Abstract
The isolation of rhizobial strains which exhibit an intrinsic tolerance to acidic conditions has been reported and has facilitated studies on the basic mechanisms underlying acid tolerance. Rhizobium tropici strain CIAT899 displays a high intrinsic tolerance to acidity and therefore was used in this work to study the molecular basis of bacterial responses to acid conditions and other environmental stresses. We generated a collection of R. tropici CIAT899 mutants affected in acid tolerance using Tn5-luxAB mutagenesis, and one mutant strain (CIAT899-13T2), which fails to grow under acid conditions, was characterized in detail. Strain CIAT899-13T2 was found to contain a single Tn5-luxAB insertion in a gene showing a high degree of similarity with the Escherichia coli gshB gene, encoding the enzyme glutathione synthetase. Intracellular potassium pools and intracellular pH levels were found to be lower in the mutant than in the parent. The glutathione-deficient mutant was shown to be sensitive to weak organic acids, osmotic and oxidative stresses, and the presence of methylglyoxal. Glutathione restores responses to these stresses almost to wild-type levels. Our data show that in R. tropici the production of glutathione is essential for growth in extreme environmental conditions. The mutant strain CIAT899-13T2 induced effective nodules; however, it was found to be outcompeted by the wild-type strain in coinoculation experiments.
Microbes are constantly challenged by a variety of stresses in their natural environments, including nutrient limitation and/or exposure to physical stresses, such as elevated temperature, acidity, high osmolarity, or oxidative shock. To adapt to these adverse conditions, microbes have evolved mechanisms to monitor the environment and to alter gene expression patterns and the activity of enzymes and transport proteins that adapt them to the new environment. Microbial differentiation into stress-resistant forms, such as endospores and cysts, constitutes one of the best-studied stress response mechanisms (18, 42). However, the majority of soil microbes are gram-negative bacteria that do not differentiate in the same fashion in response to stress conditions. Most of the present knowledge about stress survival in nondifferentiating bacteria derives from studies on enteric bacteria, such as Escherichia coli or Salmonella typhimurium, and marine bacteria, such as Vibrio spp. (27).
The responses of soil bacteria, such as Pseudomonas spp. and rhizobia, to environmental stress factors have been investigated (22, 34). For example, to identify genes which are expressed under nutrient or oxygen limitation conditions, transposon Tn5-luxAB mutagenesis has been employed to generate transcriptional fusions of lux luciferase reporter gene to genes in Pseudomona putida (28) and Sinorhizobium meliloti (33, 34). Several of the Tn5-luxAB-tagged S. meliloti genes have been shown to play a role in persistence in the soil and/or competition for nodulation of the alfalfa host plant (33). A similar approach has been used by others (40) to identify S. meliloti genes induced by alternative N or C sources such as stachydrine used under nutrient limitation conditions, which has also led to the identification of genes important for root colonization.
It is known that soil acidity, temperature, and salinity affect rhizobial persistence in the soil and rhizosphere of plants, as well as the efficiency of nodulation, especially in tropical areas (5, 9, 25). The isolation of rhizobial strains that exhibit an intrinsic acid tolerance has been described and has facilitated studies on the basic mechanisms underlying acid tolerance. Rhizobium tropici strain CIAT899 is one of the strains identified that displays a high intrinsic tolerance to acidity and therefore constitutes a suitable system to study the molecular basis of bacterial responses to acid conditions and other environmental stresses (23, 31). Different mechanisms have been proposed to be involved in the ability of microbes to survive and grow at low pH levels. The regulation of cytoplasmic pH (intracellular pH [pHi]) requires both passive and active elements (10, 11). The importance of pH homeostasis for growth of both acid-tolerant and acid-resistant Rhizobium meliloti strains has been demonstrated (36). In particular, the acid-sensitive mutants examined were unable to maintain their cytoplasmic pHs at alkaline levels under acid conditions (36).
In this work, we describe the generation and characterization of a collection of R. tropici CIAT899 mutants affected in acid tolerance using Tn5-luxAB mutagenesis. One mutant strain (CIAT899-13T2) fails to grow under acid conditions and exhibits extreme stress sensitivity. Strain CIAT899-13T2 was found to contain a single Tn5-luxAB insertion in a gene showing a high degree of similarity with the E. coli gshB gene, encoding the enzyme glutathione synthetase (Gsh). Correspondingly, glutathione essentially reverses the stress phenotype of the mutant. Although the mutant efficiently nodulates beans, it fails to compete when coinoculated with the parent strain.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth media.
R. tropici CIAT899 (Streptomycin resistant [Smr]), a bean-nodulating strain described previously (31), was grown at 28°C in TY medium (6) or minimal GTS medium (26). Strain E. coli DH5α (23) derivatives harboring the Tn5-luxAB-containing plasmid pRL1063a (46) or the helper plasmid pRK2013 (16) were grown at 37°C in Luria broth supplemented with 50 μg of kanamycin · ml−1.
Random transposon mutagenesis.
