Abstract
NixA, the high-affinity cytoplasmic membrane nickel transport protein of Helicobacter pylori, imports Ni2+ into the cell for insertion into the active site of the urease metalloenzyme, which is required for gastric colonization. NixA fractionates with the cytoplasmic membrane, and protein cross-linking studies suggest that NixA functions as a monomer. A preliminary topological model of NixA with seven transmembrane domains was previously proposed based on hydropathy, charge dispersion, and homology to other transporters. To test the proposed topology of NixA and relate critical residues to specific structural elements, a series of 21 NixA-LacZ and 21 NixA-PhoA fusions were created along the entire length of the protein. Expression of reporter fusions was confirmed by Western blotting with β-galactosidase- and alkaline phosphatase-specific antisera. The activities of reporter fusions near to and upstream of the predicted translational initiation demonstrated the presence of an additional amino-terminal transmembrane domain including a membrane localization signal. Activities of fusions immediately adjacent to motifs which have been shown to be requisite for Ni2+ transport localized these motifs entirely within transmembrane domains II and III. Fusion activities localized six additional Asp and Glu residues which reduced Ni2+ transport by >90% when mutated within or immediately adjacent to transmembrane domains II, V, VI, and VII. All fusions strongly support a model of NixA in which the amino and carboxy termini are located in the cytoplasm and the protein possesses eight transmembrane domains.
Helicobacter pylori is a well-established etiologic agent of gastritis, as well as duodenal and gastric ulceration (6, 35, 36). The development of more serious sequelae, namely, gastric carcinoma and MALT lymphoma, are also strongly associated with chronic H. pylori infection (18, 41).
H. pylori produces a battery of virulence factors, including a urease comprising up to 6% of the soluble cell protein (26). The ammonia produced by the hydrolysis of urea by this 550-kDa multimeric enzyme has been postulated to allow the bacterium to survive and colonize the low pH environment of the gastric mucosa. This is supported by the observation that H. pylori is highly sensitive to acid in the absence of urea (34). An additional role of urease in colonization has been implied by the finding that urease-negative mutants are unable to colonize animal models of infection, even when the gastric pH is maintained near neutrality by the administration of the proton pump inhibitor omeprazole (15, 48).
The divalent nickel ion is a requisite cofactor in the urease active site (23). The NixA nickel transport protein has an extremely high-affinity (KT = 11.3 nM) import mechanism (39) well suited for scavenging Ni2+ from the low concentrations found in human serum (2 to 11 nM) (45) and presumably in the gastric mucosa. Isogenic NixA-deficient H. pylori cells show predictably lower levels of Ni2+ uptake (31% of that of the wild type) and urease activity (42% of that of the wild type) (3).
Alignment of the 331-amino-acid NixA sequence with three homologous single-component Ni2+ transporters, HoxN of Alcaligenes eutrophus (50), HupN of Bradyrhizobium japonicum (19), and UreH of thermophilic Bacillus sp. strain TB-90 (30), as well as hydropathy and charge dispersion (49), suggested a preliminary model in which NixA is an integral cytoplasmic membrane protein composed of seven transmembrane domains with the amino terminus located in the periplasm (39).
Further examination of this alignment identified 12 conserved Asp, Glu, and His residues which were postulated to be involved in Ni2+ transport (20). These included the sequence motif GX2HAXDADH, which was conserved among NixA and its homologs, as well as in the NikC component of the nonhomologous Nik ATP-binding cassette transporter of Escherichia coli (20, 50). The motif GX2FX2GHSSVV, which is also shared among the four single-component Ni2+ transporters, is also present as a slight variant in the NhlF Ni2+-sensitive Co2+ transporter of Rhodococcus rhodochrous (20, 27, 50). Both sequence motifs were also predicted to be in the NicT protein, recently identified as encoded in the genome of Mycobacterium tuberculosis (10), as well as in additional putative homologs identified in the genome sequences of Salmonella typhi (4) and Schizosaccharomyces pombe (29). Initial topological predictions for NixA placed these motifs in the first transmembrane domain and across the cytoplasmic border of the first cytoplasmic loop into the second transmembrane domain, respectively (39). Site-directed mutation of two Asp and three His residues located within these motifs each resulted in the complete loss of NixA-mediated Ni2+ uptake and urease activity when each mutation was expressed in E. coli cells along with the H. pylori urease operon (20). Nonconservative mutations of six additional conserved Asp and Glu residues which were postulated to lie within transmembrane domains reduced rates of Ni2+ transport by >90%, with correlating reductions in urease activities (20). While these findings shed considerable light on the function of specific amino acid residues of NixA, a detailed analysis of the membrane topology and localization of the protein is required to assign specific location and structure to amino acid residues which appear to be critical for Ni2+ transport. We here report a new model of the structure and localization of NixA, based on the analysis of reporter fusions, membrane fractionation, and cross-linking studies.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
H. pylori ATCC 43504 and an isogenic NixA-deficient allelic exchange mutant of this strain have been described previously (3). E. coli DH5α [supE44 ΔlacU169 (ϕ80 lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1] and E. coli MC1061 [hsdR araD139 Δ(araABC-leu) ΔlacX74 galU galK rpsL thi] were used as recipients of recombinant plasmids (43). Plasmids pUEF204 [carrying the H. pylori nixA gene cloned in pBluescript II SK(+) (Stratagene)], pLKC480, and pRT733 have been described previously (39, 46, 47). Strains were maintained on Luria-Bertani agar or sheep blood agar containing the appropriate antibiotics and were stored at −70°C in Luria broth supplemented with 15% (vol/vol) glycerol or Mueller-Hinton broth supplemented with 4% horse serum and 15% (vol/vol) glycerol. Other media included Luria broth supplemented with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) and 10 mM Na2HPO4.
Recombinant DNA techniques.
Recombinant DNA methods, including restriction endonuclease digestion, ligation, and transformation, were performed according to standard protocols (2, 43). Plasmid DNA was purified by rapid alkaline lysis (5). Large-scale preparations were isolated with Qiagen DNA purification columns according to the manufacturer's instructions.
Construction of PhoA fusion plasmid pBAF.
