Abstract
Bacteria and lytic viruses (phages) engage in highly dynamic coevolutionary interactions over time, yet we have little idea of how transient selection by phages might shape the future evolutionary trajectories of their host populations. To explore this question, we generated genetically diverse phage-resistant mutants of the bacterium Pseudomonas syringae. We subjected the panel of mutants to prolonged experimental evolution in the absence of phages. Some populations re-evolved phage sensitivity, whereas others acquired compensatory mutations that reduced the costs of resistance without altering resistance levels. To ask whether these outcomes were driven by the initial genetic mechanisms of resistance, we next evolved independent replicates of each individual mutant in the absence of phages. We found a strong signature of historical contingency: some mutations were highly reversible across replicate populations, whereas others were highly entrenched. Through whole-genome sequencing of bacteria over time, we also found that populations with the same resistance gene acquired more parallel sets of mutations than populations with different resistance genes, suggesting that compensatory adaptation is also contingent on how resistance initially evolved. Our study identifies an evolutionary ratchet in bacteria–phage coevolution and may explain previous observations that resistance persists over time in some bacterial populations but is lost in others. We add to a growing body of work describing the key role of phages in the ecological and evolutionary dynamics of their host communities. Beyond this specific trait, our study provides a new insight into the genetic architecture of historical contingency, a crucial component of interpreting and predicting evolution.
Keywords: historical contingency, epistasis, experimental evolution, parallel evolution, bacteriophages, microbial evolution
Introduction
Pathogens are ubiquitous and exert strong selection on their hosts to evade infection (Stenseth and Maynard Smith 1984; Gómez et al. 2010). These selection pressures are constantly in flux, and defense-related traits are often detrimental when pathogens are not present at high levels (Clay and Kover 1996; Sheldon and Verhulst 1996). The loss of costly resistance under relaxed selection has been the focus of a plethora of theoretical and empirical studies, in large part because it helps to explain the observed coexistence of resistant and sensitive host types in many natural populations (Waterbury and Valois 1993; Stahl et al. 1999; Rodriguez-Brito et al. 2010; Lourenço et al. 2020). Whether host populations readily regress to susceptibility after escape from pathogen pressure or retain a signature of their coevolutionary history will depend on several factors, including the environmental conditions and the strength of selection. For example, resistance may persist if it is not costly to maintain; or if compensatory mutations reduce the fitness costs without reversing the trait itself, as is often observed in drug-resistant bacteria (Faria et al. 2015; Durão et al. 2018). In other cases, even when reversion to sensitivity would be favorable, it may not be possible if the host population has since acquired other mutations that would be deleterious on the wild-type genetic background (Shah et al. 2015).
In a more general sense, any mutation that affects an organism’s phenotype and is selected for (even temporarily) can alter the selection acting on subsequent mutations and can therefore shape the evolutionary trajectory of the population. For example, garter snakes that prey on tetrodotoxin-bearing newts have evolved high levels of toxin resistance, but only within lineages that already carried a prior substitution—an ancient modification to a sodium channel that took place long before the newts arose (McGlothlin et al. 2016). In a laboratory evolution experiment with Escherichia coli, different substitutions in a DNA topoisomerase enzyme were shown to have different consequences for the subsequent accumulation of other beneficial mutations. In fact, this second-order selection for evolvability was more important than the initial effects of the substitutions themselves in determining which lineage prevailed in the long term (Woods et al. 2011). Historical contingencies such as these can make it particularly challenging to interpret patterns of genetic divergence across populations or predict the lasting consequences of short-term coevolutionary interactions.
To explore how previous selection by phages can alter future bacterial evolution, we tracked phage resistance over time in experimental evolution populations of the bacterium Pseudomonas syringae. Bacteria and phages are a tractable model system frequently used for studying coevolution in the laboratory (Brockhurst et al. 2007; Dennehy 2012). Phages initiate infection by recognizing and binding to proteins on the surfaces of bacterial cells. Bacteria can evade phages by altering or deleting phage receptors, but in doing so, often compromise other fitness-related traits such as nutrient uptake, adhesion, and virulence (Dy et al. 2014; Mangalea and Duerkop 2020). In these cases, as resistance spreads in a population and phage densities decrease, resistance is predicted to be lost over time as a result of relaxed selection (Koskella 2018). Laboratory fluctuation assays show that the rates of spontaneous genetic reversion from resistance to susceptibility can be high (Chaudhry et al. 2018), suggesting that phage sensitivity can (re)emerge within resistant populations.
Despite these predicted dynamics, studies of natural bacterial communities sometimes find that bacteria remain resistant to phages that they coexisted with many months or even years in the past (Koskella and Parr 2015; LeGault et al. 2021; Dewald-Wang et al. 2022). In cases where laboratory studies propagated resistant bacteria for many generations in the absence of phages, whether phage sensitivity re-emerged has remained inconsistent and difficult to explain (Meyer et al. 2010; Avrani and Lindell 2015). For example, over 45,000 generations of relaxed selection did not reduce the observed resistance of E. coli to T6 phage (Meyer et al. 2010). In contrast, some experimental populations of Prochlorococcus became less resistant in the absence of phage, but these changes were difficult to explain in terms of compensatory adaptation, as they often occurred independently of fitness gains (Avrani and Lindell 2015).
On the basis of these observations, we hypothesized that bacteria can access many different genetic pathways to phage resistance, each with different implications for the subsequent evolutionary potential of the bacteria, including whether they compensate for fitness costs by re-evolving phage sensitivity. To test this idea, we isolated and sequenced P. syringae colonies that had evolved resistance across a panel of lytic phages and measured the fitness costs of each resistance mutation. We then used each resistant strain to seed a different population that was experimentally evolved in the absence of phages. Through a series of laboratory evolution experiments, we demonstrate that phage resistance can either be reversible or entrenched depending on the initial genetic path to resistance.
Results
Single-Step Selection for Phage Resistance
To explore whether the long-term outcomes of phage resistance were contingent on the genetic underpinnings of their initial resistance (and/or the fitness costs associated with resistance), we first generated a panel of phage-resistant bacterial strains. With the aim of generating substantial variation both in resistance mechanisms and in fitness, we used five different Pseudomonas phages to select phage-resistant colonies of P. syringae pv. tomato DC3000 through soft agar overlays. We isolated and sequenced 22 colonies to reveal 17 unique resistance mutations (positions 1184510 and 5662024 each appeared three times in the panel, and one colony had no detectable fixed genetic differences from the ancestor despite apparent phenotypic resistance).
