Abstract
A 7.5-kbp fragment of chromosomal DNA downstream of the Vibrio cholerae vibriobactin outer membrane receptor, viuA, and the vibriobactin utilization gene, viuB, was recovered from a Sau3A lambda library of O395 chromosomal DNA. By analogy with the genetic organization of the Escherichia coli enterobactin gene cluster, in which the enterobactin biosynthetic and transport genes lie adjacent to the enterobactin outer membrane receptor, fepA, and the utilization gene, fes, the cloned DNA was examined for the ability to restore siderophore synthesis to E. coli ent mutants. Cross-feeding studies demonstrated that an E. coli entF mutant complemented with the cloned DNA regained the ability to synthesize enterobactin and to grow in low-iron medium. Sequence analysis of the cloned chromosomal DNA revealed an open reading frame downstream of viuB which encoded a deduced protein of greater than 2,158 amino acids, homologous to Yersinia sp. HMWP2, Vibrio anguillarum AngR, and E. coli EntF. A mutant with an in-frame deletion of this gene, named vibF, was created with classical V. cholerae strain O395 by in vivo marker exchange. In cross-feeding studies, this mutant was unable to synthesize ferric vibriobactin but was able to utilize exogenous siderophore. Complementation of the mutant with a cloned vibF fragment restored vibriobactin synthesis to normal. The expression of the vibF promoter was found to be negatively regulated by iron at the transcriptional level, under the control of the V. cholerae fur gene. Expression of vibF was not autoregulatory and neither affected nor was affected by the expression of irgA or viuA. The promoter of vibF was located by primer extension and was found to contain a dyad symmetric nucleotide sequence highly homologous to the E. coli Fur binding consensus sequence. A footprint of purified V. cholerae Fur on the vibF promoter, overlapping the Fur binding consensus sequence, was observed using DNase I footprinting. The protein product of vibF is homologous to the multifunctional nonribosomal protein synthetases and is necessary for the biosynthesis of vibriobactin.
Iron is a required element for bacterial survival and growth. Under aerobic conditions, however, iron is present only in insoluble mineral complexes or bound to a mammalian host's own iron-binding proteins (11, 12). Vibrio cholerae, like many pathogenic bacteria, produces a siderophore—a low-molecular-weight iron chelator—which effectively competes with human iron-binding proteins, such as lactoferrin and transferrin (12, 35). The V. cholerae siderophore vibriobactin belongs to the group of catechol siderophores, of which enterobactin, a cyclic trimer of 2,3-dihydroxybenzoyl-l-serine produced by Escherichia coli, is the prototype (35). Vibriobactin contains three residues of 2,3-dihydroxybenzoic acid (2,3-DHBA) and two residues of threonine, which are in the form of oxazoline rings (21). The polyamine backbone of vibriobactin is unusual in being the rare compound norspermidine [N-(-aminopropyl)-1,3-diaminopropane] rather than spermidine, which is found in the Paracoccus denitrificans siderophore parabactin and the Agrobacterium tumefaciens siderophore agrobactin (21). The biosynthetic steps needed for vibriobactin production are not yet clearly defined.
In E. coli, the enterobactin biosynthetic (ent) and transport (fep) genes lie in a large gene cluster, with the clockwise order of the enterobactin genes on the E. coli chromosome being entD, fepA (which encodes the enterobactin outer membrane receptor), fes (which is involved in ferric enterobactin uptake and processing), entF, fepE, fepC, fepG, fepD, fepB, entC, entE, entB, and entA. Similarly, the gene encoding the vibriobactin outer membrane receptor, viuA, lies upstream of a second gene, viuB, which encodes a cytoplasmic protein analogous in function to fes (5, 6). By analogy with the enterobactin gene cluster, other genes involved in vibriobactin biosynthesis and uptake may be located near viuA and viuB. However, recent work has identified a group of vibriobactin biosynthetic (vib) and utilization (viu) genes that are not linked to the viuA-viuB region (GenBank accession no. U52150) (52). These include genes postulated to function in the synthesis of 2,3-DHBA from chorismate (vibC, vibB, and vibA), a vibriobactin synthase cluster linking the three residues of 2,3-DHBA to two residues of threonine with the norspermidine backbone (vibBDEH), and several genes with transport function (viuPDGC). The order of these genes on the V. cholerae chromosome, vibB, vibE, vibC, vibA, vibH, viuP, viuD, viuG, viuC, and vibD, is quite different from that of the enterobactin gene cluster, and no congener of entF is present in this cluster.
The production of vibriobactin is negatively regulated by iron, and many of the genes involved in vibriobactin production and uptake have been found to be negatively regulated by iron at the transcriptional level, under the control of the iron repressor protein Fur. The transcription of both viuA and viuB is derepressed under high-iron conditions in a fur mutant (5, 6), while the uptake and utilization genes vibB, vibC, vibH, viuP, and viuC all have potential Fur binding consensus sequences in their putative promoter regions (GenBank accession no. U52150) (52). In addition to the production of vibriobactin, V. cholerae possesses other iron uptake systems, which use the iron in ferric citrate, ferrichrome, hemin, or hemoglobin (44, 47). The heme transport system also depends on a gene cluster that is controlled by Fur (24, 25, 36). The gene encoding the 70-kDa major iron-regulated outer membrane protein of V. cholerae, IrgA, is negatively controlled by Fur but is also under the positive transcriptional regulation of the product of a divergently transcribed gene, irgB (17–20, 50). IrgA is a virulence factor in V. cholerae but has not yet been demonstrated to play a role in iron acquisition. The interplay of transcriptional regulators in the control of iron-regulated genes in V. cholerae remains to be investigated.
In this study, we have characterized an additional gene in the vibriobactin biosynthetic and uptake apparatus. The protein product of this gene is a congener of EntF and is homologous to a family of proteins involved in the biosynthesis of siderophores and peptide antibiotics. We describe the phenotype of a deletion mutant form of this gene, examine its regulation by iron and Fur and its interaction with other iron-regulated genes, and present studies that suggest that the protein product of this gene acts as a multifunctional nonribosomal peptide synthetase in the biosynthesis of vibriobactin.
(Portions of this work were presented at the 97th General Meeting of the American Society for Microbiology [abstr. B-235].)