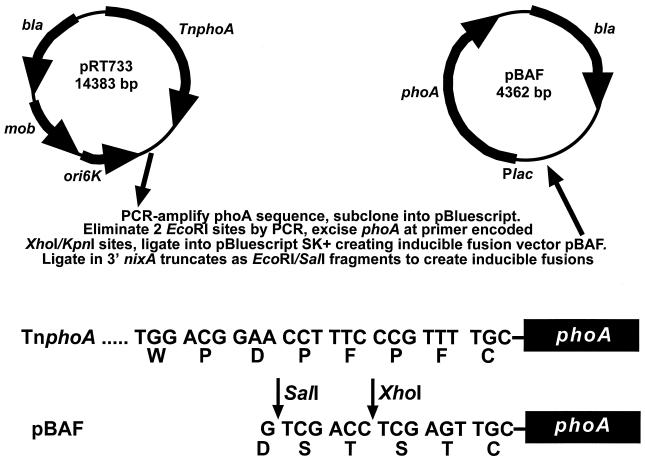
A 1,417-bp sequence encoding the mature PhoA polypeptide was PCR amplified as a XhoI-KpnI fragment from pRT733 (46) and subcloned into the EcoRV site of pBluescript II SK(+). Two EcoRI sites within the phoA sequence were eliminated by site-directed mutation by the PCR overlap extension method of Ho et al. (25). First-round PCR products were agarose gel purified to prevent the amplification of wild-type phoA. PCRs were performed with cloned Pfu DNA polymerase (Stratagene) and primers (sequences available upon request) which carried two conservative codon changes: CTG to CTC (G717C of phoA) and GAA to GAG (A1050G of phoA). The mutated phoA product was amplified as a PstI-NruI fragment and ligated into pLKC480, creating vector pAPF1. The phoA sequence was reamplified from pAPF1 as an XhoI-KpnI fragment and ligated into pBluescript II SK(+) at these sites, yielding the IPTG-inducible PhoA fusion plasmid pBAF.
Nucleotide sequencing.
Plasmid DNA was sequenced by the dideoxy chain termination method (44). Reactions were run on an Applied Biosystems Model 373A DNA sequencer.
Construction of NixA-LacZ and NixA-PhoA fusions.
A series of 21 3′ truncates of nixA were PCR amplified as EcoRI-SalI fragments (primer sequences available upon request) and ligated into vector pLKC480 (47) to create in-frame LacZ fusions or into vector pBAF to create in-frame PhoA fusions. LacZ fusion constructs were transformed into E. coli MC1061 cells and selected on Luria agar containing ampicillin (100 μg/ml), kanamycin (50 μg/ml), and X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) (40 μg/ml), PhoA fusions were transformed into E. coli DH5α cells and selected on Luria agar containing ampicillin (100 μg/ml) and XP (5-bromo-4-chloro-3-indolylphosphate) (40 μg/ml).
Enzyme assays.
β-Galactosidase activities of NixA-LacZ fusions were measured by the method of Miller (38). Alkaline phosphatase activity of NixA-PhoA fusions were measured by the method of Brickman and Beckwith (8) with the following modifications. Overnight cultures were used to inoculate fresh Luria broth containing 1 mM IPTG and 10 mM Na2HPO4, pH 7.0. Exponential-phase cells were washed with and resuspended in 50 mM Tris-HCl (pH 8.0) and spectrophotometrically assayed as described.
Expression of reporter fusions.
E. coli MC1061 cells transformed with either pLKC480 or nixA-lacZ fusions were harvested by centrifugation (5,000 × g, 10 min, 4°C) from exponential-phase Luria broth cultures. E. coli DH5α cells transformed with pBAF or nixA-phoA fusions grown in Luria broth containing 4 mM IPTG and 50 mM Na2HPO4, pH 7.0, were harvested as described above. Cells were ruptured in a French press at 20,000 lb/in2, and the lysate was centrifuged (8,000 × g, 10 min, 4°C). The cleared lysate was ultracentrifuged (170,000 × g, 90 min, 4°C). The resultant membrane pellet was resuspended in and washed with phosphate-buffered saline (PBS), pH 7.25, and then it was ultracentrifuged as before. Membranes were resuspended in PBS, and 7.5 μg of each sample was electrophoresed under denaturing conditions in a sodium dodecyl sulfate (SDS)–5% or 8% polyacrylamide gel and transferred to Immobilon P as described by Ausebel et al. (2). Blots were probed with antibodies to β-galactosidase or alkaline phosphatase (5′→3′), washed, and reprobed with a polyclonal goat anti-rabbit alkaline phosphatase conjugate (Sigma). Blots were developed in BCIP-NBT (5-bromo-4-chloro-3-indolylphosphate–nitroblue tetrazolium) substrate solution (Sigma) according to the manufacturer's instructions.
Fractionation of H. pylori.
Cells of H. pylori ATCC 43504 and its isogenic nixA mutant were grown on sheep blood agar and fractionated into periplasmic, cytosolic, outer membrane, and inner membrane fractions (2, 17). Cells were harvested by centrifugation (5,000 × g, 10 min, 4°C), cold osmotically shocked, and pelleted, and periplasmic proteins were isolated from the supernatant (2). The pelleted cells were resuspended and lysed in a French pressure cell at 20,000 lb/in2, and the lysate was cleared by centrifugation (5,000 × g, 10 min, 4°C). The lysate was further ultracentrifuged (100,000 × g, 1 h, 4°C). The supernatant (containing cytosolic components) was decanted, and the pellet (containing membrane components) was washed with PBS, pH 7.25, and pelleted again by ultracentrifugation as before. The membrane pellet was then detergent fractionated. Inner membrane components were solubilized by incubation with 0.5% (wt/vol) Sarkosyl for 20 min (17), and outer membrane components were isolated by additional ultracentrifugation (100,000 × g, 60 min, 4°C). The protein concentration of each fraction was determined by the bicinchoninic acid (BCA) assay (Pierce). Each sample was boiled for 5 min in SDS-polyacrylamide gel electrophoresis (PAGE) (2) sample buffer, and 5 μg (protein) was electrophoresed on an SDS–9% polyacrylamide gel and transferred to Immobilon P membranes which were probed (2) with rabbit polyclonal antibodies and affinity purified with an immobilized NixA peptide (20).
Cross-linking of H. pylori membrane proteins.
