Abstract
Although general physiological functions have been ascribed to the high-molecular-weight penicillin binding proteins (PBPs) of Escherichia coli, the low-molecular-weight PBPs have no well-defined biological roles. When we examined the morphology of a set of E. coli mutants lacking multiple PBPs, we observed that strains expressing active PBP 5 produced cells of normal shape, while mutants lacking PBP 5 produced cells with altered diameters, contours, and topological features. These morphological effects were visible in untreated cells, but the defects were exacerbated in cells forced to filament by inactivation of PBP 3 or FtsZ. After filamentation, cellular diameter varied erratically along the length of individual filaments and many filaments exhibited extensive branching. Also, in general, the mean diameter of cells lacking PBP 5 was significantly increased compared to that of cells from isogenic strains expressing active PBP 5. Expression of cloned PBP 5 reversed the effects observed in ΔdacA mutants. Although deletion of PBP 5 was required for these phenotypes, the absence of additional PBPs magnified the effects. The greatest morphological alterations required that at least three PBPs in addition to PBP 5 be deleted from a single strain. In the extreme cases in which six or seven PBPs were deleted from a single mutant, cells and cell filaments expressing PBP 5 retained a normal morphology but cells and filaments lacking PBP 5 were aberrant. In no case did mutation of another PBP produce the same drastic morphological effects. We conclude that among the low-molecular-weight PBPs, PBP 5 plays a principle role in determining cell diameter, surface uniformity, and overall topology of the peptidoglycan sacculus.
Of the 12 known penicillin binding proteins (PBPs) of Escherichia coli, the only enzymes with known physiological roles are four of the five high-molecular-weight PBPs: 1a, 1b, 2, and 3 (10, 21). These four proteins polymerize new peptidoglycan strands and incorporate them into the existing sacculus, which forms the rigid layer of the cell envelope. In contrast, the seven low-molecular-weight (LMW) PBPs have no well-defined physiological functions, even though the enzymological knowledge of these proteins is well advanced.
Among the LMW proteins, PBP 5 has been studied in the most depth, but its biological function remains mysterious. PBP 5 is the major cellular dd-carboxypeptidase, an enzyme that removes the terminal d-alanine residue from the pentapeptide side chain of peptidoglycan (10, 12), thereby altering the types of cross-linking that can occur between glycan chains. Even though PBP 5 is the most numerous of the PBPs, its loss is not lethal (11, 20). One possible reason that PBP 5 is dispensable is that one or more other LMW PBPs might compensate for its absence. However, E. coli survives in the absence of all known dd-carboxypeptidases (3–5) and even in the absence of all seven LMW PBPs (4). This suggests that PBP 5 has no important physiological role or that its role is invisible in the laboratory environment. On the other hand, overproduction of PBP 5 is lethal, causing E. coli to grow as spheres before dying (9, 22), suggesting that PBP 5 may have a morphological effect on growth of the cell wall.
Recently, we constructed a set of E. coli mutants lacking every possible combination of eight PBPs (4). Among these mutants are gene combinations in which PBP 5 is present or absent in genetic backgrounds that differ only in the complement of other active PBPs. Some of these mutants exhibited severe morphological defects, especially when forced to filament by inactivating PBP 3 (4, 8). The PBP patterns of these defective mutants led us to ask which of the PBPs plays the dominant role in maintaining normal bacterial shape. We report here that, in the absence of multiple LMW PBPs, deletion of PBP 5 creates E. coli mutants with cell walls of abnormal diameter and contour. Furthermore, when cells lacking PBP 5 are forced to filament, the overall topology of the bacterial cell is severely affected, resulting in gross abnormalities and branching. Therefore, PBP 5 plays an important role in the genesis and/or maintenance of bacterial shape.
MATERIALS AND METHODS
Strains and growth conditions.
The E. coli strains used in this work are listed in Table 1. pFAD38, a plasmid carrying sulA (sfiA) under araP control, was provided by J. Lutkenhaus. pBAD24-Amp and pBAD18-Cam were provided by J. Beckwith. Strains were maintained on Luria-Bertani (LB) broth or agar plates (13) with appropriate antibiotic selection, as follows: chloramphenicol (20 μg/ml) or ampicillin (100 μg/ml). When necessary, glucose (0.1%) was added to inhibit expression from the pBAD promoter. Overnight cultures were diluted 1:200 into 50 ml of fresh LB medium and were incubated with vigorous shaking at 37°C until reaching an A550 of approximately 0.2. Cells were filamented by adding 10 μg of aztreonam (Leo Pharmaceuticals, Ballerup, Denmark) per ml or 2 μg of mitomycin C per ml. All chemicals were purchased from Sigma Chemical Co. (St. Louis, Mo.), unless otherwise noted.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Genotype or characteristic | PBP(s) deleted | Source or reference |