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains and plasmids used in this study are described in Table 1.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant genotype or phenotypea | Reference or source |
|---|---|---|
| V. cholerae strains | ||
| O395 | Wild-type V. cholerae, classical biotype; Smr | 31 |
| MBG14 | O395 viuA::TnphoA; Smr Kmr | 6 |
| MBG40 | O395 irgA::TnphoA; Smr Kmr | 20 |
| CML19 | O395 Δfur; Smr | 30 |
| JRB3 | O395 ΔirgB; Smr | 50 |
| JRB15 | O395 lacZ::vibFp→phoA; Smr | This study |
| JRB16 | O395 Δfur lacZ::vibFp→phoA; Smr | This study |
| JRB17 | O395 ΔirgB lacZ::vibFp→phoA; Smr | This study |
| JRB18 | O395 ΔvibF; Smr | This study |
| JRB19 | MBG14 ΔvibF; Smr Kmr | This study |
| JRB20 | MBG40 ΔvibF; Smr Kmr | This study |
| JRB21 | O395 ΔvibF lacZ::vibFp→phoA; Smr | This study |
| JRB22 | O395 irgA lacZ::vibFp→phoA; Cmr Smr | This study |
| JRB23 | O395 viuA lacZ::vibFp→phoA; Tcr Smr | This study |
| E. coli strains | ||
| DH5α | F−deoR endA1 hsdR17 supE44 thi-1 recA1 gyrA96 relA1 Δ(argF-lacZYA)U169 (φ80d lacZ ΔM15) | BRLb |
| AB1515 | purE42 proC14 leu-6 trpE38 thi-1 fhuA23 lacY1 mtl-1 xyl-5 rpsL109 azi-6 tsx-67 | S. Payne; 45 |
| AN117 | entF derivative of AB1515 | 52 |
| SM10λpir | thi thr leu tonA lacY supE recA::RP4-2-Tc::Mu λpirR6K; Kmr | 34 |
| SY327λpir | Δ(lac-pro) nalA recA56 araD argE(Am) λpirR6K; Smr | 34 |
| Plasmids | ||
| pGEM-T | PCR cloning vector; Ampr | Promega Corp. |
| pCVD442 | Suicide vector composed of mob, ori, and bla regions of pGP704 and sacB gene of B. subtilis; Ampr | 16 |
| pBluescript II SK+ | Cloning vector; Ampr | Stratagene |
| pUJ10 | pUC18-based plasmid with multiple cloning site between divergent E. coli lacZ and phoA genes; Ampr | 15 |
| p6891MCS | pBR327 containing 8-kbp Sau3A fragment of V. cholerae DNA encoding lacZ and multiple cloning site inserted at KpnI within lacZ; Ampr | 4 |
| pJRB20 | pBluescript II SK+ with 7.5-kbp cloned SacI insert from Sau3A LambdaGEM-11 library of V. cholerae chromosomal DNA, containing 5′ end of vibF; Ampr | This study |
| pMHC4 | pGEM-T with 760-bp cloned insert of V. cholerae chromosomal DNA downstream of region cloned into pJRB20; Ampr | This study |
| pWCW1 | pBluescript II SK+ with cloned 1,552-bp HindIII fragment within vibF from pJRB20; Ampr | This study |
| pWCW2 | pWCW1 with 882-bp in-frame deletion of vibF between two internal BglII sites; Ampr | This study |
| pWCW3 | pCVD442 with SalI-SacI fragment from pWCW2 containing 882-bp in-frame deletion of vibF; Ampr | This study |
| pMHC1 | pUJ10 with 228-bp XbaI-BamHI fragment containing intergenic region of viuB-vibF, such that vibFp→phoA; Ampr | This study |
| pJRB44 | p6891MCS with 2.8-kbp NotI fragment from pMHC1 containing vibFp→phoA; Ampr | This study |
| pPAC19 | pCVD442 with 3.7-kbp SacI-SphI fragment containing irgA::Cmr; Ampr Cmr | 48 |
| pPAC20 | pCVD442 with 4.2-kbp PvuII fragment containing viuA::Tcr; Ampr Tcr | 48 |
Ampr, ampicillin resistance; Cmr, chloramphenicol resistance; Kmr, kanamycin resistance; Smr, streptomycin resistance, Tcr, tetracycline resistance.
BRL, Bethesda Research Laboratories Life Technologies, Inc.
Media.
All strains were preserved by storage at −70°C in Luria broth (LB) medium (43) containing 15% glycerol. LB medium, with or without the addition of the iron chelator 2,2-dipyridyl (final concentration, 0.2 mM), was used for growth under low- and high-iron conditions, respectively. Ampicillin (100 μg/ml), chloramphenicol (12.5 μg/ml), tetracycline (2 μg/ml), and streptomycin (100 μg/ml) were added as appropriate.
Genetic methods.
Isolation of plasmid DNA, preparation of RNA, restriction enzyme digests, agarose gel electrophoresis, plaque hybridization, and Southern hybridization of DNA separated by electrophoresis were performed by standard molecular biologic techniques (43). Bacterial chromosomal DNA was prepared using the Easy-DNA Kit (Invitrogen Corp., Carlsbad, Calif.). Genescreen Plus and Colony/PlaqueScreen hybridization transfer membranes (NEN Research Products, Boston, Mass.) were used in accordance with the manufacturer's protocols for Southern and plaque hybridizations. Primer extension analysis was performed as previously described (33); oligonucleotide primers were hybridized to RNA in 0.4 M NaCl–40 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)], pH 6.4, without formamide, at 55°C for 2 h. RNasin and avian myeloblastosis virus reverse transcriptase were obtained from Bethesda Research Laboratories Life Technologies, Inc. (Gaithersburg, Md.). DNA sequencing was performed at the Massachusetts General Hospital Department of Molecular Biology DNA Sequencing Core Facility using ABI Prism DiTerminator Cycle sequencing with AmpliTaq DNA polymerase FS and an ABI 377 DNA sequencer (Perkin-Elmer Applied Biosystems Division, Foster City, Calif.).
Plasmids were transformed into E. coli strains by standard heat shock techniques or electroporated into V. cholerae by using a Gene Pulser (Bio-Rad Laboratories, Richmond, Calif.) in accordance with the manufacturer's protocol, with modifications for electroporation into V. cholerae as previously described (19). E. coli strains DH5α and SY327λpir were used as recipients for the plasmid constructs described below. DNA restriction endonucleases and T4 DNA ligase were used in accordance with the manufacturers' specifications. Restriction enzyme-digested chromosomal and plasmid DNA fragments were separated on 1% agarose gels; fragments of interest were cut from the gel under UV illumination and purified using GenElute spin columns (Supelco Inc., Bellefonte, Pa.). DNA fragments used as probes were radiolabeled with [α-32P]dCTP using the Prime-It II random priming labeling kit (Stratagene, La Jolla, Calif.).
The PCR was carried out using a standard DNA minicycler (M. J. Research, Inc., Watertown, Mass.). All PCRs were performed using the following conditions: 94°C for 4 min; 30 cycles of 94°C for 1 min (denaturing), 50°C for 1 min (annealing), and 72°C for 1 min (extension); and 72°C for 5 min (terminal extension).
Construction of plasmids.
A 7.5-kbp fragment of V. cholerae O395 chromosomal DNA downstream of the gene encoding the vibriobactin outer membrane receptor, viuA, and the vibriobactin utilization gene, viuB, was recovered from a Sau3A LambdaGEM-11 (Promega Corp., Madison, Wis.) library of O395 chromosomal DNA (gift of Ken Peterson) as follows. Plaques were hybridized with a 1,485-bp SacI (bp 2669)-to-BglII (bp 4155) DNA fragment probe encompassing viuB (5, 6) (Fig. 1). Bacteriophage DNA was isolated from hybridizing plaques by standard techniques (43). A 7.5-kbp SacI fragment containing O395 chromosomal DNA was isolated from one of the hybridizing bacteriophages and ligated into the unique SacI site of pBluescript II SK+ (Stratagene) to create plasmid pJRB20. One of the flanking SacI sites was from O395 chromosomal DNA; the other was from the LambdaGEM-11 vector, in which a SacI site is adjacent to the BamHI site within which the Sau3A chromosomal fragments were cloned.
