Abstract
The chemical senses of olfaction and taste are well developed in fish and play a vital role in its various activities such as navigation, mate recognition, and food detection. The small teleost fish Astyanax mexicanus consists of interfertile river‐dwelling and cave‐dwelling populations, referred to as “surface fish” and “cavefish” respectively. An important anatomical feature of cavefish is the lack of eyes leading them to be referred to as blind fish and suggesting an enhanced functional role for other senses such as taste. In this study, we characterize the expression of bitter taste receptors (T2Rs or Tas2Rs) in A. mexicanus and investigate their functionality in a heterologous expression system. The genome database of A. mexicanus (ensemble and NCBI) showed 7 Tas2Rs, among these Tas2R1, Tas2R3, Tas2R4, and Tas2R114 are well characterized in humans and mice but not in A. mexicanus. Therefore, the 4 Tas2Rs were selected for further analysis and their expression in A. mexicanus was confirmed by in situ hybridization and RT‐PCR in early developmental stages. These Tas2Rs are expressed in various oral and extraoral organs (liver, fins, jaws, and gills) in A. mexicanus, and Tas2R1 has maximum expression and is localized throughout the fish body. Using the heterologous expression of A. mexicanus T2Rs in HEK293T cells coupled with cell‐based calcium mobilization assays, we show that A. mexicanus T2Rs are activated by commonly used fish food and known bitter agonists, including quinine. This study provides novel insights into the extraoral expression of T2Rs in A. mexicanus and suggests their importance in extraoral food detection.
Keywords: Astyanax mexicanus, bitter taste receptors, calcium mobilization, in situ hybridization, taste buds, teleost
1. INTRODUCTION
Taste is a fundamental chemosensory system, which plays a vital role in the identification of nutrients and regulation of food intake. Tastants stimulate specialized cells known as taste receptor cells locally organized as taste buds, consisting of signaling molecules involved in taste signal transduction. Several reports in the recent past have studied the physiology of taste receptors in extraoral tissues and have led to the assumption that these receptors could represent excellent candidates for high‐affinity chemo‐detectors in cells outside the tongue. 1 , 2 , 3 Mammals, including humans, have five basic taste modalities where sweet, bitter, and umami taste transductions are mediated through G protein‐coupled receptor (GPCR) signaling pathway. 4 , 5 In rodents, bitter taste is mediated by a family of ∼30 highly divergent bitter taste receptors (Tas2Rs or T2Rs). 4 , 5 Besides human and rodents, T2Rs are also expressed in animals such as chicken, frog, and teleost fish. 6 , 7 In fish, the chemical senses of olfaction and taste, are well developed and important for various survival cues. 8 Molecular analyses have revealed that teleosts and mammals share pivotal signaling components involved in taste signal transduction. 9
Astyanax mexicanus (Mexican tetra) is a novel animal model in evolutionary developmental biology. This fish exists in two morphological types: an eyed, pigmented surface‐dwelling form (surface fish, SF), and an eyeless unpigmented cave‐dwelling form (cavefish, CF). 10 , 11 , 12 However, both morphs share the same genome with the exception of certain polymorphisms in CF which are important for its cave adaptation. 13 A. mexicanus is an omnivorous feeder that eats flush‐out debris and leftovers from other cave‐dwelling animals, such as cave‐dwelling bats and some small organisms. The sighted SF in the wild feeds more on plant‐based materials such as water weeds and small organisms. 14 Although both forms are omnivorous, the CF lacks eyes. Over the last 1 million years, the CF has evolved mechanisms to search, identify, and consume food in these resource‐limiting environments. 14 These mechanisms include the development of constructive traits, such as large jaws to accommodate more teeth and an increased number of taste buds around the mouth and ventral surface of the head. 10 , 11 The larger jaws of the CF result in bigger mouths to maximize the opportunities to capture food. This makes more space on the lips for expansion of the gustatory system, namely tastebuds. Schemmel (1967) showed that the number of tastebuds in CF was five to sevenfold greater than compared to the sighted SF. 15
Furthermore, in SF, taste buds are mainly found in the labial epithelium, whereas in the CF, they are also found in the skin of the maxilla, lower jaw, and ventral aspect of the head. 14 Tastebuds in teleosts are typically pear‐ or onion‐shaped intraepithelial sensory structures that consist of multiple cell types. Although Schemmel (1980) showed that the morphology of the tastebuds does not differ between the two morphs, more recent evidence has shown, using immunohistochemistry, that the tastebuds in CF larvae contain more receptor cells and are innervated by more axon profiles. 16
The T2R families in teleost fish have been described as small and evolutionarily conserved, in contrast to the significant evolutionary dynamics observed for tetrapod T2Rs. The T2Rs have been identified in divergent fish species, such as zebrafish, medaka fish, and fugu fish (Takifugu rubripes). 8 , 17 , 18 , 19 A recent genomic analysis revealed an expansion of T2Rs in A. mexicanus. The cellular and molecular characteristics of specific T2Rs and their functional significance are largely unknown in fish species including A. mexicanus. 9
The aim of the present study is to investigate the expression of Tas2Rs in different organs of the A. mexicanus and to functionally characterize A. mexicanus T2Rs using heterologous systems. Quinine, a known agonist of human T2Rs, is part of the quinolone alkaloid family and is usually found in plants. 20 , 21 Therefore, the possibility that quinolone alkaloids could be present in aquatic plants and that recognition of quinolones by the fish extraoral taste receptors could be beneficial cannot be discarded. In this study, it was tested if quinine, dextromethorphan (DXM), and commonly used fish food can activate A. mexicanus T2Rs. The specificity of T2R signaling was also analyzed using blockers.
2. MATERIAL AND METHODS
2.1. Mexican tetra fish strains and maintenance
Mexican tetra (A. mexicanus) was obtained from Dr. R. Borowsky (New York University) and Dr. W. Jeffery (University of Maryland). In accordance with the Canadian council of animal care guidelines, the fish were raised at the animal care facility, University of Manitoba, Canada, with a controlled pH (7.4 ± 0.4) and conductivity (900 ± 100 μS/m) levels. Animals were maintained under a stable 12 h light and dark cycle at 24°C in a 20‐gallon glass tank. Fish were fed twice a day with flake food and to prime, for breeding they were fed with protein‐enriched foods (bloodworms, blackworms, and brine shrimp). To induce spawning, the tank temperature was increased to 26°C and one male was added to a tank containing one female. Eggs were collected the next day. Animals were raised according to the guidelines of the Canadian Council of Animal Care (CCAC) and the experiment protocols (AC 11301) were approved annually.
