Abstract
A 4.0-kb DNA fragment of Trichodesmium sp. strain WH9601 contained gene sequences encoding the nitrate reduction enzymes, nirA and narB. A third gene positioned between nirA and narB encodes a putative membrane protein with similarity to the nitrate permeases of Bacillus subtilis (NasA) and Emericella nidulans (CrnA). The gene was shown to functionally complement a ΔnasA mutant of B. subtilis and was assigned the name napA (nitrate permease). NapA was involved in both nitrate and nitrite uptake by the complemented B. subtilis cells. napA is distinct from the nrt genes that encode the nitrate transporter of freshwater cyanobacteria.
The utilization of nitrate as an essential nitrogen source for growth is a property common to many heterotrophic and photosynthetic organisms. Following the transport of nitrate into the cell, its assimilation requires the reducing activity of two enzymes, nitrate reductase and nitrite reductase. The genes required for the assimilation of nitrate in the cyanobacterium Synechococcus sp. strain PCC7942 are organized in the so-called nir operon (14). nirA, the gene encoding nitrite reductase, is the first gene of the operon, followed by the genes encoding the nitrate transporter (nrtABCD) and finally the nitrate reductase gene narB. A similar gene organization has been reported for other freshwater species: Synechocystis sp. strain PCC6803 (8), Anabaena sp. strain PCC7120 (4, 6), Plectonema boryanum (21), and Phormidium laminosum (11). Nitrate is an important N source in the ocean, and it can be readily assimilated by marine Synechococcus spp. in the absence of ammonium (7). Other marine cyanobacteria, including species of the diazotrophic, colony-forming genus Trichodesmium, grow well on nitrate as the sole N source (13). Trichodesmium filaments are incapable of N2 fixation when growing on ammonium but maintain this capacity in the presence of nitrate (13). Here we report on the identification and partial characterization of nitrate assimilation genes from axenic Trichodesmium sp. strain WH9601 (derived from original strain IMS101).
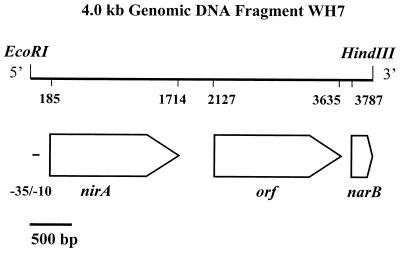
Using degenerate primers to conserved nirA regions (21), we PCR amplified a 507-bp nirA fragment using Trichodesmium sp. strain WH9601 genomic DNA as a template (PCR conditions: denaturation at 95°C for 1 min, annealing at 45°C for 1 min, and elongation at 72°C for 1 min; 40 cycles). The PCR product was used to probe EcoRI-HindIII-digested genomic DNA of Trichodesmium strain WH9601. A positive 4.0-kb fragment designated WH7 was cloned into plasmid pUC19 and transformed into Escherichia coli host strain DH5α using standard protocols (19). Sequence analysis of the WH7 fragment showed the presence of three open reading frames (ORFs) in the same orientation (Fig. 1). They are identified below as genes encoding nitrite reductase (nirA), a nitrate permease (napA), and nitrate reductase (narB). The orientation of the three genes would allow them to form part of a polycistronic message. Putative promoter elements were found at positions 27 to 32 (TTGATA) and 49 to 54 (TAAAAT). These elements show a high level of similarity to the −35 and −10 sequences for E. coli ς70. The DNA fragment WH7 contained no recognizable promoter elements further downstream.
FIG. 1.
Physical map of part of the nir operon of Trichodesmium strain WH9601. Numbers denote the positions of the first and last bases of coding regions, block arrows denote the ORFs with their direction of transcription. Double-stranded sequence was obtained by dye terminator cycle sequencing and primer hopping. Sequence reactions were analyzed on an ABI 377 Prism DNA sequencer (Perkin-Elmer Corp.) at the DNA Analysis Unit of the Life Sciences Institute, Hebrew University.
Nitrate and nitrite reductase genes.
