Abstract
The hydrogenase accessory protein HypB, or nickelin, has two functions in the N2-fixing, H2-oxidizing bacterium Bradyrhizobium japonicum. One function of HypB involves the mobilization of nickel into hydrogenase. HypB also carries out a nickel storage/sequestering function in B. japonicum, binding nine nickel ions per monomer. Here we report that the two roles (nickel mobilization and storage) of HypB can be separated in vitro and in vivo using molecular and biochemical approaches. The role of HypB in hydrogenase maturation is completely dependent on its intrinsic GTPase activity; strains which produce a HypB protein that is severely deficient in GTPase activity but that fully retains nickel-sequestering ability cannot produce active hydrogenase even upon prolonged nickel supplementation. A HypB protein that lacks the nickel-binding polyhistidine region near the N terminus lacks only the nickel storage capacity function; it is still able to bind a single nickel ion and also retains complete GTPase activity.
The maturation of nickel-containing enzymes, involving poorly described steps of nickel mobilization and insertion into metal centers, has been the subject of increasing scrutiny over the last few years with the sequencing of genes encoding “accessory” proteins required for Ni-containing-enzyme synthesis (see reference 9). For the three best understood systems—hydrogenase, urease, and carbon monoxide dehydrogenase (CODH)—interesting parallels have emerged between the properties of these accessory proteins (10). In each system, there appears to be a requirement for an accessory protein with a nucleotide-binding motif. This motif is proposed to function in a chaperone-type role for synthesis of active-site metallocenters in urease, hydrogenase, nitrous oxide reductase, and nitrogenase (9). Also conserved to varying degrees among the nickel enzymes are accessory proteins with histidine-rich areas, which in some cases have been shown to be the domains that bind nickel. Several proteins have been shown to be required for urease metallocenter biosynthesis. One of these is the histidine-rich protein UreE (Fig. 1A), and another is the nucleotide-binding protein UreG (15, 16). Similarly, CODH maturation requires the histidine-rich protein CooJ (Fig. 1A) and the nucleotide-binding protein CooC (12).
FIG. 1.
Alignments of the histidine-rich regions of known or putative HypB proteins from various organisms, including the histidine-rich regions of UreE and CooJ (A), and the G motifs from several known or putative HypB proteins (B). Also included are the nucleotide-binding P-loop residues (within the G1 region) identified in CooC and UreG. Organisms (references) are as follows: Bj, B. japonicum (7); Rl, R. leguminosarum (25); Ss, Synechocystis sp. strain PCC 6803 (11); Ac, Azotobacter chroococcum (27); Av, Azotobacter vinelandii (3); Ms, Mycobacterium smegmatis (24); Rc, Rhodobacter capsulatus (4); Ec, E. coli (17); Ka (UreE, UreG), K. aerogenes (22); Rr (CooJ, CooC), Rhodospirillum rubrum (12).
In the case of hydrogenase maturation systems, the two properties (nickel sequestering and nucleotide binding/hydrolysis) can be contained in a single protein, namely, HypB (8, 23). Bradyrhizobium japonicum HypB purified from an over-producing strain of Escherichia coli has been shown to bind up to 18 nickel ions per dimer and also to contain GTPase activity (8). In-frame mutations of hypB yield strains which are partially or completely deficient in hydrogenase activity, depending on how much of the gene is deleted. A strain which produces a truncated HypB lacking 23 of the clustered 24 histidines is still capable of producing active hydrogenase, but these activities only approach wild-type levels when very high levels of nickel are supplied to the growth medium (23). The strain expressing the truncated hypB also accumulates less nickel than the wild type under conditions when hypB is expressed (23). From these results, we have concluded that HypB has two roles in B. japonicum: (i) that of nickel binding and storage, with this function being dependent on the histidine-rich N terminus, and (ii) that of hydrogenase expression, which may require the nucleotide-binding motif and GTP hydrolysis. Due to the nickel storage role, we previously proposed the name “nickelin” for HypB (J. W. Olson, C. Fu, and R. J. Maier, Abstr. 96th Gen. Meet. Am. Soc. Microbiol. 1996, abstr. K-202, p. 570, 1996).