2.2. Materials
Human embryonic kidney (HEK) 293 T cells were purchased from American Type Culture Collection and maintained in a cell culture medium composed of DMEM‐F12 (Thermo Fisher Scientific) supplemented with 10% heat‐inactivated fetal bovine serum (Millipore Sigma) and 1% penicillin–streptomycin (Thermo Fisher Scientific). Hygromycin (Millipore Sigma) was used as the selection antibiotic for generating stable cells. The anti‐FLAG antibody used for flow cytometry was purchased from BioLegend (Cat# 637308, RRID:AB_2561497). The anti‐digoxigenin‐AP antibody used in the dot blot analysis was purchased from Roche Diagnostics (Cat# 11093274910, RRID:AB_514497). The bitter compounds (quinine hydrochloride), and dextromethorphan (DXM), were purchased from Millipore Sigma. The frozen fish food, brine shrimp (Omega One, OmegaSea LLC), and bloodworms (San Francisco Bay Brand) were purchased from a local pet shop in Winnipeg, Manitoba. Gallein was purchased from Tocris. U73122 and U73343 were purchased from Sigma–Aldrich. A commercially synthesized peptide inhibitor (AGDDAPRAVF) was acquired from GenScript. All compounds were prepared using calcium assay buffer (1X HBSS, 20 mM, 4‐(2‐hydroxyethyl)‐1‐piperazineethanesulfonic acid [HEPES]). The Fluo‐4 NW calcium assay kit was purchased from ThermoFisher Scientific. All chemicals were of analytical grade. DIG DNA Labeling Kit was purchased from Sigma–Aldrich to prepare an antisense RNA probe for in situ hybridization.
2.3. Phylogenetic analysis
The phylogenetic analysis was performed to explore the relationship of bitter taste receptor genes in A. mexicanus. Data regarding Tas2R expression in A. mexicanus were acquired from NCBI (RRID:SCR_006472), Uniprot (RRID:SCR_002380), and Ensembl (RRID:SCR_002344) databases. We used MEGA version 10 software (Molecular Evolutionary Genetics Analysis, http://www.megasoftware.net, RRID:SCR_000667) to prepare the phylogenetic tree. 22 The amino acid sequences for each T2R were obtained from the Uniprot protein search engine and included in the MEGA 10 software. As reported previously, we have selected the neighbor‐joining statistical analysis method, and the bootstrap consensus tree was inferred from 1000 replicates. 23
2.4. Reverse transcriptase‐polymerase chain reaction
Total RNA was isolated from different sections of the 30 days post‐fertilization (dpf) A. mexicanus body using an RNeasy mini kit. Total RNA was quantified using a Nanodrop spectrophotometer. cDNA synthesis: 800 ng of the total RNA was subjected to reverse transcription with iScript™ Reverse Transcription Supermix (BioRad) for cDNA synthesis, as per the manufacturer's instructions. The supermix contents were RNase H+ MMLV reverse transcriptase, RNA inhibitor, dNTPs, Oligo (dT), random primers, buffer, MgCl2, and stabilizers. Briefly, the reaction was performed at 46°C for 20 min followed by RT inactivation at 95°C for 1 min. The resulting cDNA was used for RT PCR (BioRad). The PCR primers used for the selected taste receptor genes were designed using PrimerQuest software (Integrated DNA Technologies) and validated with the NCBI primer blast tool (www.ncbi.nlm.nih.gov/tools/primer‐blast/, RRID:SCR_003095). The primer sequences, annealing temperature, and NCBI accession ID are listed in Table 1. The annealing temperature for each primer set was optimized by performing Gradient PCR (Range 50–65°C). GAPDH was used as a reference gene. Non‐template control (NTC) and no reverse transcriptase (NRT) negative control were run in parallel to assess the overall specificity of the reaction. All reactions were run in triplicate.
TABLE 1.
Primer lists for gene expression studies and antisense probe preparation for In situ hybridization experiments. bp, base pairs
| Genes | Forward primer (5′‐3′) | Reverse Primer (5′‐3′) | Amplicon (bp) | Annealing Temp (°C) | NCBI accession ID |
|---|---|---|---|---|---|
| For Gene expression | |||||
| Tas2R1 | GACCATCACAGGCATCATACA | TTGGTCAAAGTCTGCAGTAGTAA | 103 | 57.1 | XM_022680204.1 |
| Tas2R3 | CTTTGACCATGGTGCTGATTATG | GCTGCTGGTCATCTTCCTAATA | 104 | 62.8 | XM_022678463.1 |
| Tas2R4 | GAACCAACGTAACCACCAATAC | GGGCAGAGGAAGGACATATAAA | 118 | XM_007248674.2 | |
| Tas2R114 | CAAGCTGTGCAACTGTTCTATAC | ATGGTCACCCTCATCTGTTTC | 106 | XM_022679999.1 | |
| GAPDH | GTGATGAACGCTCCTCTATCTTT | GTTGCTGTAGCCAAACTCATTG | 104 | 60.2 | XM_007249798.3 |
| For in‐situ hybridization | |||||
| Tas2R1 | GTGCAACACAGCTCTTCATTT | TAATACGACTCACTATAGGGGTGTCTCCGCAGGTATGAAATA | 103 | 60 | XM_022680204.1 |
| Tas2R3 | CTCTCCTGGTGCTTGTGATTT | TAATACGACTCACTATAGGGTCAGCACCATGGTCAAAGTATC | 104 | 62.1 | XM_022678463.1 |
| Tas2R4 | GGGTGATGTGGAACTCCTTT | TAATACGACTCACTATAGGGGGCCGTGACTTTAGGAATGTA | 118 | XM_007248674.2 | |
| Tas2R114 | GTGGCTGAACGTGTTCTACTAT | TAATACGACTCACTATAGGGATGGTCACCCTCATCTGTTTC | 106 | 68 | XM_022679999.1 |
2.5. Whole‐mount in situ hybridization
The PCR products of the selected genes were used to synthesize digoxigenin‐labeled antisense RNA probes. Primers for PCR products are mentioned in Table 1. The reverse primer has an additional T7 RNA polymerase promoter. The PCR product was purified with a QIAquick PCR purification kit (Qiagen). In vitro transcription was performed to make an antisense RNA probe according to the manufacturer's instructions (Roche kit, Sigma‐Aldrich). The labeling efficiency of all probes used was confirmed using dot blot analysis (data not shown). Anti‐digoxigenin‐AP (Roche) antibody was used in the dot blot analysis.
Whole‐mount in situ hybridization was performed as described by Thisse and Thisse (2008) with the modification in proteinase K treatment. 24 , 25 Proteinase K (20 μg/ml) was used for 10 min at room temperature for 2 days post‐fertilization (dpf) old fish. A no probe control hybridization mix was used as a negative control. After the color reaction, samples were post‐fixed with 4% paraformaldehyde and dehydrated through a graded methanol series. Specimens were photographed using SteREO Discovery V8 stereomicroscope.