|---|---|---|---|
| Strains | |||
| E. coli | |||
| CS109 | W1485, λ− F−thi glnV (supE) rph-1 rpoS | None | C. Schnaitman |
| CS12-7 | CS109 dacA | 5 | 4 |
| CS211-2 | CS109 dacA dacC | 5, 6 | 4 |
| CS403-3 | CS109 mrcA dacB pbpG ampH | 1a, 4, 7, AmpH | 4 |
| CS408-1 | CS109 mrcA dacB dacC ampC | 1a, 4, 6, AmpC | 4 |
| CS502-2 | CS109 mrcA dacB pbpG ampC ampH | 1a, 4, 7, AmpC, AmpH | 4 |
| CS506-1 | CS109 mrcA dacB dacA dacC ampC | 1a, 4, 5, 6, AmpC | 4 |
| CS508-1 | CS109 mrcA dacB dacC pbpG ampC | 1a, 4, 6, 7, AmpC | 4 |
| CS509-3 | CS109 mrcA dacB dacA pbpG ampH | 1a, 4, 5, 7, AmpC | 4 |
| CS531-3 | CS109 dacB dacA dacC pbpG ampH | 4, 5, 6, 7, AmpH | 4 |
| CS533-1 | CS109 dacB dacA dacC pbpG ampC | 4, 5, 6, 7, AmpC | 4 |
| CS534-1 | CS109 dacB dacA pbpG ampC ampH | 4, 5, 7, AmpC, AmpH | 4 |
| CS535-1 | CS109 dacB dacC pbpG ampC ampH | 4, 6, 7, AmpC, AmpH | 4 |
| CS536-1 | CS109 dacA dacC pbpG ampC ampH | 5, 6, 7, AmpC, AmpH | 4 |
| CS539-1 | CS109 dacB dacA dacC ampC ampH | 4, 5, 6, AmpC, AmpH | 4 |
| CS601-3 | CS109 mrcA dacB dacA dacC pbpG ampH | 1a, 4, 5, 6, 7, AmpH | 4 |
| CS602-1 | CS109 mrcA dacB dacA dacC pbpG ampC | 1a, 4, 5, 6, 7, AmpC | 4 |
| CS603-2 | CS109 mrcA dacB dacC pbpG ampC ampH | 1a, 4, 5, 7, AmpC, AmpH | 4 |
| CS604-2 | CS109 mrcA dacB dacC pbpG ampC ampH | 1a, 4, 6, 7, AmpC, AmpH | 4 |
| CS605-4 | CS109 mrcA dacA dacC pbpG ampC ampH | 1a, 5, 6, 7, AmpC, AmpH | 4 |
| CS606-1 | CS109 mrcA dacB dacA dacC ampC ampH | 1a, 4, 5, 6, AmpC, AmpH | 4 |
| CS610-1 | CS109 mrcB dacB dacC pbpG ampC ampH | 1b, 4, 6, 7, AmpC, AmpH | 4 |
| CS612-1 | CS109 dacB dacA dacC pbpG ampC ampH | 4, 5, 6, 7, AmpC, AmpH | 4 |
| CS701-1 | CS109 mrcA dacB dacA dacC pbpG ampC ampH | 1a, 4, 5, 6, 7, AmpC, AmpH | 4 |
| CS702-1 | CS109 mrcB dacB dacA dacC pbpG ampC ampH | 1b, 4, 5, 6, 7, AmpC, AmpH | 4 |
| Plasmids | |||
| pBAD24-Amp | Arabinose-inducible cloning vector, Ampr | 7 | |
| pPJ5A | dacA cloned in MCSa of pBAD24-Amp, Ampr | This work | |
| pBAD18-Cam | Arabinose inducible cloning vector, Camr | 7 | |
| pPJ5C | dacA cloned in MCS of pBAD18-Cam, Camr | This work | |
| pFAD38 | SulA expression vector, Ampr | 1 |
MCS, multiple cloning site.
Molecular techniques.
Plasmid preparations, restriction digests, and ligations were performed as described previously (17). Competent strains were prepared by the calcium chloride technique and transformed by heat shock (17). CS109 chromosomal DNA was prepared by boiling 200 μl of overnight culture with 800 μl of distilled water for 10 min, followed by centrifugation at 14,000 × g for 1 min and collection of the supernatant. Oligonucleotide primers P1 (5′-GATCGAGAATTCGTCATGAATACCATTTTTCCGC-3′) and P2 (5′-GCATGCAAGCTTCTAGATTTTTAACCAAACCAGTGATG-3′) were purchased from Gibco Life Sciences (Grand Island, N.Y.) and were used to amplify the wild-type dacA gene from CS109 chromosomal DNA by PCR. An EcoRI-HindIII fragment of the dacA PCR product was ligated into pBAD24-Amp (7), creating the intermediate plasmid pPJ5A. An NheI-HindIII fragment was excised from pPJ5A and ligated into pBAD18-Cam (7), creating the dacA expression vector pPJ5C. The nucleotide sequence of the dacA gene in pPJ5C was confirmed by Sanger dideoxynucleotide sequencing (18). PBP 5 expression from pPJ5C was quantified by 125I-penicillin-X labeling followed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, as described previously (8). Restriction endonucleases, T4 ligase, and other molecular reagents were purchased from New England Biolabs (Beverly, Mass.).
Photography and data analysis.
Poly-l-lysine slides were prepared by briefly immersing microscope slides in poly-l-lysine (0.1% [wt/vol]; Sigma Chemical Co.) and letting them dry overnight. Bacteria were grown to exponential phase (A550 ≅ 0.2), and samples were prepared for microscopy by placing 3.5 μl of broth culture on poly-l-lysine-treated slides and covering each sample with a coverslip. Samples were viewed with a Nikon EFD-3 microscope with a 100× oil immersion objective and photographed with an attached SenSys charge-coupled device camera and capture software (Photometrics Ltd., Tucson, Ariz.) at 1,000× total magnification. Cell measurements and analyses were performed with Image Pro Plus version 3.01 software (Media Cybernetics, Silver Spring, Md.). Cell diameters of individual cells were calculated by measuring the cross-sectional area and dividing that value by the cell length. The reference measurement bar included on each photograph was calibrated using a slide-mounted 0.01-mm micrometer (Fisher Scientific, Chicago, Ill.). Statistical and graphical analyses were performed with SigmaPlot (SSPS, Chicago, Ill.) and Microsoft Excel (Microsoft Inc., Redmond, Wash.).
RESULTS
Previously, we reported that some E. coli mutants produced cells with abnormal diameters and uneven contours and cells which were prone to topological oddities such as branching (7). Therefore, we investigated several mutants lacking various PBPs to determine more specifically which of these proteins contributes most strongly to these morphological alterations.
Morphological aberrations in a mutant lacking seven PBPs.