Transposon Tn5-luxAB mutagenesis of R. tropici CIAT899 was carried out using the protocol for Tn5 mutagenesis of Rhizobium spp. described by De Bruijn and Rossbach (15). Cells of the donor strain E. coli DH5α, harboring the suicide plasmid pRL1063a or the helper plasmid pRK2013, and the recipient strain, R. tropici CIAT899, were grown in Luria broth supplemented with kanamycin and TY medium, respectively, were washed with fresh TY medium, and were concentrated 10-fold in TY medium. Equal amounts (100 μl) of donor, helper, and recipient cells were mixed and spotted on TY plates. After 24 h of incubation at 28°C, the mating mixtures were resuspended in sterile distilled water and plated on selective TY medium, supplemented with streptomycin (400 μg · ml−1) and neomycin (100 μg · ml−1).
Screen for acid-sensitive mutants.
R. tropici strains carrying Tn5-luxAB insertions were screened for acid sensitivity by using toothpicks to streak individual colonies onto GTS and GMS plates (GMS medium is similar to GTS medium, except that Tris was substituted for morpholonethanesulfonic acid and the medium was acidified to a pH of 5.0 with HCl). Transconjugants that grew poorly or not at all on GMS medium but grew well on GTS medium were selected for further analysis.
Growth in the presence and absence of glutathione.
Starter cultures were generated in GTS medium, supplemented with antibiotics, and grown to saturation. Duplicate flasks containing 100 ml of GTS or GMS medium were inoculated with an aliquot of starter culture to yield an initial optical density at 600 nm (OD600) of approximately 0.06 (equivalent to about 103 cells · ml−1). The cultures were incubated under aeration at 28°C, and growth was monitored by periodically measuring the OD600 and determining the viable cell counts on TY medium. Glutathione was added to a final concentration of 80 μM. Since methylglyoxal reacts with glutathione, cells were first grown in GTS medium supplemented with glutathione to an OD600 of 0.2 U (approximately 108 cells · ml−1). The cultures were centrifuged, and the pellet was resuspended in an equal volume of fresh medium plus 2.5 mM methylglyoxal. Incubation and growth were continued as before. For the cytoplasm acidification experiments, sodium acetate (2.5 M, pH 5.5, stock solution) was added to GTS medium to a final concentration of 20 mM (38). The experiments were replicated at least twice.
Measurement of glutathione content.
The glutathione content in rhizobia cells was measured by the method of Anderson (4). Baker yeast-glutathione reductase was supplied by Sigma-Aldrich. The protein concentration of the samples was determined by the bicinchoninic acid assay (Sigma-Aldrich).
pHi measurements.
The pHi was determined by using the distribution of a radiolabeled weak acid following the centrifugation method described previously (29), using bromodecane to separate the cell pellet from the supernatant (41). The pHi was determined using [7-14C]benzoic acid (4.5 μM; 0.1 μCi · ml−1) and [3H]-inulin (1.0 μM; 1 μCi · ml−1) as the extracellular marker. The pHi value was calculated as described previously (41).
Measurements of intracellular potassium.
Cultures were grown to mid-exponential phase (OD600 = 0.5) and 1-ml samples were used for the analyses. Levels of potassium ions were quantified by preparing the samples as described for pHi measurements. The K+ content was determined by flame photometry (Corning 420) after the samples had been boiled and allowed to cool (21).
DNA isolation and manipulations.
Plasmid DNA was prepared by the alkaline method as described by Kragelund et al. (28). Total genomic R. tropici DNA isolation, restriction enzyme digestions, ligations, and Southern blotting experiments were carried out according to procedures previously described (2, 43).
DNA sequence analysis.
Sequencing of double-stranded plasmid DNA was performed using the dideoxy method of Sanger, using Sequenase kits (U.S. Biochemicals). To determine the R. tropici DNA sequence on both sides of the transposon insertion, the procedure described by Milcamps et al. (33) was used. DNA sequence data were analyzed using the Wisconsin Package Version 9.0 program (Genetics Computer Group). Similarities were examined with the BLAST program (3).
Nodulation assays.
R. tropici strains carrying Tn5-1063 insertions were screened for their symbiotic phenotype by inoculation on beans or on leucaena seedling roots. Leucaena leucocephala seeds were first scarred by heat treatment (80°C, 10 min). Seeds were sterilized by soaking for 3 min in 95% (vol/vol) ethanol, followed by soaking for 15 min in sodium hypochloride (8.25 g · liter−1), and rinsed thoroughly with sterile distilled water. Sterilized seeds were placed on top of sterile agar-water and incubated in darkness at 28°C.
The inoculation with rhizobial suspensions was performed on roots of 5-day-old seedlings. Seedlings were transferred into pots containing sterile vermiculite at a neutral pH level and incubated in a glass house with a temperature range of 25 ± 5°C. Four to five weeks after inoculation, the plants were examined for the presence or absence of root nodules.
Nodule competition experiments.
Ten bean seedlings were inoculated with a 1:1 ratio of mutant strain CIAT899-13T2 and wild-type bacteria. Four weeks after inoculation, nodules were harvested and individually analyzed. The nodules were surface-sterilized with 95% ethanol and 30% (vol/vol) hydrogen peroxide, rinsed with sterile distilled water, and crushed in 100 μl of sterile water, and the macerate was spotted onto plates containing either streptomycin (400 μg · ml−1) or neomycin (100 μg · ml−1) to determine the ratio of wild-type versus mutant bacteria. About 100 nodules from five independent randomly selected plants were examined.