Membranes were isolated from H. pylori ATCC 43504 cells cultured for 3 days on sheep blood agar plates in anaerobe jars under microaerobic conditions generated by activated Campy paks. Bacteria were harvested and resuspended in 3 ml of PBS. Cells were ruptured in a French press at 20,000 lb/in2, and the lysate was centrifuged (8,000 × g, 10 min, 4°C). The cleared lysate was ultracentrifuged (170,000 × g, 90 min, 4°C). The resultant membrane pellet was resuspended in and washed with PBS, and then it was ultracentrifuged as before. Membranes were resuspended in PBS, and 250-μl samples were treated with dithiobis(succinimidyl proprionate) (DSP) or 3,3′-dithiobis(sulfosuccinimidyl proprionate) (DTSSP) (Pierce) for 30 min or 1 h, respectively, at room temperature at final concentrations of 0.1 to 2.5 mM with gentle mixing. Proteins in whole H. pylori cells were cross-linked in the same manner, as well as a NixA-NixA antibody control to assess cross-linker binding. Cross-linking reactions were quenched by addition of Tris-HCl (pH 7.5) to a 50 mM concentration and incubation for an additional 15 min. Each cross-linked sample, along with untreated samples (5 μg of protein), was electrophoresed under nonreducing (SDS-PAGE sample buffer without β-mercaptoethanol) conditions on an SDS–9% polyacrylamide gel and stained with Coomassie blue (Sigma) to assess reactions. Duplicate electrophoresed samples were transferred to Immobilon P membranes and immunoblotted with NixA-specific antibodies as described above.
RESULTS
Localization of NixA to the cytoplasmic membrane.
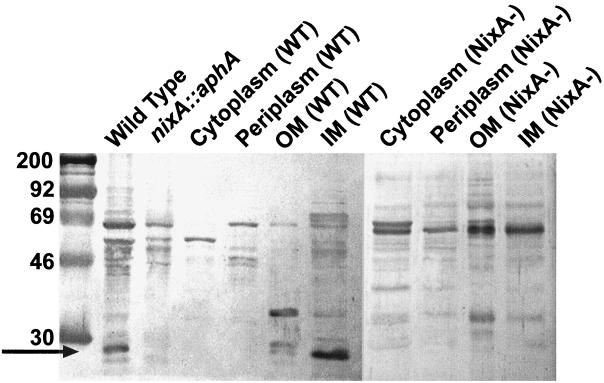
Fractionation of whole H. pylori ATCC 43504 cells into periplasmic components by osmotic shock and cytosolic and membrane components by lysis and ultracentrifugation, followed by solubilization of inner membrane components with 0.5% Sarkosyl and immunoblotting with NixA-specific antibodies, shows NixA to be an integral cytoplasmic membrane protein (Fig. 1). Some cross-reactive bands are visible in the whole-cell preparation of the wild type and the isogenic nixA mutant. The most intense of these bands also appear in either the cytoplasmic or the periplasmic fraction. An additional band with a mass of 35 kDa appears in the outer membrane fraction; its intensity is likely due to the low protein composition of the outer membrane relative to the cytoplasm, inner membrane, and periplasm. This band is also notably present in the outer membrane fraction of the isogenic nixA mutant (Fig. 1).
FIG. 1.
Localization of NixA to the cytoplasmic membrane. Whole H. pylori cells and an isogenic nixA mutant harvested from third-day growth on sheep blood agar were fractionated into periplasmic, cytoplasmic, inner membrane (IM), and outer membrane (OM) components by cold osmotic shock, lysis in a French pressure cell, ultracentrifugation, and solubilization of the inner membrane with 0.5% Sarkosyl. Whole wild-type (WT) cells, whole NixA-deficient H. pylori cells, and 5 μg of protein from each fraction were separated by SDS–9% PAGE and immunoblotted with NixA-specific affinity-purified polyclonal antibodies.
Amino-terminal structure and sequence of NixA.
Analysis of the upstream nucleotide sequence of nixA (Fig. 2) and alignment of the amino acid sequence with the homologous proteins HoxN and HupN suggested the possibility of a previously unrecognized N-terminal transmembrane helix. PCR-amplified sequences containing the upstream sequence and a putative GTG translational initiation codon only or the upstream sequence and both the GTG and ATG start codons (Fig. 2) were fused in frame to lacZY in pLKC480 (forming pNLC7 and pNLC30, respectively). A Western blot of E. coli MC1061 (Δ lac) cells expressing proteins of the fusion constructs and a vector control (see Fig. 4) probed with antiserum to β-galactosidase demonstrated significant and similar levels of synthesis of β-galactosidase subunits from both pNLC7 and pNLC30, but not from the vector control. o-Nitrophenyl-β-d-galactopyranoside (ONPG) assays of the constructs and vector control demonstrated β-galactosidase activity from both pNLC30 and pNLC7, 52 and 496 U, respectively, indicating the translation of NixA from the upstream initiation codon (see Fig. 5).
FIG. 2.
A previously unrecognized 23-amino-acid amino-terminal sequence including an additional transmembrane domain. Primers NXSTRT+, aa 7−, and aa 30− were used to amplify sequences which would include only an upstream sequence and the unreported GTG start codon or both initiation codons. PCR products were fused to β-galactosidase and alkaline phosphatase and assayed for activity (see Fig. 5B). Constructs were also assayed for expression (see Fig. 4). Amino acid 7 fusions exhibited higher β-galactosidase activity and lower alkaline phosphatase activity than did amino acid 30 fusions. ⇓, the previously reported amino terminus. Candidate promoter (−35, −10) and Shine-Dalgarno (S.D.) sequences are underlined.
FIG. 4.
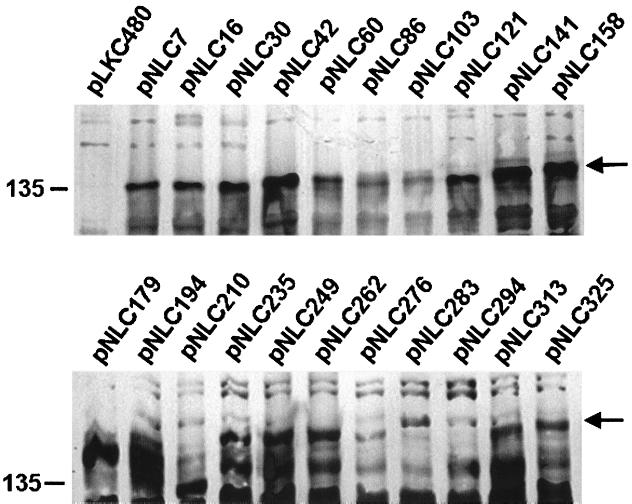
Immunodetection of NixA-LacZ fusions. E. coli MC1061 cells containing pLKC480 or each of the 21 NixA-LacZ fusions were harvested from exponential-phase cultures. Whole membranes were prepared from each by lysis in a French pressure cell and ultracentrifugation of the cleared lysate. Proteins (5 μg) from each membrane preparation were separated by SDS–5% PAGE and transferred to polyvinylidene difluoride membranes which were probed with an E. coli β-galactosidase-specific rabbit polyclonal antiserum. Arrows indicate bands corresponding to NixA-LacZ fusions.
