Abstract
Background
Neuroinflammation is emerging as an important pathological process in frontotemporal dementia (FTD), but biomarkers are lacking. We aimed to determine the value of complement proteins, which are key components of innate immunity, as biomarkers in cerebrospinal fluid (CSF) and plasma of presymptomatic and symptomatic genetic FTD mutation carriers.
Methods
We measured the complement proteins C1q and C3b in CSF by ELISAs in 224 presymptomatic and symptomatic GRN, C9orf72 or MAPT mutation carriers and non-carriers participating in the Genetic Frontotemporal Dementia Initiative (GENFI), a multicentre cohort study. Next, we used multiplex immunoassays to measure a panel of 14 complement proteins in plasma of 431 GENFI participants. We correlated complement protein levels with corresponding clinical and neuroimaging data, neurofilament light chain (NfL) and glial fibrillary acidic protein (GFAP).
Results
CSF C1q and C3b, as well as plasma C2 and C3, were elevated in symptomatic mutation carriers compared to presymptomatic carriers and non-carriers. In genetic subgroup analyses, these differences remained statistically significant for C9orf72 mutation carriers. In presymptomatic carriers, several complement proteins correlated negatively with grey matter volume of FTD-related regions and positively with NfL and GFAP. In symptomatic carriers, correlations were additionally observed with disease duration and with Mini Mental State Examination and Clinical Dementia Rating scale® plus NACC Frontotemporal lobar degeneration sum of boxes scores.
Conclusions
Elevated levels of CSF C1q and C3b, as well as plasma C2 and C3, demonstrate the presence of complement activation in the symptomatic stage of genetic FTD. Intriguingly, correlations with several disease measures in presymptomatic carriers suggest that complement protein levels might increase before symptom onset. Although the overlap between groups precludes their use as diagnostic markers, further research is needed to determine their potential to monitor dysregulation of the complement system in FTD.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12974-022-02573-0.
Keywords: Biomarker, Complement, Frontotemporal dementia, Neuroinflammation
Background
Frontotemporal dementia (FTD) is a common form of young-onset dementia and is frequently caused by autosomal dominant genetic mutations in progranulin (GRN), chromosome 9 open reading frame 72 (C9orf72) or microtubule-associated protein tau (MAPT) [1, 2]. Accumulating evidence suggests a role for neuroinflammation in FTD, although the timing and exact contribution to disease pathogenesis remains unclear [3]. Fluid biomarkers that reflect neuroinflammation in vivo could be valuable for clinical practice and therapeutic trials. Previous studies aiming to identify such biomarkers, including cytokines and microglial markers, have yielded somewhat inconsistent results [4–13]
The complement system is a key component of innate immunity and comprises a cascade of protein reactions which ultimately result in opsonisation and lysis of potential pathogens, recruitment of immune cells to create a pro-inflammatory environment, and clearance of apoptotic cells [14]. Complement proteins are also involved in microglia-mediated synaptic pruning in both the developing and adult brain [15, 16], and aberrant activation of the complement cascade is thought to play a central role in synaptic degeneration across neurodegenerative diseases [17–20]. In line with this, GRN-/- mice display excessive complement activation and synaptic pruning, whereas deletion of the complement genes C1q and C3b mitigates synapse loss and neurodegeneration [21, 22]. Complement proteins in cerebrospinal fluid (CSF) and blood are differentially regulated in Alzheimer’s disease (AD) [23–28] and other neurodegenerative diseases compared to controls [29–32], but they have not been thoroughly investigated in FTD. Promisingly, an inverse correlation was found between CSF C1q and C3b levels and Mini Mental State Examination (MMSE) score in a small series of GRN mutation carriers [21].
In the present study, we measured a range of complement proteins in CSF and plasma of presymptomatic and symptomatic genetic FTD mutation carriers participating in the international Genetic FTD Initiative (GENFI). To determine their value as disease progression biomarkers, we correlated complement levels with corresponding clinical and neuroimaging measures. Finally, we explored their relationship with biomarkers that reflect neuro-axonal degeneration (neurofilament light chain, NfL) [33, 34] and astrogliosis (glial fibrillary acidic protein, GFAP) [35, 36].
Methods
Subjects
Subjects were recruited from 19 centres collaborating in GENFI, a longitudinal cohort study which follows patients with genetic FTD due to a mutation in GRN, C9orf72 or MAPT and their 50% at-risk family members (either presymptomatic mutation carriers or non-carriers) [37]. Participants underwent an annual assessment as previously described [37], which includes a brief medical history, neurological and neuropsychological examination, magnetic resonance imaging (MRI) of the brain, and collection of blood and CSF. Clinical researchers were blinded to the genetic status of at-risk individuals unless they had undergone predictive testing. Subjects with known auto-immune diseases were excluded from the current study as complement levels could be affected [38].
CSF samples were available in 104 presymptomatic (46 GRN, 42 C9orf72, 16 MAPT) and 46 symptomatic mutation carriers (11 GRN, 28 C9orf72, 7 MAPT) and 74 healthy non-carriers. Plasma samples were available in 215 presymptomatic (88 GRN, 80 C9orf72, 47 MAPT) and 104 symptomatic mutation carriers (36 GRN, 47 C9orf72, 21 MAPT) and 112 non-carriers (Table 1). 174 subjects were included in both the CSF and plasma cohorts.
Table 1.
Subject characteristics for (a) CSF and (b) plasma measurements
| (a) CSF cohort | ||||||||
|---|---|---|---|---|---|---|---|---|
| Non-carriers | Presymptomatic carriers | Symptomatic carriersa | p | |||||
| N | 74 | 104 | 46 | |||||
| Sex, male (%) | 34 (46%) | 43 (41%) | 28 (61%) | 0.085 | ||||
|
Age at collection, years Years |
47 (39–58) | 46 (35–56) | 63 (55–69) | < 0.001 | ||||
| MMSE (n = 219) | 30 (29–30) | 30 (29–30) | 26 (24–29) | < 0.001 | ||||
| CDR® + NACC FTLD-SB (n = 185) | 0 (0–0) | 0 (0–0) | 9 (2–13) | < 0.001 | ||||
| Per genotype | GRN | C9orf72 | MAPT | GRN | C9orf72 | MAPT | ||
| N | 46 | 42 | 16 | 11 | 28 | 7 | - | |
| Age at collection, years |
54 (42–59) |
43 (33–53) |
42 (34–46) |
67 (61–70) |
60 (55–72) |
59 (52–64) |
< 0.001b | |
| Age at symptom onset, years | – | – | – | – |
64 (54–67) |
56 (49–62) |
55 (52–56) |
0.141 |
| Disease duration, years | – | – | – | – |
2.5 (1.0–4.3) |
4.1 (2.1–8.0) |
2.6 (0.4–8.0) |
0.229 |
| (b) Plasma cohort | ||||||||
|---|---|---|---|---|---|---|---|---|
| Non-carriers | Presymptomatic carriers | Symptomatic carriersc | p | |||||
| N | 112 | 215 | 104 | |||||
| Sex, male (%) | 49 (44%) | 79 (37%) | 64 (62%) | < 0.001 | ||||
|
Age at collection, years Years |
50 (39–60) | 45 (35–55) | 63 (58–69) | < 0.001 | ||||
| MMSE (n = 405) | 30 (29–30) | 30 (29–30) | 25 (20–28) | < 0.001 | ||||
| CDR® + NACC FTLD-SB (n = 329) | 0 (0–0) | 0 (0–0) | 8 (3–14) | < 0.001 | ||||
| Per genotype | GRN | C9orf72 | MAPT | GRN | C9orf72 | MAPT | ||
| N | 88 | 80 | 47 | 36 | 47 | 21 | ||
| Age at collection, years |
51 (39–59) |
44 (34–53) |
40 (33–46) |
64 (59–68) |
66 (59–72) |
58 (52–63) |
< 0.001b | |
| Age at symptom onset, years | – | – | – | – |
60 (55–66) |
59 (55–66) |
53 (47–57) |
< 0.001d |
| Disease duration, years | – | – | – | – |
2.6 (1.8–4.2) |
5 (2.6–6.6) |
5.6 (1.5–6.8) |
0.002e |
Continuous variables are expressed as median (interquartile range) and were compared between groups using Kruskal–Wallis tests. Sex distributions were compared between groups using Chi-square tests. MMSE Mini Mental State Examination, CDR Clinical Dementia Rating scale, SB sum of boxes
aPhenotypes: behavioural variant FTD (bvFTD) (n = 32), primary progressive aphasia (PPA) (n = 5), FTD with amyotrophic lateral sclerosis (ALS) (n = 3), ALS without FTD (n = 3), progressive supranuclear palsy (PSP) (n = 1), memory-predominant FTD (n = 1), dementia not otherwise specified (n = 1)
bSymptomatic mutation carriers were older than presymptomatic carriers in all genetic subgroups. cPhenotypes: bvFTD (n = 78), PPA (n = 16), FTD-ALS (n = 2), ALS without FTD (n = 5), PSP (n = 1), memory-predominant FTD (n = 1), dementia not otherwise specified (n = 1)
dSymptomatic MAPT mutation carriers were younger at symptom onset than C9orf72 (p = 0.004) and GRN mutation carriers (p = 0.002)
eSymptomatic C9orf72 mutation carriers had a longer disease duration than symptomatic GRN carriers at sample collection
Mutation carriers were considered symptomatic if they fulfilled international consensus criteria for behavioural variant FTD [39], primary progressive aphasia [40] or amyotrophic lateral sclerosis (ALS) [41]. Disease duration was defined based on when the primary caregiver first noted symptoms. Global cognition was scored using the MMSE and Clinical Dementia Rating scale® plus NACC FTLD sum of boxes (CDR® + NACC FTLD-SB) [42], collected within 6 months of CSF or plasma sampling.