When E. coli CS109 was incubated in the presence of aztreonam (which inhibits the septum-specific enzyme PBP 3), the resulting filamentous cells had a constant diameter equal to that of the original cell, producing regularly shaped elongated rods with a smooth outer contour (Fig. 1B). Somewhat surprisingly, aztreonam-induced filaments of strain CS604-2 were very similar to those of the wild-type parent even though the mutant lacked six PBPs (the deleted PBPs were 1a, 4, 6, 7, AmpC, and AmpH) (4) (Fig. 1D). In contrast, filaments of E. coli CS701-1 were grossly abnormal (Fig. 1F). The single difference between CS604-2 and CS701-1 is the deletion from the latter of the gene encoding PBP 5 (4).
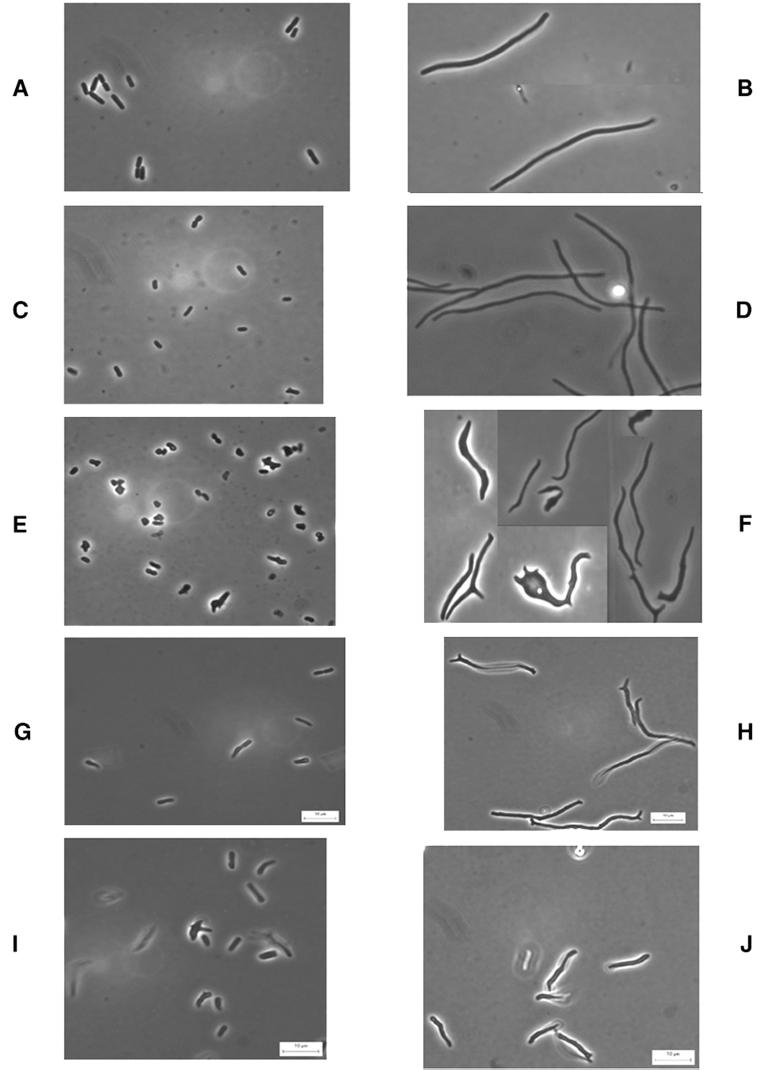
FIG. 1.
Morphology of PBP mutants before and after filamentation by aztreonam. Bacteria were grown in LB broth at 37°C until reaching an A550 of approximately 0.2, at which point 10 μg of aztreonam per ml was added. Samples were prepared for microscopy immediately before and 45 min after addition of the antibiotic. (A) CS109; (B) CS109 plus aztreonam; (C) CS604-2; (D) CS604-2 plus aztreonam; (E) CS701-1; (F) CS701-1 plus aztreonam; (G) CS601-3; (H) CS601-3 plus aztreonam; (I) CS612-1; (J) CS612-1 plus aztreonam.
Aztreonam-induced filaments of E. coli CS701-1 exhibited three types of morphological aberrations. First, filaments exhibited large cell-to-cell variations in diameter. Second, the diameter of an individual filament fluctuated along its length to a degree that was often extraordinary (Fig. 1F). We refer to this irregular diameter within an individual cell as a deviation in cell contour, to distinguish the phenotype from variations between different cells. The third aberration involved the overall shape of individual filaments. Many filaments produced one or more knobs, branches, or various outgrowths that were distinctly different from simple alterations in diameter (Fig. 1F). We refer to these types of morphological deviations as differences in cell topology. Untreated cells of CS701-1 (Fig. 1E) were also morphologically diverse compared to cells of the parent CS109 (Fig. 1A) and the mutant CS604-2 (Fig. 1C). Even though the differences were not as visually remarkable as those observed in filaments, it was apparent that CS701-1 cells had different diameters, uneven contours, and a variety of inappropriate shapes (Fig. 1E).
Severe morphological aberrations in mutants lacking PBP 5.
Because the only difference between strains CS604-2 and CS701-1 was the absence of PBP 5 from CS701-1, it was possible that this PBP was responsible for the observed morphological defects. To test this, we chose seven strains from which different sets of six PBPs were deleted. Each mutant was isogenic with CS701-1 and expressed one of the PBPs missing from CS701-1 (Table 1) (4).