Here we report that these two functions of HypB can be separated and assigned to separate domains of the protein. In vitro analysis of a truncated form of the protein missing 23 of the 24 clustered histidines shows that it retains the properties required for hydrogenase synthesis, while a mutation in the G1 domain of nickelin demonstrates that GTP hydrolysis is essential for nickelin's role in nickel donation to form an active hydrogenase.
MATERIALS AND METHODS
Purification of HypBΔ23H.
Cells of E. coli BL21(DE3) (Novagen) containing plasmid pET-HypB23H (8) were grown in baffled flasks at 37°C and 200 rpm to an optical density of ∼0.8, at which point they were induced with addition of 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) (Gold Biotechnology) for 3 h. Cells were harvested, washed, and broken by French press as described previously for purification of HypB (8). Broken cells were cleared of cell debris and membrane fractions by centrifugation at 100,000 × g for 90 min. The supernatant from the high-speed spin was then incubated at 4°C with constant stirring, while ammonium sulfate was slowly added to a final concentration of 30%. After final addition of ammonium sulfate, the solution was left stirring at 4°C for 1 h. After centrifugation at 5,000 × g for 45 min, the pellet from the ammonium sulfate precipitation was resuspended in buffer containing 10 mM Tris-Cl (pH 7.5)–25 mM NaCl (TN) and loaded directly on a 5-ml column containing DEAE-Sepharose. The column was washed with 5 column volumes of TN and eluted with a 100-ml 25 to 200 mM NaCl gradient in 10 mM Tris-Cl (pH 7.5). Fractions were assayed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and HypBΔ23H-containing fractions were pooled and dialyzed in the appropriate buffer for further assays.
Purification of HypB and HypBK119T.
Proteins were expressed from plasmid pET-HypB (8) or pET-HypBK119T in E. coli BL21(DE3) (Novagen). Proteins were purified in a single step using a nickel-loaded iminodiacetic acid-linked agarose column as described previously (8), the only difference being that a 75 mM imidazole wash was added immediately before elution of the protein by 200 mM imidazole.
Construction of a lysine 119 mutant.
Site-directed mutagenesis of the codon associated with lysine 119 was done using the Quick Change protocol (Stratagene). Primers containing the desired mutation, K119TF (5′ GCCCCGGCGCCGGTACCACCTCGCTCTTGGTC 3′) and K119TR (5′ GACCAAGAGCGAGGTGGTACCGGCGCCGGGGC 3′), were synthesized and used to introduce the mutation into the previously described plasmid pET-HypB (8). The resulting sequence introduces a silent mutation in codon 118 (GCA→GTA, glycine→glycine), changes codon 119 from AAG (lysine) to CCA (threonine), and introduces the KpnI recognition site GGTACC into the sequence. An 800-bp PstI-BamHI fragment containing the mutation was subcloned into pBluescriptKS+ (Stratagene), yielding pKSKT1. This fragment was then sequenced to verify that no errors were introduced during mutagenesis. The 800-bp fragment was then subcloned into the SacI-SalI sites of plasmid pLO-1 using the flanking SacI-SalI sites of the pBluescriptKS+ multiple cloning site, yielding pLO-KT. In-frame mutagenesis with pLO-KT was performed as previously described (23). Briefly, pLO-KT was transformed into E. coli S17-1 and mated into the parent strain JH by the biparental method, as previously described (5). Mating filters were incubated for 7 days at 30°C and then plated on MB medium plus kanamycin (75 μg/ml). Kmr colonies represented single-crossover mutants, some of which were then grown in MB medium without antibiotic selection for several days. These cultures were then plated on MB plus sucrose (5%), and any sucrose-resistant colonies were restreaked on MB-kanamycin and MB-sucrose. Kms Sucr isolates were then assayed for hydrogenase activity, and mutants were verified for the existence of the engineered KpnI restriction site by Southern hybridization (data not shown).
Hydrogenase activity.