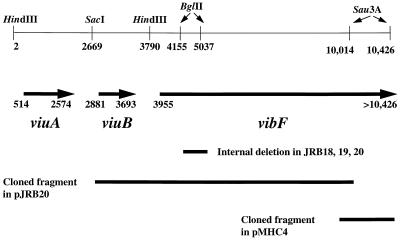
FIG. 1.
Partial restriction map of O395 chromosomal DNA with relevant restriction enzyme sites, locations of the cloned fragments in pJRB20 and pMHC4, and position of the internal deletion of vibF in JRB18, JRB19, and JRB20. The extents of the coding regions of viuA, viuB, and vibF are indicated by solid arrows. The numbering of sites corresponds to that in reference 5.
Sequence analysis of the fragment in pJRB20 revealed a single long open reading frame (see below), which we called vibF. Chromosomal DNA downstream of the insert in pJRB20 was isolated by PCR amplification of the hybridizing bacteriophages with primers complementary to the multiple cloning site of the right arm of the LambdaGEM-11 cloning vector (5′ CCATTTAGGTGACACTATGAA 3′) and to the previously sequenced downstream region of vibF (5′ AGTTTCCCTAATGCCAAG 3′). A 760-bp PCR product was amplified from one of the isolated hybridizing phages and cloned into PCR cloning vector pGEM-T (Promega) to create plasmid pMHC4 (Fig. 1). Both strands of the chromosomal DNA insert in pMHC4 were sequenced. Primers complementary to internal sequences of the pMHC4 chromosomal insert were used to amplify fragments from O395 chromosomal DNA which were then resequenced to confirm that the sequence obtained from the bacteriophage library was identical to those in the V. cholerae chromosome (data not shown).
An in-frame deletion of vibF was constructed as follows. pJRB20 was digested with HindIII, and the 1,552-bp HindIII fragment within vibF was cloned within the unique HindIII site of pBluescript II SK+ to create pWCW1. An 882-bp in-frame deletion of vibF was made between the two BglII sites in pWCW1 to create pWCW2 (Fig. 1). pWCW2 was digested with SalI and SacI; the 670-bp fragment encompassing the vibF deletion was cloned into the SalI and SacI sites of plasmid pCVD442 to create pWCW3. pCVD442 is a suicide vector containing the pir-dependent R6K replicon, the ampicillin resistance determinant, and the sacB gene from Bacillus subtilis (16).
Construction of V. cholerae vibF mutants.
The V. cholerae vibF mutant JRB18 was constructed as follows. pWCW3 was transformed into SM10λpir and then crossed with streptomycin-resistant V. cholerae O395. Streptomycin- and ampicillin-resistant merodiploids were subsequently grown overnight in LB medium without ampicillin selection, plated on LB agar containing 10% sucrose but no NaCl, and grown at 30°C for 30 h. Because the product of the sacB gene is lethal in the presence of sucrose, surviving colonies had deleted the integrated sacB gene via a second recombination event and lost the resulting plasmid (3). In particular, sucrose-resistant colonies that were ampicillin susceptible had either re-excised the original plasmid to yield the parent O395 or had resolved the merodiploid state to replace the vibF locus in O395 with the vibF deletion from pWCW3. These two possibilities were distinguished by colony blotting and Southern hybridization (data not shown). One of these colonies was purified and named JRB18. Using the above methods, vibF deletions were also made in V. cholerae strains MBG14 (O395 viuA::TnphoA) and MBG40 (O395 irgA::TnphoA) to create JRB19 and JRB20, respectively.
Construction of alkaline phosphatase reporter strains.
Reporter strains JRB15, JRB16, and JRB17 were constructed from V. cholerae strains O395, CML19 (Δfur), and JRB3 (ΔirgB) as follows. A 228-bp DNA fragment containing the intergenic region of viuB-vibF was amplified by PCR (oligonucleotides 5′-AAAATCTAGAGCGGCCGCCATTCACCTTGCCTGTTA-3′ and 5′-TTTTGGATCCTTAAACCCACAGATTCATCCC-3′) with flanking 5′ XbaI-NotI and 3′ BamHI restriction sites and inserted into the XbaI and BamHI sites within the multiple cloning site of pUJ10, a pUC18-based plasmid containing a multiple cloning site between divergent E. coli lacZ and phoA genes (15), such that the vibF promoter controls the transcription of phoA, to create pMHC1. After cloning, this insert was fully sequenced, including both junctions. A 2.8-kbp fragment containing the viuB-vibF intergenic region, as well as the flanking phoA reporter gene, was removed by NotI digestion and ligated into the unique NotI site of p6891MCS, a plasmid containing a multiple cloning site between two fragments of the V. cholerae lacZ gene (4), to create plasmid pJRB44. This construct was introduced by in vivo marker exchange into the lacZ genes of V. cholerae strains O395, CML19, and JRB3, as previously described (4), to create strains JRB15, JRB16, and JRB17, respectively. Correct insertion of the construct within lacZ was confirmed by Southern blot using the 2.1-kbp HpaI fragment of p6891MCS as a probe (data not shown).
Strains JRB21 (ΔvibF), JRB22 (irgA), and JRB23 (viuA) were constructed as follows. A vibF deletion was made in reporter strain JRB15 by in vivo marker exchange with pWCW3 as described above to create JRB21. irgA and viuA mutations were made in JRB15 by similar methods using previously described plasmids pPAC19 and pPAC20 (48) to create JRB22 and JRB23.
Bioassay.
Utilization of vibriobactin was determined by bioassay (36). Organisms (105/ml) of indicator bacterial strains were solidified in iron-depleted medium (LB agar with ethylenediamine di[o-hydroxyphenylacetic acid] [EDDA] at 75 μg/ml for V. cholerae strains or at 500 μg/ml for E. coli strains) deferrated by the method of Rogers [40]). The ability of these strains to use vibriobactin was determined by measuring the growth of the indicator strains around 10-μl spots of stationary-phase bacterial cultures (producer strains) after incubation at 37°C for 24 to 48 h. The indicator strains would not grow in the absence of usable exogenous siderophore or iron. A solution of ferrous sulfate (18 mM) was used as a positive control.
Alkaline phosphatase reporter assay.
vibFp→phoA reporter strains were grown overnight in LB with and without the iron chelator 2,2-dipyridyl. Overnight cultures were standardized for density of bacterial growth, pelleted, washed, and permeabilized as previously described (32). Measurement of the amount of hydrolysis of p-nitrophenyl phosphate (Amresco) by permeabilized cells allowed calculation of the alkaline phosphatase activity. Test samples were prepared in triplicate, and strain O395 lacking the reporter construct was used as a negative control to account for endogenous phosphatase activity.
DNase I footprinting assay.