2.6. Expression vectors and cell culture
The N‐terminal FLAG epitope‐tagged Tas2R1, Tas2R3, Tas2R4, and Tas2R114 genes were codon‐optimized for expression in mammalian cells and cloned into the KpnI‐NotI site of the pcDNA3.1/Hygro(+) expression vector. The recombinant expression vectors were acquired from commercial sources (GenScript Inc). 26 The gene constructs of Tas2Rs are listed in Table 2. The HEK293T heterologous expression system was used, similarly to previous studies that characterized ligands of fish taste receptors. 6 For recombinant expression of fish T2Rs in HEK293T cell lines, the cells were transfected with corresponding fish Tas2R expressing plasmids using Lipofectamine 2000 as per the manufacturer's instructions. Post transfection, the cells were grown in media supplemented with hygromycin (200 μg/ml). To establish the stable cell lines stably expressing the fish T2Rs, the cell was allowed to propagate in the selection media until the clones starts to appear. The clones were screened for higher expression of T2Rs using flow cytometry. The protocol used for the establishment of a stable cell line has been previously described. 27 , 28
TABLE 2.
Recombinant amino acid sequences of selected A. mexicanus T2Rs
| Gene name (NCBI accession number) | Construct |
|---|---|
|
Tas2R1 |
MDYKDDDDKFHSIHWSFSISISLKTFQSVNVPLSLFSIFMDIFFIFCLSPVSEQQRLKPPLLVLLGSLVGCNTA LHFFTLLFVLSDFANSLSETDSYSFYYFTAQCLLFTMRVSITSCLWLNVFYYCQIVPARHPFLIMLKRNIRLFVY SALIIDKFFFLEEFIVYIASYLIRLYRKPEIYNSTYTNMVTNALIVDIWLRLVYFFFSVCMMLASGCATISYLRRH MRNMEKSSRSSARLQSQLRVTITGIIQTLLYLLCSVWLILDDVAFYLTTADFDQKAYIFYTVISLYSFGTNINLG VGQTVFREQAILIWQKLFGSFLD* |
|
Tas2R3 |
MDYKDDDDKNVRQPLMTLLVLVICCTSTFQLCFIIHLSLEFMAIGGNLKQIVDTLMSYAVRVSIPASLWLN IFYCSQIIPVTYAPFIWLKKNIKTVVYFLLISTKIYFLTYFVLEILMSYGIFSRMKLPQVNSTVINGSYASVRTKFQ ISSLLDILDTLTMVLIMLGLCVMLIVTGSTGRYLYKHIRKMTSSGMPFNSQTLQNQVRVTLTSFIQGFIYLLFT VGAFIDVYCAFKSLVFDWDIVWMLHNLYSLATIINLGVGQSLFRQRVAHLCQKAVNIPSS* |
|
Tas2R4 |
MDYKDDDDKHARVHAQLTSQWINIMGLDTFILQLIAIVVLVCTGVMWNSFNLIATIRMQLKTKGIQTMIL IIFSFSFSNVILVLSCFAIVLFVTLDPVFMCTKIEAHHLILVYMWLSSSCVSFWSIALLSVLHCVKVVSFSIGCFN ALKKNISRITNIALLLICLGSFLLFIPFFTLYIPKVTATNTTIGINGTNVTTNTSNTTTKTCPLPSLSPQISTPLYTLLY MSFLCPIPLMIMMTTSLRMVIHLCQHVLSLSKNQTQVQSLDSYLFICKLNISLVGVYLITLAIVSIFFILKMLELT VSYLLIIFGFSLYCIMTAALLTASTKKLREKFWRMICCKETKKQ* |
|
Tas2R114 |
MDYKDDDDKAENTSTVGFLKMSSQAFASVNMAVASLSIFINLFFVFCMVFPSQRSEHLKQPLNILLGLLIGS SIASHVCILIFVHSGDVLFTAESPKFLINHIVEETMLFIMRTSVTSHLWLNVFYYCQIVPAQRSFLIWLKDNIRVF VYFALIMDRLFFLSSFITSILYYSEIQIISNSTTYTNTSLMDTEQNTTAAILRELSETCIIQYWLRFAYFFISLCVMLA ASCATVLYLRRHMKRMEESSKSFSSPRLQKQMRVTITGIVQLILQMICISWIISDGPLRLKLPSHFDPDRHIYST VISLYTLCSTLNLCVGQSIFRQPVINMWQNLVQFFCANSE* |
2.7. Flow cytometry
For the flow cytometry analysis 1 × 105 HEK293T cells stably expressing FLAG‐tagged T2Rs were used. The cells were stained with allophycocyanin (APC) conjugated anti‐FLAG antibody at 1:300 dilutions followed by a 1 h incubation in dark at 4°C. After incubation, the cells were washed three times with fluorescence‐activated cell sorting (FACS) buffer (0.5% BSA in 1X PBS). Next, the cell surface expression of the FLAG‐tagged T2Rs was analyzed on a BD FACSCanto II flow cytometer (BD Biosciences, RRID:SCR_018056). The percentage of cells expressing the FLAG (APC) surface marker was determined with the gating method using the BD FACSDiva (BD Biosciences Systems, RRID:SCR_001456) and FlowJo (BD Life Sciences) software. One‐way ANOVA and Holm‐Sidak's multiple comparisons tests were performed to calculate the statistical significance of three independent experiments. A p‐value of <0.05 was considered statistically significant.
2.8. Fish food preparation for calcium mobilization assay
The grinded fish food, brine shrimp, and bloodworm, were thawed on ice and 300 mg of each fish food were dissolved in 10 ml of assay buffer (1X Hanks' balanced salt solution, 20 mM HEPES). The solution was homogenized by pipetting several times, followed by three cycles of 30s sonication (W‐375 Sonicator, Heat Systems Ultrasonics Inc.) with an interval of 30s in between when the sample was placed on ice. Lastly, the solution was filtered using 0.45 μm and 0.2 μm sterile syringe filters, aliquoted, and stored at −20°C until further use.
2.9. Calcium mobilization assay
Calcium mobilization assays were performed as described previously. 26 , 28 , 29 Briefly, HEK293T cell lines stably expressing T2Rs were incubated with Fluo‐4 NW dye for 35 mins at 37°C followed by another 35 mins at room temperature. Then, the cells were treated with different concentrations of compounds by robotically adding the compounds using FlexStation 3 plate reader (Molecular Devices). The cells were treated with well‐known T2R agonists, quinine and dextromethorphan (DXM), at concentrations ranging from 0.06 to 4 mM, and commonly used fish food, brine shrimp, and blood worm, at a concentration ranging from 0.16–10 mg/ml. To characterize the canonical signaling pathway activated by T2Rs, signaling inhibition experiments were performed by pre‐treating the cells for 30 min with the Gβγ inhibitor, gallein, PLCβ inhibitor, U73122, or its inactive analog U73343 (10 μM), followed by treatment with brine shrimp. 28 , 30 For the competition‐binding assays, the cells were treated with a known T2R blocker, peptide AGDDAPRAVF (1 mM), in the presence of quinine. For elucidating the IC50 value of the peptide, the cells were treated with AGDDAPRAVF at concentrations ranging from 1 mM to 0.03 mM in the presence of a single concentration (1 mM) of quinine (Zhang et al. 2018).