FIG. 5.
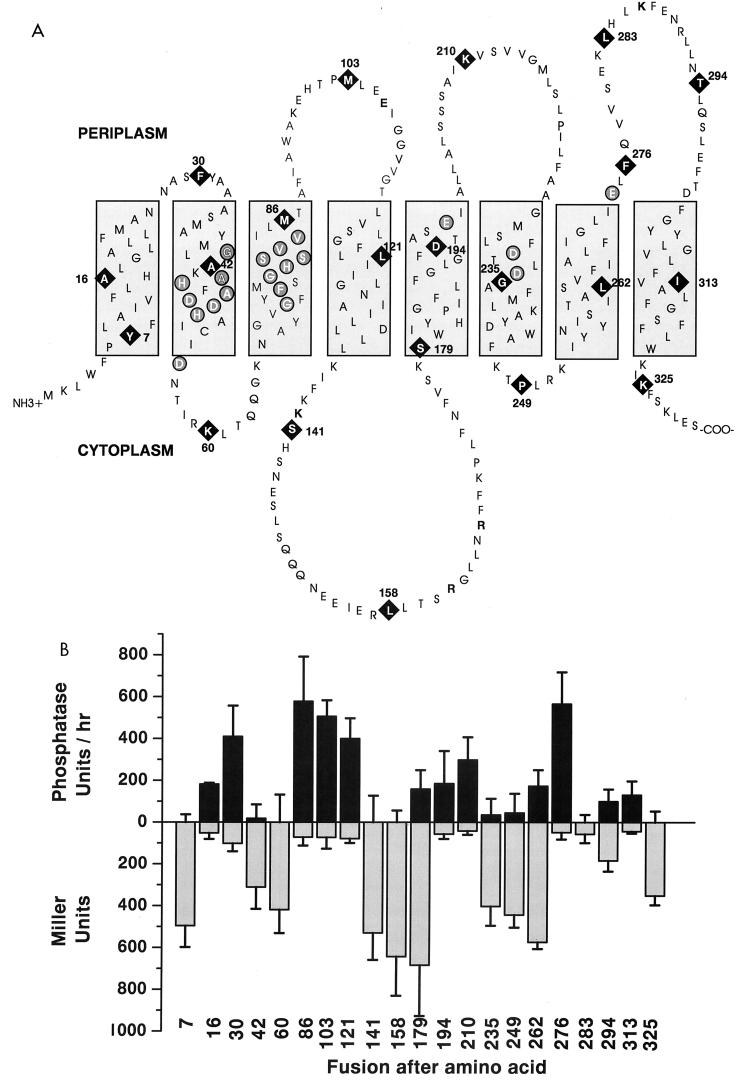
Topology of NixA and locations and activities of reporter fusions. (A) Revised topological model of NixA. Boxed regions indicate transmembrane domains. Black diamonds indicate the locations of reporter fusions by number (from the amino terminus) of the last NixA amino acid prior to the fusion junction. Circled residues indicate conserved motifs in helices II and II plus six additional transport-critical residues. (B) Graphical representation of reporter fusion activities. Values are statistical averages of at least three independent experiments ± standard deviations.
These results, along with PhoA fusion activities in the same region (Fig. 5B), indicate the presence of a previously unrecognized eighth transmembrane domain and place the amino terminus of the protein in the cytoplasm. The sequence encoding this region utilizes a GTG start codon 69 bp upstream of the originally reported initiation of translation and also now provides candidate transcriptional and translational regulatory sequences not previously identified (Fig. 2). The additional 23 amino acids encoded by this sequence also includes a Lys residue immediately after the initiation codon, a common localization feature of cytoplasmic membrane proteins, and now predicts NixA to be 331 amino acids in length with a molecular mass of 36,991 Da.
NixA-LacZ and NixA-PhoA fusions.
To confirm the prediction of eight transmembrane domains, the localization of both the amino and carboxy termini in the cytoplasm, and the location of the transport-critical motifs GX2HAXDADH and GX2FX2GHSSVV, additional β-galactosidase and alkaline phosphatase fusion proteins were constructed. Since the putative model of NixA had been designed based on hydropathy (28), charge dispersion (49), and homology to the partially solved topology of other proteins (16), reporter fusion sites were chosen based on this model and constructed with two reporter fusion vectors (Fig. 3) rather than creating random fusions by transposon mutagenesis, which often leads to multiple fusions in small regions or hot spots of recombination separated by large gaps (16).
FIG. 3.
Construction of inducible NixA-PhoA fusions. A 1,417-bp PCR fragment encoding the mature PhoA polypeptide of TnphoA was amplified from plasmid pRT733 and subcloned into the EcoRV site of pBluescript II SK(+). Two EcoRI sites within the phoA sequence were eliminated by PCR mutagenesis (G717C and A1050G). The phoA sequence was then excised at primer-encoded PstI-NruI sites and ligated into these sites in pLKC480 to create pAPF1. The phoA sequence was reamplified from pAPF1 by using primers encoding XhoI and KpnI restriction sites and ligated into pBluescript II SK(+), yielding the IPTG-inducible PhoA fusion vector pBAF. Black boxes indicate the convergence of the TnphoA sequence and the sequence encoding the mature E. coli PhoA polypeptide. The last 8 of 16 amino acids of the in-frame TnphoA insertion sequence and their nucleotide sequence are shown, along with the corresponding sequences encoded by restriction sites in pBAF. nixA truncates were ligated in EcoRI-SalI fragments.
To create strategic reporter fusions, 21 unique 3′ truncates of nixA were PCR amplified. Based on the assumption that cytoplasmic membrane-spanning helices are typically 19 to 25 amino acids long, fusion sites were chosen at sufficiently small intervals to avoid overlooking membrane spanning or associated regions. The PCR-generated fragments bearing EcoRI-SalI restriction sites were ligated in frame to lacZY in vector pLKC480 (Fig. 3) (47).
In order to construct alkaline phophatase fusions with sufficient enzymatic activity for easily measurable and reproducible results, an inducible vector was constructed (Fig. 3). The nucleotide sequence encoding the mature PhoA polypeptide from TnphoA in vector pRT733 (46) was PCR amplified as a 1,417-bp XhoI-KpnI fragment ligated into pBluescript II SK(+) (Stratagene). Two EcoRI sites present within the phoA fragment were eliminated by PCR site-directed mutagenesis (25). This allowed the nixA-PCR truncates, used to create lacZY fusions, to be used directly to create IPTG-inducible phoA fusions in the resultant vector, pBAF (Fig. 3). This redundancy of fusion sites allowed at least partial delineation of misleading fusion activities due to unusual membrane insertion of one or two fusions at the same site, as well as enhanced interpretation of mixed activities obtained from fusions within transmembrane domains (see Fig. 5).