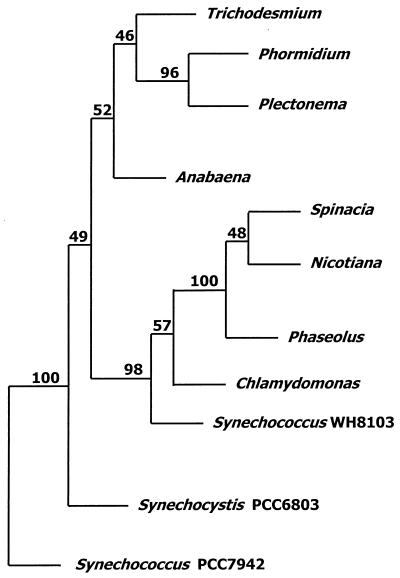
The deduced amino acid sequence of the first ORF identified it as NirA on the basis of its strong sequence identity (1) of >63% with NirA of other cyanobacterial species (4, 6, 8, 10, 11, 21) and >44% identity with eukaryotic algae and higher plants (3, 9, 17, 20). Since nirA was the only nitrate assimilation gene of Trichodesmium sp. strain WH9601 which was obtained in a full-length sequence and identified unambiguously, it was used to determine the phylogenetic relationship among the various groups of organisms. Figure 2 shows the consensus tree deduced from a parsimony analysis of available NirA sequences using the PHYLIP phylogeny package (5). The analysis was based on an aligned stretch of 376 amino acids from position 66 to position 442 in the deduced Trichodesmium NirA sequence. Our analysis places the unicellular cyanobacteria near the base of the unrooted tree with two major branches: a branch containing the sequences of filamentous, mainly N2-fixing cyanobacteria and a branch made up of higher plant and algal sequences. Interestingly, marine Synechococcus sp. strain WH8103 NirA was grouped near the eukaryotic sequences rather than with Synechococcus sp. strain PCC7942 and Synechocystis sp. strain PCC6803. This grouping occurred irrespective of the choice of parameters for construction of the tree. The narB sequence encodes an 82-amino-acid peptide with strong similarity (>55% identity) to the N terminus of cyanobacterial nitrate reductase genes. As for nirA, the highest similarity was observed for narB of filamentous cyanobacteria (4, 6, 22).
FIG. 2.
Consensus tree based on random sampling of NirA amino acid sequences of 11 photosynthetic organisms. The organisms used (and their GenBank accession numbers) are Trichodesmium sp. (AF178846), Phormidium laminosum (Q51879), Plectonema boryanum (D31732), Anabaena sp. strain PCC7120 (U61496), Synechococcus sp. strain WH8103 (AF065403), Synechococcus strain PCC7942 (P39661), Synechocystis strain PCC6803 (D64003), Chlamydomonas reinhardtii (Y08937), Spinacia oleracea (P05314), Nicotiana tabacum (S23769), and Phaseolus vulgaris (S51945). Numbers at nodes indicate the bootstrap values from 100 trees constructed using the PHYLIP software.
Nitrate and nitrite transporter gene.
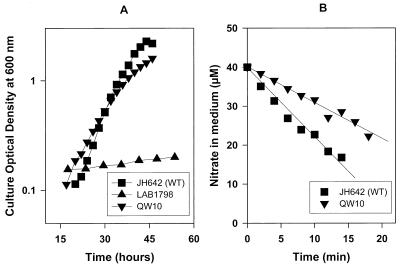
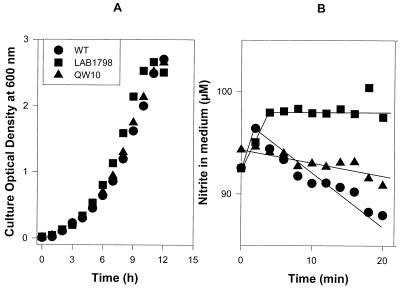
The nrtABCD genes located between nirA and narB in both unicellular and filamentous freshwater cyanobacteria are known to encode nitrate and nitrite uptake proteins which together form an ABC-type transport system (15). The nitrogen-fixing, filamentous cyanobacteria that together with Trichodesmium formed a related group based on nirA and narB sequences all possess such a nitrate transporter (4, 6, 8, 10, 11, 21). However, in Trichodesmium strain WH9601, a single ORF was found between nirA and narB. The ORF's deduced amino acid sequence predicts a 55-kDa membrane protein with 12 potential α helices and a pI of 8.1. A similarity search indicated affinity with a family of microbial permeases, especially with the nitrate permeases of Bacillus subtilis (NasA) and Emericella nidulans (CrnA), as well as the nitrite extrusion protein (NarK) of B. subtilis (Fig. 3). The similarity to each of the three proteins is relatively low (∼22% identity) and based mostly on conserved amino acid stretches in the N-terminal half of the protein. Based on the ORF's position between nirA and narB, its orientation, the characteristics of the predicted protein, and the sequence similarity to known nitrate permeases, we hypothesized that it encodes the nitrate transporter in Trichodesmium sp. strain WH9601 and tentatively named it napA. Since a genetic manipulation system