T1-weighted MRI on 3 Tesla scanners was obtained within 6 months of sample collection using a standardised GENFI protocol. T1-weighted volumetric MRI scans were parcellated into brain regions as previously described [37], using an atlas propagation and fusion strategy to generate volumes of the whole brain (WBV), frontal, temporal, parietal and occipital lobes, insula and cingulate gyrus. Brain volumes were expressed as a percentage of total intracranial volume (TIV), computed with SPM12 running under Matlab R2014b (Math Works, Natick, MA, USA) [43].
Sample collection and laboratory methods
CSF was collected by lumbar puncture in polypropylene tubes, and blood was collected by venepuncture in EDTA tubes. Samples were centrifuged and stored at -80 °C until use according to a standardised GENFI protocol.
All CSF and plasma measurements were performed in duplicate. The mean duplicate coefficient of variation (CV) was below 10% for all analytes; samples with a CV > 20% were re-measured or excluded. For sample concentrations outside of the range of quantification, we imputed the lower or upper limits of quantification (LLOQ and ULOQ) (Additional file 1: Table S1).
CSF complement proteins C1q and C3b were measured using the ELISA kits Human Complement C1q (ab170246) and Human Complement C3b (ab195461) from Abcam (Boston, MA, USA) according to the manufacturer’s instructions. Plates were read on a SpectraMax M2 plate reader (Molecular Devices, San Jose, CA). CSF NfL was measured using the Simoa NF-Light Advantage Kit from Quanterix (Billerica, MA, USA) on a Simoa HD-1 analyzer instrument according to the manufacturer’s instructions.
Plasma complement proteins were measured using the multiplex Human Complement Magnetic Bead Panel 1 (complement factors C2, C4b, C5, C5a, C9, factor D, mannose-binding lectin, and factor I) and Human Complement Magnetic Bead Panel 2 (C1q, C3, C3b, C4, factor B, factor H) (HCMP1MAG-19 K and HCMP2MAG-19 K, respectively) kits from EMD Millipore Corporation (Billerica, MA, USA) according to the manufacturer’s instructions. Plates were analysed on a Luminex MAGPIX Instrument System (Luminex Corp, Austin, TX, USA). Plasma NfL and GFAP were measured using the multiplex Neurology 4-Plex A kit from Quanterix on a Simoa HD-1 Analyzer according to the manufacturer’s instructions, as previously described [36].
Laboratory technicians were blinded to all clinical and genetic information.
Statistical analysis
Statistical analyses were performed in IBM SPSS Statistics 25 and R.
Demographic and clinical variables were compared between groups (symptomatic, presymptomatic, non-carrier) using Kruskal–Wallis tests for continuous variables and a Chi-square test for sex. Normality of biomarker data was assessed using Kolmogorov–Smirnov tests and visual inspection of Q–Q plots. All raw protein concentrations, both in CSF and plasma, were non-normally distributed. For CSF analytes, normal distributions were achieved after log-transformation, and we subsequently performed ANCOVAs with age and sex as covariates to compare protein concentrations between groups. For plasma complement proteins, normal distributions could not be achieved with conventional transformations (e.g. log transformation, Box–Cox transformation), and we therefore applied quantile regression, which is robust to non-normality and outliers, with age and sex as covariates. In comparisons between symptomatic mutation carriers, we also included disease duration as a covariate. Correlations between raw biomarker values and clinical and neuroimaging measures, as well as NfL and GFAP, were assessed using Spearman’s rho for presymptomatic and symptomatic mutation carriers separately. Correction for multiple comparisons was done with the Holm–Bonferroni method. We restricted correlative analyses between CSF and plasma measurements to subjects for whom the time interval between both sample collections was less than 6 months.
Results
Subjects
Subject characteristics of the CSF and plasma cohorts are shown in Table 1.
CSF complement levels
We excluded 7 samples from C1q analyses (5 presymptomatic carriers, 2 non-carriers) and 12 samples from C3b analyses (5 presymptomatic and 3 symptomatic carriers, 4 non-carriers) due to duplicate CV’s > 20%. CSF C1q correlated strongly with C3b levels (rs = 0.709, p < 0.001). Furthermore, C1q and C3b correlated with age at sample collection (rs = 0.359 and rs = 0.323; both p < 0.001) in the entire cohort, but not in non-carriers alone (Additional file 1: Table S2).
Group differences
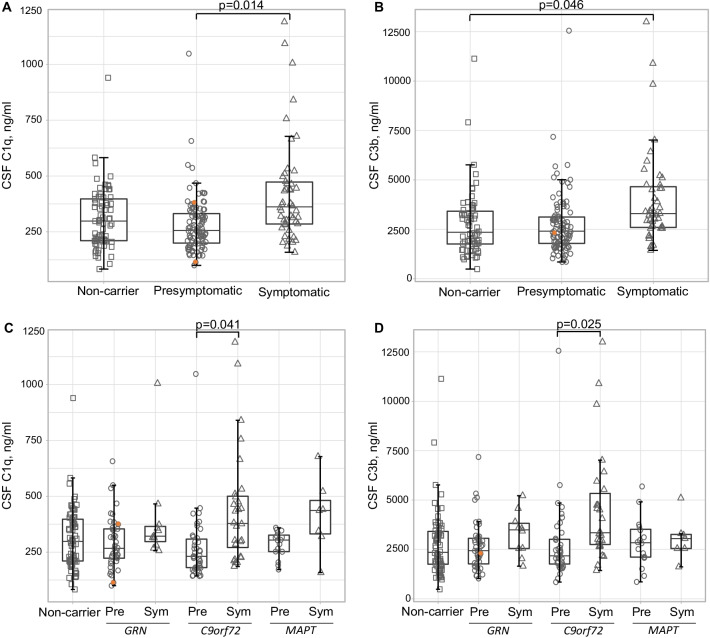
C1q levels were significantly higher in symptomatic mutation carriers (median 362 ng/ml (interquartile range 284–481)) than in presymptomatic carriers (256 ng/ml (199–337), p = 0.014), but not compared to non-carriers (298 ng/ml (210–402), p = 0.148) (Fig. 1a). Higher levels of C3b were found in symptomatic carriers (3295 ng/ml (2558–4734)) compared to non-carriers (2350 ng/ml (1730–3452), p = 0.046). However, C3b levels between symptomatic and presymptomatic carriers (2406 ng/ml (1772–3127) were not significantly different (p = 0.074) (Fig. 1b). After exclusion of extreme outliers (> median + 3*IQR; n = 1 for C1q and n = 5 for C3b), C3b levels were also elevated compared to presymptomatic carriers (p = 0.038).
Fig. 1.
Group comparisons of CSF C1q and C3b concentration. A C1q in clinical groups; B C1q in genetic subgroups; C C3b in clinical groups; D C3b in genetic subgroups. Orange circles indicate presymptomatic carriers who developed symptoms during follow-up, one of whom was excluded from C3b analyses due to a duplicate CV > 20%. P-values were derived from ANCOVAs on log-transformed biomarker data with age and sex as covariates. Pre presymptomatic, Sym symptomatic
Separated by genetic group, C1q and C3b levels were elevated in all symptomatic carriers, but after correction for age, group differences were only significant for C9orf72 mutation carriers (C1q: p = 0.041; C3b: p = 0.025) (Fig. 1c, d). C1q or C3b levels did not differ between symptomatic carriers of different genetic groups (p = 0.351).
Correlative analyses
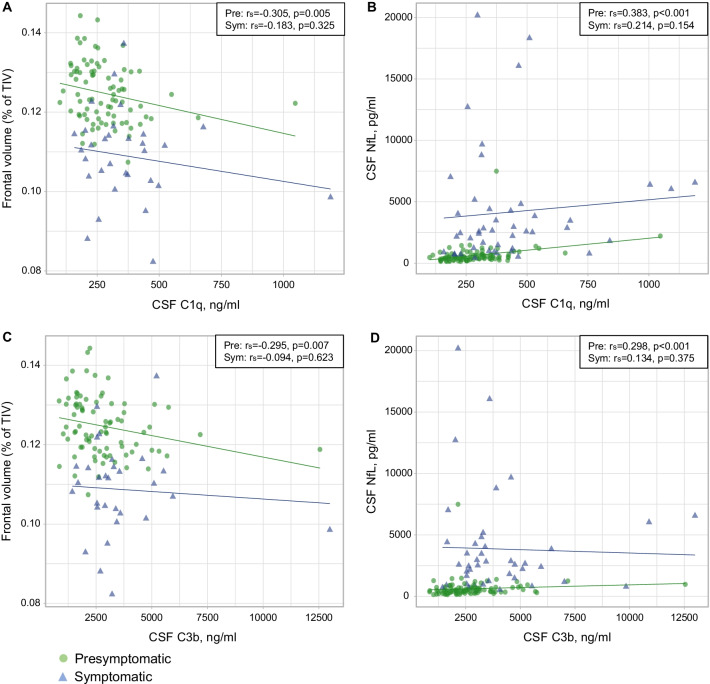
In presymptomatic mutation carriers, C1q and C3b levels correlated with NfL and inversely with frontal lobe volume (Fig. 2 and Additional file 1: Table S3a). These correlations remained significant after correction for age.
Fig. 2.
Correlations among mutation carriers between A CSF C1q and frontal lobe volume; B CSF C1q and CSF neurofilament light chain (NfL); C CSF C3b and frontal volume); and D CSF C3b and CSF NfL. P-values were derived from Spearman’s rho. NfL neurofilament light chain, TIV total intracranial volume, Pre presymptomatic, Sym symptomatic
In symptomatic carriers, we observed an inverse correlation between C1q and MMSE (rs = − 0.370, p = 0.013) (Additional file 1: Table S4a), and C3b levels—but not C1q—were correlated with disease duration (rs = 0.343, p = 0.024) (Additional file 1: Fig. S1a and 1b).