Calcium mobilized was expressed as ΔRFU after subtracting the responses of HEK293T control cells. The data from at least three independent experiments performed in triplicate were analyzed. The EC50 and IC50 values were calculated using GraphPad Prism software version 6.0 (GraphPad Software Inc., RRID:SCR_002798) and a nonlinear regression (curve‐fit) model using the function Y = a + (d−a)/(1 + 10[(logEC50 or IC50 – X)*HillSlope], where a is minimum (basal response), d is maximum (maximal response), and X is compound (agonist or inhibitor) concentration. R2 values were obtained and used to determine the goodness of the curve fit of the data.
2.10. Statistical analysis
The data shown are the ± SEM from at least three independent experiments (n). One‐way analysis of variance was performed to compare three or more groups, followed by Holm‐Sidak's multiple comparison post hoc test to check the significance. A p < 0.05 was considered statistically significant.
3. RESULTS
3.1. Phylogenetic analysis
Astyanax mexicanus is diploid with two sets of 25 chromosomes. The chromosomal locations of the seven Tas2R genes are shown in Figure 1A. Among these genes, Tas2R1, Tas2R3, Tas2R4, and Tas2R114 are well characterized in humans and mice but not in A. mexicanus, therefore they were selected for further analysis. Tas2R3 gene has two isoforms having the same location on the chromosome. The phylogenetic analysis of Tas2Rs of A. mexicanus is shown in Figure 1B. We did not include partial or pseudo Tas2R genes in the phylogenetic analysis. The amino acid sequences of all seven Tas2Rs were aligned, and the Neighbor‐Joining method with bootstrapping was performed. All ambiguous positions were removed for each sequence pair (pairwise deletion option). There was a total of 360 positions in the final dataset. The optimal tree with the sum of branch length of 4.5393 is shown in Figure 1B. The tree was drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. We observed some Tas2Rs have long branch lengths indicating a more genetic change than other Tas2Rs, Tas2R1, and Tas2R114 originate from the same branch with less genetic divergence, whereas Tas2R4 has maximum genetic changes and falls in a separate branch of the tree.
FIGURE 1.
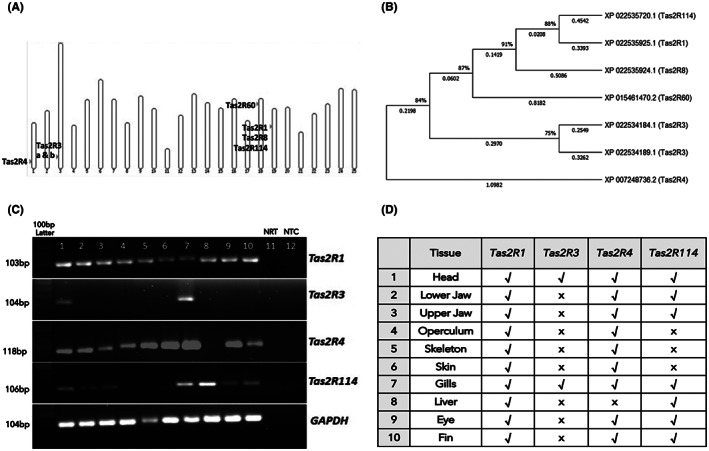
A. mexicanus bitter taste receptor genes (Tas2Rs). (A) Chromosomal location of the A. mexicanus Tas2Rs. A. mexicanus is diploid with two sets of 25 chromosomes. The source of the map is Ensembl using A. mexicanus (genome 2.0) whole genome. (B) Phylogenetic analysis of A. mexicanus Tas2Rs. We performed the analysis using the Neighbor‐Joining method. The optimal tree with the sum of branch length = 4.5393 is shown. The tree was drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. This analysis involved 7 T2Rs amino acid sequences. Evolutionary analyses were conducted in MEGA software version X. (C,D) Tas2Rs are expressed in different tissues of A. mexicanus. 10 μl of PCR product was loaded in 2% agarose gel and stained specific product bands were visualized using a UV transilluminator (BioRad). GAPDH was used as a reference gene. Samples in lanes 1 to 10 represent different tissues and are mentioned in Table (D). NRT (lane 11) is negative control with no reverse transcriptase and NTC (lane 12) is a non‐template control. 100 bp DNA ladder was used. Present: √; Absent: X
3.2. Expression analysis of Tas2Rs
We performed RT PCR analysis to examine the presence of Tas2R1, Tas2R3, Tas2R4, and Tas2R114 and observed that Tas2R1 is expressed in all body parts of A. mexicanus (Figure 1C,D). The four Tas2Rs were expressed in the head and gills and showed varying levels of expression in different body parts (Figure 1C). Next, we checked the expression of these Tas2Rs at early developmental stages from 1 to 5 dpf and found Tas2R1 is constantly expressed at all early stages compared to other Tas2Rs (Figure S1A). However, 30 dpf fish showed the presence of all the Tas2R genes (Figure S1B).
3.3. Whole‐mount in situ hybridization
After confirming the presence of Tas2Rs in early developmental stages, 2 dpf old embryos were analyzed to see the mRNA expression in craniofacial and body region. The whole‐mount in situ hybridization analysis showed that Tas2Rs were present in the developing pharyngeal region, around the spinal cord, and above the eye (Figure 2). Tas2R1 has strong expression throughout the embryo at 2 dpf. At this stage, Tas2R4 has moderate expression and Tas2R3 and Tas2R114 have low expression in similar regions (Figure 2).
FIGURE 2.