Hydrogenase activities of whole cells which had been derepressed for hydrogenase were measured amperometrically by hydrogen electrode with O2 as the final electron acceptor, as previously described (20).
HypB and hydrogenase detection.
Proteins were detected from whole-cell extracts by immunoblotting, using antibody directed toward the hydrogenase large subunit or HypB as described previously (23), except that bands were visualized using an ECL kit (NEB) and autoradiography.
GTPase assays.
GTPase activities were determined as described previously (8).
Nickel-binding assays.
HypBΔ23H, HypB, and HypBK119T nickel binding was assayed by equilibrium dialysis and atomic absorption spectrophotometry, as described previously (8).
hup promoter activity.
Hydrogenase promoter activity was measured by introduction of the hup-lacZ fusion plasmid pSY7 (13). Cells carrying pSY7 were derepressed for hydrogenase as described previously (26), and β-galactosidase activity was measured on permeabilized cells as described previously (14, 21).
Complementation of JHK119T.
Plasmid pCF1 (6), which contains a 6.4-kb BglII fragment of B. japonicum DNA encoding the hypB promoter but no genes downstream of hypB, was introduced via triparental mating (23).
RESULTS AND DISCUSSION
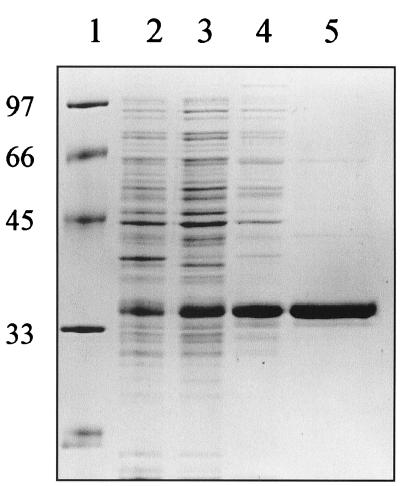
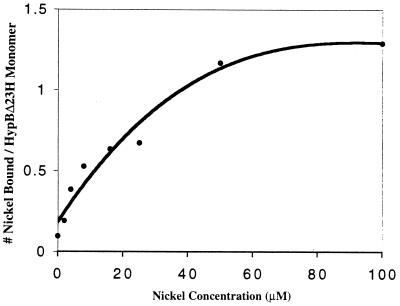
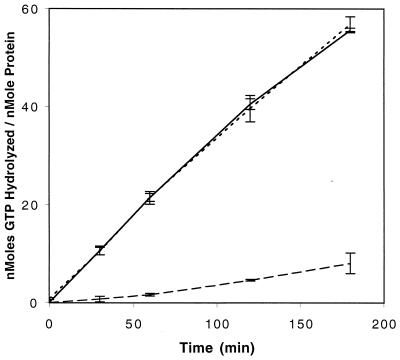
We have previously described the properties of an in-frame mutant strain lacking 23 of the clustered 24 histidines near the N terminus of HypB (23). The strain produced active Ni-containing hydrogenase, although the strain was deficient in its ability to store nickel for later hydrogenase expression. The properties of the altered (His-truncated) form of HypB were thus of interest, especially its ability to bind nickel, as the His-deleted protein can still function in mobilization of nickel in vivo. The histidine-truncated version of HypB (HypBΔ23H) was purified to near homogeneity (Fig. 2) from E. coli harboring plasmid pET-HypBΔ23 (8). In contrast to the wild-type protein, the His-truncated version was unable to bind to a nickel-loaded metal chelate affinity chromatography (MCAC) column, a qualitative example of its significantly reduced nickel-binding capacity. Nevertheless, purified protein was obtained by ammonium sulfate precipitation and DEAE chromatography, and as shown in Fig. 3, HypBΔ23H retains a modest nickel-binding capacity that saturates at 1.19 ± 0.12 atoms of nickel per monomer, with an apparent Kd of 14.8 ± 4.6 μM. Although the residue(s) within HypBΔ23H responsible for the remaining nickel binding has not been identified, it should be noted that the truncated protein still contains three histidines. GTP hydrolysis has been implicated for the proposed nickel mobilization role of HypB in E. coli (8). Consistent with the GTPase domain playing such a role in B. japonicum, the B. japonicum HypBΔ23H protein retains full (like wild-type) GTPase activity (Fig. 4). Therefore, HypB, even when lacking the His-rich nickel storage domain, still contains characteristics of nickel binding and GTP hydrolysis that correlate with active (Ni mobilization) function. The role of the His-rich area seems to be primarily in nickel storage/sequestering.