Plasma complement levels
C3b, C5a and C9 were excluded from analyses as concentrations were below the LLOQ in 80–100% of samples; the samples with levels above the LLOQ were from all clinical groups. For some analytes, a small number of samples was excluded due to CVs > 20% (Additional file 1: Table S1).
Moderate correlations were found between most plasma analytes (Additional file 1: Table S5). Furthermore, we found positive correlations between age and almost all analytes (Additional file 1: Table S2); several of these remained significant when analyses were limited to non-carriers.
Group comparisons
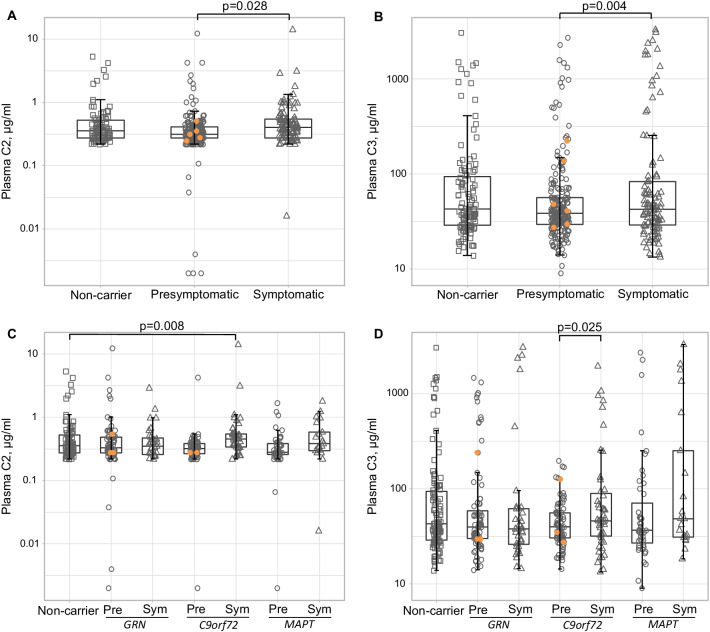
Symptomatic mutation carriers had significantly higher levels of plasma C2 and C3 than presymptomatic carriers (Table 2, Fig. 3a, b).
Table 2.
Plasma complement levels per clinical group
| Non-carriers | Presymptomatic carriers | Symptomatic carriers | p | |
|---|---|---|---|---|
| C2 | 0.358 (0.274–0.532) | 0.332 (0.274–0.431) | 0.411 (0.319–0.548) | 0.006* |
| C4b | 9.97 (8.00–12.3) | 9.92 (8.15–12.8) | 10.8 (8.88–14.3) | 0.913 |
| C5 | 29.1 (21.6–35.8) | 27.0 (21.7–35.2) | 31.0 (25.1–38.3) | 0.358 |
| Factor D | 3.57 (2.99–4.37) | 3.46 (2.69–4.15) | 4.19 (3.50–5.28) | 0.481 |
| MBL | 2.44 (0.841–4.20) | 2.01 (0.860–4.12) | 1.71 (0.763–4.41) | 0.710 |
| Factor I | 39.6 (32.9–46.6) | 37.9 (32.8–45.2) | 40.9 (35.0–47.0) | 0.863 |
| C1q | 71.4 (64.5–81.2) | 71.2 (60.6–80.1) | 68.7 (62.2–76.2) | 0.636 |
| C3 | 44.8 (29.9–98.2) | 39.9 (29.9–58.8) | 45.5 (30.5–102) | 0.047** |
| C4 | 293 (254–360) | 297 (252–348) | 294 (254–349) | 0.577 |
| Factor B | 168 (147–211) | 167 (146–205) | 170 (143–202) | 0.772 |
| Factor H | 250 (213–295) | 250 (212–288) | 254 (228–285) | 0.849 |
All concentrations are expressed as medians (interquartile range) in µg/ml. P-values are derived from quantile regression models with age and sex as covariates. MBL mannose-binding lectin. *Pairwise comparisons: symptomatic vs presymptomatic carriers: p = 0.028. **Pairwise comparisons: symptomatic vs presymptomatic carriers: p = 0.004
Fig. 3.
Group comparisons of plasma C2 and C3 concentration. A Plasma C2 in clinical groups; B Plasma C3 in clinical groups; C Plasma C2 in genetic subgroups; D Plasma C3 in genetic subgroups. Protein concentrations were plotted on a logarithmic scale for ease of visualisation. Orange circles indicate presymptomatic carriers who developed symptoms during follow-up, one of whom was excluded from C2 analyses due to a duplicate CV > 20%. P-values were derived from quantile regression models with age and sex as covariates. Pre presymptomatic, Sym symptomatic
Separated by genetic subgroup, elevated levels of C2 and C3 were observed in symptomatic C9orf72 and MAPT—but not GRN—mutation carriers, reaching statistical significance in C9orf72 (Fig. 3c, d). No significant differences were observed in C2 or C3 levels between symptomatic carriers of different genetic subgroups (C2: p = 0.425; C3: p = 0.512).
Correlative analyses
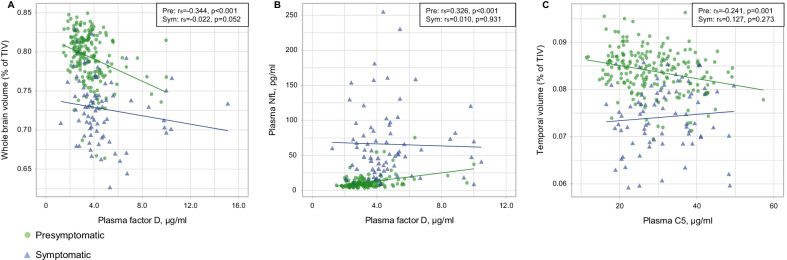
In the presymptomatic stage, inverse correlations were observed between several complement proteins and regional grey matter volume. The strongest correlations were observed for factor D with WBV (rs = − 0.344, p < 0.001), temporal volume (rs = − 0.271, p < 0.001) and volume of the cingulate gyrus (rs = − 0.262, p < 0.001), and for C5 with temporal volume (rs = − 0.241, p = 0.001), which remained significant after correction for age. Furthermore, C4b, C5 and factor D were positively correlated with NfL and GFAP (Fig. 4, Additional file 1: Tables S3b, 6).
Fig. 4.
Correlations among mutation carriers between A plasma factor D and whole brain volume; B plasma factor D and plasma neurofilament light chain (NfL); and C plasma C5 and temporal lobe volume. P-values were derived from Spearman’s rho. NfL neurofilament light chain, TIV total intracranial volume, Pre presymptomatic, Sym symptomatic
In the symptomatic stage, C2, C3 and factor D were inversely correlated with WBV and volume of the temporal and parietal lobes, cingulate gyrus and insula, and C2, C3, factor D, factor I and factor H were correlated with CDR® + NACC FTLD-SB score (Additional file 1: Tables S3b, 4b). Furthermore, we found positive correlations with disease duration for C2 (rs = 0.279, p = 0.006), factor D (rs = 0.239, p = 0.015) and factor I (rs = 0.202, p = 0.039).
Correlation between CSF and plasma C1q levels
CSF and plasma C1q levels were not correlated among 147 subjects with matched CSF and plasma samples (rs = 0.092, p = 0.266; mean time interval between CSF and plasma: 13 days). Restricting analyses to samples collected on the same day similarly revealed no correlation (rs = 0.065, p = 0.543, n = 91).
CSF and plasma complement levels in converters
In the seven presymptomatic carriers who were diagnosed with FTD during follow-up (‘converters’), no relationship between CSF or plasma complement levels and time to symptom onset was observed (Additional file 1: Table S7).
Discussion
This large, international study demonstrated elevated levels of several complement proteins in CSF and plasma in the symptomatic stages of genetic FTD, as well as correlations with various measures of disease severity. Our findings provide in vivo evidence of an inflammatory component in FTD and could aid therapeutic trials aimed at modulation of the immune response.
The elevated levels of C1q and C3b in CSF of symptomatic mutation carriers probably reflect increased local synthesis of complement proteins by glial cells and neurons, as has previously been reported in neurodegeneration [18, 44–47]. C1q is the initiator molecule of the classical pathway, and its binding to immune complexes, apoptotic cells and various other stimuli triggers a cascade of protein reactions to generate C3b [14]. C3b is one of the primary complement opsonins, and its accumulation on synapses and subsequent recognition by phagocytic microglia is thought to underlie the synapse loss observed early in the neurodegenerative process [18–20]. If direct associations can be confirmed, CSF C1q and C3b might provide a means to monitor complement-mediated synaptic pruning and measure the effect of complement-directed therapeutics [44]. C3b also elicits generation of the cytotoxic terminal C5b-C9 complex (TCC) [14]. It would be interesting to expand on our results by measuring CSF complement proteins directly implicated in the TCC, as well as regulatory factors, a decrease of which might further amplify aberrant complement activation [48].
In symptomatic mutation carriers, plasma measurements revealed elevated levels of C2, a component of the classical pathway, and C3. Rather than reflecting overflow from the central nervous system (CNS), these findings might reflect a systemic immune response, which could in turn contribute to neuroinflammation by passing through the (compromised) blood–brain barrier [44, 49]. The lack of a correlation between CSF and plasma C1q suggests that systemic and local complement activation might not occur simultaneously and indicates that plasma complement measurement is not a suitable surrogate for CSF. Investigation of CSF–plasma associations of other complement proteins besides C1q might confirms this, in which case brain-derived extracellular vesicles could provide a better peripheral measure of CNS complement activation [25, 28]. Furthermore, since consumption of intact complement components (e.g. C3, C4 and C5) can paradoxically reduce plasma levels during strong complement activation [38], future measurements of activated fragments (e.g. C3a, C3b and components of the TCC) in FTD might provide more robust measures of peripheral complement activation.