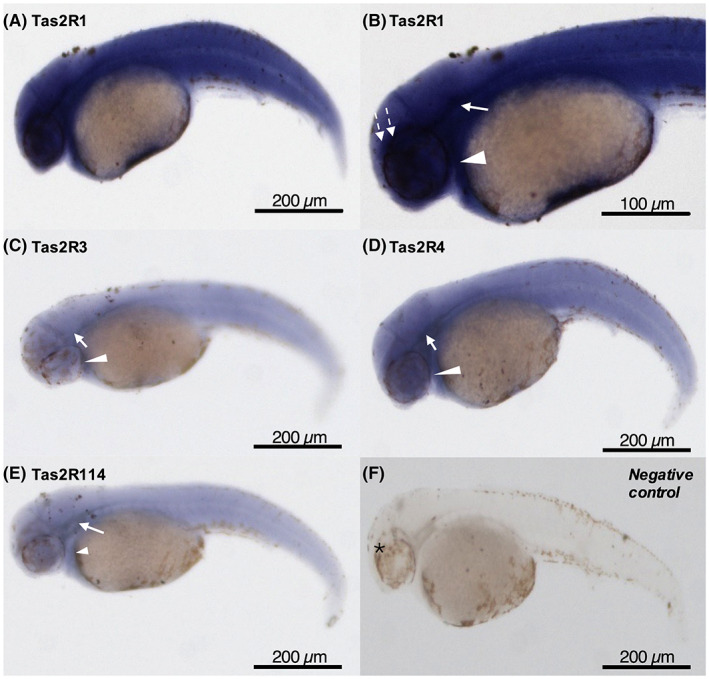
Whole‐mount In‐situ hybridization of Tas2Rs expression of 2‐day post‐fertilization (dpf) A. mexicanus embryos. (A,B) Tas2R1, (C) Tas2R3, (D) Tas2R4, (E) Tas2R114, and (F) no probe negative control. Expression was present in the developing pharyngeal region (arrowhead), around the spinal cord (arrow), and above the eye (dash arrow). Tas2R1 has strong expression throughout the embryo at this stage (A). Tas2R4 has moderate expression (D) and Tas2R3 and Tas2R114 have lower expression in similar regions. *, eye. Scale bar is shown
3.4. Functional characterization of A. mexicanus T2Rs
HEK293T cells stably expressing T2R1, T2R114, T2R3, and T2R4 were used for the functional calcium mobilization assays. The stable expression of T2Rs in these cell lines was confirmed using flow cytometry (Figure S2). The intracellular calcium mobilization was measured after T2R stable cells were treated with quinine (0.06–4 mM) and DXM (0.06–4 mM), which are known to activate multiple human T2Rs. 31 , 33 , 34 Next, to evaluate whether the expressed T2Rs can recognize commonly used fish food, we treated the cells with brine shrimp and bloodworm (0.16–10 mg/ml). Similar to human T2Rs, the selected A. mexicanus T2Rs were activated by multiple compounds. Increased calcium mobilization was observed when these T2Rs were treated with quinine (EC50 0.42 to 1.11 mM) and DXM (EC50 1.43 to 2.04 mM) (Figure 3A,B). On the other hand, the fish food activated only T2R1 (EC50(brine shrimp) 4.99 ± 0.8 mg/ml, EC50(bloodworm) 5.23 ± 1.79 mg/ml) and T2R3 (EC50(brine shrimp) 4.63 ± 0.72 mg/ml, EC50(bloodworm) 4.8 ± 0.2 mg/ml). T2R4 and T2R114 failed to show significant calcium mobilization upon treatment with brine shrimp (Figure 3C). No saturation in calcium mobilization was observed for T2R4 with bloodworm treatment (Figure 3D). As T2R1 was highly expressed through the A. mexicanus body and activated by all selected compounds in this study, it was further selected to characterize the canonical Gβγ‐PLC signaling pathway activated by T2Rs. Upon pretreatment of the cells with the Gβγ inhibitor gallein and PLCβ inhibitor U73122 followed by treatment with brine shrimp, a significant decrease in calcium mobilization was observed. As expected, no significant differences were observed with the U73122 inactive analog U73343 (Figure 3E,F). The inhibition of quinine‐dependent T2R activation by the peptide AGDDAPRAVF previously demonstrated with human T2Rs, 32 was also tested. The results showed that the peptide inhibited quinine response upon activation of fish T2R1 in a concentration‐dependent manner (IC50 = 176.92 ± 31.01 μM, Figure 4).
FIGURE 3.
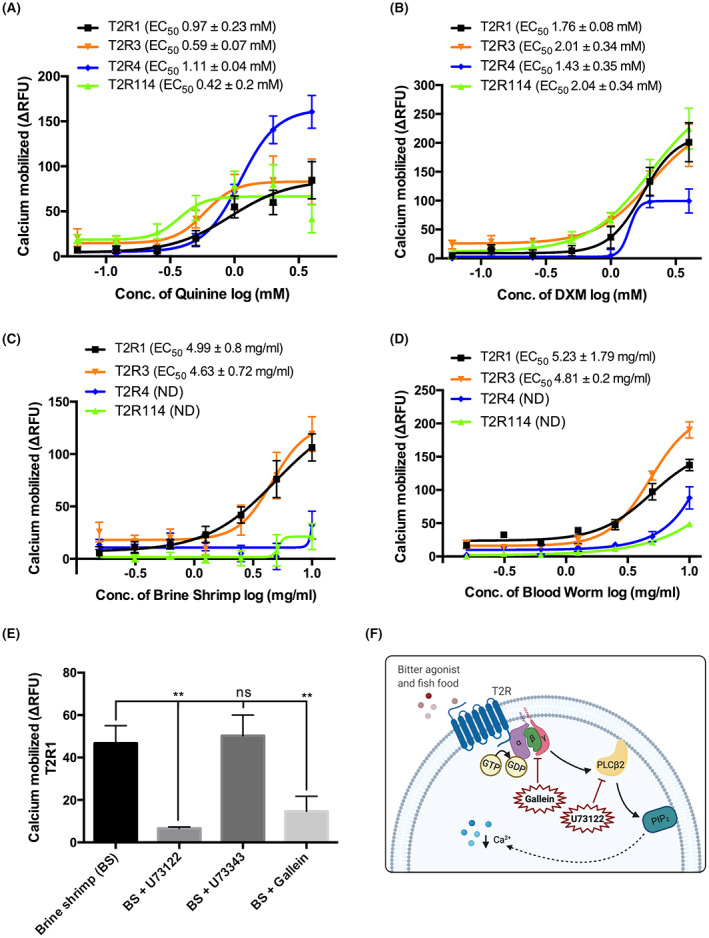
Functional analysis of A. mexicanus T2R1, T2R3, T2R4, and T2R114 in response to quinine, DXM, fish food, and signaling blockers. T2Rs stably expressed in HEK293T cell lines were treated with (A) quinine and (B) dextromethorphan (DXM) at a concentration ranging from 0.06 to 4 mM, and (C) brine shrimp and (D) bloodworm at a concentration ranging from 0.16 mg/ml–10 mg/ml. (E) The graph shows the decrease in calcium mobilization when the cells were treated with the signaling blockers U73122 (10 μM) and gallein (10 μM) in combination with the brine shrimp (5 mg/ml). No significant differences were observed with the negative control U73343 (10 μM). Calcium mobilization assay was performed using Fluo‐4NW dye, and baseline subtraction was carried out with HEK293T control cells and plotted as ΔRFU or Relative Fluorescence Units. The data represent SEM of at least three independent experiments performed in triplicate. (F) Schematic display of the signaling steps inhibited by gallein (Gβγ inhibitor) and U73122 (PLCβ inhibitor). Created with BioRender.com. **, p ≤ 0.01
FIGURE 4.