Various amounts of purified Fur (50, 51) were incubated with a biotin end-labeled 228-bp DNA fragment containing the intergenic region of viuB-vibF described above. The binding buffer contained 20 mM Tris (pH 7.5), 5 mM MgCl2, 50 mM NaCl, bovine serum albumin at 100 μg/ml, salmon sperm DNA at 5 μg/ml, 100 μl of MnCl2, and 10% glycerol. One picomole of DNA and the designated amount of purified Fur were added to a 95-μl sample volume and allowed to incubate at room temperature for 30 min. Samples were subsequently incubated with bovine pancreatic DNase I (Worthington Biochemical Corporation, Freehold, N.J.) for exactly 30 s. The reaction was stopped by addition of 10 μl of a solution of 0.125 M EDTA and tRNA at 2.5 mg/ml, and DNA fragments were precipitated with ethanol. Samples were resuspended in a loading buffer consisting of 8 M urea, 40 mM Tris (pH 8.45), 40 mM borate, 0.05% bromophenol blue, and 0.05% xylene cyanol and stored at −80°C until use. A Maxam-and-Gilbert G + A reaction was performed on the same DNA fragment used for footprinting, and this was used as a position marker (30a). Samples were run on a 5% polyacrylamide gel, transferred to a Sequenase Images nylon membrane (United States Biochemical Corp.) by contact blotting, and developed using the Sequenase Images Detection Kit.
DNA and protein database searches.
Nucleotide and derived amino acid sequences were assembled and initially analyzed using DNA Strider software version 1.0 (Christian Marck, Commissariat a l'Energie Atomique, Paris, France). Database comparisons were performed with the Basic Local Alignment Search Tool programs (1) using the server at the National Center for Biotechnology Information. Preliminary sequence data was obtained from the website of The Institute for Genomic Research (http://www/tigr.org). Protein homology searches were performed using the FASTA program, version 3.26, using the server at the European Bioinformatics Institute (37). A homology comparison to a database of conserved protein motifs was performed with the BLOCKS program (26). The hydropathicity index profile was calculated by using the Kyte-Doolittle formula (28).
Nucleotide sequence accession number.
The GenBank accession number for the vibF sequence presented here is AF030977.
RESULTS
Complementation of an E. coli entF mutant with the V. cholerae chromosomal insert in pJRB20.
V. cholerae chromosomal DNA downstream of the gene encoding the vibriobactin outer membrane receptor, viuA, and the utilization gene, viuB, was recovered as a 7.5-kbp SacI insert in pJRB20. In E. coli, entF, which encodes a cytoplasmic protein required for enterobactin biosynthesis, is adjacent to fepA, encoding the enterobactin outer membrane receptor, and fes, which encodes a protein involved in utilization of ferric enterobactin. To investigate whether the cloned V. cholerae chromosomal DNA might have EntF-like activity, the ability of pJRB20 to complement an E. coli entF mutant, AN117, was investigated. Wild-type parent E. coli strain AB1515, when complemented with the cloning vector pBluescript II SK+, grew to an optical density at 600 nm (OD600) of 0.598 ± 0.117 (mean ± standard deviation of triplicate cultures) following overnight growth in LB medium with added 2,2-dipyridyl. The entF mutant AN117(pBluescript II SK+) grew poorly under iron stress (OD600 of 0.036 ± 0.003). pJRB20 partially restored the growth of AN117 in low-iron medium to the level of the parent strain (OD600 of 0.338 ± 0.002).
The ability of the cloned V. cholerae chromosomal DNA on pJRB20 to complement the mutation in AN117 was also evaluated with a bioassay. In this assay, indicator strains do not grow in the absence of usable exogenous siderophore or iron; when siderophore or iron is present, a visible zone of growth can be seen around the siderophore-producing strains or iron compounds. The indicator strain used was the entF deletion mutant AN117, which is unable to synthesize ferric enterobactin but utilizes the siderophore normally. The producer strains tested were wild-type E. coli strain AB1515, the entF deletion mutant AN117, and AN117 complemented with pJRB20. The indicator strain grew when cross-fed by wild-type producer strain AB1515 (zone of growth, 18 mm) but had no zone of growth in the presence of the entF deletion mutant. The complemented mutant AN117(pJRB20) regained the ability to cross-feed the indicator strain (zone of growth, 15 mm), demonstrating that complementation of the entF mutant with pJRB20 restored enterobactin synthesis to normal.
Nucleotide sequence of vibF.
The 7.5-kbp V. cholerae chromosomal insert in pJRB20 and the overlapping 760-bp insert in pMHC4 were cloned and sequenced as described above. One large open reading frame, which was named vibF, was identified (Fig. 1), beginning at bp 3955 and extending beyond the recovered chromosomal DNA, and therefore potentially encoding a deduced protein of greater than 2,158 amino acids. Three potential starting methionines were present, but a strongly conserved Shine-Dalgarno sequence was not identified.
Identification of components of a multifunctional nonribosomal peptide synthetase in VibF.
The hydropathicity plot of VibF revealed no signal sequence or transmembrane domains (data not shown), suggesting that VibF is not a periplasmic or membrane protein but may be a cytoplasmic protein involved in ferric vibriobactin biosynthesis.
The FASTA algorithm for protein homology was used to compare VibF with other proteins in the SWISS-PROT database. VibF was highly homologous to a number of polypeptides involved in the biosynthesis of siderophores and peptide antibiotics, including Yersinia sp. HMWP2, a virulence factor involved in yersiniabactin synthesis (SWISS-PROT accession no. P48633; 24.9% identity in a 1,747-amino-acid overlap) (2, 22); Vibrio anguillarum AngR, a positive transcriptional regulator involved in anguibactin synthesis (accession no. P19828; 25.0% identity in a 984-amino-acid overlap) (9, 49); E. coli EntF, which is required for enterobactin synthesis (accession no. P11454; 24.0% identity in a 987-amino-acid overlap) (41); Bacillus brevis gramicidin S synthetases (accession no. P14688 and P14687); and B. subtilis surfactin synthetases (accession no. P27206, Q08787, and Q04747) (10), serine-activating enzyme (accession no. P45745), and peptide synthetases (accession no. P39846 and P39845).
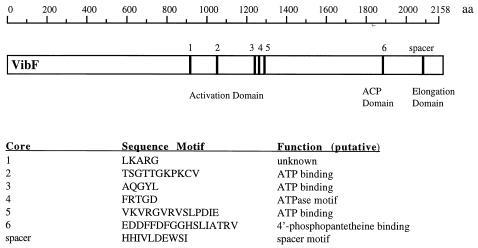
Using the BLOCKS program to search for conserved domains, motifs suggestive of components of one module of a multifunctional nonribosomal peptide synthetase were detected in the deduced amino acid sequence of VibF (13, 29, 38, 46). The organization of these sequence motifs within the N terminus of VibF is shown in Fig. 2. An activation or AMP-binding domain, belonging to the family of adenylate-forming enzymes, extends from amino acid 922 to amino acid 1325 of the predicted protein and contains five core sequence motifs. A phosphopantetheinyl attachment site is present within the acyl carrier protein domain, with a conserved sequence beginning at amino acid 1882 containing the core motif GGHSL. A spacer motif begins at amino acid 2083, within the presumptive elongation domain.
FIG. 2.
Organization of domains and core sequence motifs within the peptide synthetase module identified from the deduced amino acid sequence of VibF. The relative locations of the core sequences (cores 1 to 6) and spacer motif, their amino acid sequences in one-letter code, and their putative functions are indicated. The locations of the activation, acyl carrier protein (ACP), and elongation domains are marked.
Bioassay of ferric vibriobactin production and utilization.