In genetic subgroup analyses, the elevated complement protein levels in CSF and plasma remained statistically significant only in C9orf72 mutation carriers. Interestingly, C9orf72−/− mice have been shown to have upregulated interferon-β expression, increased microglial activation, and excessive synaptic pruning compared to wild-type mice [50]. In vivo administration of interferon-β drives microglial activation and complement C3-dependent synapse elimination [51]. C9orf72 deficiency might thus promote microglial activation through interferon-β, in turn leading to synaptic elimination by complement activation. Alternatively, since complement activation has also been reported in cell and animal models of GRN and MAPT mutations [17, 21, 22, 52], the lack of significant differences in GRN- and MAPT-related FTD might instead reflect a lack of statistical power given the smaller sample size of these genetic subgroups. The elevated levels of complement proteins in various other neurodegenerative diseases similarly point towards a general rather than gene- or disease-specific upregulation of the complement system [21–30]. Future studies comparing complement levels in genetic and sporadic forms of FTD and associated clinical subtypes might further elucidate potential gene-specific effects.
In presymptomatic mutation carriers, CSF and plasma complement levels correlated with regional grey matter volume and NfL. These correlations remained significant even after correction for age, and suggest that complement activation might occur in the late-presymptomatic stage in conjunction with early brain atrophy. Accordingly, elevated complement levels have been observed in presymptomatic genetic AD [27, 53] and mild cognitive impairment [26, 28]. In AD mouse models, complement aggregation is observed prior to plaque formation [17, 18, 52]. Despite applying statistical correction for age, the lack of group differences in complement levels between presymptomatic carriers and non-carriers could partly be due to including carriers of all ages, and thus time to symptom onset was highly variable.
CSF and plasma complement levels showed substantial overlap between groups, which has also been reported in AD [26, 27, 54] and precludes their use as diagnostic biomarkers. NfL may be a more powerful tool to distinguish symptomatic from presymptomatic mutation carriers [34]. The large variability in complement levels, which was observed even among non-carriers, suggests that within-individual changes in complement levels may be more informative for disease monitoring than single measurements. The lack of correlation in symptomatic mutation carriers between CSF complement factors and most disease severity measures, including brain atrophy, NfL, GFAP and MMSE, indicates that complement levels probably do not increase linearly as the disease progresses. Instead, in line with the dynamic nature of neuroinflammation [3], they might fluctuate depending on the disease stage [55]. Longitudinal studies of CSF and plasma complement factors, including a larger number of converters, might elucidate their dynamics over the course of FTD.
Strengths of this study include the very large, well-characterised genetic FTD cohort with corresponding clinical and neuroimaging data. In plasma, we measured a broad range of complement proteins covering all three activation pathways as well as various regulatory molecules. The strong correlations between CSF C1q and C3b, as well as between the various plasma complement factors, support the validity of our results.
The findings presented in this study must be viewed in light of some limitations. Our plasma complement measurements could have been affected by various confounding factors, including body mass index, hypertension, diabetes mellitus and (asymptomatic or low-grade) inflammatory processes [38, 56]. Although we excluded subjects with known auto-immune diseases, we cannot rule out the presence of other inflammatory conditions, such as infections. Future research should include a blood panel to check for infectious parameters at the time of sample collection. Furthermore, complement proteins are sensitive to variability in pre-analytical parameters [57], which could have affected our results, despite following standardised protocols for sample collection and processing. Finally, we were unable to quantify plasma C3b, C5a and C9 levels, presumably due to very low concentrations, highlighting the need for more sensitive assays.
Conclusions
In conclusion, we provide in vivo evidence of complement activation in genetic FTD, which might already occur in late-presymptomatic stages in conjunction with neuronal loss. Future longitudinal studies could elucidate at which stage of disease complement levels start to change, and might reveal their potential value as monitoring biomarkers [44].
Supplementary Information
Additional file 1: Table S1. Number of samples for each of the analytes in CSF and plasma. Table S2. Correlations between complement proteins and age. Table S3. Correlations between grey matter volume and (a) CSF and (b) plasma complement protein concentration. Table S4. Correlations between clinical measures of disease severity and (a) CSF and (b) plasma complement proteins. Table S5. Correlations between plasma complement factors. Table S6. Correlations between plasma complement proteins, neurofilament light chain (NfL) and glial fibrillary acidic protein (GFAP). Table S7. Complement protein levels of seven presymptomatic carriers who became symptomatic during follow-up (‘converters’). Figure S1. Correlations between CSF C1q, C3b and disease duration. P-values were derived from Spearman’s rho.
Acknowledgements
We thank all participants and their family members for taking part in this study. Several authors of this publication are members of the European Reference Network for Rare Neurological Diseases—Project ID no. 739510.
GENFI consortium author list: Sónia Afonso, Instituto Ciencias Nucleares Aplicadas a Saude, Universidade de Coimbra, Coimbra, Portugal; Maria Rosario Almeida, Faculty of Medicine, University of Coimbra, Coimbra, Portugal; Sarah Anderl-Straub, Department of Neurology, University of Ulm, Ulm, Germany; Christin Andersson, Department of Clinical Neuroscience, Karolinska Institutet, Stockholm, Sweden; Anna Antonell, Alzheimer’s disease and Other Cognitive Disorders Unit, Neurology Service, Hospital Clínic, Barcelona, Spain; Silvana Archetti, Biotechnology Laboratory, Department of Diagnostics, ASST Brescia Hospital, Brescia, Italy; Andrea Arighi, Fondazione IRCCS Ca’ Granda Ospedale Maggiore Policlinico, Neurodegenerative Diseases Unit, Milan, Italy; University of Milan, Centro Dino Ferrari, Milan, Italy; Mircea Balasa, Alzheimer’s disease and Other Cognitive Disorders Unit, Neurology Service, Hospital Clínic, Barcelona, Spain; Myriam Barandiaran, Cognitive Disorders Unit, Department of Neurology, Donostia University Hospital, San Sebastian, Gipuzkoa, Spain; Neuroscience Area, Biodonostia Health Research Insitute, San Sebastian, Gipuzkoa, Spain; Nuria Bargalló, Imaging Diagnostic Center, Hospital Clínic, Barcelona, Spain; Robart Bartha, Department of Medical Biophysics, The University of Western Ontario, London, Ontario, Canada; Centre for Functional and Metabolic Mapping, Robarts Research Institute, The University of Western Ontario, London, Ontario, Canada; Benjamin Bender, Department of Diagnostic and Interventional Neuroradiology, University of Tübingen, Tübingen, Germany; Alberto Benussi, Centre for Neurodegenerative Disorders, Department of Clinical and Experimental Sciences, University of Brescia, Italy; Luisa Benussi, Istituto di Ricovero e Cura a Carattere Scientifico Istituto Centro San Giovanni di Dio Fatebenefratelli, Brescia, Italy; Valentina Bessi, Department of Neuroscience, Psychology, Drug Research and Child Health, University of Florence, Florence, Italy ; Giuliano Binetti, Istituto di Ricovero e Cura a Carattere Scientifico Istituto Centro San Giovanni di Dio Fatebenefratelli, Brescia, Italy; Sandra Black, Sunnybrook Health Sciences Centre, Sunnybrook Research Institute, University of Toronto, Toronto, Canada; Martina Bocchetta, Department of Neurodegenerative Disease, Dementia Research Centre, UCL Institute of Neurology, Queen Square, London, UK; Sergi Borrego-Ecija, Alzheimer’s disease and Other Cognitive Disorders Unit, Neurology Service, Hospital Clínic, Barcelona, Spain; Jose Bras, Center for Neurodegenerative Science, Van Andel Institute, Grand Rapids, Michigan, MI 49503, USA; Rose Bruffaerts, Laboratory for Cognitive Neurology, Department of Neurosciences, KU Leuven, Leuven, Belgium; Marta Cañada, CITA Alzheimer, San Sebastian, Gipuzkoa, Spain; Valentina Cantoni, Centre for Neurodegenerative Disorders, Neurology Unit, Department of Clinical and Experimental Sciences, University of Brescia, Brescia, Italy; Paola Caroppo, Fondazione IRCCS Istituto Neurologico Carlo Besta, Milano, Italy; David Cash, Department of Neurodegenerative Disease, Dementia Research Centre, UCL Institute of Neurology, Queen Square, London, UK; Miguel Castelo-Branco, Faculty of Medicine, University of Coimbra, Coimbra, Portugal; Rhian Convery, Department of Neurodegenerative Disease, Dementia Research Centre, UCL Institute of Neurology, Queen Square, London, UK; Thomas Cope, Department of Clinical Neuroscience, University of Cambridge, Cambridge, UK; Giuseppe Di Fede, Fondazione IRCCS Istituto Neurologico Carlo Besta, Milano, Italy; Alina Díez, Neuroscience Area, Biodonostia Health Research Insitute, San Sebastian, Gipuzkoa, Spain; Diana Duro, Faculty of Medicine, University of Coimbra, Coimbra, Portugal; Chiara Fenoglio, Fondazione IRCCS Ca’ Granda Ospedale Maggiore Policlinico, Neurodegenerative Diseases Unit, Milan, Italy; University of Milan, Centro Dino Ferrari, Milan, Italy; Camilla Ferrari, Department of Neuroscience, Psychology, Drug Research and Child Health, University of Florence, Florence, Italy; Catarina B. Ferreira, Laboratory of Neurosciences, Institute of Molecular Medicine, Faculty of Medicine, University of Lisbon, Lisbon, Portugal; Nick Fox, Department of Neurodegenerative Disease, Dementia