FIG. 2.
Purification of HypBΔ23H. Five micrograms (each) of crude extract (lane 2), membrane-free (S100 supernatant) extract (lane 3), ammonium sulfate-precipitated protein (lane 4), and pooled DEAE fractions (lane 5) were subjected to SDS-PAGE (12.5% polyacrylamide) and stained with Coomassie blue. Molecular weight markers are given in lane 1, and Mrs (in thousands) are shown at left.
FIG. 3.
Nickel-binding saturation curve of purified HypBΔ23H. Nickel binding was determined by equilibrium dialysis of HypBΔ23H versus increasing concentrations of nickel, followed by atomic absorption spectrophotometry. The best-fit curve was generated as a 3rd order polynominal equation.
FIG. 4.
GTPase assays of HypB (solid line), HypBΔ23H (dotted line), and HypBK119T (dashed line). Error bars indicate standard deviations of three separate determinations.
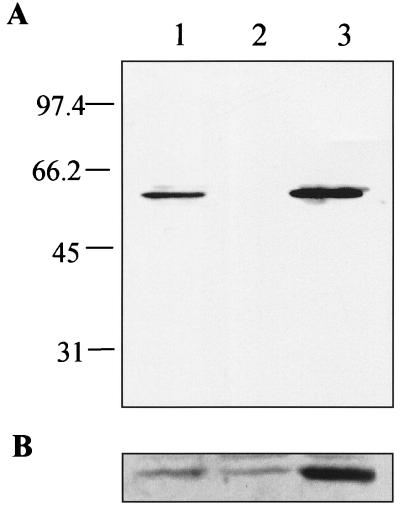
The role of the GTPase region was directly addressed by site-directed mutation of the codon which encodes the conserved lysine residue in the G1 domain of HypB (Fig. 1B). The resulting protein, HypBK119T (lysine changed to threonine), binds to the Ni-charged MCAC column and elutes from the column at the same imidazole concentration as the wild-type protein, indicating a normal affinity for nickel. Nickel-saturated HypB and HypBK119T were shown to bind the same amount of nickel when assayed by equilibrium dialysis and atomic absorption spectrophotometry (8.5 ± 0.6 nickel atoms per monomer for HypB K119T and 8.7 ± 1.8 nickel atoms per monomer for HypB, based on the average ± standard deviation for three replicates). HypBK119T retained a low (about 7% of wild type) GTPase activity (Fig. 4); when this site-directed change was introduced back into wild-type B. japonicum (via in-frame mutagenesis), a hydrogenase-negative phenotype was observed. The phenotype was not cured by adding high levels of nickel (up to 100 μM), in contrast to the same type of mutant in the E. coli system (18). Immunoblots from extracts using antibodies directed against the large subunit of hydrogenase revealed that the GTPase-deficient strain lacked hydrogenase protein (Fig. 5A). Hydrogenase protein synthesis (Fig. 5A) and activity (data not shown) can be restored by plasmid pCF1, indicating that these phenotypes are due only to the mutation within hypB and not to polar mutations on downstream genes. It should also be noted that JHK119T accumulates nearly wild-type levels of the mutant version of HypB (Fig. 5B). The fact that no hydrogenase (not even the nickel-free “apo” form) is produced is likely due to the fact that, unlike any of the other hydrogenase systems, HypB in B. japonicum plays a role in transcriptional regulation of the hydrogenase structural genes (23). We previously attributed this to a likely role for HypB as a nickel source for HupV, a protein that contains the Ni-binding motif of the hydrogenase large subunit and is necessary for the nickel-dependent transcription of B. japonicum hydrogenase (23). β-Galactosidase activities from JHK119T carrying the hup-lacZ fusion plasmid pSY7 (13) confirm that JHK119T is transcriptionally silent from the hydrogenase promoter at all nickel concentrations tested (up to 100 μM) (data not shown). These results indicate that GTP hydrolysis by HypB in B. japonicum is also required for transcriptional regulation of hydrogenase.