To further assess the role of VibF in vibriobactin synthesis, an internal deletion of vibF was made in classical V. cholerae strain O395; this mutant strain, JRB18, was then complemented with the vibF fragment on plasmid pJRB20. Analysis of the ability of JRB18 to synthesize or utilize exogenous ferric vibriobactin was evaluated in a bioassay. JRB18 was used as the indicator strain, while the parent O395, vibF mutant JRB18, and JRB18 complemented with pJRB20 were the producer strains. The indicator strain grew when cross-fed by O395 (zone of growth, 40 mm) but did not grow in the presence of mutant JRB18. Complementation of the vibF mutant with the portion of vibF cloned on pJRB20 restored growth of the indicator strain (zone of growth, 20 mm).
Regulation of the vibF promoter.
To investigate the regulation of vibF transcription in more detail, chromosomal vibFp-phoA operon fusions were constructed. The role of iron in vibF expression was determined by measuring the alkaline phosphatase activity of reporter strain JRB15 (O395 lacZ::vibFp→phoA) after overnight growth in medium with or without the iron chelator 2,2-dipyridyl. Reporter strains JRB16 and JRB17 were used to test whether vibF expression was influenced by the regulatory proteins Fur and IrgB, while strains JRB21, -22, and -23 were used to evaluate the influence of VibF, IrgA, and ViuA on vibF expression. As shown in Table 2, vibF was negatively regulated by iron at the transcriptional level, under the control of the V. cholerae fur gene. In addition, vibF expression was reduced approximately twofold in an irgB mutant (P < 0.02 by Student's t test). However, vibF expression was not autoregulatory and neither affected nor was affected by the expression of irgA or viuA.
TABLE 2.
Alkaline phosphatase activities in high- and low-iron media
| Strain | Genotype | Alkaline phosphatase activity (U/A600 of cells)a
|
|
|---|---|---|---|
| High ironb | Low ironc | ||
| O395 | Wild type | 1 ± 0 | 1 ± 0 |
| MBG14 | viuA::TnphoA | 1 ± 0 | 26 ± 8 |
| MBG40 | irgA::TnphoA | 2 ± 0 | 572 ± 316 |
| JRB15 | lacZ::vibFp→phoA | 2 ± 0 | 112 ± 7 |
| JRB16 | lacZ::vibFp→phoA fur | 413 ± 8 | 151 ± 46 |
| JRB17 | lacZ::vibFp→phoA irgB | 2 ± 0 | 47 ± 19 |
| JRB18 | O395 vibF | 1 ± 0 | 2 ± 0 |
| JRB19 | MBG14 vibF | 1 ± 0 | 23 ± 3 |
| JRB20 | MBG40 vibF | 1 ± 0 | 832 ± 96 |
| JRB21 | lacZ::vibFp→phoA vibF | 2 ± 0 | 138 ± 5 |
| JRB22 | lacZ::vibFp→phoA irgA | 2 ± 0 | 159 ± 8 |
| JRB23 | lacZ::vibFp→phoA viuA | 2 ± 0 | 117 ± 32 |
Units of alkaline phosphatase activity reflect the mean ± the standard deviation of triplicate samples (rounded to the nearest unit) and include a normalization factor for the density of culture growth. V. cholerae O395 and JRB18 were used as negative controls to account for endogenous phosphatase activity.
High iron, LB medium.
Low iron, LB medium with 0.2 mM 2,2-dipyridyl.
Localization of the transcriptional start site and promoter of vibF by primer extension.
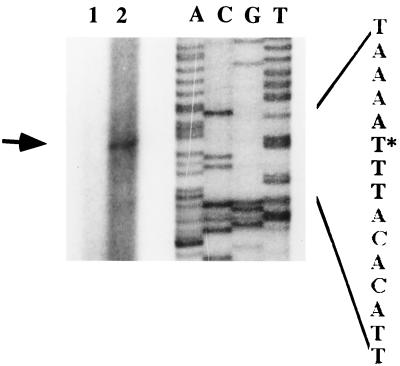
Primer extension analysis with RNA from strain O395 grown in low-iron medium was performed with a 21-mer synthetic oligonucleotide, 5′ CAAATTCGGCGTAGAGATGCG 3′, complementary to the DNA sequence located 76 bp downstream of the methionine start codon of vibF. As shown in Fig. 3, primer extension analysis revealed a potential transcriptional start site 65 bp upstream of the most proximal translational start site (marked with an asterisk in Fig. 4). Possible −10 and −35 boxes of the vibF promoter, homologous to the E. coli consensus sequences, are indicated in Fig. 4.
FIG. 3.
Primer extension analysis of RNA from strain O395 grown in low-iron medium using a 21-mer oligonucleotide at an annealing temperature of 55°C. Lanes A, C, G, and T are the corresponding lanes of the DNA sequencing ladder. Lane 1 is the control without added primer, and lane 2 is the primer extension reaction. The identified transcriptional start site (arrow) is shown with an asterisk in Fig. 4.
FIG. 4.
Nucleotide sequence of the putative promoter region of vibF. The deduced amino acid sequence of VibF is shown in single-letter code beneath the DNA sequence. The likely transcriptional start site (∗), −10 box (−10), and −35 box (−35) are indicated above the sequence. Three potential starting methionine codons are shown. An inverted repeat homologous to the Fur binding consensus sequence of E. coli is underlined. The binding site for Fur deduced from DNase I protection (see Fig. 5) is boxed.
Homology of the vibF promoter region to the Fur binding consensus sequence.
The postulated promoter region of vibF was examined for homology to the Fur binding consensus sequence of E. coli. As shown in Fig. 4, the vibF promoter region has an area of dyad symmetry that shares 16 of 19 bp with the E. coli Fur binding consensus sequence GATAATGATTATTATTAAC (7, 8, 14, 23).
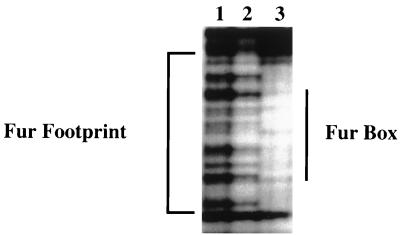
DNase I footprinting assays.
Footprinting studies were performed to document binding of Fur to the predicted vibF promoter. When Fur was titrated into the binding reaction mixture, a DNase I footprint was seen overlapping the Fur binding consensus sequence within the vibF promoter (Fig. 4 and 5). The footprint was dependent on the presence of metal in the binding mixture (data not shown).
FIG. 5.
Various amounts of purified Fur were incubated with a DNA fragment containing the viuB-vibF intergenic region (10 nM concentration) and subjected to DNase I digestion. Lanes: 1, DNA fragment alone; 2, DNA fragment incubated with 10 nM Fur; 3, DNA fragment incubated with 50 nM Fur. The DNase I footprint is indicated, as is the location of the Fur binding consensus sequence shown in Fig. 4.