As the SalI and XhoI sites are immediately adjacent in pBluescript, primer-encoded amino acid changes in the carboxy terminus of NixA and the amino terminus of PhoA should have little effect on membrane insertion of the fusion proteins. The adjacent sites in pBAF encode the sequence Ser-Thr-Ser-Ser, replacing the sequence Pro-Phe-Pro-Phe in TnphoA (Fig. 3), which is followed by a Cys encoded by TnphoA and the mature PhoA amino acid sequence. In the native TnphoA sequence, two Ser residues, a Thr residue, two Asp residues, and a Glu residue are included within the 16 amino acids encoded by the in-frame insertion sequence immediately upstream of the mature PhoA polypeptide. This sequence also includes three Pro residues. As these negatively charged Asp or Glu residues, Ser or Thr residues, or Pro residues do not interfere with or confer any localization on TnphoA mutagenized proteins, their replacement by a short four-amino-acid Ser-Thr sequence does not appear to alter the activities of pBAF-encoded fusions.
Western blots of the 21 NixA-LacZ fusion proteins demonstrate the synthesis of the full-length fusion proteins and their presence in the bacterial membrane (Fig. 4), although certain fusions in transmembrane and periplasmic domains appear to be present at lower levels. This may likely be due to proteolysis and the instability of locked translocation intermediates. Similarly, Western blotting of the 21 NixA-PhoA fusion proteins (data not shown) confirmed the synthesis of fusion proteins of the expected size. Notably, bands corresponding to PhoA fusions in the cytoplasmic domains are significantly less prominent. It has been previously noted that the non-disulfide-linked monomeric form of the alkaline phosphatase precursor is highly unstable in the cytoplasm (13) and likely accounts for discrepancies in signal intensities. Likewise, LacZ fusions to regions of NixA which localize to the periplasmic space, e.g., pNLC103, pNLC210, and pNLC276, appear to be less stable (Fig. 4), as evidenced by immunoblotting with β-galactosidase-specific antiserum. There also appears to be a general trend toward lower expression and/or stability of fusions closer to the carboxy terminus of NixA.
As predicted, LacZY fusions near the amino and carboxy termini (amino acids 7 and 325) produced strong reproducible β-galactosidase activities and no alkaline phosphatase activity above the vector control, localizing both to the cytoplasm (Fig. 5). With the exception of fusions at amino acid 179, all fusions to predicted cytoplasmic loops yielded high β-galactosidase activities and no significant alkaline phosphatase activity. Fusions at amino acid 179, which was initially proposed to be located near the carboxy-terminal end of the large cytoplasmic loop, possessed both significant β-galactosidase and alkaline phosphatase activities. Two Lys residues located within 10 amino acids upstream of amino acid 179 likely act as membrane anchors (21). It has been previously shown that positively charged Lys and Arg residues commonly act as stop transfer sequences, lying at or near the junction of cytosolic and transmembrane domains and preventing further insertion of the amino acid sequence into the membrane (21). Additionally, the next fusion site (amino acid 194) demonstrated high alkaline phophatase-to-β-galactosidase activity, indicating that although it was predicted to be nearly in the center of the following transmembrane domain, it is more likely to be near to or within the following periplasmic loop (Fig. 5).
Significantly, reporter fusions (amino acids 30, 42, 60, and 86) within and near the highly conserved, transport-critical motifs GX2HAXDADH and GX2FX2GHSSVV (20, 50) place these motifs entirely within transmembrane domains II and III. Six additional residues which have been shown to reduce nickel transport by ≥90% when mutated (20) localized within or immediately adjacent to transmembrane domains II, V, VI, and VII (Fig. 5).
The activities of fusions in the predicted final periplasmic loop and transmembrane domain VIII are difficult to interpret as strictly periplasmic or transmembrane (Fig. 5). Amino acid 262 fusions produced significant levels of both β-galactosidase and alkaline phophatase activities and predict amino acid 262 to lie within transmembrane domain VII. The following fusion site, amino acid 276, yielded high alkaline phosphatase activity and only basal β-galactosidase activity, indicating periplasmic exposure. Fusions at amino acid 283 had no alkaline phosphatase activity and only basal β-galactosidase activity. Fusions at amino acid 294 produced low but significant activity with each reporter, while predicted transmembrane helix amino acid 313 fusions have alkaline phosphatase activities similar to those at amino acid 294 (100 versus 131 U/h, respectively) but only basal β-galactosidase activity. The sole following fusion is near the carboxy terminus and is clearly cytoplasmic. These findings indicate that the protein clearly spans the cytoplasmic membrane between amino acids 276 and 325, but the intervening region is difficult to interpret, possibly due to protein instability (Fig. 4) or misincorporation into the membrane due to the number of positive charges in the region. The region between amino acids 276 and 325 is clearly not long enough to include two additional transmembrane domains, and addition of only a ninth helix would place the carboxy terminus in the periplasm, which is clearly inconsistent with the fusion activities.
NixA appears to function as a monomer.
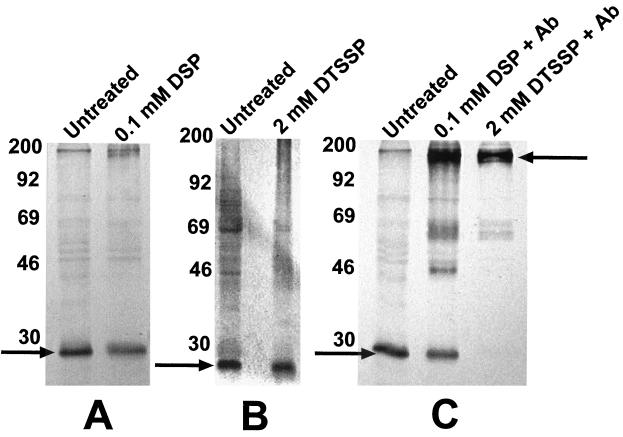
To detect possible multimerization of NixA, whole H. pylori cells and membrane preparations were subjected to protein cross-linking with the membrane-soluble 12-Å cross-linker DSP and its water-soluble analog DTSSP over a concentration range of 0.1 to 2.5 mM. Coomassie-stained SDS-polyacrylamide gel-separated samples confirmed the general effectiveness of the cross-linking reactions, as did control cross-linking of NixA to NixA-specific polyclonal antibodies, which resulted in intense bands with masses of approximately 175 kDa, representative of the combined electrophoretic mobility of immunoglobulin G and NixA. However, immunoblotting with NixA-specific antiserum (Fig. 6) demonstrated that NixA remained present only as a monomer in cross-linked membrane samples, and no significant loss of monomeric-sized NixA or addition of multimeric-sized bands was observed in Western blots of cross-linked whole-cell proteins or cross-linked membrane protein preparations.