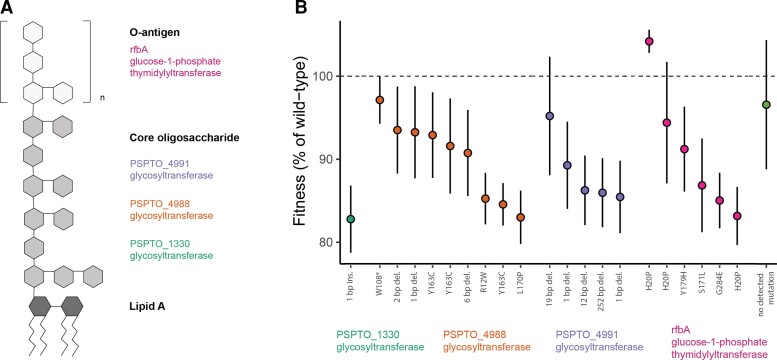
In contrast to the diversity of exact mutations, there was relatively high convergence in the genes in which they were found. The mutations in this study were all located in one of four genes, all involved in biosynthesis of outer membrane lipopolysaccharide molecules: PSPTO_4988 (RfaB family glycosyltransferase), PSPTO_4991 (GlcII glycosyltransferase), and PSPTO_1330 (Glycosyltransferase α-L-Rha), involved in assembling the LPS core structure, and rfbA (glucose-1-phosphate thymidylyltransferase) responsible for the O-antigen (Kutschera et al. 2021) (fig. 1A). Resistance mutations included single-nucleotide missense mutations, single-nucleotide nonsense mutations, and insertions and deletions of varying sizes. When resistant mutants were tested in the absence of phages, these strains grew more slowly than their phage-sensitive ancestor, indicating trade-offs between resistance and other aspects of fitness (one-sample t-test, t = −8.405, P < 0.001, fig. 1B). Variation in growth rates could not be explained by the genes in which those mutations occurred (ANOVA, F = 0.673, df = 3, P = 0.580), indicating that different resistance mutations in the same gene could have different impacts on fitness. This lack of systematic differences in fitness costs across genes underlying resistance meant that we could tease apart the effects of underlying genetic mechanisms and fitness costs on the maintenance of phage resistance, as these two predictors were not confounded with each other.
Fig. 1.
Genetic mechanisms and fitness costs of phage resistance. (A) Schematic illustration of the lipopolysaccharide structure, with the genes implicated in phage resistance in this study matched to their corresponding substructures. (B) Population growth rates of resistant strains in the absence of phage, based on a logistic model fitted to a 40 h growth curve (n = 22 strains). The error bars represent standard error across technical replicates. Bacterial colonies with resistance mutations in the same gene are denoted by a color scheme. The location and the nature of the mutation is indicated along the x-axis; for example, R12W indicates a change from arginine to tryptophan at the 12th position, and W108* indicates a change from tryptophan to a stop codon.
Of note, there was no effect of the identity of the selecting phage on either resistance mechanism or fitness of the bacteria; in fact, different phages often selected for resistance mutations in the same gene or even the exact same resistance mutation (table 1). Further, resistance to one phage typically conferred cross-resistance to all other phages in the study, suggesting that the phages in this study used the same receptor. We therefore aggregated data across selecting phages for the analyses in this study.
Table 1.
Genetic Mechanisms and Evolutionary Outcomes of Phage-resistant Populations.
| Population | Resistance gene | Position of resistance mutation | Resistance mutation | Outcome in original evolution experiment |
|---|---|---|---|---|
| FMS6 | rfbA | 1183718 | SNP (G284E) | Remained resistant |
| FMS4 | rfbA | 1184034 | SNP (Y179H) | Remained resistant |
| VCM4 | rfbA | 1184057 | SNP (S171L) | Re-evolved sensitivity |
| MS10 | rfbA | 1184510 | SNP (H20P) | Remained resistant |
| MS2 | rfbA | 1184510 | SNP (H20P) | Remained resistant |
| QAC4 | rfbA | 1184510 | SNP (H20P) | Remained resistant |
| QAC5 | PSPTO_1330 | 1461031 | 1 bp ins. | Re-evolved sensitivity |
| VCM19 | PSPTO_4988 | 5661617 | 2 bp del. | Remained resistant |
| MS12 | PSPTO_4988 | 5661621 | 1 bp del. | Remained resistant |
| FMS13 | PSPTO_4988 | 5661925 | 6 bp del. | Remained resistant |
| SNK12 | PSPTO_4988 | 5662003 | SNP (L170P) | Remained resistant |
| QAC3 | PSPTO_4988 | 5662024 | SNP (Y163C) | Remained resistant |
| SNK11 | PSPTO_4988 | 5662024 | SNP (Y163C) | Remained resistant |
| VCM17 | PSPTO_4988 | 5662024 | SNP (Y163C) | Remained resistant |
| SNK6 | PSPTO_4988 | 5662188 | SNP (W108*) | Remained resistant |
| MS1 | PSPTO_4988 | 5662478 | SNP (R12W) | Remained resistant |
| FMS12 | PSPTO_4991 | 5664934 | 19 bp del. | Remained resistant |
| FMS9 | PSPTO_4991 | 5665124 | 252 bp del. | Remained resistant |
| SNK7 | PSPTO_4991 | 5665501 | 1 bp del. | Re-evolved sensitivity |
| FMS11 | PSPTO_4991 | 5665743 | 1 bp del. | Remained resistant |
| FMS16 | PSPTO_4991 | 5665890 | 12 bp del. | Remained resistant |
indicates a stop codon.
Multiple Evolutionary Paths to Recover Fitness Costs
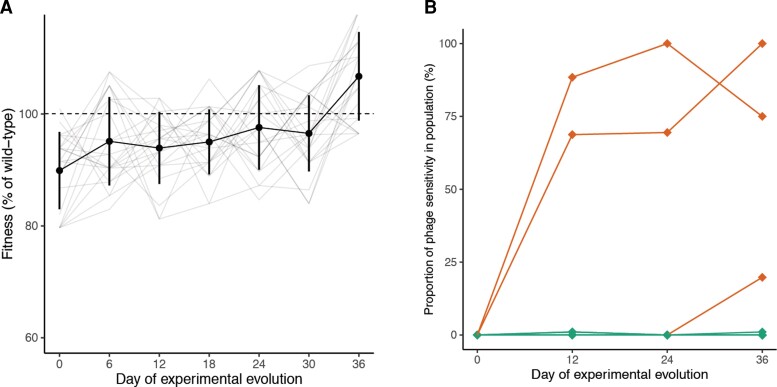
To test whether phage sensitivity tends to re-evolve in the absence of phages, we inoculated 22 experimental microcosms of King’s Broth media each with one of the resistant colonies. We also inoculated six microcosms with the phage-sensitive ancestor to assess changes due to overall adaptation to the environment, for a total of 28 experimentally evolving populations. Populations were transferred to fresh media every 3 days for a total of 36 days. Over the course of the experiment, the difference in growth rates between the populations initiated with phage-resistant bacteria and those initiated with phage-sensitive (ancestral) bacteria gradually narrowed, and by the end of the experiment there was no observed fitness difference between the two groups (fig. 2A). Changes in fitness followed a diminishing returns pattern of adaptation: populations with the lowest initial fitness made the greatest fitness gains over time (ANOVA, F = 33.226, P < 0.001; supplementary fig. S1, Supplementary Material online).