RESULTS
Isolation and characterization of the acid-sensitive mutant CIAT899-13T2.
Acid sensitivity was defined experimentally as the inability to grow on minimal medium buffered to a pH of 5.0. A total of 6,000 Tn5-luxAB-containing strains were analyzed, of which three mutants were found to be unable to grow on acidified GMS medium. One of the mutant strains (CIAT899-13T2), was further characterized. Using Southern hybridization analysis, it was shown that strain CIAT899-13T2 carries a single Tn5-luxAB insertion (data not shown).
In order to identify the gene carrying the Tn5-luxAB insertion in strain CIAT899-13T2, the tagged locus was cloned from the rhizobial genome. This was facilitated by the presence of an origin of replication within the Tn5-luxAB transposon (46). Total genomic DNA of strain CIAT899-13T2 was digested with EcoRI, self-ligated, and introduced into E. coli via transformation. This plasmid was used to confirm that the Tn5-luxAB insertion correlates with the phenotype found in mutant CIAT899-13T2. The mutated region was recombined in wild-type CIAT899 by marker exchange. Randomly chosen exchange mutants were assayed for growth on GTS and GMS media, and found to be unable to grow at a pH of 5.0. Finally, the DNA sequence of the Tn5-luxAB target junctions was determined and an open reading frame of 945 nucleotides was identified. The deduced amino acid sequence of this open reading frame was compared to sequences in the databases. It was found that the Tn5-luxAB-tagged locus shares a high degree of similarity with glutathione synthetase (Gsh), which is encoded by the gshB gene, of E. coli (39% identity, 72% similarity) (8) and other bacteria, such as a Sinechococcus sp. (40% identity, 73% similarity) (37), a Synechocystis sp. (37% identity, 72% similarity) (35), an Anabaena sp. (36% identity, 73% similarity) (17), and Anaplasma centrale (35% identity, 68% similarity) (39). Glutathione synthetase catalyzes the second step of glutathione biosynthesis and adds glycine to the C-terminal site of γ-glutamylcysteine to form glutathione.
In order to confirm that strain CIAT899-13T2 is a glutathione-deficient mutant, we measured the content of glutathione in the cell extracts of the wild-type strain and the mutant strain. In the cell extracts prepared from the wild-type strain, the glutathione content was 14 nmol · mg of protein−1, whereas in the cell extracts prepared from the mutant strain the glutathione content was very low, about 3% of the wild-type level.
Glutathione is important for acid tolerance.
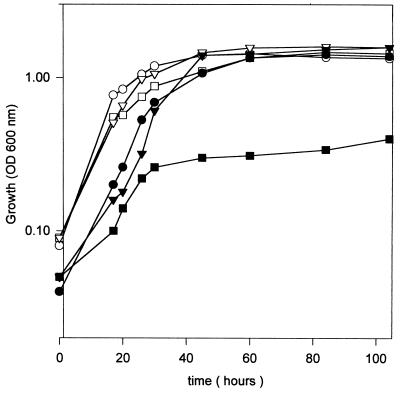
Strain CIAT899-13T2 was found to grow at the same rate as the parent strain in liquid media buffered to a pH of 7.5, with a mean generation time of approximately 3 h. However, after dilution in fresh GTS medium from late-exponential-phase cultures, strain CIAT899-13T2 exhibited a slower adaptation to the new medium before achieving a growth rate similar to that of the parent. Contrarily, the growth yield of the mutant was substantially reduced in acidic medium GMS, with a cessation of growth after four generations (Fig. 1). Growth was also affected, but to a lesser extent, in the pH range of 5.0 to 6.0.
FIG. 1.
Recovery of acid tolerance by R. tropici CIAT899-13T2. At time zero, rhizobia cells were diluted with GTS or GMS medium to an OD600 of 0.08. Growth of wild-type strain CIAT899 (open symbols) and mutant strain CIAT 899-13T2 (closed symbols) in GTS at a pH of 7.5 (circles), in GMS at a pH of 5.0 (squares), and in GMS (pH 5.0) supplemented with 80 μM glutathione at time zero (triangles) was monitored by measuring the OD600. The data are the means from three replicate experiments, with standard deviations of less than 10%.
To determine whether endogenous glutathione was involved in providing protection against low external pH stress, cells of R. tropici CIAT 899-13T2 were diluted in fresh GMS medium (pH 5.0) supplemented with 80 μM glutathione, and bacterial growth was monitored (Fig. 1). It was found that mutant CIAT899-13T2 recovered the ability to grow at a pH of 5.0. In addition, strain CIAT899-13T2 exhibited an extended lag before growth was initiated at a pH of 7.5 and this could be shortened by incubation with glutathione (data not shown), which is similar to observations made for E. coli glutathione-deficient mutants (19).