FIG. 6.
Immunodetection of NixA in cross-linker-treated whole cells and membranes. Whole H. pylori cells were subjected to cross-linking with the membrane-soluble cross-linker DSP at a final concentration of 0.1 mM, and membranes were isolated from treated cultures. H. pylori membrane preparations were treated with the water-soluble cross-linker DTSSP at a final concentration of 2.0 mM. As a control, whole-cell lysates were similarly treated with DSP and DTSSP in the presence of affinity-purified NixA-specific polyclonal antibodies (Ab) to demonstrate cross-linker binding to NixA. Nonreduced cross-linked samples were analyzed along with nonreduced untreated membranes and cross-linked NixA-antibody fusions by SDS-PAGE. Immunoblots were probed with NixA-specific antibodies. Arrows indicate monomeric NixA (A and B) and the cross-linked NixA-NixA-immunoglobulin (C).
DISCUSSION
Assembly of catalytically active urease, central to the virulence of H. pylori, is dependent upon the incorporation of Ni2+ ions into the active site of the apoenzyme (40). The NixA Ni2+ transport protein of this organism is a potent mediator of this requirement, as demonstrated by direct measurement of NixA-dependent 63Ni2+ uptake and urease activity (39) and the reduction of Ni2+ uptake and urease activity seen in isogenic nixA mutants (3). NixA is representative of a small family of high-affinity Ni2+ transporters (4, 10, 12, 19, 27, 29, 30, 39, 50) which have not been extensively studied and whose structures and functions are not well understood.
We have recently reported that His, Asp, and Glu residues contained within two distinct sequence motifs, GX2HAXDADH and GX2FX2GHSSVV, and putative transmembrane domains are critical for Ni2+ transport, based on site-directed mutation of these residues (20). However, the relationship of these residues to any specific structural determinant or the cellular location of these residues was largely speculative, based on a model derived from hydropathy predictions (28), the “positive inside rule” (11, 49) with a net cytoplasmic charge of +12 and a periplasmic charge of −1, and homology to other transporters, such as HoxN (50).
Although the high substrate affinity, single-component nature, and charge dispersion strongly suggested that NixA is an integral cytoplasmic membrane protein, this had not been demonstrated for NixA or any homologous transporter. We have shown here that NixA is recognizable by immunoblotting with NixA-specific antibodies only in the Sarkosyl-solubilized inner membrane components as expected. The somewhat low observed molecular weight of NixA on Western blots is typical of hydrophobic integral membrane proteins and may be the result of increased SDS binding in hydrophobic regions (42). The technique of fractionation is somewhat limited in H. pylori cells by the altruistic lysis of cells, and the binding of cytoplasmic, inner membrane, and periplasmic proteins to the surface of live H. pylori cells, where they may act in vivo as a means of host immune avoidance. Nevertheless, fractionation and immunoblotting results combined with the criteria of cytoplasmic membrane proteins strongly support its localization in the cytoplasmic membrane.
An eighth amino-terminal transmembrane domain has also been added here to the previous model, based on the observation that in-frame reporter fusions to the region upstream of the reported translational initiation were expressed equally as well as reporter fusions placed downstream of the originally reported start. The lower β-galactosidase activity and higher alkaline phosphatase activity of the downstream reporter fusions relative to those of the upstream fusions further suggest that the amino terminus is located in the cytoplasm, not in the periplasm as previously reported. This now yields a model of NixA with eight putative transmembrane domains.
Additional LacZ and PhoA reporter fusions confirmed the eight membrane-spanning domains model. A total of 42 reporter fusions were constructed such that the distance between fusions would be too small to allow membrane-spanning regions to be overlooked. All reporter fusions except those in the carboxy-terminal region of predicted periplasmic loop IV (amino acids 283 and 294) and transmembrane domain VIII (amino acid 313) expressed strong complementary fusion activities and were clearly indicative of an eight-transmembrane-domain topology.
Relative to the originally proposed working model, fusions within transmembrane domains II through VII demonstrated enzymatic activities reflective of their relative degrees of insertion into the membrane. Fusions to the first three periplasmic loops demonstrated higher relative alkaline phosphatase activities, as expected, though each possessed some residual β-galactosidase activity, presumably due to tetramerization of the β-galactosidase subunits prior to membrane insertion in some small population of molecules.
The precise structures of periplasmic domain IV and transmembrane domain VIII cannot be discerned entirely from reporter fusion activities. While fusions at amino acid 276 clearly demonstrate periplasmic exposure and the near-carboxy-terminal amino acid 325 is clearly cytoplasmic, three fusions placed between these residues have perplexing activities. Despite the general agreement of the proposed and resolved NixA models with the positive inside rule (49), having a net cytoplasmic charge of at least +12 and a net periplasmic charge of −1 or possibly lower, there are four positively charged residues between amino acids 276 and 294. It has been observed that the loss of Lys or Arg residues in truncated fusion proteins can lead to erroneous results (7). Fusions in this region, such as those in pNLC294 and pNLC313, also appear to be unstable or weakly expressed (Fig. 4). So, while it is clear that NixA spans the membrane between amino acids 276 and 325, the precise nature of this region is not clear. There is, however, sufficient sequence to allow for only a transmembrane domain VIII consistent with observed reporter activities, as addition of a transmembrane IX helix would place the carboxy terminus in the periplasm. Protease protection studies or epitope mapping may be necessary to define the precise topology of this region.
Conserved residues which were shown to be absolutely requisite for Ni2+ transport (Fig. 5) in the motif GX2HAXDADH (20) localized within transmembrane domain II. Similarly, the requisite conserved motif GX2FX2GHSSVV (20) now appears to lie entirely within transmembrane domain III. Four of six additional Asp and Glu residues which reduced transport by >90% when mutated (20) were localized within transmembrane domains V and VI. The remaining two transport-critical residues localized near the cytoplasmic interface of transmembrane domain II and the periplasmic interface of transmembrane domain VII.