Fig. 2.
Phenotypic changes in bacterial populations during experimental evolution in the absence of phages. (A) Population growth rates of populations inoculated with phage-resistant colonies over experimental evolution in the absence of phages, calculated based on optical density measurements in a microplate reader over 40 h of growth (n = 22). Lines represent the trajectories of individual populations over time, whereas points indicate the mean fitness among populations sampled at the same day and error bars indicate SD. Values are normalized to the growth rates of the phage-sensitive control populations sampled at the same point in experimental evolution. (B) Proportion of colonies in each population that were scored as phage-sensitive over time, as indicated by disruption in bacterial growth upon encountering phage on an agar plate (n = 22 populations with 96 colonies sampled per population). Orange lines indicate populations from which sensitive colonies were isolated and verified in a microplate assay.
Bacterial populations evolved lower levels of resistance to their original selecting phages under relaxed selection, but only rarely. A total of five populations (of 22 initially resistant populations) contained colonies with sensitive, partially resistant, or ambiguous phenotypes on agar plates. We tracked the growth of these clones over time in liquid culture in the presence or absence of the selecting phage, revealing that three of these five populations contained phage-sensitive bacteria (fig. 2B). Interestingly, the overall increase in bacterial fitness over time could not be attributed to the re-evolution of phage sensitivity alone. Although there was a main effect of the day of experimental evolution on bacterial fitness, the rate of change did not differ among populations that re-evolved sensitivity and those that remained resistant (ANOVA, F = 2.146 for the interaction term, P = 0.145). This observation suggested that the resistant populations may be able to access compensatory mutations that lessen the costs of phage resistance without reversing it.
We isolated and sequenced the genomes of phage-sensitive colonies to determine how phage sensitivity re-evolved. The population “SNK7”, which originally acquired resistance through a single base-pair deletion in the glycosyltransferase-encoding gene PSPTO_4991, later acquired a single base-pair insertion five bases away that restored the reading frame. In contrast, the other two populations acquired mutations that fell outside of their initial resistance genes. VCM4, which had become phage-resistant through a substitution in the rfbA gene, evolved additional mutations in two other membrane transport proteins. QAC5, which had evolved resistance through a single base-pair insertion in the glycosyltransferase-encoding gene PSPTO_1330, acquired a mutation in a membrane transport protein as well as several intergenic mutations whose function (if any) is unknown.
We had predicted that populations with particularly costly resistance would be most likely to re-evolve sensitivity due to strong selection. However, we did not observe a relationship between initial costs of resistance and the evolution of phage sensitivity (Welch’s unequal variances t-test, t = 1.818, P = 0.138). We also asked whether phage sensitivity would be more likely to re-emerge among populations with particular resistance genes. Although the unequal distribution of phage resistance outcomes makes it difficult to entirely rule out this possibility, the three populations that re-evolved sensitivity in this study did so from three different initial resistance genes, providing no evidence that trait reversion is historically contingent at the gene level (table 1). However, we noticed two important observations that warranted further study.
First, our study included four populations that had originally acquired resistance through large deletions in oligosaccharide synthesis genes, spanning from 6 to 252 bp in length. None of these populations re-evolved sensitivity, whether by re-insertion of the missing sequence or through any other means. Second, by chance our study included two sets of three populations each with identical resistance mutations. In both cases, these genetic triplets all followed the same evolutionary outcome (remaining resistant). Given that re-evolving phage sensitivity was generally rare in this study, it was not apparent whether these two observations were simply probabilistic, or whether they pointed to historical contingency at the level of the individual mutation rather than the resistance gene as a whole. As this experiment was not originally set up to test this possibility, we developed an experimental design that would test the effects of exact resistance mutations on phage resistance outcomes.
Replay of Experimental Evolution
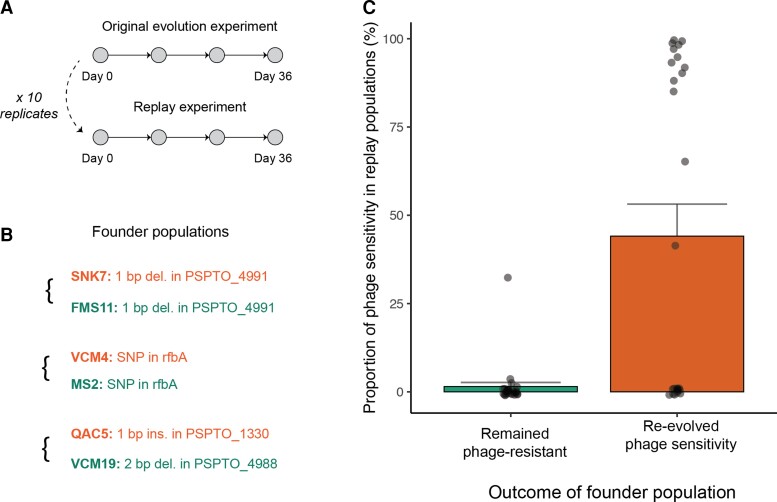
The initial evolution experiment revealed that the identity of the gene conferring phage resistance did not explain whether bacteria remained resistant or re-evolved sensitivity. To ask whether phage resistance outcomes were contingent on the exact resistance mutation instead, we returned to the progenitor frozen stocks of the three resistant colonies that had re-evolved sensitivity during experimental evolution. We also included three resistant colonies that were never observed to re-evolve sensitivity, selecting the closest possible genetic matches to the colonies that did. For example, colonies VCM4 and MS2 had both originally acquired resistance through point mutations in the rfbA gene, yet VCM4 had re-evolved phage sensitivity, whereas MS2 had not.
We used each of these six resistant mutants (“founders”) to seed ten replicate populations, resulting in a total of 60 experimentally evolving populations. As described previously, these populations were maintained in experimental microcosms of King’s Broth and transferred every 3 days for 36 days. At the end of the replay experiment, founder identity accounted for the majority of variance among populations in the prevalence of phage sensitivity, with evolutionary stochasticity playing a smaller role. In fact, populations derived from a founder that had re-evolved sensitivity in the original experiment were far more likely to re-evolve sensitivity in the replay experiment as well (Welch’s unequal variances t-test, t = 4.744, P < 0.001; fig. 3).