When mutants lacking six PBPs were filamented in the presence of aztreonam, the only strain that produced filaments of relatively normal length, diameter, contour, and shape was the mutant that expressed active PBP 5, CS604-2 (Fig. 1D). The five mutants that expressed one of the other LMW PBPs (PBPs 4, 6, 7, AmpC, and AmpH) produced filaments with aberrant morphology. The same general effect was observed in untreated cells: mutants lacking PBP 5 and any combination of five other PBPs were much more variable in size and shape. For example, strain CS601-3 (expressing AmpC) exhibited morphological irregularities in untreated cells (Fig. 1G) and in aztreonam-induced filaments (Fig. 1H). Similar results were observed in untreated cells of other sextuple mutants lacking PBP 5, as follows: CS605-4 (expressing PBP 4), CS606-1 (expressing PBP 7), and CS602-1 (expressing AmpH) (data not shown). Thus, an active PBP 5 gene reversed the morphological oddities seen in E. coli CS701-1, whereas expression of any one of the other LMW PBPs had little or no effect.
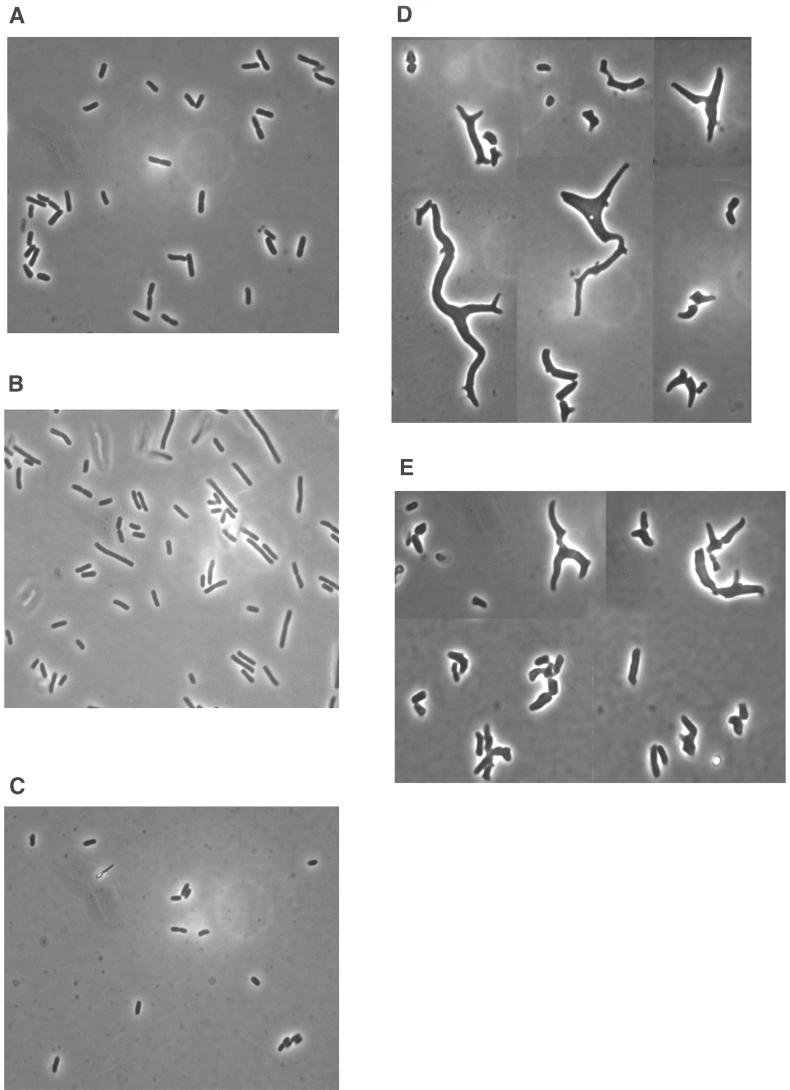
The same PBP 5-dependent morphological characteristics were observed in mutants having fewer than six PBPs deleted. For example, strains producing PBP 5, such as CS502-2 (lacking PBPs 1a, 4, 7, AmpC, and AmpH) (Fig. 2A) and CS508-1 (lacking PBPs 1a, 4, 6, 7, and AmpC) (Fig. 2B), produced cells that looked quite normal, though some were longer than usual. On the other hand, strains lacking PBP 5, such as CS506-1 (lacking PBPs 1a, 4, 5, 6, and AmpC) (Fig. 2D) and CS509-3 (lacking PBPs 1a, 4, 5, 7, and AmpH) (Fig. 2E), produced a high percentage of cells whose morphology was extremely unusual. Once again, the morphological differences were reversed in isogenic mutants expressing PBP 5. For example, CS403-3 (lacking PBPs 1a, 4, 7, and AmpH) is an isogenic relative of CS509-3 that produces active PBP 5 (4), and this one difference returned the cells to normal size and shape (Fig. 2C). The same result was observed for the isogenic partner of CS506-1, strain CS408-1, which expresses active PBP 5 (data not shown). (Note: the examples shown in Fig. 2 were selected from photographs of many cells and are not meant to imply that every cell was equally aberrant. Nonetheless, the photographs do reflect the high degree of morphological variation among cells of these mutants.)
FIG. 2.
Morphology of untreated PBP mutants. Bacteria were grown in LB broth at 37°C until reaching log phase and then photographed. (A) CS502-1; (B) CS508-1; (C) CS403-3; (D) CS506-1; (E) CS509-3.
Although deletion of PBP 5 was associated with significant changes in cell morphology, the degree of morphological variation increased in frequency and extent as the total number of LMW PBPs was reduced. For example, in the mutant in which PBP 5 was the only gene deleted (strain CS12-7) there were only very slight morphological changes visible in filaments formed after treatment with aztreonam (data not shown). This was also true for all but one of the mutants in which two PBPs were deleted (data not shown). The exception to this rule was the double mutant CS211-2 (PBPs 5 and 6 deleted) in which a few irregularities, such as minor branching at the ends of fewer than 1% of the filaments, appeared (data not shown). Deletion of PBP 5 produced the greatest morphological defects in mutants that were missing a total of three or more PBPs.