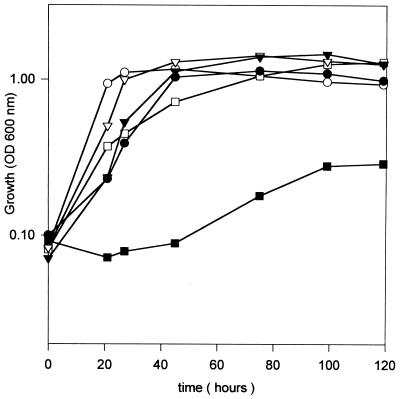
In order to examine if glutathione played a role in the growth response of R. tropici CIAT899 to changes in pHi, the effect of adding sodium acetate (NaAc) to cultures of strains CIAT899-13T2 and CIAT899 was examined. Acetic acid in equilibrium with NaAc is a weak acid capable of crossing the bacterial membrane in its undissociated form, but it dissociates and liberates protons once inside the cells, causing cytoplasmatic acidification (38, 41). The wild-type strain CIAT899 was found to display a reduced growth rate at a pH of 7.5 in the presence of NaAc, but it eventually achieved the same final growth yield (Fig. 2). Supplementation of the growth medium with glutathione enhanced the growth of the wild-type strain (Fig. 2). Incubation of mutant strain CIAT 899-13T2 with NaAc provoked a 40-h lag prior to the initiation of exponential growth (Fig. 2). The growth rate was much slower but could be restored to normal by the addition of glutathione to the growth medium. These results suggest that glutathione synthesis in R. tropici CIAT899 is essential for growth at acid pH levels and in the presence of weak organic acids that provoke perturbations of cytoplasmic pH levels.
FIG. 2.
Acidification of cytoplasm and protection by glutathione in R. tropici CIAT899. At time zero, cells of CIAT899 (open symbols) and mutant CIAT 899-13T2 (closed symbols) were diluted in fresh GTS medium. Growth in GTS medium (circles), in GTS medium supplemented with 20 mM sodium acetate (squares), and in GTS supplemented with 20 mM sodium acetate plus 80 μM glutathione (triangles) was monitored by measuring the OD600. The data are the means from at least two replicate experiments, with standard deviations of less than 12%.
pHi and potassium levels in the CIAT 899-13T2 mutant strain.
We investigated the ability of mutant strain CIAT899-13T2 to control pHi levels and to maintain potassium pools when exposed to acid medium. At a pH of 7.5 the pHi values for the mutant and the wild type were identical, but 60 min after the cells were transferred from a pH of 7.5 to a pH of 5.0, the acid-sensitive CIAT 899-13T2 mutant strain was found to have a lower pHi level than the wild-type strain had (Table 1).
TABLE 1.
Intracellular pH and potassium in glutathione-deficient R. tropici mutanta
| Strain | pHi
|
[Ki] (mmol K+/mg of protein)
|
||
|---|---|---|---|---|
| pH 7.0 | pH 5.0 | pH 7.4 | pH 5.0 | |
| Wild-type CIAT899 | 7.52 ± 0.04 | 7.23 ± 0.04 | 1.69 ± 0.05 | 2.60 ± 0.05 |
| Mutant CIAT899-13T2 | 7.52 ± 0.04 | 7.01 ± 0.04 | 1.58 ± 0.06 | 1.90 ± 0.03 |
Cells were grown in GTS medium, centrifuged, and resuspended in fresh prewarmed GTS medium buffered to a pH of 7.0 or 7.4 or GMS medium (pH 5.0). After 60 min of incubation at 28°C, samples were removed for determination of pHi and K+ levels according to procedures described in Materials and Methods. Ki, intracellular potassium.
Since potassium efflux activity, mediated by KefB- and KefC-generated channels, involves glutathione (19), we measured the intracellular K pool in the CIAT899-13T2 mutant strain under neutral and acid conditions (Table 1). The level of potassium was found to be higher in wild-type cells than in the mutant CIAT899-13T2 cells incubated in either neutral or acid media, and in each case the potassium level was found to be higher under acidic conditions, indicating that exposure to acidity stimulated potassium accumulation (Table 1). This result indicates that the observed acid sensitivity of mutant strain CIAT899-13T2 may be related to the inability to regulate the transport of ion potassium.
Glutathione is important for osmotic stress tolerance.
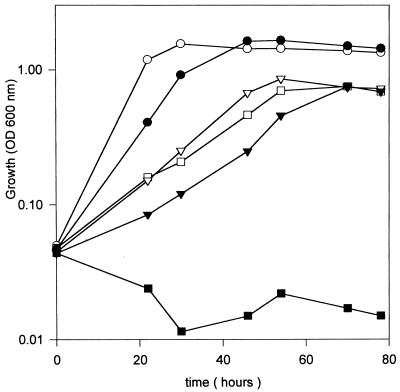
In order to examine whether glutathione was important for the response of R. tropici CIAT899 to other environmental stresses, we tested the effect of osmotic shock (addition of 0.3 M NaCl) on the growth characteristics of strains CIAT899 and CIAT899-13T2. In the presence of 0.3 M NaCl, growth of the wild-type CIAT899 strain was found to be significantly reduced compared to growth in normal GTS medium (Fig. 3). Addition of glutathione to the medium only slightly enhanced growth of CIAT899. On the other hand, growth of the mutant strain CIAT899-13T2 was completely inhibited in the presence of NaCl and the OD600 declined. This inhibition was relieved by glutathione and the growth rate of the mutant approached that of the parent strain (Fig. 3). Similarly, the mutant strain CIAT899-13T2 was found to be more sensitive to incubation in the presence of the oxidative stress agent H2O2; after 20 min of exposure in the presence of 1.5 mM H2O2, the number of viable cells was reduced one order of magnitude from 105 to 104 cells · ml−1, whereas the parent strain was found to be unaffected. These results suggest that glutathione is important for both acid tolerance and responses to other environmental stresses in R. tropici CIAT899.