Fig. 3.
Phage resistance outcomes in the replay experiment mirror the outcomes of their founding population. (A) Populations in the replay experiment were founded by frozen stocks of resistant colonies prior to their evolution in the absence of phages. Each founder was used to generate ten replicate populations from genetically identical starting points. (B) Resistance mutations of the six founders. Each colony that had re-evolved sensitivity in the original experiment (orange) was paired with a genetically similar colony that had remained resistant (green). Of note, QAC5 was the only colony in the study with a mutation in the gene PSPTO_1330, so it was matched with a population that also had a small frameshift mutation but in a different glycosyltransferase-encoding gene. (C) Proportion of colonies in each population that were scored as phage-sensitive after 36 days in the absence of phages, as indicated by disruption in bacterial growth upon encountering phage on an agar plate (n = 54 populations with 96 colonies sampled per population).
Historical Contingency in Genome Evolution
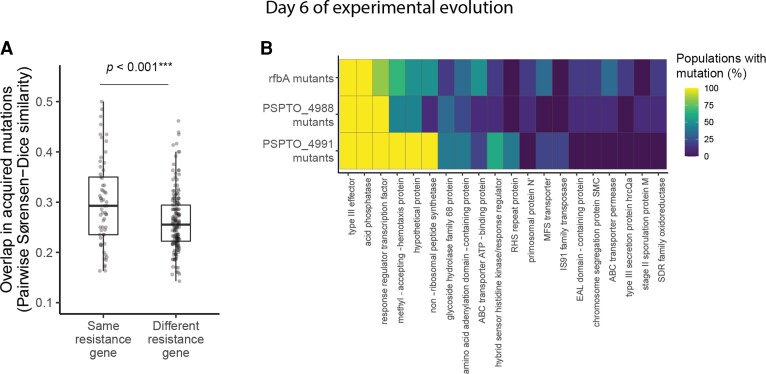
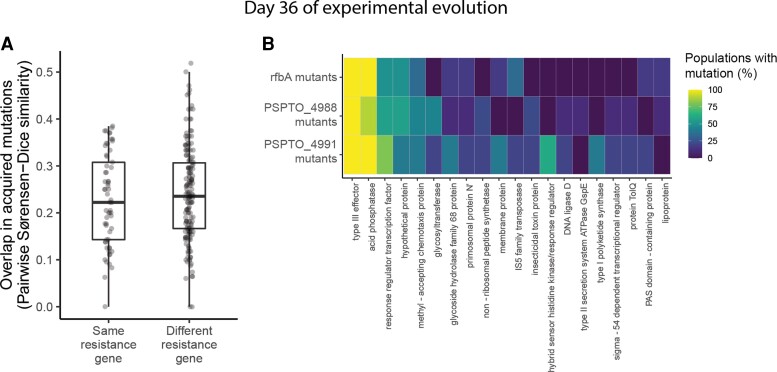
We pool-sequenced each of the populations of the original evolution experiment after 6 days of evolution in the absence of phages. To characterize the molecular basis of compensatory evolution in phage-resistant bacteria, we first considered that compensatory mutations in other systems are often related to biochemical protein stability or specialized protein-protein interactions (Bridgham et al. 2009; Gong et al. 2013). We therefore predicted that different mechanisms of resistance would likely require different compensatory mutations to ameliorate their costs. We would then expect populations with the same resistance gene to exhibit greater evolutionary parallelism than populations with different resistance genes. We calculated the Sørenson–Dice similarity coefficient, which describes the proportion of mutated genes that two populations have in common, controlling for their initial genetic backgrounds. We found that populations with the same resistance mechanisms had acquired more similar sets of mutations than populations with different resistance mechanisms (randomization test, P < 0.001, fig. 4A). For example, populations that had originally acquired phage resistance through a mutation in the glycosyltransferase-encoding gene PSPTO_4991 evolved particularly similarly to one another, with many genes acquiring mutations in either all or none of the populations (see fig. 4B for a diagram of genetic differences between evolved and ancestral populations organized by initial resistance gene).
Fig. 4.
Genetic contingency in identity of mutated genes during experimental evolution. (A) Pairwise similarity coefficients among pairs of evolved populations at day 6 of experimental evolution (n = 22). Statistical significance was assessed by randomizing whether pairs were labeled as having the same or different resistance genes and recalculating their similarity coefficients for 10,000 permutations. (B) Heatmap depicting the relationship between initial resistance genes and mutations acquired during experimental evolution for rfbA mutants (n = 6), PSPTO_4988 mutants (n = 9), and PSPTO_4991 mutants (n = 5). Rows represent all populations with resistance mutations in the same gene (note that resistance genes represented by fewer than two populations are not pictured, as there was no way to assess parallelism in these cases). Columns represent the top 20 genes that were most frequently mutated across populations by day 6 of experimental evolution. Colors indicate the percentage of populations of each resistance gene that had acquired one or more mutations by day 6 of experimental evolution.
Effect of History on Genome Evolution Diminishes Over Time
To examine adaptation and historical contingency in the longer term, we pool-sequenced each of the populations at the end of the evolution experiment (day 36). By the end of the experiment, the evolving populations had fixed relatively few mutations in protein-coding genes [mean = 0.71 ± 0.66 standard deviation (SD)] but acquired and maintained polymorphisms at many loci (mean = 22.61 ± 7.20 SD). Across all populations, the most frequent mutations occurred in genes coding for effector proteins, membrane-bound enzymes, and transcription factors (supplementary table S1, Supplementary Material online). Additionally, several populations had acquired substitutions in the DNA mismatch repair proteins mutS or mutL during experimental evolution. Although none of these mutations fixed within their respective populations, populations with a mutation in mutS or mutL acquired more fixed mutations or polymorphisms overall than the other populations by the end of the experiment (Student’s t-test, t = 2.746, P = 0.005), suggesting that they may contain “hypermutator” lineages with elevated mutation rates (Shaver et al. 2002; Harris et al. 2021).
After populations had evolved for a total of 36 days in identical environments, there was no remaining signature of the initial resistance gene on pairwise similarity coefficients (randomization test, P = 0.710, fig. 5).
Fig. 5.