Multimerization of transporters with eight transmembrane domains has not been extensively studied, as few examples are known. Dimerization has been suggested in the KefC protein of E. coli (14) while the transmembrane components of the CopA and CopB copper-translocating P-type ATPases function as monomers (22, 37). Integral membrane transport proteins with 10 to 13 membrane-spanning domains have been extensively studied and commonly function as monomers (31). Smaller transport proteins including the vast number of ATP-binding cassette transporters with six membrane-spanning domains function nearly exclusively as multimers, commonly dimers, hence the 6 times 2 rule (1, 24). Chemical cross-linking is commonly used to detect the presence or absence of protein interaction and multimerization in vivo and in vitro (9). Cross-linking of H. pylori proteins in membrane preparations or whole live cells using the homobifunctional membrane soluble cross-linker DSP or its water-soluble analog DTSSP gave no indication of any multimerization of NixA at cross-linker concentrations of 0.1 to 2.5 mM (Fig. 6).
In conclusion, we can now present an empirically derived model of NixA as a monomerically functioning, integral cytoplasmic membrane protein with a mass of 36,991 Da, eight membrane-spanning domains, and the amino and carboxy termini located in the cytoplasm. Furthermore, we have now also localized two requisite sequence motifs common to all Ni2+-specific transporters of this type, GX2HAXDADH and GX2FX2GHSSVV (20, 50), to transmembrane domains II and III. Finally, six additional Glu and Asp residues which also reduced transport by >90% when mutated in previous studies (20) all localized within or immediately adjacent to transmembrane domains II, V, VI, and VII.
ACKNOWLEDGMENT
This work was supported by Public Health Service grant AI25567 from the NIH.
REFERENCES
- 1.Aquila H, Link T, Klingenberg M. Solute carriers involved in energy transfer of mitochondria form a homologous protein family. FEBS Lett. 1987;212:1–9. doi: 10.1016/0014-5793(87)81546-6. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Smith J A, Seidman J G, Struhl K. Current protocols in molecular biology. New York, N.Y: Greene Publishing Associates and Wiley Interscience; 1987. [Google Scholar]
- 3.Bauerfeind P, Garner R M, Mobley H L T. Allelic exchange mutagenesis of nixA in Helicobacter pylori results in reduced nickel transport and urease activity. Infect Immun. 1996;64:2877–2880. doi: 10.1128/iai.64.7.2877-2880.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baumler A J, Heffman F. Direct Genbank submission. Accession no. AF029846.1. 1997. [Google Scholar]
- 5.Birnboim H C, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blaser M J. Helicobacter pylori and the pathogenesis of gastroduodenal inflammation. J Infect Dis. 1990;161:626–633. doi: 10.1093/infdis/161.4.626. [DOI] [PubMed] [Google Scholar]
- 7.Boyd D, Beckwith J. Positively charged amino acid residues can act as topogenic determinants in membrane proteins. Proc Natl Acad Sci USA. 1989;86:9446–9450. doi: 10.1073/pnas.86.23.9446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brickman E, Beckwith J. Analysis of the regulation of Escherichia coli alkaline phosphatase synthesis using deletions and Φ80 transducing phages. J Mol Biol. 1975;96:307–316. doi: 10.1016/0022-2836(75)90350-2. [DOI] [PubMed] [Google Scholar]
- 9.Burgess A J, Matthews I, Grimes E A, Mata A M, Mukonge F M, Lee A J, East J M. Chemical crosslinking and enzyme kinetics provide no evidence for a regulatory role for the 53 kDa glycoprotein of sarcoplasmic reticulum in calcium transport. Biochim Biophys Acta. 1991;1064:139–144. doi: 10.1016/0005-2736(91)90420-d. [DOI] [PubMed] [Google Scholar]
- 10.Cole S T, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon S V, Eiglmeier K, Gas S, Barry C E, 3rd, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Barrell B G. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 11.Dalbey R E. Positively charged residues are important determinants of membrane protein topology. Trends Biochem Sci. 1990;15:353–357. doi: 10.1016/0968-0004(90)90047-f. [DOI] [PubMed] [Google Scholar]
- 12.Degen O, Kobayashi M, Shimizu S, Eitinger T. Selective transport of divalent cations by transition metal permeases: the Alcaligenes eutrophus HoxN and the Rhodococcus rhodochrous NhlF. Arch Microbiol. 1999;171:139–145. doi: 10.1007/s002030050691. [DOI] [PubMed] [Google Scholar]
- 13.Derman A I, Beckwith J. Escherichia coli alkaline phosphatase fails to aquire disulfide bonds when retained in the cytoplasm. J Bacteriol. 1991;173:7719–7722. doi: 10.1128/jb.173.23.7719-7722.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Douglas R M, Ritchie G Y, Munro A W, McLaggan D, Booth I R. The K+-efflux system of Escherichia coli: genetic evidence for oligomeric structure. Mol Membr Biol. 1994;11:55–61. doi: 10.3109/09687689409161030. [DOI] [PubMed] [Google Scholar]
- 15.Eaton K A, Krakowka S. Effect of gastric pH on urease dependent colonization of gnotobiotic piglets by Helicobacter pylori. Infect Immun. 1994;62:3604–3607. doi: 10.1128/iai.62.9.3604-3607.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eitinger T, Friedrich B. A topological model for the high-affinity nickel transporter of Alcaligenes eutrophus. Mol Microbiol. 1994;12:1025–1032. doi: 10.1111/j.1365-2958.1994.tb01090.x. [DOI] [PubMed] [Google Scholar]
- 17.Filip C, Fletcher G, Wulff J L, Earhart C F. Solubilization of the cytoplasmic membrane of Escherichia coli by the ionic detergent sodium-lauryl sarcosinate. J Bacteriol. 1973;115:717–722. doi: 10.1128/jb.115.3.717-722.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Forman D, Newell D G, Fullerton F, Yarnell J W G, Stacey A R, Wald N, Sitas F. Association between Helicobacter pylori infection and risk of gastric cancer: evidence from a prospective study. Br Med J. 1991;302:1302–1305. doi: 10.1136/bmj.302.6788.1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fu C, Javedan S, Moshiri F, Maier R. Bacterial genes involved in incorporation of nickel into a hydrogenase enzyme. Proc Natl Acad Sci USA. 1994;91:5099–5103. doi: 10.1073/pnas.91.11.5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fulkerson J F, Jr, Garner R M, Mobley H L T. Conserved residues and motifs in the NixA protein of Helicobacter pylori are critical for the high affinity transport of nickel ions. J Biol Chem. 1998;273:235–241. doi: 10.1074/jbc.273.1.235. [DOI] [PubMed] [Google Scholar]