FIG. 3.
Growth inhibition by osmotic shock is relieved by glutathione. At time zero, cells of CIAT899 (open symbols) and mutant CIAT 899-13T2 (closed symbols) were diluted in fresh GTS medium. Growth in GTS medium (circles), GTS medium supplemented with 0.3 M sodium chloride (squares), and GTS medium supplemented with 0.3 M sodium chloride plus 80 μM glutathione (triangles) was monitored by measuring the OD600. The data are the means from two replicate experiments, with standard deviations of less than 10%.
Glutathione is important for protection against methylglyoxal toxicity.
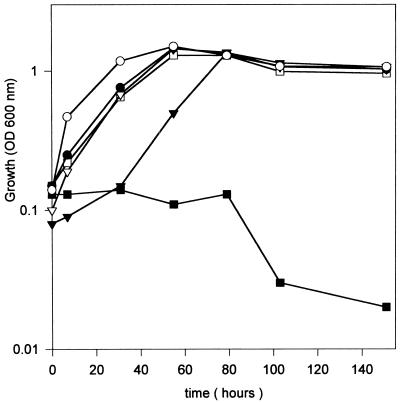
Methylglyoxal is a naturally occurring toxic electrophilic compound, and it has been shown previously that in E. coli glutathione is involved in its detoxification (19, 20, 21). In order to examine if glutathione was also involved in preventing methylglyoxal toxicity in R. tropici CIAT899, the effect of adding methylglyoxal to the growth medium of both CIAT899 and CIAT899-13T2 cultures was assessed.
The addition of methylglyoxal did not substantially alter the growth pattern of the wild-type strain CIAT899 (Fig. 4). However, growth of the mutant CIAT 899-13T2 strain was found to be completely inhibited in the presence of 2.5 mM methylglyoxal (Fig. 4). The effect of the addition of glutathione was also examined. Since methylglyoxal reacts with glutathione to form hemithiolacetal (19), we assessed the effect of adding glutathione by growing the rhizobia in the presence of glutathione alone, prior to resuspension in a medium containing methylglyoxal. The results are shown in Fig. 4 and demonstrate that glutathione prevents the inhibitory effect of methylglyoxal, allowing restoration of growth of CIAT899-13T2 cells. However, the lag period before active growth was observed was found to be greater than that for the parent strain (Fig. 4). These data suggest that R. tropici CIAT899 may require glutathione for the detoxification of methylglyoxal, by a mechanism probably similar to that found in E. coli (19).
FIG. 4.
Effect of methylglyoxal and protection by glutathione. Cells of strain CIAT899 (closed symbols) and mutant CIAT899-13T2 (open symbols) were grown in fresh GTS medium (circles) and in fresh GTS medium supplemented with 80 μM glutathione (triangles). After outgrowth for about 5 h, cells were resuspended in GTS medium with 2.5 mM methylglyoxal (squares). The data are the means from at least three replicate experiments, with standard deviations of less than 10%.
Symbiotic properties of mutant strain CIAT899-13T2.
Strain CIAT899-13T2 was examined for its symbiotic properties and found to form effective nodules on beans and leucaena. The appearance of plants inoculated with the mutant strain was found to be similar to that of plants inoculated with the wild-type strain. Control plants without rhizobial inoculation showed clear symptoms of nitrogen deficiency (data not shown). This result suggests that a mutation in the glutathione synthetase gene (gshB) does not affect the symbiotic ability of CIAT899 to form effective nodules. However, in a coinoculation experiment, in which bean plants were coinoculated with a combination of wild-type and mutant CIAT899-13T2 cells in a 1:1 ratio, the total number of nodules harvested from five independent bean plants were found to be occupied exclusively by the wild-type strain. Therefore, the gshB mutation appears to affect significantly the ability to compete during the process of bean nodulation.
DISCUSSION
A Tn5-luxAB insertion mutant of R. tropici CIAT899 that is unable to grow under acidic conditions was isolated and characterized. The mutation in this strain was shown to be due to a single Tn5-luxAB insertion in the gshB gene, based on the following evidence. Firstly, Southern analysis of EcoRI-restricted DNA from strain CIAT899-13T2 with a Tn5-luxAB probe revealed a single hybridizing EcoRI fragment. Secondly, DNA sequence analysis of the Tn5-luxAB flanking region revealed a high degree of similarity with the gshB gene of several bacteria. Finally, exogenous glutathione allowed mutant CIAT899-13T2 to recover the wild-type tolerance to low pH levels, high osmolarity, and the effects of weak organic acids, hydrogen peroxide, and methylglyoxal.