Genetic contingency is no longer detectable after extended evolution in the absence of phages. (A) Pairwise similarity coefficients among pairs of evolved populations at day 36 of experimental evolution. Statistical significance was assessed by randomizing whether pairs were labeled as having the same or different resistance genes and recalculating their similarity coefficients for 10,000 permutations. (B) Heatmap depicting the relationship between initial resistance genes and mutations acquired during experimental evolution for rfbA mutants (n = 6), PSPTO_4988 mutants (n = 9), and PSPTO_4991 mutants (n = 5). Rows represent all populations with resistance mutations in the same gene (note that resistance genes represented by fewer than two populations are not pictured, as there was no way to assess parallelism in these cases). Columns represent the top 20 genes that were most frequently mutated across populations by day 36 of experimental evolution. Note that these are not necessarily identical to the top 20 genes identified in figure 4. Colors indicate the percentage of populations of each resistance gene that had acquired one or more mutations by day 36 of experimental evolution.
Discussion
Phages are ubiquitous in microbial communities and are expected to play a central role in bacterial evolution, with critical implications for bacteria–bacteria and bacteria–host interactions (Thurber 2009; Koskella and Brockhurst 2014). Predation by phages can maintain population and community diversity in their bacterial hosts (Weinbauer and Rassoulzadegan 2004), regulate the dissemination of antibiotic resistance genes (Burmeister, Fortier, et al. 2020; LeGault et al. 2021), select for hypermutator strains (Pal et al. 2007), and alter competitive outcomes among bacterial species (Bohannan and Lenski 2000). Here, we show that even transient exposure to phages can have lasting consequences for the evolutionary trajectories of bacterial populations. Through a series of laboratory evolution experiments, we demonstrate that phage resistance in the bacterium P. syringae can be reversible or entrenched depending on the original genetic path to resistance.
Phage resistance in our study primarily occurred through mutations in lipopolysaccharide biosynthesis genes, which have been previously implicated in phage resistance in this bacterial species and others (Picken and Beacham 1977; Evans et al. 2010; Meaden et al. 2015; Kulikov et al. 2019; Holtappels et al. 2020). One population did not appear to have any fixed genetic differences from the ancestor despite its resistant phenotype. Resistance in this case may be conferred by mutations that are not adequately resolved by short-read sequencing, such as copy number variation or sequence region inversions, or through unstable genetic changes such as phase variations (Kircher and Kelso 2010; Bull et al. 2014). Of note, this population remained phenotypically resistant throughout the evolution experiment, possibly because it did not experience detectable fitness costs relative to its phage-sensitive ancestor.
Even though the resistance mutations in our study were concentrated within a handful of genes, they occurred at many unique positions and altered the amino acid sequences in a variety of ways, including missense and nonsense substitutions and frameshift mutations of vastly different sizes. Such convergence in resistance mechanisms at the gene level, and diversity at the sequence level, is consistent with the expectation that phage infection relies on specific receptor structures and that recognition can easily be disrupted by modifying or deleting these structures in one of many ways (Dy et al. 2014). However, resistance was generally costly in the absence of phages, as is often observed in this system and others (Koskella et al. 2012; Meaden et al. 2015; Scanlan et al. 2015; Burmeister, Fortier, et al. 2020; Burmeister, Sullivan, et al. 2020).
Costs of resistance may stem from alterations to lipopolysaccharide molecules that destabilize the bacterial membrane or reduce surface adhesion (Mangalea and Duerkop 2020). The commonly observed costs of phage resistance suggest that resistance might be selected against in the absence of phage pressure, yet previous studies of this question have produced inconsistent results (Meyer et al. 2010; Avrani and Lindell 2015; Wielgoss et al. 2016). When we propagated the bacterial populations for an extended period in the absence of phages, we observed that phage sensitivity re-appeared and swept to high frequencies in several populations but that the majority of populations remained resistant. In the case of one population, this reversion to sensitivity was due to a mutation that restored the reading frame of the original sequence, but in other cases, populations re-evolved phage sensitivity without reversing the original mutation.
The diversity of phage-resistant mutants in this study allowed us to ask whether the re-evolution of phage sensitivity was contingent on the mechanisms and/or costs of phage resistance. Costs of trait maintenance are often expected to predict trait loss under relaxed selection (Lahti et al. 2009; Meyer et al. 2010), yet we did not observe a relationship between the magnitude of fitness costs and the re-evolution of phage sensitivity in our study. This observation, along with the fact that populations that did not re-evolve sensitivity nevertheless improved their fitness to match their phage-sensitive counterparts, suggests the existence of compensatory mutations that reduce the costs of resistance. Compensatory mutations could eventually restore fitness levels to a point where phage sensitivity is no longer advantageous (Teotónio and Rose 2007; McCandlish et al. 2016; Pennings et al. 2022), or may even be disadvantageous or lethal on the ancestral background, further discouraging reversion of the original trait (Rojas Echenique et al. 2019). Therefore, it appears that trade-offs between phage resistance and other aspects of fitness can be strong, yet bacteria are able to access two evolutionary pathways (reversion and compensation) in response.
We hypothesized that the probabilities of these two pathways could be contingent on the genetic mechanism underlying phage resistance. Some traits might have a greater supply of reversion mutations than others; for example, there may be more mutations that restore the ancestral expression levels of a gene than those that reconstitute the exact 3D structure of a receptor (Wielgoss et al. 2016), or more mutations that reverse duplications than point mutations (Chaudhry et al. 2018). Similarly, resistance mechanisms may impact bacterial fitness in different ways, thus requiring different sets of compensatory mutations to restore their costs (Rojas Echenique et al. 2019). We did not find an overall correlation between the initial resistance gene and whether phage sensitivity re-evolved but with two important caveats. First, when resistance was acquired through a large deletion in a receptor biosynthesis gene, it was never reversed, whether through a complementary insertion or other mutations. This suggests that the mutations that would reverse such a large change are so vanishingly rare that other compensatory mutations are much more likely to appear first.
Our second observation was that the genetic replicates in the mutant panel (i.e., the populations that independently acquired resistance via the same mutation) all followed the same phenotypic trajectories. Specifically, they all remained resistant throughout experimental evolution in the absence of phage. This suggested that historical contingencies—if they existed here—might be generated not from the identity of the resistance gene but from the exact genetic sequence. Even within the same resistance gene, some mutations might be more reversible than others. As the original experiment was not explicitly focused on this possibility, we designed a follow-up experiment that “replayed” experimental evolution ten times per founding genotype (inspired by Stephen Jay Gould’s famous thought experiment about replaying the tape of life, [Gould 1990]). Strikingly, we found that the evolutionary outcomes of the populations in our replay experiment closely mirrored those of their founders. Populations whose founders had re-evolved phage sensitivity also tended to re-evolve phage sensitivity at high rates, whereas populations whose founders had remained resistant tended to remain resistant as well.