DISCUSSION
The identification of two genes in the vibriobactin uptake apparatus, viuA, the ferric vibriobactin outer membrane receptor gene, and viuB, a vibriobactin utilization gene, suggested that other genes involved in vibriobactin biosynthesis and uptake might be clustered in the same region of the V. cholerae chromosome. A large DNA fragment downstream of viuB was recovered from V. cholerae O395. Sequence analysis of the cloned region revealed a single large open reading frame which extended beyond the recovered chromosomal DNA, suggesting that the entire coding region is very large and encodes a protein greater than 2,158 amino acids in length. Complementation studies demonstrated that the cloned fragment, despite encoding a truncated protein, restored the ability of an E. coli entF mutant to grow in low-iron medium. Given the complementation of entF function, the open reading frame was named vibF.
The construction of a stable, in-frame vibF mutant by in vivo marker exchange and the effect of introduction of the amino-terminal 2,158 amino acids of VibF in trans on the phenotype of the mutant provided evidence that vibF encodes a protein involved in ferric vibriobactin biosynthesis. Cross-feeding studies demonstrated that V. cholerae vibF mutant strain JRB18 was unable to synthesize vibriobactin but utilized the exogenous siderophore normally; the clone of the 5′ region of vibF in trans restored the ability of the mutant to produce vibriobactin. The mutant phenotype of JRB18 is unlikely to be caused by polar effects on other genes downstream of vibF in an operon, since the deletion of vibF in JRB18 is in frame and the complementing chromosomal fragment on pJRB20 contains only viuB in addition to the cloned fragment of vibF.
Many genes encoding proteins involved in siderophore biosynthesis and uptake are themselves regulated by iron, and the suggested role of vibF in vibriobactin biosynthesis is supported by the observation that vibF is negatively regulated by iron at the transcriptional level. Analysis of vibF promoter activity in different mutant strains, using an operon fusion of the vibF promoter to E. coli phoA, revealed that vibF expression is under the control of the fur gene but is unaffected by the expression of irgA or viuA. The biological significance of the twofold reduction of vibF expression observed in a strain mutant in the positive transcriptional regulator IrgB is unclear. In prior studies, gel shift assays demonstrated that IrgB does not bind to the vibF promoter region (50), suggesting at most an indirect role of IrgB in vibF expression. Analysis of the promoter region of vibF identified a region homologous to the E. coli repressor protein Fur binding consensus sequence. The footprint of purified V. cholerae Fur overlapping this consensus sequence in the vibF promoter further supported the hypothesis that the transcriptional regulation of vibF by iron is under the control of Fur.
The very large size of the deduced VibF protein is unusual, as it is twice the size of the previous largest open reading frame reported for V. cholerae (27); such large open reading frames are not common in bacterial genomes in general. One class of proteins that are very large are the multifunctional nonribosomal peptide synthetases, which are multisubunit enzymes ranging in size from 100 to over 1,600 kDa (46). These proteins, produced mainly by soil bacteria and filamentous fungi, synthesize bioactive peptides “nonribosomally” on a protein template. Evaluation of the deduced amino acid sequence of VibF revealed significant homology with many members of this protein class.
Peptide synthetases are multienzyme complexes comprising one or more modules in a single, large polypeptide chain; each module represents a functional unit and acts as an independent enzyme to catalyze the sequential activation and condensation of one amino or hydroxy acid into the peptide product. The specific linear order of the modules determines the final sequence of the linked amino acids in the product. Each module shares homology with the others in specific functional domains (46). The most common module characterized in bacteria contains about 600 amino acids, with an approximately 500-amino-acid acyladenylation domain, essential for amino acid recognition and activation, and a 100-amino-acid domain involved in thioester formation. Up to nine core sequence motifs have been identified in the acyladenylation domain, with an additional core motif within the thioester domain (29, 38, 46). The serine residue of the thioester motif (LGGXSI) is the binding site for the cofactor 4′-phosphopantetheine and is essential for thioester formation. Between modules are elongation domains, which contain spacer motifs (HHXXXDG), involved in acyl transfer. In the multiple-carrier model, each amino acid is activated as an aminoacyl adenylate and linked to the enzyme as a thioester with a phosphopantetheinyl group. Elongation then occurs by transfer of the activated carboxyl to the amino group of the next amino acid, allowing stepwise condensation (13).
Six core sequence motifs, along with a spacer motif, are present in the VibF sequence (Fig. 2), suggesting that the truncated protein encoded on pJRB20 comprises one complete functional module of a peptide synthetase. The enterobactin synthetases EntE (2,3-dihydroxybenzoate AMP ligase) (42) and EntF (a serine-activating enzyme) (39) also belong to the family of adenylate-forming enzymes. We speculate that the identified module in VibF may act to link 2,3-DHBA to threonine in the production of vibriobactin in V. cholerae, but this proposed function does not explain the way in which pJRB20 complements the entF mutation in AN117, as it seems unlikely that VibF would activate serine to form enterobactin in E. coli. Wyckoff et al. were able to complement the entF mutation in AN117 with a cosmid clone containing portions of the different vibriobactin gene cluster they identified; however, the siderophore produced was clearly novel, as it did not comigrate with either enterobactin or vibriobactin in chromatographic analysis (52). This gene cluster contains a gene, vibH, which encodes a protein similar to the amino-terminal region of EntF (GenBank accession no. U52150). It may be that both VibH and VibF complement an entF mutation in E. coli by directing the synthesis of variant siderophores.
The linkage of viuA-viuB-vibF to vibB-vibE-vibC-vibA-vibH-viuP-viuD-viuG-viuC-vibD has been previously investigated (52). A probe from the viuA gene did not hybridize with cosmid clones containing the vibB-vibD region, and the cosmids did not complement a viuA mutant strain. We believed that the vibB-vibD region was unlikely to lie upstream of viuA, as 3 kbp of chromosomal DNA upstream of viuA has been recovered from V. cholerae O395 and found to contain the genes gltX, ompA, and orfA, a gene without database homology (GenBank accession no. AF030977) (W. J. Liao, M. H. Choi, and J. R. Butterton, 33rd Joint Conf. Cholera Relat. Diarrheal Dis., p. 179, 1997). However, since several members of the family of nonribosomal peptide synthetases are extremely large, reaching over 10,000 bp in length, it remained possible that vibF was linked downstream to vibB-vibD. However, preliminary sequence data from The Institute for Genomic Research suggests that these regions, although both on the V. cholerae large chromosome, are widely separated. The observation that a vibF mutation eliminates siderophore biosynthesis argues against the possibility that V. cholerae possesses two separate but redundant siderophore production and utilization clusters.
The identified extent of VibF represents one of the longest proteins identified in gram-negative enteric pathogens, being similar in size to HMWP2 of Yersinia spp. (2,035 amino acids), which also likely functions as a nonribosomal peptide synthetase in siderophore biosynthesis (2, 22). Analysis of preliminary sequence data from The Institute for Genomic Research of a different V. cholerae strain, O1 El Tor N16961, suggests that the complete vibF open reading frame encodes a 2,413-residue protein, and the complete open reading frame contains only one identified biosynthetic module. There are no additional open reading frames in the region downstream of vibF that encode proteins with homology to nonribosomal peptide synthetases or siderophore biosynthetic enzymes. If the biosynthetic module identified in VibF acts to link a single 2,3-DHBA molecule to threonine in the production of vibriobactin, other proteins, perhaps encoded by the vibB-vibD region, may complete the synthesis of vibriobactin by linking another molecule of 2,3-DHBA to norspermidine via a threonine residue, along with a third 2,3-DHBA molecule attached directly to the norspermidine backbone. Further characterization of the vibriobactin biosynthetic genes will allow a detailed understanding of the biosynthetic steps that are needed for production of this siderophore.