The morphological effects of losing PBP 5 did not depend on the simultaneous absence of PBP 1a. For example, PBP 1a was active in the Δ5 mutant, CS612-1, but untreated cells and filaments of this strain still exhibited abnormal morphologies (Fig. 1I and J). In addition, we examined five quintuple mutants related to CS535-1, each of which contained a deletion of PBP 5 in place of one of the five genes missing from CS535-1 (strains CS531-3, CS533-1, CS534-1, CS536-1, and CS539-1). In these five isogenic mutants, filaments formed in the presence of aztreonam were significantly abnormal, indicating that no one of the other five LMW PBPs could substitute completely for the loss of PBP 5 (data not shown). Because PBP 1a was expressed in these five quintuple mutants, the alterations in morphology did not depend on the simultaneous absence of PBPs 1a and 5.
Loss of PBP 5 increases the average diameter of E. coli.
The morphological results of deleting PBP 5 were visible by qualitative microscopic inspection. In an effort to obtain a more quantitative idea of the effect of PBP 5 on cell morphology, photographs of cells were subjected to image analysis to measure cell length, cross-sectional area, and derived diameter (Materials and Methods).
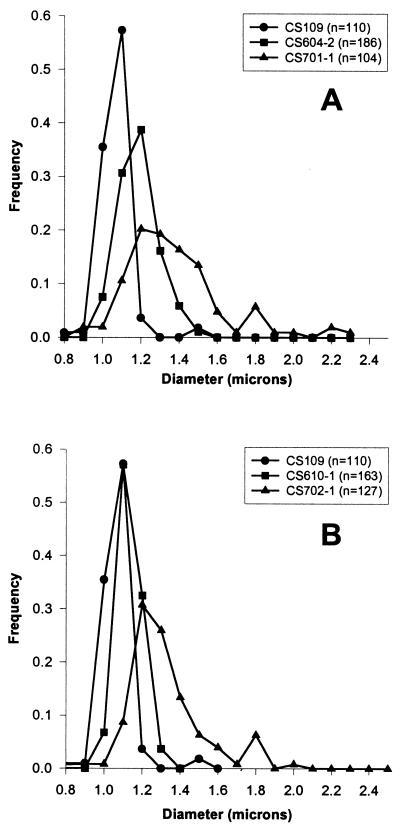
The derived diameters of 92% of untreated wild-type E. coli CS109 cells clustered between 1.0 and 1.1 μm (Fig. 3A), and only 2% of the cells had diameters greater than 1.2 μm (Table 2). In strain CS604-2, which lacks six PBPs but in which PBP 5 is active, 68% of the cells were 1.1 to 1.2 μm in diameter (Fig. 3A) and 23% had diameters that exceeded 1.2 μm (Table 2). Thus, even in the presence of active PBP 5, the average diameter increased slightly in this mutant. However, what is not reflected in these figures is cell shape. As stated above, cells containing active PBP 5 were unbranched and uniform in width along the length of individual cells (i.e., they had a uniform topology and contour). Untreated CS701-1 cells (which lack PBP 5) had much larger derived diameters than did CS604-2 cells (Fig. 3A and Table 2). The diameters of CS701-1 were spread over a greater range, reaching as high as 2.3 μm (Fig. 3A), and 65% of the cells had diameters greater than normal (>1.2 μm) (Table 2). When CS109, CS604-2, and CS701-1 were filamented in the presence of aztreonam for 45 min, the resulting filaments were long enough to exhibit visible differences in morphology but short enough so that each cell could be photographed in a single plane. The same relationships were observed among the diameters of these filaments (Table 2 and data not shown).
FIG. 3.
Differences in cellular diameter of strains isogenic for dacA. Strains were grown in LB broth at 37°C until reaching an A550 of approximately 0.2, at which point samples were prepared for microscopy. “Frequency” indicates the proportion of the population with a particular diameter. n, the number of cells measured for each population.
TABLE 2.
Distribution of cell diameters of PBP mutants
| Strain | PBP(s) deleteda | No treatment
|
Aztreonamc
|
Mitomycin Cd
|
Arabinosee
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| nb | No. (%) of cells with diameters that were:
|
n | No. (%) of cells with diameters that were:
|
n | No. (%) of cells with diameters that were:
|
n | No. (%) of cells with diameters that were:
|
||||||
| <1.2 μm | >1.2 μm | <1.2 μm | >1.2 μm | <1.2 μm | >1.2 μm | <1.2 μm | >1.2 μm | ||||||
| CS109 | None | 110 | 98 | 2 | 92 | 99 | 1 | 182 | 98 | 2 | —f | — | — |
| CS604-2 | 1a | 186 | 77 | 23 | 130 | 82 | 18 | 132 | 92 | 8 | — | — | — |
| CS610-1 | 1b | 163 | 96 | 4 | — | — | — | 120 | 93 | 7 | — | — | — |
| CS701-1 | 1a, 5 | 104 | 35 | 65 | 91 | 38 | 62 | 178 | 39 | 61 | — | — | — |
| CS702-1 | 1b, 5 | 127 | 41 | 59 | — | — | — | 158 | 76 | 24 | — | — | — |
| CS604-2(pFAD38)g | 1a | 144 | 72 | 28 | — | — | — | — | — | — | 135 | 89 | 11 |
| CS610-1(pFAD38)g | 1b | 144 | 97 | 3 | — | — | — | — | — | — | 128 | 98 | 2 |
| CS701-1(pFAD38)g | 1a, 5 | 122 | 8 | 92 | — | — | — | — | — | — | 131 | 21 | 79 |
| CS702-1(pFAD38)g | 1b, 5 | 198 | 95 | 5 | — | — | — | — | — | — | 59 | 100 | 0 |
| CS604-2(pBAD18)h | 1a | 71 | 41 | 59 | 62 | 47 | 53 | 71 | 58 | 42 | — | — | — |
| CS701-1(pBAD18)h | 1a, 5 | 151 | 20 | 80 | 131 | 35 | 65 | 122 | 12 | 88 | — | — | — |
| CS701-1(pPJ5)i | 1a, 5 | 101 | 40 | 60 | 128 | 19 | 81 | 146 | 30 | 70 | — | — | — |
In addition to PBPs 4, 6, 7, AmpC, and AmpH.