Different genera of rhizobia show significant variability in their ability to grow under low pH conditions, although the molecular basis for the observed differences is not clear. A correlation between the bacterial cell surface, including the amount of exopolysaccharides, and lipopolysaccharide composition and acid tolerance has been reported (1, 23, 44, 45). Metabolic changes upon shifting cultures to acid media have been reported, including the accumulation of cellular polyamines and elevated levels of glutamate (23). Overall, these responses seem to be a consequence of pH stress, rather than factors required for survival. Here, we demonstrate that metabolic production of glutathione is essential to protect R. tropici against environmental stresses that are frequently found in nature, such as acidity and osmotic or oxidative shock. Soil acidity and other acid-related toxicity factors, including aluminum and manganese toxicity, have previously been implicated to be major environmental constraints affecting the symbiotic performance of legumes (9).
An important role for glutathione has been demonstrated in E. coli. Glutathione is involved in the binding, transformation, and detoxification of a wide variety of compounds. One recognized component of this process of detoxification is the formation of a glutathione adduct, which in turns activates potassium efflux systems (KefB and KefC) (19). The KefB and KefC channels offer the cell a means to lower the cytoplasmic pH level to effect protection against cytotoxic chemicals such as methylglyoxal (19). Glutathione also provides protection against chlorine compounds in E. coli and against oxidants in E. coli and Rhizobium leguminosarum bv. phaseoli (13, 14). The behavior of the glutathione-deficient CIAT899-13T2 strain of R. tropici reported here appears to be similar to that reported for equivalent mutant strains of E. coli and other bacteria. Therefore, the role of glutathione in bacterial stress protection seems to be shared by a diversity of bacteria, although the molecular basis of its action mechanism remains unknown. Furthermore, our finding of glutathione involvement in the acid tolerance adds a novel aspect to the observed protection of cells against environmental stresses.
pHi homeostasis is not fully understood, but it had been proposed to involve the circulation of Na+ and K+ ions (10, 11, 30). Higher potassium levels in CIAT899 following exposure to acid pH levels were indeed detected (1). Other studies have shown an increased expression of the E. coli Kdp system, a high-affinity potassium transporter found in many bacteria, under low pH conditions (6, 30). In E. coli the potassium channels KefB and KefC are inhibited by glutathione, and in the absence of glutathione, K+ leaks out of the cells (11). In addition, it was found that the pHi level in a glutathione-deficient E. coli strain was lower than that in the parent strain in medium containing a low level of K+ (19). Therefore, we hypothesized that the CIAT899-13T2 mutant strain might be affected in its ability to maintain K+ levels. Furthermore, if the pHi homeostasis of R. tropici also involves the control of cytoplasmic potassium levels, then the essential requirement of glutathione may result from its involvement in a process associated with acid tolerance rather than the movement of H+ ions, per se. Consistent with this hypothesis we observed that the intracellular K+ pool of the wild-type strain indeed increased when cells were exposed to acidic conditions and that the K+ level in the acid-sensitive mutant CIAT899-13T2 was lower than that in the wild-type strain. Therefore, it is reasonable to conclude that in order to tolerate external acidity, bacterial cells respond to adjust their intracellular potassium levels and that the capacity to generate a pH gradient, associated with potassium uptake, may be disturbed in the CIAT899-13T2 mutant (10). Under acidic conditions, the available mechanisms for potassium uptake, such as Kdp and Trk found in E. coli (32), may not be sufficient in Gsh− mutant strains to reverse the effect of potassium leakage via the glutathione-dependent Kef system, resulting in an inadequate intracellular potassium level.
With regard to the observed impairment in nodule occupancy efficiency in our R. tropici Gsh-deficient strain CIAT899-13T2, we do not know which of the different stages of infection (e.g., root colonization, infection thread, bacterial release, etc.) is affected in the glutathione-deficient mutant. Competition for nodulation of legumes is a very complex process. Both environmental influences, such as temperature and soil characteristics, and specific rhizobial genes impact competition (12). Clearly, glutathione is important for the symbiotic process in R. tropici, since a glutathione-deficient strain is outcompeted by the wild-type strain.
In conclusion, the data presented here demonstrate the essential nature of glutathione biosynthesis for R. tropici under acidic conditions and other environmental stresses. Our results provide new evidence for the proposed role for glutathione in bacterial physiology in general and in optimal symbiotic performance of rhizobia in particular.
ACKNOWLEDGMENTS
This work was supported by grants from Agencia de Promoción Científica y Tecnológica, Argentina (PICT-97 No. 0628), from the NSF Center of Microbial Ecology, and from the Department of Energy. O.M.A. is member of the research career of the National Research Council-CONICET, Argentina. P.M.R. was supported by CIC, Buenos Aires, Argentina.