FIG. 5.
Immunoblot analysis of whole-cell extracts of JH (lane 1), JHK119T (lane 2), and JHK119T(pCF1) (lane 3). (A) Ten micrograms of each extract was subjected to SDS-PAGE (12.5% polyacrylamide), transferred to nitrocellulose, and blotted with antibody directed to the hydrogenase large subunit. Mrs (in thousands) are shown at left. (B) A 50-μg extract similarly treated was blotted with antibody directed to HypB.
Taken together, these data are in agreement with the conclusion that the HypB protein can be considered to possess two “domains” with different roles. These roles can be studied in vivo by phenotypic analysis of mutants and biochemically by characterizing the pure proteins. The core of the protein is the GTPase, which is highly conserved in all HypB sequences found to date (Fig. 1B). This GTPase core is obviously central to the role of all HypB proteins, but some organisms have evolved a second function for HypB, that of nickel storage/sequestering via addition of a region high in histidine residues. This His-rich domain, with its associated Ni-binding function, is most evident in the protein from B. japonicum; however, HypB proteins from other organisms also have clustered histidines near the N terminus to various extents. By “dissecting” the histidine-rich area of HypB, we have shown that the histidine-truncated protein is capable of supporting hydrogenase expression but only at dramatically increased nickel availability. This phenotype is consistent with the metal-sequestering role of the His-rich domain. The histidine-truncated HypB strain also was impaired in its ability to store nickel (23). A case can be made that maintaining an intracellular nickel reservoir, even in a Ni-poor environment, could influence the survivability of an H2-oxidizing organism, meaning that the degree of histidine residue association with HypB could be critical to survival. B. japonicum and Rhizobium leguminosarum display the most dramatic histidine-containing span (Fig. 1A), and both of these organisms express hydrogenase when in symbiosis with plants. It could be that the root nodule is a nickel-poor environment which requires the bacteroids to compete with plant enzymes for nickel. In the case of the soybean, the nickel-containing enzyme urease is ubiquitously produced (28). Also, nickel availability to the pea is a limiting factor for hydrogenase expression in R. leguminosarum bv. vicae in symbiosis (2).
An interesting parallel to the hydrogenase system is the urease accessory protein UreE. Although UreE proteins from most organisms contain the histidine-rich motifs, some do not. Organisms which do not have histidine-rich UreE proteins contain nickel-specific permeases (1). UreE from Klebsiella aerogenes normally binds six nickel ions. When its histidine-rich C terminus was deleted (the 15 amino acids shown in Fig. 1A), the strain retained reduced urease activity and it was demonstrated that the truncated UreE protein could still bind two nickel ions. A role in nickel storage was consequently proposed for the histidine-rich region (1). The structural characterization of Ni-binding sites that play metal storage or catalytic roles in enzymes is bringing about a new appreciation for the importance of nickel in metallobiochemistry (10, 19).
ACKNOWLEDGMENT
This work was supported the Department of Energy (grant DE-FG02-99ER20321).