Research Centre, UCL Institute of Neurology, Queen Square, London, UK; Morris Freedman, Baycrest Health Sciences, Rotman Research Institute, University of Toronto, Toronto, Canada; Giorgio Fumagalli, Fondazione IRCCS Ca’ Granda Ospedale Maggiore Policlinico, Neurodegenerative Diseases Unit, Milan, Italy; University of Milan, Centro Dino Ferrari, Milan, Italy; Alazne Gabilondo, Neuroscience Area, Biodonostia Health Research Insitute, San Sebastian, Gipuzkoa, Spain; Roberto Gasparotti, Neuroradiology Unit, University of Brescia, Brescia, Italy; Serge Gauthier, Alzheimer Disease Research Unit, McGill Centre for Studies in Aging, Department of Neurology & Neurosurgery, McGill University, Montreal, Québec, Canada; Stefano Gazzina, Neurology, ASST Brescia Hospital, Brescia, Italy; Giorgio Giaccone, Fondazione IRCCS Istituto Neurologico Carlo Besta, Milano, Italy; Ana Gorostidi, Neuroscience Area, Biodonostia Health Research Insitute, San Sebastian, Gipuzkoa, Spain; Caroline Greaves, Department of Neurodegenerative Disease, Dementia Research Centre, UCL Institute of Neurology, Queen Square, London, UK; Rita Guerreiro, Center for Neurodegenerative Science, Van Andel Institute, Grand Rapids, Michigan, MI 49503, USA; Tobias Hoegen, Neurologische Klinik, Ludwig-Maximilians-Universität München, Munich, Germany; Begoña Indakoetxea, Cognitive Disorders Unit, Department of Neurology, Donostia University Hospital, San Sebastian, Gipuzkoa, Spain; Neuroscience Area, Biodonostia Health Research Insitute, San Sebastian, Gipuzkoa, Spain; Vesna Jelic, Division of Clinical Geriatrics, Karolinska Institutet, Stockholm, Sweden; Hans-Otto Karnath, Division of Neuropsychology, Hertie-Institute for Clinical Brain Research and Center of Neurology, University of Tübingen, Tübingen, Germany; Ron Keren, The University Health Network, Toronto Rehabilitation Institute, Toronto, Canada; Tobias Langheinrich, Manchester Centre for Clinical Neurosciences, Department of Neurology, Salford Royal NHS Foundation Trust, Manchester, UK;; Manchester Centre for Clinical Neurosciences, Department of Neurology, Salford Royal NHS Foundation Trust, Manchester, UK; Maria João Leitão, Centre of Neurosciences and Cell Biology, Universidade de Coimbra, Coimbra, Portugal; Albert Lladó, Alzheimer’s disease and Other Cognitive Disorders Unit, Neurology Service, Hospital Clínic, Barcelona, Spain; Gemma Lombardi, Department of Neuroscience, Psychology, Drug Research and Child Health, University of Florence, Florence, Italy; Sandra Loosli, Neurologische Klinik, Ludwig-Maximilians-Universität München, Munich, Germany; Carolina Maruta, Laboratory of Language Research, Centro de Estudos Egas Moniz, Faculty of Medicine, University of Lisbon, Lisbon, Portugal; Simon Mead, MRC Prion Unit, Department of Neurodegenerative Disease, UCL Institute of Neurology, Queen Square, London, UK; Gabriel Miltenberger, Faculty of Medicine, University of Lisbon, Lisbon, Portugal; Rick van Minkelen, Department of Clinical Genetics, Erasmus Medical Center, Rotterdam, Netherlands; Sara Mitchell, Sunnybrook Health Sciences Centre, Sunnybrook Research Institute, University of Toronto, Toronto, Canada; Katrina Moore, Department of Neurodegenerative Disease, Dementia Research Centre, UCL Institute of Neurology, Queen Square, London, UK; Benedetta Nacmias, Department of Neuroscience, Psychology, Drug Research and Child Health, University of Florence, Florence, Italy; Jennifer Nicholas, Department of Medical Statistics, London School of Hygiene and Tropical Medicine, London, UK; Linn Öijerstedt, Center for Alzheimer Research, Division of Neurogeriatrics, Department of Neurobiology, Care Sciences and Society, Bioclinicum, Karolinska Institutet, Solna, Sweden; Unit for Hereditary Dementias, Theme Aging, Karolinska University Hospital, Solna, Sweden ; Jaume Olives, Alzheimer’s disease and Other Cognitive Disorders Unit, Neurology Service, Hospital Clínic, Barcelona, Spain; Sebastien Ourselin, School of Biomedical Engineering & Imaging Sciences, King's College London, London, UK; Alessandro Padovani, Centre for Neurodegenerative Disorders, Department of Clinical and Experimental Sciences, University of Brescia, Italy; Georgia Peakman, Department of Neurodegenerative Disease, Dementia Research Centre, UCL Institute of Neurology, Queen Square, London, UK; Michela Pievani, Istituto di Ricovero e Cura a Carattere Scientifico Istituto Centro San Giovanni di Dio Fatebenefratelli, Brescia, Italy; Cristina Polito, Department of Biomedical, Experimental and Clinical Sciences “Mario Serio”, Nuclear Medicine Unit, University of Florence, Florence, Italy; Enrico Premi, Stroke Unit, ASST Brescia Hospital, Brescia, Italy; Sara Prioni, Fondazione IRCCS Istituto Neurologico Carlo Besta, Milano, Italy; Catharina Prix, Neurologische Klinik, Ludwig-Maximilians-Universität München, Munich, Germany; Rosa Rademakers, Department of Neurosciences, Mayo Clinic, Jacksonville, Florida, USA; Veronica Redaelli, Fondazione IRCCS Istituto Neurologico Carlo Besta, Milano, Italy; Tim Rittman, Department of Clinical Neurosciences, University of Cambridge, Cambridge, UK; Ekaterina Rogaeva, Tanz Centre for Research in Neurodegenerative Diseases, University of Toronto, Toronto, Canada; Pedro Rosa-Neto, Translational Neuroimaging Laboratory, McGill Centre for Studies in Aging, McGill University, Montreal, Québec, Canada; Giacomina Rossi, Fondazione IRCCS Istituto Neurologico Carlo Besta, Milano, Italy; Martin Rosser, Department of Neurodegenerative Disease, Dementia Research Centre, UCL Institute of Neurology, Queen Square, London, UK; Beatriz Santiago, Neurology Department, Centro Hospitalar e Universitario de Coimbra, Coimbra, Portugal; Elio Scarpini, Fondazione IRCCS Ca’ Granda Ospedale Maggiore Policlinico, Neurodegenerative Diseases Unit, Milan, Italy; University of Milan, Centro Dino Ferrari, Milan, Italy; Sonja Schönecker, Neurologische Klinik, Ludwig-Maximilians-Universität München, Munich, Germany; Elisa Semler, Department of Neurology, University of Ulm, Ulm; Rachelle Shafei, Department of Neurodegenerative Disease, Dementia Research Centre, UCL Institute of Neurology, Queen Square, London, UK; Christen Shoesmith, Department of Clinical Neurological Sciences, University of Western Ontario, London, Ontario, Canada; Miguel Tábuas-Pereira, Neurology Department, Centro Hospitalar e Universitario de Coimbra, Coimbra, Portugal; Mikel Tainta, Neuroscience Area, Biodonostia Health Research Insitute, San Sebastian, Gipuzkoa, Spain; Ricardo Taipa, Neuropathology Unit and Department of Neurology, Centro Hospitalar do Porto—Hospital de Santo António, Oporto, Portugal; David Tang-Wai, The University Health Network, Krembil Research Institute, Toronto, Canada; David L Thomas, Neuroimaging Analysis Centre, Department of Brain Repair and Rehabilitation, UCL Institute of Neurology, Queen Square, London, UK; Paul Thompson, Division of Neuroscience and Experimental Psychology, Wolfson Molecular Imaging Centre, University of Manchester, Manchester, UK; Hakan Thonberg, Center for Alzheimer Research, Division of Neurogeriatrics, Karolinska Institutet, Stockholm, Sweden; Carolyn Timberlake, Department of Clinical Neurosciences, University of Cambridge, Cambridge, UK; Pietro Tiraboschi, Fondazione IRCCS Istituto Neurologico Carlo Besta, Milano, Italy; Emily Todd, Department of Neurodegenerative Disease, Dementia Research Centre, UCL Institute of Neurology, Queen Square, London, UK; Philip Van Damme, Neurology Service, University Hospitals Leuven, Belgium; Laboratory for Neurobiology, VIB-KU Leuven Centre for Brain Research, Leuven, Belgium; Mathieu Vandenbulcke, Geriatric Psychiatry Service, University Hospitals Leuven, Belgium; Neuropsychiatry, Department of Neurosciences, KU Leuven, Leuven, Belgium; Michele Veldsman, Nuffield Department of Clinical Neurosciences, Medical Sciences Division, University of Oxford, Oxford, UK; Ana Verdelho, Department of Neurosciences and Mental Health, Centro Hospitalar Lisboa Norte—Hospital de Santa Maria & Faculty of Medicine, University of Lisbon, Lisbon, Portugal; Jorge Villanua, OSATEK, University of Donostia, San Sebastian, Gipuzkoa, Spain; Jason Warren, Department of Neurodegenerative Disease, Dementia Research Centre, UCL Institute of Neurology, Queen Square, London, UK; Carlo Wilke, Department of Neurodegenerative Diseases, Hertie-Institute for Clinical Brain Research and Center of Neurology, University of Tübingen, Tübingen, Germany; Center for Neurodegenerative Diseases (DZNE), Tübingen, Germany; Ione Woollacott, Department of Neurodegenerative Disease, Dementia Research Centre, UCL Institute of Neurology, Queen Square, London, UK; Elisabeth Wlasich, Neurologische Klinik, Ludwig-Maximilians-Universität München, Munich, Germany; Miren Zulaica, Neuroscience Area, Biodonostia Health Research Insitute, San Sebastian, Gipuzkoa, Spain
Abbreviations
- AD
Alzheimer’s disease
- ALS
Amyotrophic lateral sclerosis
- bvFTD
Behavioural variant FTD
- C9orf72
Chromosome 9 open reading frame 72
- CDR® + NACC FLTD-SB
Clinical Dementia Rating scale plus NACC frontotemporal lobar degeneration—sum of boxes
- CNS
Central nervous system
- CSF
Cerebrospinal fluid
- CV
Coefficient of variation
- FTD
Frontotemporal dementia
- GENFI
Genetic frontotemporal dementia initiative
- GFAP
Glial fibrillary acidic protein
- GRN
Granulin
- LLOQ
Lower limit of quantification
- MAPT
Microtubule-associated protein tau
- MBL
Mannose-binding lectin
- MMSE
Mini Mental State Examination
- NfL
Neurofilament light chain
- PPA
Primary progressive aphasia
- PSP
Progressive supranuclear palsy
- TCC
Terminal complement complex
- TIV
Total intracranial volume
- ULOQ
Upper limit of quantification
- WBV
Whole brain volume
Author contributions
ELvdE, CH, ASE, IJS, DM and EH performed biomarker measurements. ELvdE, CH, EH, JCvS, JDR and HS contributed to data acquisition, conception and design of the study. ELvdE and HS contributed to statistical analyses. ELvdE, HS, JCvS and JDR contributed to drafting of the manuscript and figures. The remaining authors recruited patients and collected data. All authors critically reviewed the manuscript, read and approved the final manuscript.