REFERENCES
- 1.Aarons S R, Graham P H. Response of Rhizobium leguminosarum bv. phaseoli to acidity. Plant Soil. 1991;134:145–151. [Google Scholar]
- 2.Aguilar O M, Grasso D H. The product of the Rhizobium meliloti ilvC gene is required for isoleucine and valine synthesis and nodulation of alfalfa. J Bacteriol. 1991;173:7756–7764. doi: 10.1128/jb.173.24.7756-7764.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 4.Anderson M E. Determination of glutathione and glutathione disulfide in biological samples. Methods Enzymol. 1985;113:548–555. doi: 10.1016/s0076-6879(85)13073-9. [DOI] [PubMed] [Google Scholar]
- 5.Anyango B, Wilson K J, Beynon J L, Giller K E. Diversity of rhizobia nodulating Phaseolus vulgaris L. in two Kenyan soils with contrasting pHs. Appl Environ Microbiol. 1995;61:4016–4021. doi: 10.1128/aem.61.11.4016-4021.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Asha H, Gowrishankar J. Regulation of kdp operon expression in Escherichia coli: evidence against turgor as signal for transcriptional control. J Bacteriol. 1993;175:4528–4537. doi: 10.1128/jb.175.14.4528-4537.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beringer J E. R factor transfer in Rhizobium leguminosarum. J Gen Microbiol. 1974;84:188–198. doi: 10.1099/00221287-84-1-188. [DOI] [PubMed] [Google Scholar]
- 8.Blattner F R, Plunkett III G, Bloch C A, Perna N T, Burland V, Riley M, et al. The complete genome sequence of Escherichia coli K12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 9.Bohlool B B, Ladha J K, Garrity D P, George T. Biological nitrogen fixation for sustainable agriculture: a perspective. Plant Soil. 1992;141:1–11. [Google Scholar]
- 10.Booth I R. Regulation of cytoplasmic pH in bacteria. Microbiol Rev. 1985;49:359–378. doi: 10.1128/mr.49.4.359-378.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Booth I R. Bacterial responses to pH. Novartis Foundation Symposium 221. Chichester, United Kingdom: Wiley; 1999. The regulation of intracellular pH in bacteria; pp. 19–37. [DOI] [PubMed] [Google Scholar]
- 12.Buttery B R, Park S J, Hume D J. Potential for increasing nitrogen fixation in grain legumes. Can J Plant Sci. 1992;72:323–349. [Google Scholar]
- 13.Chesney J A, Eaton J W, Mahoney J R., Jr Bacterial glutathione: a sacrificial defense against chlorine compounds. J Bacteriol. 1996;178:2131–2135. doi: 10.1128/jb.178.7.2131-2135.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Crockford A J, Behncke C, Williams H D. The adaptation of Rhizobium leguminosarum bv. phaseoli to oxidative stress and its overlap with other environmental stress responses. Microbiology. 1998;142:331–336. doi: 10.1099/13500872-142-2-331. [DOI] [PubMed] [Google Scholar]
- 15.De Bruijn F J, Rossbach S. Transposon mutagenesis. In: Gerhardt P, Murray R G E, Wood W A, Krieg N R, editors. Methods for general and molecular biology. Washington, D.C.: American Society for Microbiology; 1994. pp. 387–405. [Google Scholar]
- 16.Ditta G, Stanfield S, Corbin D, Helinski D R. Broad host range DNA cloning system for Gram negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc Natl Acad Sci USA. 1980;77:7347–7351. doi: 10.1073/pnas.77.12.7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Doherty H M, Adams D G. Cloning and sequence of ftsZ and flanking regions from the cyanobacterium Anabaena PCC 7120. Gene. 1995;163:93–96. doi: 10.1016/0378-1119(95)00416-4. [DOI] [PubMed] [Google Scholar]
- 18.Errington J. Bacillus subtilis sporulation: regulation of gene expression and control of morphogenesis. Microbiol Rev. 1993;57:1–33. doi: 10.1128/mr.57.1.1-33.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ferguson G P, Booth I R. Importance of glutathione for growth and survival of Escherichia coli cells: detoxification of methylglyoxal and maintenance of intracellular K+ J Bacteriol. 1998;180:4314–4318. doi: 10.1128/jb.180.16.4314-4318.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ferguson G P, Munro A W, Douglas R M, McLaggan D, Booth I R. Activation of potassium channels during metabolite detoxification in Escherichia coli. Mol Microbiol. 1993;9:1297–1303. doi: 10.1111/j.1365-2958.1993.tb01259.x. [DOI] [PubMed] [Google Scholar]
- 21.Ferguson G P, McLaggan D, Booth I R. Potassium channel activation by glutathione-S-conjugates in Escherichia coli: protection against methylglyoxal is mediated by cytoplasmic acidification. Mol Microbiol. 1995;17:1025–1033. doi: 10.1111/j.1365-2958.1995.mmi_17061025.x. [DOI] [PubMed] [Google Scholar]
- 22.Givskov M, Eberl L, Molin S. Responses to nutrient starvation in Pseudomonas putida KT2442L two-dimensional electrophoretic analysis of starvation- and stress-induced proteins. J Bacteriol. 1994;176:4816–4824. doi: 10.1128/jb.176.16.4816-4824.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Graham P H, Draeger K J, Ferrey M L, Conroy M J, Hamer B E, Martinez E, et al. Acid pH tolerance in strains of Rhizobium and Bradyrhizobium, and initial studies on the basis for acid tolerance of Rhizobium tropici UMR 1899. Can J Microbiol. 1994;40:198–207. [Google Scholar]