REFERENCES
- 1.Brayman T, Hausinger R. Purification, characterization, and functional analysis of a truncated Klebsiella aerogenes UreE urease accessory protein lacking the histidine-rich carboxyl terminus. J Bacteriol. 1996;178:5410–5416. doi: 10.1128/jb.178.18.5410-5416.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brito B, Palacios J-M, Hidalgo E, Imperial J, Ruiz-Argüeso T. Nickel availability to pea (Pisum sativum L.) plants limits hydrogenase activity of Rhizobium leguminosarum bv. viciae bacteroids by affecting the processing of the hydrogenase structural subunits. J Bacteriol. 1994;176:5297–5303. doi: 10.1128/jb.176.17.5297-5303.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen J C, Mortenson L E. Identification of six open reading frames from a region of the Azotobacter vinelandii genome likely involved in dihydrogen metabolism. Biochim Biophys Acta. 1992;1131:199–202. doi: 10.1016/0167-4781(92)90077-d. [DOI] [PubMed] [Google Scholar]
- 4.Colbeau A, Richaud P, Toussaint B, Caballero F J, Elster C D, Smith R L, Jacqueline C, Vignais P M. Organization of the genes necessary for hydrogenase expression in Rhodobacter capsulatus. Sequence analysis and identification of two hyp regulatory mutants. Mol Microbiol. 1993;8:15–29. doi: 10.1111/j.1365-2958.1993.tb01199.x. [DOI] [PubMed] [Google Scholar]
- 5.Fu C, Maier R J. Identification of a locus within the hydrogenase gene cluster involved in intracellular nickel metabolism in Bradyrhizobium japonicum. Appl Environ Microbiol. 1991;57:3502–3510. doi: 10.1128/aem.57.12.3502-3510.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fu C, Maier R J. A genetic region downstream of the hydrogenase structural genes of Bradyrhizobium japonicum that is required for hydrogenase processing. J Bacteriol. 1993;175:295–298. doi: 10.1128/jb.175.1.295-298.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fu C, Maier R J. Nucleotide sequences of two hydrogenase-related genes (hypA and hypB) from Bradyrhizobium japonicum, one of which (hypB) encodes an extremely histidine-rich region and guanine nucleotide domains. Biochim Biophys Acta. 1994;1184:135–138. doi: 10.1016/0005-2728(94)90163-5. [DOI] [PubMed] [Google Scholar]
- 8.Fu C, Olson J W, Maier R J. HypB protein of Bradyrhizobium japonicum is a metal-binding GTPase capable of binding 18 divalent nickel ions per dimer. Proc Natl Acad Sci USA. 1995;92:2333–2337. doi: 10.1073/pnas.92.6.2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hausinger R P. General principles and mechanisms of metallocenter assembly. In: Hausinger R P, Eichhorn G L, Marzilli L G, editors. Mechanisms of metallocenter assembly. New York, N.Y: VCH Publishers, Inc.; 1996. pp. 1–18. [Google Scholar]
- 10.Hausinger R P. Metallocenter assembly in nickel-containing enzymes. J Biol Inorg Chem. 1997;2:279–286. [Google Scholar]
- 11.Kaneko T, Sato S, Kotani H, Tanaka A, Asamizu E, Nakamura Y, Miyajima N, Hirosawa M, Sugiura M, Sasamoto S, Kimura T, Hosouchi T, Matsuno A, Muraki A, Nakazaki N, Naruo K, Okumura S, Shimpo S, Takeuchi C, Wada T, Watanabe A, Yamada M, Yasuda M, Tabata S. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 1996;3:109–136. doi: 10.1093/dnares/3.3.109. [DOI] [PubMed] [Google Scholar]
- 12.Kerby R L, Ludden P W, Roberts G P. In vivo nickel insertion into the carbon monoxide dehydrogenase of Rhodospirillum rubrum: molecular and physiological characterization of cooCTJ. J Bacteriol. 1997;179:2259–2266. doi: 10.1128/jb.179.7.2259-2266.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim H, Maier R J. Transcriptional regulation of hydrogenase synthesis by nickel in Bradyrhizobium japonicum. J Biol Chem. 1990;265:18729–18732. [PubMed] [Google Scholar]