for Trichodesmium does not exist, we studied napA function by complementing ΔnasA strain LAB1798 of B. subtilis, which is impaired in nitrate uptake (12). The napA coding sequence was fused to the nitrate-specific B. subtilis promoter PnasA. Both the PnasA region and the napA region were obtained by PCR amplification using primers nas-f (5′-CCT CCT TTG GCG AAG CTT GTA AG-3′); nas-r (5′-CAG TTC CGA CAG CTT CAT AGA ATT CCC-3′), nap-F (5′-GGA CAA TTT CTG CAA TTA TCA AGA ATT CTA TGT TAAC-3′), and nap-r 5′-CCT CAA GGA TCC AGC ATA ACC GAG-3′) (underlining denotes engineered restriction sites, and boldface denotes base replacements). Primers were designed such that the products ligated at an engineered EcoRI site 3 bases upstream of the ATG start codon. The modification involved a minimal number of bases and did not affect obvious regulatory sequence or alter the length of the upstream sequence. The PnasA-napA construct was introduced into plasmid pHT304, a suitable shuttle vector for Bacillus spp. (2), to yield plasmid pQW10. In order to obtain a nonmethylated plasmid, pQW10 was amplified in dam dcm mutant E. coli strain JM110 using ampicillin at 100 μg · ml−1 as a selective marker. Competent B. subtilis LAB1798 cells were then transformed by electroporation (two pulses at 200 Ω, 1.25 kV, and 25 μF [GenePulse II; Bio-Rad Inc.] for 6 × 108 cells with 0.5 μg of plasmid). Transformants were allowed to recover in Luria-Bertani medium at 37°C for 3 h and subsequently transferred to solid MMG medium (12) containing nitrate as the nitrogen source and erythromycin at 25 μg · ml−1. Of 25 erythromycin-resistant colonies, 7 were capable of sustained growth on liquid minimal medium with nitrate. One of these transformants, QW10, was chosen for further analysis. QW10 attained growth rates that approached those of wild-type B. subtilis JH642 grown in minimal MMG medium with 20 mM nitrate (Fig. 4A). Nitrate removal was studied in nitrate-grown B. subtilis strains that were collected by centrifugation for 5 min at 8,000 rpm and resuspended in MMG medium lacking combined nitrogen. Nitrate was measured as described in reference 16 after Cd-catalyzed reduction of nitrate to nitrite. Following 5 min of acclimation, cells received a 40 μM nitrate addition. QW10 cells assimilated nitrate at 57% of the rate of wild-type B. subtilis (Fig. 4B). LAB1798 cells did not take up any nitrate from the medium (data not shown). Figure 5A shows the growth on 20 mM nitrite of wild-type B. subtilis alongside mutant strain LAB1978 and strain QW10, napA-complemented LAB1798. All three strains grew equally well on nitrite, and apparently the presence of NapA is not required to meet the N demands of B. subtilis when it is growing on nitrite. Nitrite assimilation by B. subtilis was studied as described above for nitrate. Nitrite utilization by B. subtilis LAB1798 cells was undetectably low during the 20-min assay (Fig. 5B). These cells probably satisfy their N requirement from the slow diffusion of nondissociated HNO2 over the cytoplasmic membrane at the elevated NO2− concentration in the growth medium. JH642 cells readily assimilated nitrite, albeit at a rate slower than that of nitrate assimilation (0.53 ± 0.05 μmol of NO2− · OD600 U−1 · min−1 versus 2.73 ± 0.05 μmol of NO3− · OD600 U−1 · min−1). The napA-complemented QW10 cells removed nitrite from the medium at 0.20 ± 0.04 μmol of NO2− · OD600 U−1 · min−1, approximately 50% of the rate found for wild-type JH642. It was concluded that napA encodes an essential component of the nitrate transport system in Trichodesmium strain WH9601 and that this component also facilitates uptake of nitrite. Both functions showed an apparent efficiency of 50% compared to wild-type B. subtilis.
FIG. 3.