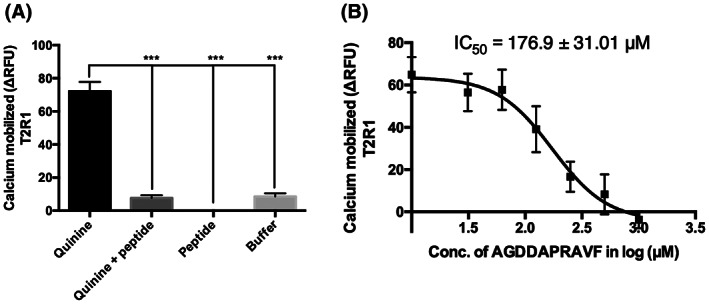
Competition calcium mobilization assay of quinine‐dependent T2R1 activation with peptide AGDDAPRAVF. (A) A. mexicanus T2R1 stably expressed in HEK293T cell lines were co‐treated with quinine (1 mM) and a known T2R blocker, the peptide AGDDAPRAVF (1 mM). One‐way analysis of variance was performed followed by Tukey's multiple comparison post hoc tests to check the significance. (B) To obtain the inhibitory concentrations (IC50), peptide AGDDAPRAVF, at a concentration ranging from 0.03 mM to 1 mM, was co‐incubated with quinine (1 mM). Calcium mobilization assay was performed using Fluo‐4NW dye and baseline subtraction was carried out with HEK293T control cells and plotted as ΔRFU or Relative Fluorescence Units. The IC50 values were calculated using Graph Pad Prism 6.0. Data were collected from at least three independent experiments performed in triplicate. ***p < 0.001
4. DISCUSSION
Although much research has been conducted on the loss of eyes and the effect of lens degeneration on associated or surrounding eye structures on CF, 35 , 36 , 37 , 38 few studies have focused on molecular mechanisms of other sensory functions such as olfaction and taste. Thus, we attempted to explore the T2R biology on teleost using A. mexicanus as an animal model.
In animals, bitter taste perception is important to detect and reject potentially toxic substances including foods. 39 , 40 Humans can tolerate mild or moderate bitter compounds which are important for survival. 41 Several studies showed the presence of human T2Rs in oral and extraoral tissues. 3 , 42 However, the existence and function of extraoral T2Rs in A. mexicanus remained to be explored. The evolution of T2Rs is very dynamic on a genomic scale where frequent pseudogene and gene duplication happened which generated a large variation of T2Rs repertoire within bony vertebrates. 43 This was the first study to show that Tas2Rs are expressed in different body parts of A. mexicanus, an early derived teleost fish like Zebrafish. 44
The genome of 15 fish species was previously analyzed and the presence of 21 intact and 3 pseudogenes from the Tas2R family in the A. mexicanus genome was reported. 7 Here, analyzing more recent transcriptomics data of A. mexicanus from Gene Expression Omnibus (GEO)‐NCBI and Ensembl databases, we found seven Tas2Rs and analyzed the gene expression of four Tas2Rs in oral and extraoral tissues. Our data showed in A. mexicanus, Tas2R1 has maximum expression followed by Tas2R4. Mainly, the head region that showed expression of all selected Tas2Rs. Similarly, zebrafish, a phylogenetically close fish species, also expresses Tas2Rs in taste receptor cells in the lips, gills, and pharynx. 6 , 18 , 45
In order to facilitate the analysis of the functional role of T2Rs in A. mexicanus, we used the established HEK293T heterologous expression system. 28 , 31 We selected the bitter agonists' quinine and DXM, which we have previously studied in human T2Rs. 31 , 34 , 46 Other studies have also performed characterization of known bitter agonists in zebrafish and common carp (Cyprinus carpio) T2Rs. 6 , 47 In comparison to what we observed in previous studies on human T2R4 (hT2R4), A. mexicanus T2R4 showed an EC50 value of 1.1 mM with quinine, which was similar to that usually obtained with hT2R4 quinine treatments. 28 , 31 We have previously shown that quinine acts intracellularly to enhance hT2R4 expression, acting as a pharmacological chaperone for hT2R4. 26 This suggests that it is possible that quinine treatment may also cause an increase in cell surface trafficking of A. mexicanus T2R4. More studies are required to investigate the possible chaperone activity of quinine in A. mexicanus T2Rs.
Our results also suggest differences in the ligand recognition profiles between human and A. mexicanus T2Rs. For example, the A. mexicanus T2R4 showed a response to DXM (EC50 1.43 mM), which does not activate the hT2R4. 33 Interestingly, A. mexicanus T2R1 was also activated by quinine, which does not activate hT2R1. 33 Our results confirm A. mexicanus T2R1 specificity for quinine, as the peptide blocker inhibited quinine activated T2R1 (Figure 4). The observed differential functional responses between human and A. mexicanus T2Rs could be partially related to the small sequence homology between human and A. mexicanus T2Rs.
Live or frozen brine shrimp and bloodworm are commonly used food for A. mexicanus (https://diszhal.info/english/characins/en_Astyanax_mexicanus.php, accessed on December 27, 2021). Interestingly, we observed that even though the brine shrimp is a heterogeneous mix, up to 86% of the calcium response elicited by brine shrimp was blocked by T2R signaling inhibitors, suggesting that fish food can activate the canonical bitter signal transduction pathway. Future studies targeting the behavioral analysis of A. mexicanus can be performed to confirm the fish's preferences for bitter agonists. Additional assessment of the fish food, brine shrimp, and bloodworm, can also be performed to identify which specific constituents activate T2Rs. These studies can assist with modulating the composition of fish food to enhance its acceptance by cultured fish or as an aquarium fish food.
5. CONCLUSION
Our findings suggest that A. mexicanus has functional T2Rs that are expressed in many extraoral tissues and might be involved in food detection. In humans, T2Rs elicit distinctive effects in different tissues. 2 , 48 , 49 Therefore, it is highly likely that fish T2Rs may also be involved in many other physiological responses. Future studies are required to clarify the physiological effects mediated by the T2Rs in A. mexicanus and other fish species.
AUTHOR CONTRIBUTIONS
V.B., V.C.J., F.A.S., A.J., and N.S. conducted the experiments and analyzed the data. V.B. and V.C.J. wrote the manuscript. P.C. and D.A. contributed to the conceptualization, design, and data interpretation. D.A. directed the study. All authors critically revised and edited the manuscript.
CONFLICT OF INTEREST
The authors declare no competing financial interests.
Supporting information
Figure S1
Figure S2
ACKNOWLEDGMENTS
We acknowledge the RFHS, University of Manitoba for supporting the Fish Culture Facility. We would also like to thank Dr. Richard Borowsky (New York University) and Dr. William Jeffery (University of Maryland) for providing us with A. mexicanus breeding stock and the University of Manitoba fish care personnel for their ongoing support.
Bhatia V, de Jesus VC, Shaik FA, et al. Extraoral expression and characterization of bitter taste receptors in Astyanax mexicanus (Mexican tetra fish). FASEB BioAdvances. 2022;4(9):574‐584. doi: 10.1096/fba.2022-00032
Vikram Bhatia and Vivianne Cruz de Jesus have contributed equally to this work and share the first authorship.