Number of bacterial cells measured.
Cells were incubated for 45 min with 10 μg of aztreonam per ml.
Cells were incubated for 45 min with 2 μg of mitomycin C per ml.
Cells were incubated for 45 min with 0.1% (wt/vol) arabinose per ml.
—, no measurements made.
Strain characterized by SulA overproduction.
Strain characterized with control vector.
Strain characterized by PBP 5 complementation.
The effect of PBP 5 was also evident in untreated cells from which PBP 1b had been deleted instead of PBP 1a. In CS610-1 (lacking PBP 1b and five LMW PBPs), the derived diameters of cells were virtually indistinguishable from those of the wild-type strain CS109 (Fig. 3B and Table 2). However, in CS702-1, from which PBP 5 is deleted in addition to those PBPs absent in CS610-1, the derived diameters increased (Fig. 3B and Table 2).
Cloned PBP 5 reverses the morphological defects in dacA deletion mutants.
The PBP 5 gene, dacA, was placed under control of the arabinose promoter, creating plasmid pPJ5C (Materials and Methods). When this plasmid was transformed into CS701-1, a small amount of PBP 5 was produced by cells grown in LB medium in the absence of inducer, protein expression rose with increasing concentrations of arabinose, and no PBP 5 was produced when glucose was present (data not shown). Complementation studies were performed in the absence of arabinose or by inducing PBP 5 by adding 0.005% arabinose.
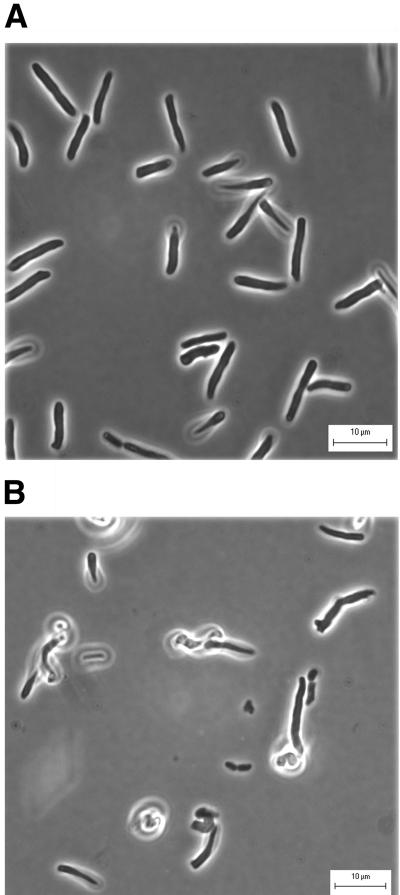
Expression of cloned PBP 5 complemented the major morphological defects in E. coli CS701-1. pPJ5C and the control vector pBAD18-Cam were introduced into E. coli strains CS701-1 and CS604-2, and the cells were filamented by addition of aztreonam. CS701-1 cells containing the vector pBAD18-Cam had diameters and morphologies as abnormal as those exhibited by CS701-1 without the plasmid (Table 2 and data not shown). However, aztreonam-induced filaments of CS701-1 pPJ5C returned to a uniform size and shape (Fig. 4A). Cloned PBP 5 complemented the morphological defects in CS701-1 even if arabinose was omitted from the medium, indicating that the low level of basal expression from the arabinose promoter produced sufficient PBP 5 to counteract the dacA deletion (data not shown). When PBP 5 expression was inhibited by adding glucose to the medium, CS701-1(pPJ5C) cells had the same types of aberrant diameters and shapes as the mutant without the plasmid (Fig. 4B). Thus, visual inspection established that controlled expression of PBP 5 eliminated the gross morphological defects associated with deletion of the dacA gene. In addition, PBP 5 expression in CS701-1(pPJ5C) reduced the range of untreated cell diameters to that exhibited by strain CS604-2(pBAD18), the isogenic strain in which the dacA gene was intact (Table 2).
FIG. 4.
Restoration of normal morphology by cloned PBP 5. E. coli CS701-1 pPJ5C was grown at 37°C in LB broth supplemented with 20 μg of chloramphenicol and either 0.005% arabinose or 0.1% glucose until reaching an A550 of 0.2, at which point 10 μg of aztreonam per ml was added. Samples were prepared for microscopy 45 min after addition of the antibiotic. (A) PBP 5 expression induced by 0.005% arabinose; (B) inhibition of PBP 5 expression by 0.1% glucose.
Loss of PBP 5 produces morphological defects in SulA-induced filaments.
In the preceding experiments the most obvious morphological defects were produced in filaments induced by the addition of aztreonam, which binds to PBP 3 and inhibits septation. It was therefore possible that some of the morphological changes associated with the loss of PBP 5 might require the simultaneous inactivation of PBP 3. To test this, cells were filamented by controlled expression of the SulA protein from plasmid pFAD38 (1). Because SulA inhibits the activity of FtsZ and halts septation at its earliest identified stage (14, 24), this method allowed us to produce filamentous cells without directly inactivating PBP 3.