- 24.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 25.Hungría M, Franco A A, Sprent J I. New sources of high-temperature tolerant rhizobia for Phaseolus vulgaris L. Plant Soil. 1993;149:103–109. [Google Scholar]
- 26.Kiss G B, Vincze É, Kálman Z, Forrai T, Kondorasi Á. Genetic and biochemical analysis of mutants affects nitrate reduction in Rhizobium meliloti. J Gen Microbiol. 1979;113:105–118. [Google Scholar]
- 27.Kjelleberg S. Starvation in bacteria. New York, N.Y: Plenum Press; 1993. [Google Scholar]
- 28.Kragelund L, Christoffsen B, Nybroe O, de Bruijn F J. Isolation of lux reporter gene fusions in Pseudomonas fluorescens DF57 inducible by nitrogen or phosphorus starvation. FEMS Microbiol Ecol. 1995;17:95–106. [Google Scholar]
- 29.Kroll R G, Booth I R. The role of potassium transport in the generation of a pH gradient in Escherichia coli. Biochem J. 1981;198:691–698. doi: 10.1042/bj1980691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Malli R, Epstein W. Expression of the Kdp ATPase is consistent with regulation by turgor pressure. J Bacteriol. 1998;180:5102–5108. doi: 10.1128/jb.180.19.5102-5108.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martínez Romero E, Segovia L, Mercante F M, Franco A A, Graham P, Pardo M A. Rhizobium tropici, a novel species nodulating Phaseolus vulgaris L beans and Leucaena sp. trees. Int J Syst Bacteriol. 1991;41:417–426. doi: 10.1099/00207713-41-3-417. [DOI] [PubMed] [Google Scholar]
- 32.Meury J, Kepes A. Glutathione and the gated potassium channels of Escherichia coli. EMBO J. 1982;1:339–343. doi: 10.1002/j.1460-2075.1982.tb01171.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Milcamps A, Ragatz D M, Lim P, Berger K A, de Bruijn F J. Isolation of carbon- and nitrogen-deprivation induced loci of Sinorhizobium meliloti 1021 by Tn5-luxAB mutagenesis. Microbiology. 1998;144:3205–3218. doi: 10.1099/00221287-144-11-3205. [DOI] [PubMed] [Google Scholar]
- 34.Milcamps A, de Bruijn F J. Identification of a novel nutrient-deprivation induced Sinorhizobium meliloti gene (hmgA) involved in the degradation of tyrosine. Microbiology. 1999;145:935–947. doi: 10.1099/13500872-145-4-935. [DOI] [PubMed] [Google Scholar]
- 35.Miyajima N, Hirosawa M, Sugiura M, Sasamoto S, Kimura T, Hosouchi T, et al. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 1996;3:109–136. doi: 10.1093/dnares/3.3.109. [DOI] [PubMed] [Google Scholar]
- 36.O'Hara G W, Gross T J, Dilworth M J, Glenn A R. Maintenance of intracellular pH and acid tolerance in Rhizobium meliloti. Appl Environ Microbiol. 1989;55:1870–1876. doi: 10.1128/aem.55.8.1870-1876.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Okumura N, Masamoto K, Wada H. The gshB gene in the cyanobacterium Sinechococcus sp. PCC 7942 encodes a functional glutathione synthetase. Microbiology. 1997;143:2883–2890. doi: 10.1099/00221287-143-9-2883. [DOI] [PubMed] [Google Scholar]
- 38.Perez-Galdona R, Kahn M L. Effects of organic acids and low pH on Rhizobium meliloti 104A14. Microbiology. 1994;140:1231–1235. doi: 10.1099/13500872-140-5-1231. [DOI] [PubMed] [Google Scholar]
- 39.Peters J M, Dalrymple B P, Jorgensen W K. Sequence of a putative glutathione synthetase-II gene and flanking regions from Anaplasma centrale. Biochem Biophys Res Commun. 1992;182:1040–1046. doi: 10.1016/0006-291x(92)91836-f. [DOI] [PubMed] [Google Scholar]
- 40.Phillips D A, Sande E S, Vriezen J A C, de Bruijn F J, Le Rudulier D, Joseph C M. A new genetic locus in Sinorhizobium meliloti is involved in stachydrine utilization. Appl Environ Microbiol. 1998;64:3954–3960. doi: 10.1128/aem.64.10.3954-3960.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roe A J, McLaggan D, Davidson I, O'Byrne C, Booth I R. Perturbation of anion balance during inhibition of growth of Escherichia coli by weak acids. J Bacteriol. 1998;180:767–772. doi: 10.1128/jb.180.4.767-772.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 43.Watson N, Dunyak D S, Rosey E L, Stoneczewski J L, Olson E R. Identification of elements involved in the transcriptional regulation of the Escherichia coli operon by external pH. J Bacteriol. 1992;174:530–540. doi: 10.1128/jb.174.2.530-540.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wayne G R, Tiwari R P, Wong C M, Dilworth M J, Glenn A R. The transcriptional regulator gene phR in Sinorhizobium meliloti WSM419 is regulated by low pH and other stresses. Microbiology. 1998;144:3335–3342. doi: 10.1099/00221287-144-12-3335. [DOI] [PubMed] [Google Scholar]
- 45.Wolk P C, Cai Y, Panoff J M. Use of transposon with luciferase as a reporter to identify environmentally responsive genes in a cyanobacterium. Proc Natl Acad Sci USA. 1991;88:5355–5359. doi: 10.1073/pnas.88.12.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]