Funding
This study was supported in the Netherlands by Memorabel grants from Deltaplan Dementie (ZonMw and Alzheimer Nederland; grant numbers 733050813, 733050103, 733050513), the Bluefield Project to Cure Frontotemporal Dementia, the Dioraphte foundation (grant number 1402 1300), and the European Joint Programme—Neurodegenerative Disease Research and the Netherlands Organisation for Health Research and Development (PreFrontALS: 733051042, RiMod-FTD: 733051024); in Belgium by the Mady Browaeys Fonds voor Onderzoek naar Frontotemporale Degeneratie; in the UK by the MRC UK GENFI grant (MR/M023664/1) and the JPND GENFI-PROX grant (2019-02248); JDR is supported by an MRC Clinician Scientist Fellowship (MR/M008525/1) and has received funding from the NIHR Rare Disease Translational Research Collaboration (BRC149/NS/MH); ASE supported by the UK Dementia Research Institute which receives its funding from DRI Ltd, funded by the UK Medical Research Council, Alzheimer’s Society and Alzheimer’s Research UK; IJS is supported by the Alzheimer’s Association; JBR is supported by the Wellcome Trust (103838); in Spain by the Fundació Marató de TV3 (20143810 to RSV); in Germany by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under Germany’s Excellence Strategy within the framework of the Munich Cluster for Systems Neurology (EXC 2145 SyNergy—ID 390857198) and by grant 779357 “Solve-RD” from the Horizon 2020 Research and Innovation Programme (to MS); in Sweden by grants from the Swedish FTD Initiative funded by the Schörling Foundation, grants from JPND PreFrontALS Swedish Research Council (VR) 529–2014-7504, Swedish Research Council (VR) 2015–02926, Swedish Research Council (VR) 2018–02754, Swedish Brain Foundation, Swedish Alzheimer Foundation, Stockholm County Council ALF, Swedish Demensfonden, Stohnes foundation, Gamla Tjänarinnor, Karolinska Institutet Doctoral Funding, and StratNeuro. HZ is a Wallenberg Scholar.
Availability of data and materials
The raw data of this project are part of GENFI. De-identified patient data can be accessed upon reasonable request to genfi@ucl.ac.uk.
Declarations
Ethics approval and consent to participate
Local ethics committees at each site approved the study, and all participants provided written informed consent.
Consent for publication
Not applicable.
Competing interests
JL reports speaker fees from Bayer Vital and Roche, consulting fees from Axon Neuroscience, author fees from Thieme medical publishers and W. Kohlhammer GmbH medical publishers, non-financial support from Abbvie and compensation for duty as part-time CMO from MODAG, outside the submitted work. HZ has served at scientific advisory boards for Alector, Eisai, Denali, Roche Diagnostics, Wave, Samumed, Siemens Healthineers, Pinteon Therapeutics, Nervgen, AZTherapies and CogRx, has given lectures in symposia sponsored by Cellectricon, Fujirebio, Alzecure and Biogen, and is a co-founder of Brain Biomarker Solutions in Gothenburg AB (BBS), which is a part of the GU Ventures Incubator Program. The other authors report no competing interests relevant to this study.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Emma L. van der Ende and Carolin Heller contributed equally
Jonathan D. Rohrer and Harro Seelaar contributed equally
Contributor Information
Emma L. van der Ende, Email: e.l.vanderende@amsterdamumc.nl
Carolin Heller, Email: c.heller@ucl.ac.uk.
Aitana Sogorb-Esteve, Email: a.sogorb-esteve@ucl.ac.uk.
Imogen J. Swift, Email: i.swift.17@ucl.ac.uk
David McFall, Email: david.mcfall@ucsf.edu.
Georgia Peakman, Email: georgia.peakman.18@ucl.ac.uk.
Arabella Bouzigues, Email: arabella.bouzigues.18@ucl.ac.uk.
Jackie M. Poos, Email: j.m.poos@erasmusmc.nl
Lize C. Jiskoot, Email: l.c.jiskoot@erasmusmc.nl
Jessica L. Panman, Email: panman@essb.eur.nl
Janne M. Papma, Email: j.papma@erasmusmc.nl
Lieke H. Meeter, Email: h.meeter@erasmusmc.nl
Elise G. P. Dopper, Email: e.dopper@erasmusmc.nl
Martina Bocchetta, Email: m.bocchetta@ucl.ac.uk.
Emily Todd, Email: emily.todd.18@ucl.ac.uk.
David Cash, Email: d.cash@ucl.ac.uk.
Caroline Graff, Email: caroline.graff@ki.se.
Matthis Synofzik, Email: matthis.synofzik@uni-tuebingen.de.
Fermin Moreno, Email: fermin.morenoizco@gmail.com.
Elizabeth Finger, Email: elizabeth.finger@lhsc.on.ca.
Raquel Sanchez-Valle, Email: rsanchez@clinic.cat.
Rik Vandenberghe, Email: rik.vandenberghe@uzleuven.be.
Robert Laforce, Jr, Email: robert.laforce@fmed.ulaval.ca.
Mario Masellis, Email: mario.masellis@utoronto.ca.
Maria Carmela Tartaglia, Email: carmela.tartaglia@uhn.ca.
James B. Rowe, Email: james.rowe@mrc-cbu.cam.ac.uk
Chris Butler, Email: chris.butler@ndcn.ox.ac.uk.
Simon Ducharme, Email: simon.ducharme@mcgill.ca.
Alexander Gerhard, Email: Alex.Gerhard@manchester.ac.uk.
Adrian Danek, Email: adrian.danek@med.uni-muenchen.de.
Johannes Levin, Email: johannes.levin@med.uni-muenchen.de.
Yolande A. L. Pijnenburg, Email: yal.pijnenburg@amsterdamumc.nl
Markus Otto, Email: markus.otto@uni-ulm.de.
Barbara Borroni, Email: bborroni@inwind.it.
Fabrizio Tagliavini, Email: fabrizio.tagliavini@istituto-besta.it.
Alexandre de Mendonça, Email: mendonca@medicina.ulisboa.pt.
Isabel Santana, Email: isabeljsantana@gmail.com.
Daniela Galimberti, Email: daniela.galimberti@unimi.it.
Sandro Sorbi, Email: sandro.sorbi@unifi.it.
Henrik Zetterberg, Email: henrik.zetterberg@clinichem.gu.se.
Eric Huang, Email: eric.huang2@ucsf.edu.
John C. van Swieten, Email: j.c.vanswieten@erasmusmc.nl
Jonathan D. Rohrer, Email: j.rohrer@ucl.ac.uk
Harro Seelaar, Email: h.seelaar@erasmusmc.nl.