- 14.Kim H, Yu C, Maier R J. Common cis-acting region responsible for transcriptional regulation of Bradyrhizobium japonicum hydrogenase by nickel, oxygen, and hydrogen. J Bacteriol. 1991;173:3993–3999. doi: 10.1128/jb.173.13.3993-3999.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee M H, Mulrooney S B, Renner M J, Markowicz Y, Hausinger R P. Klebsiella aerogenes urease gene cluster: sequence of ureD and demonstration that four accessory genes (ureD, ureE, ureF, and ureG) are involved in nickel metallocenter biosynthesis. J Bacteriol. 1992;174:4324–4330. doi: 10.1128/jb.174.13.4324-4330.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee M H, Pankratz H S, Wang S, Scott R A, Finnegan M G, Johnson M K, Ippolito J A, Christianson D W, Hausinger R P. Purification and characterization of Klebsiella aerogenes UreE protein: a nickel-binding protein that functions in urease metallocenter assembly. Protein Sci. 1993;2:1042–1052. doi: 10.1002/pro.5560020617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lutz S, Jacobi A, Schlensog V, Böhm R, Sawers G, Böck A. Molecular characterization of an operon (hyp) necessary for the activity of the three hydrogenase isoenzymes in Escherichia coli. Mol Microbiol. 1991;5:123–135. doi: 10.1111/j.1365-2958.1991.tb01833.x. [DOI] [PubMed] [Google Scholar]
- 18.Maier T, Lottspeich F, Böck A. GTP hydrolysis by HypB is essential for nickel insertion into hydrogenases of Escherichia coli. Eur J Biochem. 1995;230:133–138. [PubMed] [Google Scholar]
- 19.Maroney M J. Structure/function relationships in nickel metallobiochemistry. Curr Opin Chem Biol. 1999;3:188–199. doi: 10.1016/S1367-5931(99)80032-5. [DOI] [PubMed] [Google Scholar]
- 20.Merberg D, O'Hara E B, Maier R J. Regulation of hydrogenase in Rhizobium japonicum: analysis of mutants altered in regulation by carbon substrates and oxygen. J Bacteriol. 1983;156:1236–1242. doi: 10.1128/jb.156.3.1236-1242.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 22.Mulrooney S B, Hausinger R P. Sequence of the Klebsiella aerogenes urease genes and evidence for accessory proteins facilitating nickel incorporation. J Bacteriol. 1990;172:5837–5843. doi: 10.1128/jb.172.10.5837-5843.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Olson J W, Fu C, Maier R J. The HypB protein from Bradyrhizobium japonicum can store nickel and is required for the nickel-dependent transcriptional regulation of hydrogenase. Mol Microbiol. 1997;24:119–128. doi: 10.1046/j.1365-2958.1997.3251690.x. [DOI] [PubMed] [Google Scholar]
- 24.Papavinasasundaram K G, Movahedzadeh F, Keer J T, Stoker N G, Colston M J, Davis E O. Mycobacterial recA is cotranscribed with a potential regulatory gene called recX. Mol Microbiol. 1997;24:141–153. doi: 10.1046/j.1365-2958.1997.3441697.x. [DOI] [PubMed] [Google Scholar]
- 25.Rey L, Murillo J, Hernando Y, Hidalgo E, Cabrera E, Imperial J, Ruiz-Argüeso T. Molecular analysis of a microaerobically induced operon required for hydrogenase synthesis in Rhizobium leguminosarum biovar vicae. Mol Microbiol. 1993;8:471–481. doi: 10.1111/j.1365-2958.1993.tb01591.x. [DOI] [PubMed] [Google Scholar]
- 26.Stults L W, Moshiri F, Maier R J. Aerobic purification of hydrogenase from Rhizobium japonicum by affinity chromatography. J Bacteriol. 1986;166:795–800. doi: 10.1128/jb.166.3.795-800.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tibelius K H, Du L, Tito D, Stejskal F. The Azotobacter chroococcum hydrogenase gene cluster: sequences and genetic analysis of four accessory genes, hupA, hupB, hupY, and hupC. Gene. 1993;127:53–61. doi: 10.1016/0378-1119(93)90616-b. [DOI] [PubMed] [Google Scholar]
- 28.Torisky R S, Griffin J D, Yenofsky R L, Polacco J C. A single gene (Eu4) encodes the tissue-ubiquitous urease of soybean. Mol Gen Genet. 1994;242:404–414. doi: 10.1007/BF00281790. [DOI] [PubMed] [Google Scholar]