Variation in the reversibility of different resistance mutations may occur if different sets of compensatory mutations are required to restore their costs. To explore this possibility, we compared the similarity of acquired mutations among pairs of the experimentally evolving populations. We found that populations with the same initial resistance gene evolved more in parallel with one another than populations with different resistance genes, suggesting that compensatory adaptation in phage-resistant bacteria also depends on evolutionary history. Several other studies have observed greater genomic parallelism among experimental evolution populations with similar starting genotypes for other traits, including in antibiotic resistance evolution (Card et al. 2021) and in compensatory adaptation after gene deletion (Rojas Echenique et al. 2019). And outside of the laboratory, convergence in sequence evolution appears to be more common among populations with a recent common ancestor than a distant one (Conte et al. 2012; Goldstein et al. 2015). Notably, the effect we observed was strong when the phage selection event was recent but was no longer detectable after populations had spent an extended period of time in the same environment. Thus, recent historical differences appear to be more important than distant historical differences in shaping subsequent evolution (Travisano et al. 1995; Yen and Papin 2017; Santos-Lopez et al. 2021).
Our study provides evidence for an evolutionary ratchet in bacteria–phage coevolution, where compensatory adaptation enables the persistence of certain resistance mutations even after the original selection pressure has ceased to operate. Why did history constrain evolution in this study but not in some others (Weinreich et al. 2006; Feldman et al. 2012; Toledo et al. 2016)? One important clue may lie in the underlying genetic structure of phage resistance. The phage-sensitive ancestor in this study could mutate to phage resistance through any one of at least 17 individual mutations, including nonsense mutations and large deletions. Phage adsorption relies on highly specific molecular interactions (Sharma et al. 2008), suggesting that there are many ways to change lipopolysaccharide structure to avoid phage recognition, and that at least some of these mutations are not lethal to the cell. In other cases, traits can evolve in parallel across lineages despite their historical differences. For example, several distantly related groups of animals have identical substitutions conferring tetrodotoxin resistance (Toledo et al. 2016). In this case, mutagenic screens have identified additional possible mutations that confer toxin resistance, but these additional mutations are so detrimental to sodium channel performance—an essential function—that they are not observed in nature (Feldman et al. 2012). The role of history in evolution is therefore likely related to the target size of mutations that confer novel functions yet are not overly disruptive to the original trait.
Our findings add to a growing body of work that selection by phages plays a key role in the ecological and evolutionary dynamics of bacterial communities. Further, many of the bacterial populations in our study acquired mutations in genes known to interact with eukaryotic immune systems. Phage resistance was directly mediated in many cases by changes to lipopolysaccharide molecules, which are recognized by both animal and plant immune systems (Triantafilou and Triantafilou 2005; Newman et al. 2007), and many populations also acquired mutations in Type III effector proteins that underlie bacterial virulence. It will thus be important to characterize whether and how phages are indirectly responsible for shaping coevolution in between bacteria and eukaryotes as well (Wahida et al. 2021).
Materials and Methods
Selection and Validation of Phage Resistance
Pseudomonas syringae pv. tomato DC3000 was obtained from Gail Preston at Oxford. The phage strains FMS, VCM, M5.1, QAC, and SNK were obtained from OmniLytics, Inc. and are considered potential biocontrol agents for plant-pathogenic Pseudomonas bacteria. Information on which mutant was selected from which phage is available in table 1. The population name indicates the selecting phage (e.g., “FMS”) and the position of the resistant colony in the original assay for selection of resistance (e.g., “6”).
Phage-resistant colonies were selected through soft agar overlays. Briefly, we amplified P. syringae in King’s B Medium overnight and then mixed bacterial cells with soft agar (King’s Broth supplemented with 0.6% agar). The mixture was spread evenly on petri dishes and allowed to dry. Droplets of high titer phage were pipetted on top. Plates were incubated for 48 h at 28°C. Large clearing zones (plaques) appeared and expanded from the phage droplet sites, and resistant colonies were picked from plaques.
To verify resistance, each bacterial colony was streaked on hard agar plates (King’s B Medium supplemented with 1.2% agar), across a high titer line of the phage strain it was isolated on (Burlage et al. 1998). Ancestral DC3000 was also streaked against all phages as a phage-sensitive negative control. Colonies were verified as phage-resistant if bacterial growth was uninterrupted at the phage line (supplementary fig. S2, Supplementary Material online).
To ensure that colonies were entirely free of phage particles prior to experimental evolution, each colony was streaked on hard agar, and cells were sampled at the end of the streak to seed an overnight culture. The next day, the overnight culture was passed through a 0.22 μm filter. The filtrate was spotted on a mixture of ancestral DC3000 and soft agar, as described above. Plates were incubated for 48 h at 28°C and checked for plaques. This entire process was repeated twice, at which point none of the filtrates produced phage plaques.
Experimental Evolution in the Absence of Phage
Phage-resistant colonies were passaged in King’s Broth media in cell culture plates. Six phage-sensitive populations founded from ancestral DC3000 were passaged alongside phage-resistant colonies to serve as controls for adaptation to the lab environment, and each transfer also included a media-only negative control. Every 3 days, 75 µl of each population (∼106 cells) were transferred to a new well with 4 ml of media. At every other transfer (every 6 days), a subset of each population was combined with a 50:50 mixture of King’s Broth and glycerol for storage at −80°C. The experiment lasted for 36 days (12 transfers), at which point there were no longer any detectable fitness differences between populations founded from phage-resistant colonies and populations founded from ancestral DC3000. According to previous work, the average doubling time of P. syringae at this temperature is approximately 1.27 h (Young et al. 1977).
Resistance and Fitness Measurements
On days 12, 24, and 36 of experimental evolution, 100 µl from each culture was sampled, serially diluted, and plated on hard agar. From the dilution level at which individual colonies were visible, 96 colonies were picked from each population. Each colony was streaked against the phage strain that the population was originally isolated on. After 2 days of incubation at 28°C, colony phenotypes were scored as resistant (uninterrupted bacterial growth across the plate), moderate (partly interrupted bacterial growth), or sensitive (fully interrupted bacterial growth at the intersection with the phage line).
It is possible that populations could contain phage-sensitive cells, yet appear partially resistant on agar due to habitat structure and/or biofilm growth (Testa et al. 2019; Simmons et al. 2020). We took two measures to combat this possibility. First, while phage-sensitive sub-populations may be able to survive among a larger population of resistant cells, separately streaking 96 colonies from each population should ensure that colonies of entirely sensitive cells are visibly impacted by phages, even in a structured environment. Second, in any populations with colonies scored as moderate or sensitive on agar, resistance was assessed in greater quantitative resolution by measuring bacterial population growth in liquid culture in the presence and absence of phages (supplementary fig. S3, Supplementary Material online).