ClustalW (version 1.74) alignment of the predicted NapA protein of Trichodesmium strain WH9601 with nitrate permeases of B. subtilis (NasA) and E. nidulans (CrnA), as well as the nitrite extrusion protein of B. subtilis (NarK). Asterisks indicate positions of identical amino acids; colons indicate positions with conserved replacements, and periods indicate semiconserved positions.
FIG. 4.
Growth (A) and nitrate utilization (B) of B. subtilis wild-type (WT) strain JH642, ΔnasA strain LAB1798, and strain QW10 (LAB1798/PnasA-napA) at 37°C on minimal MMG medium with nitrate as the N source.
FIG. 5.
Growth (A) and nitrite utilization (B) of B. subtilis wild-type (WT) strain JH642, ΔnasA strain LAB1798, and strain QW10 (LAB1798/PnasA-napA) at 37°C on minimal MMG medium with nitrite as the N source.
The napA gene encountered in marine Trichodesmium strain WH9601 has not been described as part of the nir operon in any cyanobacterium. Genes with high similarity to napA have recently been found in euryhaline Synechococcus strain PCC7002 (18) and in oceanic Synechococcus sp. strain WH7803 (23). These observations suggest that nitrate utilization by cyanobacteria in saline environments is facilitated by NapA rather than by NrtABCD. The two transporters have not been found to coexist in cyanobacteria. The delineation of the two nitrate transport systems along low-salt and high-salt cyanobacteria is interesting, especially considering that (i) NirA and NarB sequences of both marine and freshwater filamentous cyanobacteria group together in phylogenetic analyses and (ii) the delineation occurs within marine and freshwater representatives of the genus Synechococcus. These observations suggest that both environments exert considerable selective pressure on cyanobacteria with respect to their nitrate and nitrite acquisition systems.
Nucleotide sequence accession number.
The sequence of the WH7 fragment was deposited in the GenBank database and assigned accession no. AF178846.
Acknowledgments
The study presented here received financial support from “Red Sea Program” grant 03F0151A provided by the German Ministry for Education, Science, Research and Technology (BMBF). The study was further supported by the “Moshe Shilo” Minerva Center for Marine Biogeochemistry, Minerva Stiftung Gesellschaft fuer die Forschung, Muenchen, Germany.
Axenic Trichodesmium sp. strain WH 9601 was kindly supplied by J. Waterbury. We are grateful to M. M. Nakano for B. subtilis strains LAB1798 and JH642 and to D. Lindell for critical remarks on the manuscript.
REFERENCES
- 1.Altschul S F, Madden T L, Schaeffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arantes O, Dereclus D. Construction of cloning vectors for Bacillus thuringiensis. Gene. 1991;108:115–119. doi: 10.1016/0378-1119(91)90495-w. [DOI] [PubMed] [Google Scholar]
- 3.Back E, Dunne W, Schneiderbauer A, de Framond A, Rastogi R, Rothstein S J. Isolation of the spinach nitrite reductase gene promoter which confers nitrate inducibility on GUS gene expression in transgenic tobacco. Plant Mol Biol. 1991;17:9–18. doi: 10.1007/BF00036801. [DOI] [PubMed] [Google Scholar]
- 4.Cai Y, Wolk C P. Nitrogen deprivation of Anabaena sp. strain PCC 7120 elicits rapid activation of a gene cluster that is essential for uptake and utilization of nitrate. J Bacteriol. 1997;179:258–266. doi: 10.1128/jb.179.1.258-266.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Felsenstein J. PHYLIP (Phylogeny Interference Package) version 3.5c manual. Seattle: University of Washington; 1993. [Google Scholar]
- 6.Frías J E, Flores E, Herrero A. Nitrate assimilation gene cluster from the heterocyst-forming cyanobacterium Anabaena sp. strain PCC 7120. J Bacteriol. 1997;179:477–486. doi: 10.1128/jb.179.2.477-486.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Glibert P M, Ray R T. Different patterns of growth and nitrogen uptake in two clones of marine Synechococcus spp. Mar Biol. 1990;107:273–280. [Google Scholar]