the Genetic Frontotemporal Dementia Initiative (GENFI):
Sónia Afonso, Maria Rosario Almeida, Sarah Anderl-Straub, Christin Andersson, Anna Antonell, Silvana Archetti, Andrea Arighi, Mircea Balasa, Myriam Barandiaran, Nuria Bargalló, Robart Bartha, Benjamin Bender, Alberto Benussi, Luisa Benussi, Valentina Bessi, Giuliano Binetti, Sandra Black, Martina Bocchetta, Sergi Borrego-Ecija, Jose Bras, Rose Bruffaerts, Marta Cañada, Valentina Cantoni, Paola Caroppo, David Cash, Miguel Castelo-Branco, Rhian Convery, Thomas Cope, Giuseppe Di Fede, Alina Díez, Diana Duro, Chiara Fenoglio, Camilla Ferrari, Catarina B. Ferreira, Nick Fox, Morris Freedman, Giorgio Fumagalli, Alazne Gabilondo, Roberto Gasparotti, Serge Gauthier, Stefano Gazzina, Giorgio Giaccone, Ana Gorostidi, Caroline Greaves, Rita Guerreiro, Tobias Hoegen, Begoña Indakoetxea, Vesna Jelic, Hans-Otto Karnath, Ron Keren, Tobias Langheinrich, Maria João Leitão, Albert Lladó, Gemma Lombardi, Sandra Loosli, Carolina Maruta, Simon Mead, Gabriel Miltenberger, Rick van Minkelen, Sara Mitchell, Katrina Moore, Benedetta Nacmias, Jennifer Nicholas, Linn Öijerstedt, Jaume Olives, Sebastien Ourselin, Alessandro Padovani, Georgia Peakman, Michela Pievani, Cristina Polito, Enrico Premi, Sara Prioni, Catharina Prix, Rosa Rademakers, Veronica Redaelli, Tim Rittman, Ekaterina Rogaeva, Pedro Rosa-Neto, Giacomina Rossi, Martin Rosser, Beatriz Santiago, Elio Scarpini, Sonja Schönecker, Elisa Semler, Rachelle Shafei, Christen Shoesmith, Miguel Tábuas-Pereira, Mikel Tainta, Ricardo Taipa, David Tang-Wai, David L. Thomas, Paul Thompson, Hakan Thonberg, Carolyn Timberlake, Pietro Tiraboschi, Emily Todd, Philip Van Damme, Mathieu Vandenbulcke, Michele Veldsman, Ana Verdelho, Jorge Villanua, Jason Warren, Carlo Wilke, Ione Woollacott, Elisabeth Wlasich, and Miren Zulaica
References
- 1.Lashley T, Rohrer JD, Mead S, Revesz T. Review: an update on clinical, genetic and pathological aspects of frontotemporal lobar degenerations. Neuropathol Appl Neurobiol. 2015;41(7):858–881. doi: 10.1111/nan.12250. [DOI] [PubMed] [Google Scholar]
- 2.Seelaar H, Rohrer JD, Pijnenburg YA, Fox NC, van Swieten JC. Clinical, genetic and pathological heterogeneity of frontotemporal dementia: a review. J Neurol Neurosurg Psychiatry. 2011;82(5):476–486. doi: 10.1136/jnnp.2010.212225. [DOI] [PubMed] [Google Scholar]
- 3.Bright F, Werry EL, Dobson-Stone C, Piguet O, Ittner LM, Halliday GM, et al. Neuroinflammation in frontotemporal dementia. Nat Rev Neurol. 2019;15(9):540–555. doi: 10.1038/s41582-019-0231-z. [DOI] [PubMed] [Google Scholar]
- 4.Bossu P, Salani F, Alberici A, Archetti S, Bellelli G, Galimberti D, et al. Loss of function mutations in the progranulin gene are related to pro-inflammatory cytokine dysregulation in frontotemporal lobar degeneration patients. J Neuroinflamm. 2011;8:65. doi: 10.1186/1742-2094-8-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Galimberti D, Bonsi R, Fenoglio C, Serpente M, Cioffi SM, Fumagalli G, et al. Inflammatory molecules in Frontotemporal Dementia: cerebrospinal fluid signature of progranulin mutation carriers. Brain Behav Immun. 2015;49:182–187. doi: 10.1016/j.bbi.2015.05.006. [DOI] [PubMed] [Google Scholar]
- 6.Galimberti D, Schoonenboom N, Scheltens P, Fenoglio C, Venturelli E, Pijnenburg YA, et al. Intrathecal chemokine levels in Alzheimer disease and frontotemporal lobar degeneration. Neurology. 2006;66(1):146–147. doi: 10.1212/01.wnl.0000191324.08289.9d. [DOI] [PubMed] [Google Scholar]
- 7.Galimberti D, Venturelli E, Fenoglio C, Guidi I, Villa C, Bergamaschini L, et al. Intrathecal levels of IL-6, IL-11 and LIF in Alzheimer's disease and frontotemporal lobar degeneration. J Neurol. 2008;255(4):539–544. doi: 10.1007/s00415-008-0737-6. [DOI] [PubMed] [Google Scholar]
- 8.Gibbons L, Rollinson S, Thompson JC, Robinson A, Davidson YS, Richardson A, et al. Plasma levels of progranulin and interleukin-6 in frontotemporal lobar degeneration. Neurobiol Aging. 2015;36(3):1603 e1-4. doi: 10.1016/j.neurobiolaging.2014.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oeckl P, Weydt P, Steinacker P, Anderl-Straub S, Nordin F, Volk AE, et al. Different neuroinflammatory profile in amyotrophic lateral sclerosis and frontotemporal dementia is linked to the clinical phase. J Neurol Neurosurg Psychiatry. 2019;90(1):4–10. doi: 10.1136/jnnp-2018-318868. [DOI] [PubMed] [Google Scholar]
- 10.Rentzos M, Paraskevas GP, Kapaki E, Nikolaou C, Zoga M, Rombos A, et al. Interleukin-12 is reduced in cerebrospinal fluid of patients with Alzheimer's disease and frontotemporal dementia. J Neurol Sci. 2006;249(2):110–114. doi: 10.1016/j.jns.2006.05.063. [DOI] [PubMed] [Google Scholar]
- 11.Van der Ende EL, Morenas-Rodríguez E, McMillan CT, Grossman M, Irwin D, Sanchez-Valle R, et al. CSF sTREM2 is elevated in a subset in GRN-related frontotemporal dementia. Neurobiol Aging. 2021. [DOI] [PubMed]
- 12.Woollacott IOC, Nicholas JM, Heller C, Foiani MS, Moore KM, Russell LL, et al. Cerebrospinal fluid YKL-40 and chitotriosidase levels in frontotemporal dementia vary by clinical, genetic and pathological subtype. Dement Geriatr Cogn Disord. 2020;49(1):56–76. doi: 10.1159/000506282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Woollacott IOC, Nicholas JM, Heslegrave A, Heller C, Foiani MS, Dick KM, et al. Cerebrospinal fluid soluble TREM2 levels in frontotemporal dementia differ by genetic and pathological subgroup. Alzheimers Res Ther. 2018;10(1):79. doi: 10.1186/s13195-018-0405-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rutkowski MJ, Sughrue ME, Kane AJ, Mills SA, Fang S, Parsa AT. Complement and the central nervous system: emerging roles in development, protection and regeneration. Immunol Cell Biol. 2010;88(8):781–786. doi: 10.1038/icb.2010.48. [DOI] [PubMed] [Google Scholar]
- 15.Lee JD, Coulthard LG, Woodruff TM. Complement dysregulation in the central nervous system during development and disease. Semin Immunol. 2019;45:101340. doi: 10.1016/j.smim.2019.101340. [DOI] [PubMed] [Google Scholar]
- 16.Shi Q, Colodner KJ, Matousek SB, Merry K, Hong S, Kenison JE, et al. Complement C3-deficient mice fail to display age-related hippocampal decline. J Neurosci. 2015;35(38):13029–13042. doi: 10.1523/JNEUROSCI.1698-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dejanovic B, Huntley MA, De Mazière A, Meilandt WJ, Wu T, Srinivasan K, et al. Changes in the synaptic proteome in tauopathy and rescue of Tau-induced synapse loss by C1q antibodies. Neuron. 2018;100(6):1322–36 e7. doi: 10.1016/j.neuron.2018.10.014. [DOI] [PubMed] [Google Scholar]
- 18.Hong S, Beja-Glasser VF, Nfonoyim BM, Frouin A, Li S, Ramakrishnan S, et al. Complement and microglia mediate early synapse loss in Alzheimer mouse models. Science. 2016;352(6286):712–716. doi: 10.1126/science.aad8373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krukowski K, Chou A, Feng X, Tiret B, Paladini MS, Riparip LK, et al. Traumatic brain injury in aged mice induces chronic microglia activation, synapse loss, and complement-dependent memory deficits. Int J Mol Sci. 2018 doi: 10.3390/ijms19123753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schafer DP, Lehrman EK, Kautzman AG, Koyama R, Mardinly AR, Yamasaki R, et al. Microglia sculpt postnatal neural circuits in an activity and complement-dependent manner. Neuron. 2012;74(4):691–705. doi: 10.1016/j.neuron.2012.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lui H, Zhang J, Makinson SR, Cahill MK, Kelley KW, Huang HY, et al. Progranulin deficiency promotes circuit-specific synaptic pruning by microglia via complement activation. Cell. 2016;165(4):921–935. doi: 10.1016/j.cell.2016.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang J, Velmeshev D, Hashimoto K, Huang YH, Hofmann JW, Shi X, et al. Neurotoxic microglia promote TDP-43 proteinopathy in progranulin deficiency. Nature. 2020;588(7838):459–465. doi: 10.1038/s41586-020-2709-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bonham LW, Desikan RS, Yokoyama JS, Alzheimer’s Disease Neuroimaging I The relationship between complement factor C3, APOE ε4, amyloid and tau in Alzheimer's disease. Acta Neuropathol Commun. 2016;4(1):65. doi: 10.1186/s40478-016-0339-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Daborg J, Andreasson U, Pekna M, Lautner R, Hanse E, Minthon L, et al. Cerebrospinal fluid levels of complement proteins C3, C4 and CR1 in Alzheimer's disease. J Neural Transm (Vienna) 2012;119(7):789–797. doi: 10.1007/s00702-012-0797-8. [DOI] [PubMed] [Google Scholar]