To measure population growth rates over the course of experimental evolution, time series growth data was collected using a Molecular Devices VersaMax Microplate Reader. An overnight culture of each population was initially diluted to 0.001 OD600 in 200 ml King’s Broth. Colonies were randomized with respect to spatial layout, and media-only wells were included as negative controls. The plate was incubated at 28°C for 40 h with continuous shaking, with optical density readings taken at 600 nm every 5 min. These readings were used to fit a logistic growth model with the R package growthrates and estimate the intrinsic growth rate μmax and the final population size ODmax. To express these values as a proportion of wild-type fitness while controlling for technical effects, the fitted values were extracted from a model that included the day of experimental evolution, the population type (phage-resistant colony or phage-sensitive control), and the location of the well within the plate. While µmax and ODmax were modestly correlated across populations (r = 0.689, P < 0.001, supplementary fig. S4, Supplementary Material online), the final population sizes of resistant populations were close to that of the phage-sensitive ancestor, resulting in only marginally detectable costs of resistance for ODmax (one-sample t-test, t = −1.382, P = 0.091). This suggests that phage resistance is the most costly during periods of exponential growth; thus, for subsequent analyses we focused on variation in µmax over time.
Whole-Genome Sequencing
To identify mutations contributing to phage resistance, bacterial DNA was extracted from each phage-resistant clone using the DNeasy Blood and Tissue Kit. Concentrations were measured using a Qubit 3.0 Fluorometer, and samples were concentrated if necessary using an ethanol precipitation to obtain a minimum of 10 ng/µl of DNA per sample. DNA was sequenced at a depth of 300 Mb (estimated coverage of 45.9×) on the Illumina NextSeq 2000 platform at the Microbial Genome Sequencing Center (Pittsburgh, PA, USA). To identify and track the frequencies of mutations that arose during experimental evolution, bacterial DNA was extracted, quantified, and concentrated as described previously from populations at day 6 and day 36 of the experiment. DNA was sequenced at a depth of 625 Mb (estimated coverage of 95.6×) as described previously.
Paired-end reads were filtered and trimmed using Trimmomatic (Bolger et al. 2014). Reads shorter than 25 bp were discarded, and reads with an average quality score below 20 within a 4 bp sliding window were discarded. Pairs of reads that both passed filtering (>95% of total reads per sample) were retained. Reads were mapped to the P. syringae pv. tomato DC3000 genome (BioSample accession SAMN02604017 from the Pseudomonas Genome Database) and variants were identified using breseq (Deatherage and Barrick 2014), a pipeline for identifying genetic variation within microbial populations. Any sites at which the genome of the ancestral strain differed from the reference genome were removed from subsequent analyses. To avoid false positive calls from repetitive regions, mutations were filtered to exclude regions of high polymorphism (five or more mutations in a 50 bp sliding window within a population at a single time-point).
Analysis of Genomic Parallelism
To quantify the genome-wide parallelism of experimentally evolved populations, a matrix was generated of all genes with newly acquired mutations in each population (i.e., those not already fixed at day 0 of the experiment). The Sørenson–Dice similarity coefficient was calculated for each pair of populations as follows, where G1 represents genes with newly acquired mutations in Population 1, and G2 represents genes with newly acquired mutations in Population 2.
Mutations were included if they appeared at any population frequency (they were not required to be fixed), but synonymous and intergenic mutations were excluded. The analysis, therefore, focused on nonsynonymous point mutations or indels within coding regions.
Following previous work (Card et al. 2021), the distribution of pairwise Sørenson–Dice values was analyzed using a randomization test. The labels annotating pairs of populations as having the same or different resistance genes were shuffled, and the mean difference was calculated between pairs with the same resistance gene and pairs with different resistance genes. This process was repeated 10,000 times to generate a null distribution. The true difference in means in the observed data was compared with the null distribution, and the observed difference was considered significant if it was more extreme than the upper 5% of permuted values.
Replay of Experimental Evolution
To assess the repeatability of phage resistance outcomes in the initial evolution experiment, six populations were identified for further study. This list included three populations that re-evolved phage sensitivity in the initial experiment, each paired with the closest possible genetic match that maintained phage resistance. Ten replicates of each population were seeded from samples taken at day 0 of the initial evolution experiment (i.e., before they eventually lost or maintained phage resistance). The resulting 60 populations were maintained in King’s Broth media and passaged as described previously for 36 days. At this point, 96 colonies were picked from each population, individually streaked against phage, and scored as resistant or sensitive. Resistance was analyzed as a proportion of colonies within each population. Since many populations were either completely resistant or completely sensitive, Welch’s t-test for unequal variances was used to compare resistance outcomes in the replay experiment according to their founder populations.
Supplementary Material
Acknowledgments
The authors thank Tadashi Fukami, Kyle Card, Catherine Hernandez, and members of the Koskella lab for helpful discussion and methods advice. This work was supported by the National Science Foundation (award number 1650114 to R.D.; award number 1942881 to B.K.), the Society for the Study of Evolution (award 047408 to R.D.), and the Amgen Foundation (Amgen Scholars Fellowship to N.D.L.).
Contributor Information
Reena Debray, Department of Integrative Biology, University of California, Berkeley, Berkeley, CA, USA.
Nina De Luna, Department of Immunology, Pennsylvania State University, State College, PA, USA.
Britt Koskella, Department of Integrative Biology, University of California, Berkeley, Berkeley, CA, USA; Chan Zuckerberg BioHub, San Francisco, CA, USA.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Data Availability
The data and code supporting the figures in this paper can be found in the online repository https://github.com/reenadebray/loss-of-resistance.