Funding information
This work was supported by Discovery grants (RGPIN‐2019‐05364 to DA and RGPIN‐2020‐05670 to PC) from the Natural Sciences and Engineering Research Council of Canada (NSERC).
Contributor Information
Prashen Chelikani, Email: prashen.chelikani@umanitoba.ca.
Devi Atukorallaya, Email: devi.atukorallaya@umanitoba.ca.
REFERENCES
- 1. Finger TE, Kinnamon SC. Taste isn't just for taste buds anymore. F1000 Biol Rep. 2011;3:20. doi: 10.3410/b3-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Shaik FA, Singh N, Arakawa M, Duan K, Bhullar RP, Chelikani P. Bitter taste receptors: extraoral roles in pathophysiology. Int J Biochem Cell Biol. 2016;77:197‐204. doi: 10.1016/j.biocel.2016.03.011 [DOI] [PubMed] [Google Scholar]
- 3. Jaggupilli A, Singh N, Upadhyaya J, et al. Analysis of the expression of human bitter taste receptors in extraoral tissues. Mol Cell Biochem. 2017;426(1):137‐147. doi: 10.1007/s11010-016-2902-z [DOI] [PubMed] [Google Scholar]
- 4. Chandrashekar J, Mueller KL, Hoon MA, et al. T2Rs function as bitter taste receptors. Cell. 2000;100(6):703‐711. [DOI] [PubMed] [Google Scholar]
- 5. Mueller KL, Hoon MA, Erlenbach I, Chandrashekar J, Zuker CS, Ryba NJ. The receptors and coding logic for bitter taste. Nature. 2005;434(7030):225‐229. doi: 10.1038/nature03352 [DOI] [PubMed] [Google Scholar]
- 6. Oike H, Nagai T, Furuyama A, et al. Characterization of ligands for fish taste receptors. J Neurosci. 2007;27(21):5584‐5592. doi: 10.1523/JNEUROSCI.0651-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Shiriagin V, Korsching SI. Massive expansion of bitter taste receptors in blind cavefish, Astyanax mexicanus. Chem Senses. 2019;44:23‐32. doi: 10.1093/chemse/bjy062 [DOI] [PubMed] [Google Scholar]
- 8. Okada S. The taste system of small fish species. Biosci Biotechnol Biochem. 2015;79(7):1039‐1043. doi: 10.1080/09168451.2015.1023251 [DOI] [PubMed] [Google Scholar]
- 9. Yasuoka A, Abe K. Gustation in fish: search for prototype of taste perception. Results Probl Cell Differ. 2009;47:239‐255. doi: 10.1007/400_2008_6 [DOI] [PubMed] [Google Scholar]
- 10. Jeffery WR. Cavefish as a model system in evolutionary developmental biology. Dev Biol. 2001;231(1):1‐12. doi: 10.1006/dbio.2000.0121 [DOI] [PubMed] [Google Scholar]
- 11. Jeffery WR. Emerging model systems in evo‐devo: cavefish and microevolution of development. Evol Dev. 2008;10(3):265‐272. doi: 10.1111/j.1525-142X.2008.00235.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rohner N. Cavefish as an evolutionary mutant model system for human disease. Dev Biol. 2018;441:355‐357. doi: 10.1016/j.ydbio.2018.04.013 [DOI] [PubMed] [Google Scholar]
- 13. Hinaux H, Pottin K, Chalhoub H, et al. A developmental staging table for Astyanax mexicanus surface fish and Pachón cavefish. Zebrafish. 2011;8(4):155‐165. doi: 10.1089/zeb.2011.0713 [DOI] [PubMed] [Google Scholar]
- 14. Keene A, Yoshizawa M, McGaugh S. Biology and Evolution of the Mexican Cavefish. Academic Press; 2015. [Google Scholar]
- 15. Schemmel C. Studies on the genetics of feeding behaviour in the cave fish Astyanax mexicanus f. anoptichthys. An example of apparent monofactorial inheritance by polygenes. Z Tierpsychol. 1980;53(1):9‐22. doi: 10.1111/j.1439-0310.1980.tb00730.x [DOI] [PubMed] [Google Scholar]
- 16. Varatharasan N, Croll RP, Franz‐Odendaal T. Taste bud development and patterning in sighted and blind morphs of Astyanax mexicanus. Dev Dyn. 2009;238(12):3056‐3064. doi: 10.1002/dvdy.22144 [DOI] [PubMed] [Google Scholar]
- 17. Venkatesh B, Gilligan P, Brenner S. Fugu: a compact vertebrate reference genome. FEBS Lett. 2000;476(1–2):3‐7. [DOI] [PubMed] [Google Scholar]
- 18. Ishimaru Y, Okada S, Naito H, et al. Two families of candidate taste receptors in fishes. Mech Dev. 2005;122(12):1310‐1321. doi: 10.1016/j.mod.2005.07.005 [DOI] [PubMed] [Google Scholar]
- 19. Pfister P, Rodriguez I. Olfactory expression of a single and highly variable V1r pheromone receptor‐like gene in fish species. Proc Natl Acad Sci U S A. 2005;102(15):5489‐5494. doi: 10.1073/pnas.0402581102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sinha S, Mishra P, Amin H, et al. A new cytotoxic quinolone alkaloid and a pentacyclic steroidal glycoside from the stem bark of Crataeva nurvala: study of anti‐proliferative and apoptosis inducing property. Eur J Med Chem. 2013;60:490‐496. doi: 10.1016/j.ejmech.2012.12.017 [DOI] [PubMed] [Google Scholar]
- 21. Yang S, Tian M, Yuan L, et al. Analysis of E. rutaecarpa alkaloids constituents in vitro and in vivo by UPLC‐Q‐TOF‐MS combined with diagnostic fragment. J Anal Methods Chem. 2016;2016:e4218967. doi: 10.1155/2016/4218967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hall BG. Building phylogenetic trees from molecular data with MEGA. Mol Biol Evol. 2013;30(5):1229‐1235. doi: 10.1093/molbev/mst012 [DOI] [PubMed] [Google Scholar]
- 23. Monteiro Ferreira A, Tomás Marques A, Bhide M, et al. Sequence analysis of bitter taste receptor gene repertoires in different ruminant species. PLoS ONE. 2015;10(6):e0124933. doi: 10.1371/journal.pone.0124933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Thisse C, Thisse B. High‐resolution in situ hybridization to whole‐mount zebrafish embryos. Nat Protoc. 2008;3(1):59‐69. doi: 10.1038/nprot.2007.514 [DOI] [PubMed] [Google Scholar]
- 25. Atukorala ADS, Franz‐Odendaal TA. Spatial and temporal events in tooth development of Astyanax mexicanus. Mech Dev. 2014;134:42‐54. doi: 10.1016/j.mod.2014.09.002 [DOI] [PubMed] [Google Scholar]