ACKNOWLEDGMENTS
This work was supported by a Howard Hughes Medical Institute Postdoctoral Research Fellowship for Physicians (J.R.B.) and grant RO1 AI34968 (S.B.C.). Sequencing of V. cholerae was accomplished with support from the National Institute of Allergy and Infectious Diseases.
We are grateful to C. W. Wang for his generous technical help, Ken Peterson for his gift of the O395 DNA library, and Shelley Payne for providing E. coli AB1515 and AN117. Preliminary sequence data was obtained from The Institute for Genomic Research website at http://www/tigr.org.
REFERENCES
- 1.Altschul A F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Bearden S W, Fetherston J D, Perry R D. Genetic organization of the yersiniabactin biosynthetic region and construction of avirulent mutants in Yersinia pestis. Infect Immun. 1997;65:1659–1668. doi: 10.1128/iai.65.5.1659-1668.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blomfield I C, Vaughn V, Rest R F, Eisenstein B I. Allelic exchange in Escherichia coli using the Bacillus subtilis sacB gene and a temperature-sensitive pSC101 replicon. Mol Microbiol. 1991;5:1447–1457. doi: 10.1111/j.1365-2958.1991.tb00791.x. [DOI] [PubMed] [Google Scholar]
- 4.Butterton J R, Beattie D T, Gardel C L, Carroll P A, Hyman T, Killeen K P, Mekalanos J J, Calderwood S B. Heterologous antigen expression in Vibrio cholerae vector strains. Infect Immun. 1995;63:2689–2696. doi: 10.1128/iai.63.7.2689-2696.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Butterton J R, Calderwood S B. Identification, cloning, and sequencing of a gene required for ferric vibriobactin utilization by Vibrio cholerae. J Bacteriol. 1994;176:5631–5638. doi: 10.1128/jb.176.18.5631-5638.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Butterton J R, Stoebner J A, Payne S M, Calderwood S B. Cloning, sequencing, and transcriptional regulation of viuA, the gene encoding the ferric vibriobactin receptor of Vibrio cholerae. J Bacteriol. 1992;174:3729–3738. doi: 10.1128/jb.174.11.3729-3738.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calderwood S B, Mekalanos J J. Iron regulation of Shiga-like toxin expression in Escherichia coli is mediated by the fur locus. J Bacteriol. 1987;169:4759–4764. doi: 10.1128/jb.169.10.4759-4764.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Calderwood S B, Mekalanos J J. Confirmation of the Fur operator site by insertion of a synthetic oligonucleotide into an operon fusion plasmid. J Bacteriol. 1988;170:1015–1017. doi: 10.1128/jb.170.2.1015-1017.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen Q, Wertheimer A M, Tolmasky M E, Crosa J H. The AngR protein and the siderophore anguibactin positively regulate the expression of iron-transport genes in Vibrio anguillarum. Mol Microbiol. 1996;22:127–134. doi: 10.1111/j.1365-2958.1996.tb02662.x. [DOI] [PubMed] [Google Scholar]
- 10.Cosmina P, Rodriguez F, de Ferra F, Perego G G M, Venema G, van Sinderen D. Sequence and analysis of the genetic locus responsible for surfactin synthesis in Bacillus subtilis. Mol Microbiol. 1993;8:821–831. doi: 10.1111/j.1365-2958.1993.tb01629.x. [DOI] [PubMed] [Google Scholar]
- 11.Crosa J H. The relationship of plasmid-mediated iron transport and bacterial virulence. Annu Rev Microbiol. 1984;38:69–89. doi: 10.1146/annurev.mi.38.100184.000441. [DOI] [PubMed] [Google Scholar]
- 12.Crosa J H. Genetics and molecular biology of siderophore-mediated iron transport in bacteria. Microbiol Rev. 1989;53:517–530. doi: 10.1128/mr.53.4.517-530.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Crecy-Lagard V, Blanc V, Gil P, Naudin L, Lorenzon S, Famechon A, Bamas-Jacques N, Crouzet J, Thibaut D. Pristinamycin I biosynthesis in Streptomyces pristinaespiralis: molecular characterization of the first two structural peptide synthetase genes. J Bacteriol. 1997;179:705–713. doi: 10.1128/jb.179.3.705-713.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Grandis S, Ginsberg J, Toone M, Climie S, Friesen J, Brunton J. Nucleotide sequence and promoter mapping of the Escherichia coli Shiga-like toxin operon of bacteriophage H-19B. J Bacteriol. 1987;169:4313–4319. doi: 10.1128/jb.169.9.4313-4319.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Lorenzo V, Herrero M, Jakubzik U, Timmis K N. Mini-Tn5 transposon derivatives for insertional mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J Bacteriol. 1990;172:6568–6572. doi: 10.1128/jb.172.11.6568-6572.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Donnenberg M S, Kaper J B. Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect Immun. 1991;59:4310–4317. doi: 10.1128/iai.59.12.4310-4317.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goldberg M B, Boyko S A, Butterton J R, Stoebner J A, Payne S M, Calderwood S B. Characterization of a Vibrio cholerae virulence factor homologous to the family of TonB-dependent proteins. Mol Microbiol. 1992;6:2407–2418. doi: 10.1111/j.1365-2958.1992.tb01415.x. [DOI] [PubMed] [Google Scholar]
- 18.Goldberg M B, Boyko S A, Calderwood S B. Transcriptional regulation by iron of a Vibrio cholerae virulence gene and homology of the gene to the Escherichia coli Fur system. J Bacteriol. 1990;172:6863–6870. doi: 10.1128/jb.172.12.6863-6870.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goldberg M B, Boyko S A, Calderwood S B. Positive transcriptional regulation of an iron-regulated virulence gene in Vibrio cholerae. Proc Natl Acad Sci USA. 1991;88:1125–1129. doi: 10.1073/pnas.88.4.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goldberg M B, DiRita V J, Calderwood S B. Identification of an iron-regulated virulence determinant in Vibrio cholerae, using TnphoA mutagenesis. Infect Immun. 1990;58:55–60. doi: 10.1128/iai.58.1.55-60.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Griffiths G L, Sigel S P, Payne S M, Neilands J B. Vibriobactin, a siderophore from Vibrio cholerae. J Biol Chem. 1984;259:383–385. [PubMed] [Google Scholar]
- 22.Guilvout I, Mercereau-Puijalon O, Bonnefoy S, Pugsley A P, Carniel E. High-molecular-weight protein 2 of Yersinia enterocolitica is homologous to AngR of Vibrio anguillarum and belongs to a family of proteins involved in nonribosomal peptide synthesis. J Bacteriol. 1993;175:5488–5504. doi: 10.1128/jb.175.17.5488-5504.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hantke K. Cloning of the repressor protein gene of iron-regulated systems in Escherichia coli K12. Mol Gen Genet. 1984;197:337–341. doi: 10.1007/BF00330982. [DOI] [PubMed] [Google Scholar]
- 24.Henderson D P, Payne S M. Cloning and characterization of the Vibrio cholerae genes encoding the utilization of iron from hemin and hemoglobin. Mol Microbiol. 1993;7:461–469. doi: 10.1111/j.1365-2958.1993.tb01137.x. [DOI] [PubMed] [Google Scholar]