E. coli CS604-2 pFAD38 exhibited nearly wild-type morphology both before and after induction of SulA with arabinose, with only a small fraction of cells having minor terminal branching (not shown). In contrast, the isogenic mutant lacking PBP 5, CS701-1(pFAD38), exhibited extremely distorted contours along the length of individual filaments (data not shown). In addition, the derived diameters of these populations were increased in cells lacking PBP 5. For CS701-1(pFAD38) (lacking PBP 5), 79% of the filaments had diameters greater than 1.2 μm, whereas for CS604-2 pFAD38 (with active PBP 5), only 11% of the filaments were larger than 1.2 μm in diameter (Table 2). Because the morphological effects were similar to those observed in filaments produced by addition of aztreonam, the effect of PBP 5 does not appear to require the simultaneous inactivation of PBP 3. However, we cannot exclude the possibility that disruption of FtsZ activity and the cytokinetic ring may have affected the activity of PBP 3 indirectly by interfering with normal septal structure.
DISCUSSION
There are two major difficulties in determining the biological roles of the LMW PBPs. First, because multiple PBPs have similar enzymatic activities there is always the question of whether or not the loss of one protein might be compensated for by the presence of another PBP. The second problem is that no one LMW PBP nor any combination of LMW PBPs is essential for normal laboratory growth of E. coli (4). Thus, if one of these proteins plays a significant physiological role, then that role either is subtle or is expressed only under specific circumstances. Previously, we addressed the first of these problems by creating a set of mutants that encompasses all possible combinations of deletion mutations among eight PBPs (4). The premise was that the function of one or more PBPs might become visible in genetic backgrounds from which collaborating PBPs had been removed. By examining a subset of these multiple mutants, we now report that PBP 5 plays an important role in determining the diameter, topology, and proper proportions of the cell wall in E. coli.
PBP 5 and shape control.
The deletion of PBP 5 was the major determining factor in producing cells with abnormal morphology. In fact, mutants from which all LMW PBPs were deleted still formed cells with virtually wild-type morphology as long as PBP 5 remained active. No other individual PBP could substitute for PBP 5 in this regard. However, even in the absence of PBP 5, major morphological alterations did not appear until at least three PBPs had been deleted from a single strain. Therefore, it is likely that some combination of high-molecular-weight and/or LMW PBPs can substitute for the relevant activity of PBP 5. As yet, we do not know the exact combination of PBPs that can do this, and it is possible that several different PBP combinations may be able to obscure the loss of PBP 5. This would explain why the morphological role of PBP 5 was not discovered until now.
In addition to producing cells with branching and gross morphological alterations, a more general effect of the loss of PBP 5 was to increase the overall diameter of cells in a mutant population. However, although PBP 5 clearly affects diameter, the protein does not appear to determine diameter, at least not by itself. In the absence of PBP 5 the cells were still rod-like, and filamentous versions of PBP 5 mutants remained within a fairly narrow range of increased diameters. Even so, the contour of single cells or filaments was not uniform. Thus, although some mechanism imparts to E. coli a rough cylindrical form, the uniformity of individual cells requires the operation of PBP 5.
Recently, Gullbrand et al. reported that an E. coli strain with an altered form of chromosomal replication forms branches at a fairly high rate in a minimal medium (6; see also reference 2). Of particular interest is that the branching frequency increased in the presence of β-lactam antibiotics that bound multiple PBPs but was not affected by β-lactams specific for one PBP, nor did it depend on defects in the cell division proteins PBP 3 or FtsZ (6). These results are consistent with those reported here: branching and other morphological aberrations were enhanced in cells lacking multiple PBPs, and filamentation by aztreonam or SulA expression demonstrated that neither PBP 3 nor FtsZ was required for the effect. What is not apparent in the previous studies is the proximal cause of branching. In light of our current results, the simplest extrapolation is that the reported medium- and strain-dependent morphologies are mediated indirectly by an effect on the level or activity of PBP 5.
Phenotypes associated with loss of PBP 5 homologues in other bacteria.
Unfortunately, no other gram-negative PBP 5 homologues have been studied to any degree. However, an extensive set of studies has been performed in the gram-positive organism Bacillus subtilis (15, 16). Even though numerous mutants have been constructed from which multiple PBPs were deleted, there is no reported phenotype in vegetative cells that accompanies the loss of on of the PBP 5 homologues, either alone or in combination with one or two other LMW PBPs (15, 23). Therefore, either such a PBP does not exert a major influence on the morphology of B. subtilis or other proteins continue to substitute for that role in these multiple mutants.
Two gram-positive cocci, Streptococcus pneumoniae and Staphylococcus aureus, each contain only one known LMW PBP that acts as a d,d-carboxypeptidase (19, 25). Each species is viable in the absence of its single LMW PBP, but morphological effects have been observed. Cells of a mutant S. pneumoniae lacking the LMW PBP are irregularly shaped with septa at inappropriate positions or at multiple sites, the daughter cells have difficulty separating, and the peptidoglycan surface is laid down in peculiar fashion and is of variable thickness (19). Although the results were not reported in detail, a similar PBP mutant of S. aureus may exhibit a similar phenotype in that the strain grows as enlarged spheres in certain media (25). Both of these proteins are homologous to PBP 5 from E. coli, though neither is as strongly similar as the PBPs from B. subtilis (data not shown). In short, homologues of E. coli PBP 5 may influence cell morphology in bacteria other than E. coli.
Summary.
The genesis of specific bacterial shapes remains mysterious. From the data presented here, we cannot derive a detailed picture of how E. coli determines its diameter or shape except to say that PBP 5 is required for constructing a uniform peptidoglycan cylinder. Because PBP 5 is a carboxypeptidase, the evidence suggests that the length of the peptide side chains may play a significant role in how these morphological parameters are determined, although the exact mechanism is left undecided for now.
ACKNOWLEDGMENTS
We especially thank Tom Henderson, Fran Sailer, and Bernadette Meberg for technical advice, Joe Lutkenhaus for pFAD38, and Gayle Streier for graphics.