- 8.Kaneko T, Sato S, Kotani H, Tanaka A, Asamizu E, Nakamura Y, Miyajima N, Hirosawa M, Sugiura M, Sasamoto S, Kimura T, Hosouchi T, Matsuno A, Muraki A, Nakazaki N, Naruo K, Okumura S, Shimpo S, Takeuchi C, Wada T, Watanabe A, Yamada M, Yasuda M, Tabata S. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC 6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 1996;3:109–136. doi: 10.1093/dnares/3.3.109. [DOI] [PubMed] [Google Scholar]
- 9.Kronenberger J, Lepingle A, Caboche M, Vaucheret H. Cloning and expression of distinct nitrite reductases in tobacco leaves and roots. Mol Gen Genet. 1993;236:203–208. doi: 10.1007/BF00277113. [DOI] [PubMed] [Google Scholar]
- 10.Luque I, Flores E, Herrero A. Nitrite reductase gene from Synechococcus sp. PCC7942: homology between cyanobacterial and higher-plant nitrite reductases. Plant Mol Biol. 1993;21:1201–1205. doi: 10.1007/BF00023618. [DOI] [PubMed] [Google Scholar]
- 11.Merchand F, Kindle K L, Llana M J, Serra J L, Fernandez E. Cloning and sequencing of the nitrate transport system from the thermophilic filamentous cyanobacterium Phormidium laminosum: comparative analysis with the homologous system from Synechococcus sp. PCC 7942. Plant Mol Biol. 1995;28:759–766. doi: 10.1007/BF00021199. [DOI] [PubMed] [Google Scholar]
- 12.Ogawa K-I, Akagawa E, Yamane K, Sun Z-W, LaCelle M, Zuber P, Nakano M M. The nasB operon and nasA gene are required for nitrate/nitrite assimilation in Bacillus subtilis. J Bacteriol. 1995;177:1409–1413. doi: 10.1128/jb.177.5.1409-1413.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ohki K, Zehr J P, Falkowski P G, Fujita Y. Regulation of nitrogen-fixation by different nitrogen sources in the marine non-heterocystous cyanobacterium Trichodesmium sp. NIBB1067. Arch Microbiol. 1991;156:335–337. [Google Scholar]
- 14.Omata T. Structure, function and regulation of the nitrate transport system of the cyanobacterium Synechococcus sp. PCC7942. Plant Cell Physiol. 1995;36:207–213. doi: 10.1093/oxfordjournals.pcp.a078751. [DOI] [PubMed] [Google Scholar]
- 15.Omata T, Andriesse X, Hirano A. Identification and characterization of a gene cluster involved in nitrate transport in the cyanobacterium Synechococcus sp. PCC7942. Mol Gen Genet. 1993;236:193–202. doi: 10.1007/BF00277112. [DOI] [PubMed] [Google Scholar]
- 16.Parsons T R, Maita Y, Lalli C M. A manual of chemical and biological methods of sea water analysis. Elmsford, N.Y: Pergamon Press Inc.; 1984. [Google Scholar]
- 17.Quesada A, Gomez I, Fernandez E. Clustering of the nitrite reductase gene and a light-regulated gene with nitrate assimilation loci in Chlamydomonas reinhardtii. Planta. 1998;206:259–265. doi: 10.1007/s004250050398. [DOI] [PubMed] [Google Scholar]
- 18.Sakamoto T, Inoue-Sakamoto K, Bryant D A. A novel nitrate/nitrite permease in the marine cyanobacterium Synechococcus sp. strain PCC 7002. J Bacteriol. 1999;181:7363–7372. doi: 10.1128/jb.181.23.7363-7372.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 20.Sander L, Jensen P E, Back L F, Stummann B M, Henningsen K W. Structure and expression of a nitrite reductase gene from bean (Phaseolus vulgaris) and promoter analysis in transgenic tobacco. Plant Mol Biol. 1995;27:165–177. doi: 10.1007/BF00019188. [DOI] [PubMed] [Google Scholar]
- 21.Suzuki I, Kikuchi H, Nakanishi S, Fujita Y, Sugiyama T, Omata T. A novel nitrite reductase gene from the cyanobacterium Plectonema boryanum. J Bacteriol. 1995;177:6137–6143. doi: 10.1128/jb.177.21.6137-6143.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Unthan M, Klipp W, Schmid G H. Nucleotide sequence of the narB gene encoding assimilitory nitrate reductase from the cyanobacterium Oscillatoria chalybea. Biochim Biophys Acta. 1996;1305:19–24. doi: 10.1016/0167-4781(95)00210-3. [DOI] [PubMed] [Google Scholar]
- 23.Wang, Q., and A. F. Post. Unpublished data.