- 25.Henderson D P, Payne S M. Characterization of the Vibrio cholerae outer membrane heme transport protein HutA: sequence of the gene, regulation of expression, and homology to the family of TonB-dependent proteins. J Bacteriol. 1994;176:3269–3277. doi: 10.1128/jb.176.11.3269-3277.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Henikoff S, Henikoff J G. Protein family classification based on searching a database of blocks. Genomics. 1994;19:97–107. doi: 10.1006/geno.1994.1018. [DOI] [PubMed] [Google Scholar]
- 27.Karaolis D K R, Johnson J A, Bailey C C, Boedeker E C, Kaper J B, Reeves P R. A Vibrio cholerae pathogenicity island associated with epidemic and pandemic strains. Proc Natl Acad Sci USA. 1998;95:3134–3139. doi: 10.1073/pnas.95.6.3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kyte J, Doolittle R F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 29.Lambalot R H, Gehring A M, Flugel R S, Zuber P, LaCelle M, Marahiel M A, Reid R, Khosla C, Walsh C T. A new enzyme superfamily—the phosphopantetheinyl transferases. Chem Biol. 1996;3:923–936. doi: 10.1016/s1074-5521(96)90181-7. [DOI] [PubMed] [Google Scholar]
- 30.Litwin C M, Calderwood S B. Analysis of the complexity of gene regulation by Fur in Vibrio cholerae. J Bacteriol. 1994;176:240–248. doi: 10.1128/jb.176.1.240-248.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30a.Maxam A M, Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65:499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- 31.Mekalanos J J, Swartz D J, Pearson G D, Harford N, Groyne F, de Wilde M. Cholera toxin genes: nucleotide sequence, deletion analysis and vaccine development. Nature. 1983;306:551–557. doi: 10.1038/306551a0. [DOI] [PubMed] [Google Scholar]
- 32.Michaelis S, Inouye H, Oliver D, Beckwith J. Mutations that alter the signal sequence of alkaline phosphatase in Escherichia coli. J Bacteriol. 1983;154:366–374. doi: 10.1128/jb.154.1.366-374.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miller S I, Landfear S M, Wirth D F. Cloning and characterization of a Leishmania gene encoding a RNA spliced leader sequence. Nucleic Acids Res. 1986;14:7341–7360. doi: 10.1093/nar/14.18.7341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miller V L, Mekalanos J J. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol. 1988;170:2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Neilands J B. Microbial iron compounds. Annu Rev Biochem. 1981;50:715–731. doi: 10.1146/annurev.bi.50.070181.003435. [DOI] [PubMed] [Google Scholar]
- 36.Occhino D A, Wyckoff E E, Henderson D P, Wrona T J, Payne S M. Vibrio cholerae iron transport: haem transport genes are linked to one of two sets of tonB, exbB, exbB genes. Mol Microbiol. 1998;29:1493–1507. doi: 10.1046/j.1365-2958.1998.01034.x. [DOI] [PubMed] [Google Scholar]
- 37.Pearson W R, Lipman D J. Improved tools for biological sequence comparison. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pfeifer E, Pavela-Vrancic M, von Dohren H, Kleinkauf H. Characterization of tyrocidine synthetase 1 (TY1): requirement of posttranslational modification for peptide biosynthesis. Biochemistry. 1995;34:7450–7459. doi: 10.1021/bi00022a019. [DOI] [PubMed] [Google Scholar]
- 39.Reichert J, Sakaitani M, Walsh C T. Characterization of EntF as a serine-activating enzyme. Protein Sci. 1992;1:549–556. doi: 10.1002/pro.5560010410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rogers H J. Iron-binding catechols and virulence in Escherichia coli. Infect Immun. 1973;7:445–456. doi: 10.1128/iai.7.3.445-456.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rusnak F, Sakaitani M, Drueckhammer D, Reichert J, Walsh C T. Biosynthesis of the Escherichia coli siderophore enterobactin: sequence of the entF gene, expression and purification of EntF, and analysis of covalent phosphopantetheine. Biochemistry. 1991;30:2916–2927. doi: 10.1021/bi00225a027. [DOI] [PubMed] [Google Scholar]
- 42.Rusnak F W, Faraci S, Walsh C T. Subcloning, expression and purification of the enterobactin biosynthetic enzyme 2,3-dihydroxybenzoate-AMP ligase: demonstration of enzyme-bound (2,3-dihydroxybenzoyl)adenylate product. Biochemistry. 1989;28:6827–6835. doi: 10.1021/bi00443a008. [DOI] [PubMed] [Google Scholar]
- 43.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 44.Sigel S P, Stoebner J A, Payne S M. Iron-vibriobactin transport system is not required for virulence of Vibrio cholerae. Infect Immun. 1985;47:360–362. doi: 10.1128/iai.47.2.360-362.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Staab J F, Earhart C F. EntG activity of Escherichia coli enterobactin synthetase. J Bacteriol. 1990;172:6403–6410. doi: 10.1128/jb.172.11.6403-6410.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stachelhaus T, Marahiel M M. Modular structure of genes encoding multifunctional peptide synthetases required for non-ribosomal peptide synthesis. FEMS Microbiol Lett. 1995;125:3–14. doi: 10.1111/j.1574-6968.1995.tb07328.x. [DOI] [PubMed] [Google Scholar]
- 47.Stoebner J A, Payne S M. Iron-regulated hemolysin production and utilization of heme and hemoglobin by Vibrio cholerae. Infect Immun. 1988;56:2891–2895. doi: 10.1128/iai.56.11.2891-2895.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tashima K T, Carroll P A, Rogers M B, Calderwood S B. Relative importance of three iron-regulated outer membrane proteins for in vivo growth of Vibrio cholerae. Infect Immun. 1996;64:1756–1761. doi: 10.1128/iai.64.5.1756-1761.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tolmasky M E, Actis L A, Crosa J H. A single amino acid change in AngR, a protein encoded by pJM1-like virulence plasmids, results in hyperproduction of anguibactin. Infect Immun. 1993;61:3228–3233. doi: 10.1128/iai.61.8.3228-3233.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Watnick P I, Butterton J R, Calderwood S B. The interaction of the Vibrio cholerae transcription factors, Fur and IrgB, with the overlapping promoters of two virulence genes, irgA and irgB. Gene. 1998;209:65–70. doi: 10.1016/s0378-1119(98)00018-3. [DOI] [PubMed] [Google Scholar]
- 51.Watnick P I, Eto T, Takahashi T, Calderwood S B. Purification of Vibrio cholerae Fur and estimation of its intracellular abundance by antibody sandwich enzyme-linked immunosorbent assay. J Bacteriol. 1997;179:243–247. doi: 10.1128/jb.179.1.243-247.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wyckoff E E, Stoebner J A, Reed K E, Payne S M. Cloning of a Vibrio cholerae vibriobactin gene cluster: identification of genes required for early steps in siderophore biosynthesis. J Bacteriol. 1997;179:7055–7062. doi: 10.1128/jb.179.22.7055-7062.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]