- 26. Upadhyaya JD, Chakraborty R, Shaik FA, Jaggupilli A, Bhullar RP, Chelikani P. The Pharmacochaperone activity of quinine on bitter taste receptors. PLoS ONE. 2016;11(5):e0156347. doi: 10.1371/journal.pone.0156347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chakraborty R, Xu B, Bhullar RP. Chelikani P. expression of G protein‐coupled receptors in mammalian cells. Methods in Enzymology. Vol 556. Elsevier; 2015:267‐281. doi: 10.1016/bs.mie.2014.12.013 [DOI] [PubMed] [Google Scholar]
- 28. Jaggupilli A, Singh N, de Jesus VC, Gounni MS, Dhanaraj P, Chelikani P. Chemosensory bitter taste receptors (T2Rs) are activated by multiple antibiotics. FASEB J. 2019;33(1):501‐517. doi: 10.1096/fj.201800521RR [DOI] [PubMed] [Google Scholar]
- 29. Jaggupilli A, Singh N, Jesus VCD, Duan K, Chelikani P. Characterization of the binding sites for bacterial acyl homoserine lactones (AHLs) on human bitter taste receptors (T2Rs). ACS Infect Dis. 2018;4(7):1146‐1156. doi: 10.1021/acsinfecdis.8b00094 [DOI] [PubMed] [Google Scholar]
- 30. Medapati MR, Singh N, Bhagirath AY, et al. Bitter taste receptor T2R14 detects quorum sensing molecules from cariogenic Streptococcus mutans and mediates innate immune responses in gingival epithelial cells. FASEB J. 2021;35(3):e21375. doi: 10.1096/fj.202000208R [DOI] [PubMed] [Google Scholar]
- 31. Pydi SP, Sobotkiewicz T, Billakanti R, Bhullar RP, Loewen MC, Chelikani P. Amino acid derivatives as bitter taste receptor (T2R) blockers. J Biol Chem. 2014;289(36):25054‐25066. doi: 10.1074/jbc.M114.576975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhang C, Alashi AM, Singh N, Liu K, Chelikani P, Aluko RE. Beef protein‐derived peptides as bitter taste receptor T2R4 blockers. J Agric Food Chem. 2018;66(19):4902‐4912. doi: 10.1021/acs.jafc.8b00830 [DOI] [PubMed] [Google Scholar]
- 33. Meyerhof W, Batram C, Kuhn C, et al. The molecular receptive ranges of human TAS2R bitter taste receptors. Chem Senses. 2010;35(2):157‐170. doi: 10.1093/chemse/bjp092 [DOI] [PubMed] [Google Scholar]
- 34. Upadhyaya JD, Singh N, Sikarwar AS, et al. Dextromethorphan mediated bitter taste receptor activation in the pulmonary circuit causes vasoconstriction. PLoS One. 2014;9(10):e110373. doi: 10.1371/journal.pone.0110373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yamamoto Y, Jeffery WR. Central role for the lens in cave fish eye degeneration. Science. 2000;289(5479):631‐633. doi: 10.1126/science.289.5479.631 [DOI] [PubMed] [Google Scholar]
- 36. Yamamoto Y, Espinasa L, Stock DW, Jeffery WR. Development and evolution of craniofacial patterning is mediated by eye‐dependent and ‐independent processes in the cavefish Astyanax. Evol Dev. 2003;5(5):435‐446. doi: 10.1046/j.1525-142X.2003.03050.x [DOI] [PubMed] [Google Scholar]
- 37. Jeffery WR. Adaptive evolution of eye degeneration in the Mexican blind cavefish. J Hered. 2005;96(3):185‐196. doi: 10.1093/jhered/esi028 [DOI] [PubMed] [Google Scholar]
- 38. Protas M, Conrad M, Gross JB, Tabin C, Borowsky R. Regressive evolution in the Mexican cave tetra, Astyanax mexicanus. Curr Biol. 2007;17(5):452‐454. doi: 10.1016/j.cub.2007.01.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mura E, Taruno A, Yagi M, Yokota K, Hayashi Y. Innate and acquired tolerance to bitter stimuli in mice. PLoS ONE. 2018;13(12):e0210032. doi: 10.1371/journal.pone.0210032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Behrens M, Di Pizio A, Redel U, Meyerhof W, Korsching SI. At the root of T2R gene evolution: recognition profiles of coelacanth and zebrafish bitter receptors. Genome Biol Evol. 2021;13(1):evaa264. doi: 10.1093/gbe/evaa264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Breslin PAS. An evolutionary perspective on food and human taste. Curr Biol. 2013;23(9):R409‐R418. doi: 10.1016/j.cub.2013.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Adler E, Hoon MA, Mueller KL, Chandrashekar J, Ryba NJ, Zuker CS. A novel family of mammalian taste receptors. Cell. 2000;100(6):693‐702. doi: 10.1016/s0092-8674(00)80705-9 [DOI] [PubMed] [Google Scholar]
- 43. Shi P, Zhang J, Yang H, Zhang Y. Adaptive diversification of bitter taste receptor genes in mammalian evolution. Mol Biol Evol. 2003;20(5):805‐814. doi: 10.1093/molbev/msg083 [DOI] [PubMed] [Google Scholar]
- 44. Betancur‐R R, Broughton RE, Wiley EO, et al. The tree of life and a new classification of bony fishes. PLoS Curr. 2013;5. doi: 10.1371/currents.tol.53ba26640df0ccaee75bb165c8c26288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ohmoto M, Okada S, Nakamura S, Abe K, Matsumoto I. Mutually exclusive expression of Gαia and Gα14 reveals diversification of taste receptor cells in zebrafish. J Comp Neurol. 2011;519(8):1616‐1629. doi: 10.1002/cne.22589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Singh N, Chakraborty R, Bhullar RP, Chelikani P. Differential expression of bitter taste receptors in non‐cancerous breast epithelial and breast cancer cells. Biochem Biophys Res Commun. 2014;446(2):499‐503. doi: 10.1016/j.bbrc.2014.02.140 [DOI] [PubMed] [Google Scholar]
- 47. Shimizu T, Kubozono T, Asaoka R, Toda Y, Ishimaru Y. Expression profiles and functional characterization of common carp (Cyprinus carpio) T2Rs. Biochem Biophys Rep. 2021;28:101123. doi: 10.1016/j.bbrep.2021.101123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lee RJ, Xiong G, Kofonow JM, et al. T2R38 taste receptor polymorphisms underlie susceptibility to upper respiratory infection. J Clin Invest. 2012;122(11):4145‐4159. doi: 10.1172/JCI64240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Singh N, Shaik FA, Myal Y, Chelikani P. Chemosensory bitter taste receptors T2R4 and T2R14 activation attenuates proliferation and migration of breast cancer cells. Mol Cell Biochem. 2020;465(1–2):199‐214. doi: 10.1007/s11010-019-03679-5 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1
Figure S2