- 21.Gafvelin G, von Heijne G. Topological “frustration” in multispanning E. coli inner membrane proteins. Cell. 1994;77:401–412. doi: 10.1016/0092-8674(94)90155-4. [DOI] [PubMed] [Google Scholar]
- 22.Glynn I M, Karlisch S J D. Occluded cations in active transport. Annu Rev Biochem. 1990;59:171–205. doi: 10.1146/annurev.bi.59.070190.001131. [DOI] [PubMed] [Google Scholar]
- 23.Hawtin P R, Delves H T, Newell D G. The demonstration of nickel in urease of Helicobacter pylori by atomic absorption spectroscopy. FEMS Microbiol Lett. 1991;77:51–54. doi: 10.1016/0378-1097(91)90012-y. [DOI] [PubMed] [Google Scholar]
- 24.Higgins C F. ABC transporters: from microorganisms to man. Annu Rev Cell Biol. 1992;8:67–113. doi: 10.1146/annurev.cb.08.110192.000435. [DOI] [PubMed] [Google Scholar]
- 25.Ho S, Hunt H D, Horton R M, Pullen J K, Pease L R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 26.Hu L-T, Mobley H L T. Purification and N-terminal analysis of urease from Helicobacter pylori. Infect Immun. 1990;58:992–998. doi: 10.1128/iai.58.4.992-998.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Komeda H, Kobayashi M, Shimizu S. A novel transporter involved in cobalt uptake. Proc Natl Acad Sci USA. 1997;94:36–41. doi: 10.1073/pnas.94.1.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kyte J, Doolittle R F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 29.Lyne M, Rajandream M A, Barrel B G, Bothe G, Pohl T. Direct Genbank submission. Accession no. AL031787.1. 1998. [Google Scholar]
- 30.Maeda M, Hidaka M, Nakamura A, Masaki S, Vozumi T. Cloning, sequencing, and expression in thermophilic Bacillus sp. strain TB90 of a urease gene complex. J Bacteriol. 1994;176:432–442. doi: 10.1128/jb.176.2.432-442.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maloney P C, Wilson T H. Ion-coupled transport and transporters. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 1130–1148. [Google Scholar]
- 32.Manoil C. Analysis of protein localization by use of gene fusions with complementary activities. J Bacteriol. 1990;172:1035–1042. doi: 10.1128/jb.172.2.1035-1042.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Manoil C, Mekalanos J J, Beckwith J. Alkaline phosphatase fusions: sensors of subcellular location. J Bacteriol. 1990;172:515–518. doi: 10.1128/jb.172.2.515-518.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marshall B J, Barett L J, Prakesh C, McCallum R W, Guerrant R L. Campylobacter IV. Goteberg, Sweden: University of Goteberg; 1988. pp. 402–403. [Google Scholar]
- 35.Marshall B J, McGechie D B, Rogers P A, Glancy R J. Pyloric Campylobacter infection and gastroduodenal disease. Med J Aust. 1985;142:439–444. doi: 10.5694/j.1326-5377.1985.tb113444.x. [DOI] [PubMed] [Google Scholar]
- 36.Marshall B J, Warren J R. Unidentified curved bacilli on gastric epithelium in active chronic gastritis. Lancet. 1984;i:1311–1315. [PubMed] [Google Scholar]
- 37.Melchers K, Weitzenegger T, Buhmann A, Steinbiller W, Sachs G, Schafer K P. Cloning and membrane topology of a P type ATPase from Helicobacter pylori. J Biol Chem. 1996;271:446–457. doi: 10.1074/jbc.271.1.446. [DOI] [PubMed] [Google Scholar]
- 38.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. pp. 352–355. [Google Scholar]
- 39.Mobley H L T, Garner R M, Bauerfeind P G. Helicobacter pylori nickel transport gene nixA: synthesis of catalytically active urease in E. coli independent of growth conditions. Mol Microbiol. 1995;16:97–109. doi: 10.1111/j.1365-2958.1995.tb02395.x. [DOI] [PubMed] [Google Scholar]
- 40.Mobley H L T, Island M D, Hausinger R P. Molecular biology of microbial ureases. Microbiol Rev. 1995;59:451–480. doi: 10.1128/mr.59.3.451-480.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Parsonnet J, Hansen S, Rodriguez L, Gelb A, Warnke R, Jellum E, Orentreich N, Vogelman J, Friedman G. Helicobacter pylori infection and gastric lymphoma. N Engl J Med. 1994;330:1267–1271. doi: 10.1056/NEJM199405053301803. [DOI] [PubMed] [Google Scholar]
- 42.Pigeon R P, Silver R P. Topological and mutational analysis of KpsM, the hydrophobic component of the ABC-transporter involved in the export of polysialic acid in E. coli K1. Mol Microbiol. 1994;14:871–881. doi: 10.1111/j.1365-2958.1994.tb01323.x. [DOI] [PubMed] [Google Scholar]
- 43.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 44.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sundermann F W., Jr Biological monitoring of nickel in humans. Scand J Work Environ Health. 1993;19(Suppl. 1):34–38. [PubMed] [Google Scholar]
- 46.Taylor R K, Manoil C, Mekalanos J J. Broad-host-range vectors for delivery of TnphoA: use in genetic analysis of secreted virulence determinants in Vibrio cholorae. J Bacteriol. 1989;171:1870–1878. doi: 10.1128/jb.171.4.1870-1878.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tiedeman A A, Smith J M. lacZY cassettes with KanR resistance. Nucleic Acids Res. 1988;16:3587. doi: 10.1093/nar/16.8.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tsuda M, Karita M, Moreshed M G, Okita K, Nakazawa T. A urease negative mutant of Helicobacter pylori constructed by allelic exchange mutagenesis lacks the ability to colonize the nude mouse stomach. Infect Immun. 1994;62:3586–3589. doi: 10.1128/iai.62.8.3586-3589.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Von Heijne G. Membrane protein structure prediction: hydrophobicity analysis and the positive inside rule. J Mol Biol. 1992;225:487–494. doi: 10.1016/0022-2836(92)90934-c. [DOI] [PubMed] [Google Scholar]
- 50.Wolfram L, Friedrich B, Eitinger T. The Alcaligenes eutrophus protein HoxN mediates nickel transport in Escherichia coli. J Bacteriol. 1995;177:1840–1843. doi: 10.1128/jb.177.7.1840-1843.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]