REFERENCES
- 1.Addinall S G, Lutkenhaus J. FtsA is localized to the septum in an FtsZ-dependent manner. J Bacteriol. 1996;178:7167–7172. doi: 10.1128/jb.178.24.7167-7172.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Åkerlund T, Nordström K, Bernander R. Branched Escherichia coli cells. Mol Microbiol. 1993;10:849–858. doi: 10.1111/j.1365-2958.1993.tb00955.x. [DOI] [PubMed] [Google Scholar]
- 3.Baquero M-R, Bouzon M, Quintela J C, Ayala J A, Moreno F. dacD, an Escherichia coli gene encoding a novel penicillin-binding protein (PBP6b) with dd-carboxypeptidase activity. J Bacteriol. 1996;178:7106–7111. doi: 10.1128/jb.178.24.7106-7111.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Denome S A, Elf P K, Henderson T A, Nelson D E, Young K D. Escherichia coli mutants lacking all possible combinations of eight penicillin binding proteins: viability, characteristics, and implications for peptidoglycan synthesis. J Bacteriol. 1999;181:3981–3993. doi: 10.1128/jb.181.13.3981-3993.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Edwards D H, Donachie W D. Construction of a triple deletion of penicillin-binding proteins 4, 5, and 6 in Escherichia coli. In: de Pedro M A, Höltje J-V, Löffelhardt W, editors. Bacterial growth and lysis. New York, N.Y: Plenum Press; 1993. pp. 369–374. [Google Scholar]
- 6.Gullbrand B, Åkerlund T, Nordström K. On the origin of branches in Escherichia coli. J Bacteriol. 1999;181:6607–6614. doi: 10.1128/jb.181.21.6607-6614.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guzman L-M, Belin D, Carson M J, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Henderson T A, Young K D, Denome S A, Elf P K. AmpC and AmpH, proteins related to the class C β-lactamases, bind penicillin and contribute to the normal morphology of Escherichia coli. J Bacteriol. 1997;179:6112–6121. doi: 10.1128/jb.179.19.6112-6121.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Markiewicz Z, Broome-Smith J K, Schwarz U, Spratt B G. Spherical E. coli due to elevated levels of D-alanine carboxypeptidase. Nature. 1982;297:702–704. doi: 10.1038/297702a0. [DOI] [PubMed] [Google Scholar]
- 10.Matsuhashi M. Utilization of lipid-precursors and the formation of peptidoglycan in the process of cell growth and division: membrane enzymes involved in the final steps of peptidoglycan synthesis and the mechanism of their regulation. In: Ghuysen J-M, Hakenbeck R, editors. Bacterial cell wall. Amsterdam, The Netherlands: Elsevier Science B.V.; 1994. pp. 55–71. [Google Scholar]
- 11.Matsuhashi M, Maruyama I N, Takagaki Y, Tamaki S, Nishimura Y, Hirota Y. Isolation of a mutant of Escherichia coli lacking penicillin-sensitive D-alanine carboxypeptidase 1A. Proc Natl Acad Sci USA. 1978;75:2631–2635. doi: 10.1073/pnas.75.6.2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matsuhashi M, Tamaki S, Curtis S J, Strominger J L. Mutational evidence for identity of penicillin-binding protein 5 in Escherichia coli with major d-alanine carboxypeptidase 1A activity. J Bacteriol. 1979;137:644–647. doi: 10.1128/jb.137.1.644-647.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 14.Mukherjee A, Cao C N, Lutkenhaus J. Inhibition of FtsZ polymerization by SulA, an inhibitor of septation in Escherichia coli. Proc Natl Acad Sci USA. 1998;95:2885–2890. doi: 10.1073/pnas.95.6.2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Popham D I, Gilmore M E, Setlow P. Roles of low-molecular-weight penicillin-binding proteins in Bacillus subtilis spore peptidoglycan synthesis and spore properties. J Bacteriol. 1999;181:126–132. doi: 10.1128/jb.181.1.126-132.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Popham D L, Setlow P. Phenotypes of Bacillus subtilis mutants lacking multiple class A high-molecular-weight penicillin-binding proteins. J Bacteriol. 1996;178:2079–2085. doi: 10.1128/jb.178.7.2079-2085.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 18.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schuster C, Dobrinski B, Hakenbeck R. Unusual septum formation in Streptococcus pneumoniae mutants with an alteration in the d,d-carboxypeptidase penicillin-binding protein 3. J Bacteriol. 1990;172:6499–6505. doi: 10.1128/jb.172.11.6499-6505.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spratt B G. Deletion of the penicillin-binding protein 5 gene of Escherichia coli. J Bacteriol. 1980;144:1190–1192. doi: 10.1128/jb.144.3.1190-1192.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Spratt B G. Distinct penicillin binding proteins involved in the division, elongation, and shape of Escherichia coli K12. Proc Natl Acad Sci USA. 1975;72:2999–3003. doi: 10.1073/pnas.72.8.2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stoker N G, Broome-Smith J K, Edelman A, Spratt B G. Organization and subcloning of the dacA-rodA-pbpA cluster of cell shape genes in Escherichia coli. J Bacteriol. 1983;155:847–853. doi: 10.1128/jb.155.2.847-853.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Todd J A, Roberts A N, Johnstone K, Piggot P J, Winter G, Ellar D J. Reduced heat resistance of mutant spores after cloning and mutagenesis of the Bacillus subtilis gene encoding penicillin-binding protein 5. J Bacteriol. 1986;167:257–264. doi: 10.1128/jb.167.1.257-264.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trusca D, Scott S, Thompson C, Bramhill D. Bacterial SOS checkpoint protein SulA inhibits polymerization of purified FtsZ cell division protein. J Bacteriol. 1998;180:3946–3953. doi: 10.1128/jb.180.15.3946-3953.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wyke A W, Ward J B, Hayes M V, Curtis N A. A role in vivo for penicillin-binding protein 4 of Staphylococcus aureus. Eur J Biochem. 1981;119:389–393. doi: 10.1111/j.1432-1033.1981.tb05620.x. [DOI] [PubMed] [Google Scholar]