- 25.Goetzl EJ, Schwartz JB, Abner EL, Jicha GA, Kapogiannis D. High complement levels in astrocyte-derived exosomes of Alzheimer disease. Ann Neurol. 2018;83(3):544–552. doi: 10.1002/ana.25172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morgan AR, Touchard S, Leckey C, O'Hagan C, Nevado-Holgado AJ, Consortium N, et al. Inflammatory biomarkers in Alzheimer's disease plasma. Alzheimers Dement. 2019;15(6):776–787. doi: 10.1016/j.jalz.2019.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muenchhoff J, Poljak A, Thalamuthu A, Gupta VB, Chatterjee P, Raftery M, et al. Changes in the plasma proteome at asymptomatic and symptomatic stages of autosomal dominant Alzheimer's disease. Sci Rep. 2016;6:29078. doi: 10.1038/srep29078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Winston CN, Goetzl EJ, Schwartz JB, Elahi FM, Rissman RA. Complement protein levels in plasma astrocyte-derived exosomes are abnormal in conversion from mild cognitive impairment to Alzheimer's disease dementia. Alzheimers Dement (Amst) 2019;11:61–66. doi: 10.1016/j.dadm.2018.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ganesalingam J, An J, Shaw CE, Shaw G, Lacomis D, Bowser R. Combination of neurofilament heavy chain and complement C3 as CSF biomarkers for ALS. J Neurochem. 2011;117(3):528–537. doi: 10.1111/j.1471-4159.2011.07224.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goldknopf IL, Sheta EA, Bryson J, Folsom B, Wilson C, Duty J, et al. Complement C3c and related protein biomarkers in amyotrophic lateral sclerosis and Parkinson's disease. Biochem Biophys Res Commun. 2006;342(4):1034–1039. doi: 10.1016/j.bbrc.2006.02.051. [DOI] [PubMed] [Google Scholar]
- 31.Gonzalez-Garza MT, Martinez HR, Cruz-Vega DE, Hernandez-Torre M, Moreno-Cuevas JE. Adipsin, MIP-1b, and IL-8 as CSF biomarker panels for ALS diagnosis. Dis Markers. 2018;2018:3023826. doi: 10.1155/2018/3023826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kitamura Y, Kojima M, Kurosawa T, Sasaki R, Ichihara S, Hiraku Y, et al. Proteomic profiling of exosomal proteins for blood-based biomarkers in Parkinson's disease. Neuroscience. 2018;392:121–128. doi: 10.1016/j.neuroscience.2018.09.017. [DOI] [PubMed] [Google Scholar]
- 33.Petzold A. Neurofilament phosphoforms: surrogate markers for axonal injury, degeneration and loss. J Neurol Sci. 2005;233(1–2):183–198. doi: 10.1016/j.jns.2005.03.015. [DOI] [PubMed] [Google Scholar]
- 34.van der Ende EL, Meeter LH, Poos JM, Panman JL, Jiskoot LC, Dopper EGP, et al. Serum neurofilament light chain in genetic frontotemporal dementia: a longitudinal, multicentre cohort study Lancet Neurol. 2019;18(12):1103–11. [DOI] [PubMed]
- 35.Colangelo AM, Alberghina L, Papa M. Astrogliosis as a therapeutic target for neurodegenerative diseases. Neurosci Lett. 2014;565:59–64. doi: 10.1016/j.neulet.2014.01.014. [DOI] [PubMed] [Google Scholar]
- 36.Heller C, Foiani MS, Moore K, Convery R, Bocchetta M, Neason M, et al. Plasma glial fibrillary acidic protein is raised in progranulin-associated frontotemporal dementia. J Neurol Neurosurg Psychiatry. 2020. [DOI] [PubMed]
- 37.Rohrer JD, Nicholas JM, Cash DM, van Swieten J, Dopper E, Jiskoot L, et al. Presymptomatic cognitive and neuroanatomical changes in genetic frontotemporal dementia in the Genetic Frontotemporal dementia Initiative (GENFI) study: a cross-sectional analysis. Lancet Neurol. 2015;14(3):253–262. doi: 10.1016/S1474-4422(14)70324-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ekdahl KN, Persson B, Mohlin C, Sandholm K, Skattum L, Nilsson B. Interpretation of serological complement biomarkers in disease. Front Immunol. 2018;9:2237. doi: 10.3389/fimmu.2018.02237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rascovsky K, Hodges JR, Knopman D, Mendez MF, Kramer JH, Neuhaus J, et al. Sensitivity of revised diagnostic criteria for the behavioural variant of frontotemporal dementia. Brain. 2011;134(Pt 9):2456–2477. doi: 10.1093/brain/awr179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gorno-Tempini ML, Hillis AE, Weintraub S, Kertesz A, Mendez M, Cappa SF, et al. Classification of primary progressive aphasia and its variants. Neurology. 2011;76(11):1006–1014. doi: 10.1212/WNL.0b013e31821103e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brooks BR, Miller RG, Swash M, Munsat TL, World Federation of Neurology Research Group on Motor Neuron D El Escorial revisited: revised criteria for the diagnosis of amyotrophic lateral sclerosis. Amyotroph Lateral Scler Other Motor Neuron Disord. 2000;1(5):293–299. doi: 10.1080/146608200300079536. [DOI] [PubMed] [Google Scholar]
- 42.Miyagawa T, Brushaber D, Syrjanen J, Kremers W, Fields J, Forsberg LK, et al. Use of the CDR(R) plus NACC FTLD in mild FTLD: data from the ARTFL/LEFFTDS consortium. Alzheimers Dement. 2019 doi: 10.1016/j.jalz.2019.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Malone IB, Leung KK, Clegg S, Barnes J, Whitwell JL, Ashburner J, et al. Accurate automatic estimation of total intracranial volume: a nuisance variable with less nuisance. Neuroimage. 2015;104:366–372. doi: 10.1016/j.neuroimage.2014.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Carpanini SM, Torvell M, Morgan BP. Therapeutic Inhibition of the complement system in diseases of the central nervous system. Front Immunol. 2019;10:362. doi: 10.3389/fimmu.2019.00362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liddelow SA, Guttenplan KA, Clarke LE, Bennett FC, Bohlen CJ, Schirmer L, et al. Neurotoxic reactive astrocytes are induced by activated microglia. Nature. 2017;541(7638):481–487. doi: 10.1038/nature21029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Luchena C, Zuazo-Ibarra J, Alberdi E, Matute C, Capetillo-Zarate E. Contribution of neurons and glial cells to complement-mediated synapse removal during development, aging and in Alzheimer's disease. Mediators Inflamm. 2018;2018:2530414. doi: 10.1155/2018/2530414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Veerhuis R. Histological and direct evidence for the role of complement in the neuroinflammation of AD. Curr Alzheimer Res. 2011;8(1):34–58. doi: 10.2174/156720511794604589. [DOI] [PubMed] [Google Scholar]
- 48.Cong Q, Soteros BM, Wollet M, Kim JH, Sia GM. The endogenous neuronal complement inhibitor SRPX2 protects against complement-mediated synapse elimination during development. Nat Neurosci. 2020;23(9):1067–1078. doi: 10.1038/s41593-020-0672-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dionisio-Santos DA, Olschowka JA, O'Banion MK. Exploiting microglial and peripheral immune cell crosstalk to treat Alzheimer's disease. J Neuroinflamm. 2019;16(1):74. doi: 10.1186/s12974-019-1453-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lall D, Lorenzini I, Mota TA, Bell S, Mahan TE, Ulrich JD, et al. C9orf72 deficiency promotes microglial-mediated synaptic loss in aging and amyloid accumulation. Neuron. 2021;109(14):2275–91 e8. doi: 10.1016/j.neuron.2021.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Roy ER, Wang B, Wan YW, Chiu G, Cole A, Yin Z, et al. Type I interferon response drives neuroinflammation and synapse loss in Alzheimer disease. J Clin Invest. 2020;130(4):1912–1930. doi: 10.1172/JCI133737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Litvinchuk A, Wan YW, Swartzlander DB, Chen F, Cole A, Propson NE, et al. Complement C3aR inactivation attenuates tau pathology and reverses an immune network deregulated in tauopathy models and Alzheimer's disease. Neuron. 2018;100(6):1337–1535. doi: 10.1016/j.neuron.2018.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ringman JM, Schulman H, Becker C, Jones T, Bai Y, Immermann F, et al. Proteomic changes in cerebrospinal fluid of presymptomatic and affected persons carrying familial Alzheimer disease mutations. Arch Neurol. 2012;69(1):96–104. doi: 10.1001/archneurol.2011.642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hakobyan S, Harding K, Aiyaz M, Hye A, Dobson R, Baird A, et al. Complement biomarkers as predictors of disease progression in Alzheimer's disease. J Alzheimers Dis. 2016;54(2):707–716. doi: 10.3233/JAD-160420. [DOI] [PubMed] [Google Scholar]
- 55.Liu D, Cao B, Zhao Y, Huang H, McIntyre RS, Rosenblat JD, et al. Soluble TREM2 changes during the clinical course of Alzheimer's disease: a meta-analysis. Neurosci Lett. 2018;686:10–16. doi: 10.1016/j.neulet.2018.08.038. [DOI] [PubMed] [Google Scholar]
- 56.Nilsson B, Hamad OA, Ahlström H, Kullberg J, Johansson L, Lindhagen L, et al. C3 and C4 are strongly related to adipose tissue variables and cardiovascular risk factors. Eur J Clin Invest. 2014;44(6):587–596. doi: 10.1111/eci.12275. [DOI] [PubMed] [Google Scholar]
- 57.Morgan AR, O'Hagan C, Touchard S, Lovestone S, Morgan BP. Effects of freezer storage time on levels of complement biomarkers. BMC Res Notes. 2017;10(1):559. doi: 10.1186/s13104-017-2885-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Number of samples for each of the analytes in CSF and plasma. Table S2. Correlations between complement proteins and age. Table S3. Correlations between grey matter volume and (a) CSF and (b) plasma complement protein concentration. Table S4. Correlations between clinical measures of disease severity and (a) CSF and (b) plasma complement proteins. Table S5. Correlations between plasma complement factors. Table S6. Correlations between plasma complement proteins, neurofilament light chain (NfL) and glial fibrillary acidic protein (GFAP). Table S7. Complement protein levels of seven presymptomatic carriers who became symptomatic during follow-up (‘converters’). Figure S1. Correlations between CSF C1q, C3b and disease duration. P-values were derived from Spearman’s rho.
Data Availability Statement
The raw data of this project are part of GENFI. De-identified patient data can be accessed upon reasonable request to genfi@ucl.ac.uk.