References
- Avrani S, Lindell D. 2015. Convergent evolution toward an improved growth rate and a reduced resistance range in Prochlorococcus strains resistant to phage. Proc Natl Acad Sci U S A. 112:E2191–E2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohannan BJM, Lenski RE. 2000. Linking genetic change to community evolution: insights from studies of bacteria and bacteriophage. Ecol Lett. 3:362–377. [Google Scholar]
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridgham JT, Ortlund EA, Thornton JW. 2009. An epistatic ratchet constrains the direction of glucocorticoid receptor evolution. Nature 461:515–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockhurst MA, Morgan AD, Fenton A, Buckling A. 2007. Experimental coevolution with bacteria and phage. The Pseudomonas fluorescens–Phi2 model system. Infect Genet Evol. 7:547–552. [DOI] [PubMed] [Google Scholar]
- Bull JJ, Vegge CS, Schmerer M, Chaudhry WN, Levin BR. 2014. Phenotypic resistance and the dynamics of bacterial escape from phage control. PLoS One 9:e94690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burlage RS, Atlas R, Stahl D, Sayler G, Geesey G. 1998. Techniques in microbial ecology. New York, NY: Oxford University Press on Demand. [Google Scholar]
- Burmeister AR, Fortier A, Roush C, Lessing AJ, Bender RG, Barahman R, Grant R, Chan BK, Turner PE. 2020a. Pleiotropy complicates a trade-off between phage resistance and antibiotic resistance. Proc Natl Acad Sci U S A. 117:11207–11216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burmeister AR, Sullivan RM, Lenski RE. 2020b. Fitness costs and benefits of resistance to phage lambda in experimentally evolved Escherichia coli. In: Banzhaf W, Cheng BHC, Deb K, Holekamp KE, Lenski RE, Ofria C, Phennock RT, Punch WF, Whittaker DJ, editors. Evolution in action: past, present and future: a festschrift in honor of Erik D. Goodman. Springer Nature Switzerland. p. 123–143. [Google Scholar]
- Card KJ, Thomas MD, Graves JL Jr, Barrick JE, Lenski RE. 2021. Genomic evolution of antibiotic resistance is contingent on genetic background following a long-term experiment with Escherichia coli. Proc Natl Acad Sci U S A. 118(5):e2016886118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhry WN, Pleska M, Shah NN, Weiss H, McCall IC, Meyer JR, Gupta A, Guet CC, Levin BR. 2018. Leaky resistance and the conditions for the existence of lytic bacteriophage. PLoS Biol. 16(7):e2005971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clay K, Kover PX. 1996. The Red Queen Hypothesis and plant/pathogen interactions. Annu Rev Phytopathol. 34:29–50. [DOI] [PubMed] [Google Scholar]
- Conte GL, Arnegard ME, Peichel CL, Schluter D. 2012. The probability of genetic parallelism and convergence in natural populations. Proc Biol Sci. 279:5039–5047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deatherage DE, Barrick JE. 2014. Identification of mutations in laboratory-evolved microbes from next-generation sequencing data using breseq. Methods Mol Biol. 1151:165–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennehy JJ. 2012. What can phages tell us about host-pathogen coevolution? Int J Evol Biol. 2012:396165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewald-Wang EA, Parr N, Tiley K, Lee A, Koskella B. 2022. Multiyear time-shift study of bacteria and phage dynamics in the phyllosphere. Am Nat. 199(1):126–140. [DOI] [PubMed] [Google Scholar]
- Durão P, Balbontín R, Gordo I. 2018. Evolutionary mechanisms shaping the maintenance of antibiotic resistance. Trends Microbiol. 26:677–691. [DOI] [PubMed] [Google Scholar]
- Dy RL, Richter C, Salmond GPC, Fineran PC. 2014. Remarkable mechanisms in microbes to resist phage infections. Annu Rev Virol. 1:307–331. [DOI] [PubMed] [Google Scholar]
- Evans TJ, Ind A, Komitopoulou E, Salmond GPC. 2010. Phage-selected lipopolysaccharide mutants of Pectobacterium atrosepticum exhibit different impacts on virulence. J Appl Microbiol. 109:505–514. [DOI] [PubMed] [Google Scholar]
- Faria VG, Martins NE, Paulo T, Teixeira L, Sucena É, Magalhães S. 2015. Evolution of Drosophila resistance against different pathogens and infection routes entails no detectable maintenance costs. Evolution 69:2799–2809. [DOI] [PubMed] [Google Scholar]
- Feldman CR, Brodie ED Jr, Brodie ED 3rd, Pfrender ME. 2012. Constraint shapes convergence in tetrodotoxin-resistant sodium channels of snakes. Proc Natl Acad Sci U S A. 109(12):4556–4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein RA, Pollard ST, Shah SD, Pollock DD. 2015. Nonadaptive amino acid convergence rates decrease over time. Mol Biol Evol. 32:1373–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez JM, Verdú M, Perfectti F. 2010. Ecological interactions are evolutionarily conserved across the entire tree of life. Nature 465:918–921. [DOI] [PubMed] [Google Scholar]
- Gong LI, Suchard MA, Bloom JD. 2013. Stability-mediated epistasis constrains the evolution of an influenza protein. Elife 2:e00631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould SJ. 1990. Wonderful life: the burgess shale and the nature of history: New York, NY: W.W. Norton & Company. [Google Scholar]
- Harris KB, Flynn KM, Cooper VS. 2021. Polygenic adaptation and clonal interference enable sustained diversity in experimental Pseudomonas aeruginosa populations. Mol Biol Evol. 38(12):5359–5375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtappels D, Kerremans A, Busschots Y, Van Vaerenbergh J, Maes M, Lavigne R, Wagemans J. 2020. Preparing for the KIL: receptor analysis of Pseudomonas syringae pv. Porri phages and their impact on bacterial virulence. Int J Mol Sci. 21(2930):1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher M, Kelso J. 2010. High-throughput DNA sequencing—concepts and limitations. Bioessays 32:524–536. [DOI] [PubMed] [Google Scholar]
- Koskella B. 2018. Resistance gained, resistance lost: an explanation for host-parasite coexistence. PLoS Biol. 16(9):e3000013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koskella B, Brockhurst MA. 2014. Bacteria-phage coevolution as a driver of ecological and evolutionary processes in microbial communities. FEMS Microbiol Rev. 38:916–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koskella B, Lin DM, Buckling A, Thompson JN. 2012. The costs of evolving resistance in heterogeneous parasite environments. Proc Biol Sci. 279:1896–1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koskella B, Parr N. 2015. The evolution of bacterial resistance against bacteriophages in the horse chestnut phyllosphere is general across both space and time. Phil Trans R Soc B. 370:20140297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulikov EE, Golomidova AK, Prokhorov NS, Ivanov PA, Letarov AV. 2019. High-throughput LPS profiling as a tool for revealing of bacteriophage infection strategies. Sci Rep. 9:2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutschera A, Schombel U, Schwudke D, Ranf S, Gisch N. 2021. Analysis of the structure and biosynthesis of the lipopolysaccharide core oligosaccharide of Pseudomonas syringae pv. tomato DC3000. Int J Mol Sci. 22(6):3250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahti DC, Johnson NA, Ajie BC, Otto SP, Hendry AP, Blumstein DT, Coss RG, Donohue K, Foster SA. 2009. Relaxed selection in the wild. Trends Ecol Evol. 24:487–496. [DOI] [PubMed] [Google Scholar]
- LeGault KN, Hays SG, Angermeyer A, McKitterick AC, Johura F-T, Sultana M, Ahmed T, Alam M, Seed KD. 2021. Temporal shifts in antibiotic resistance elements govern phage-pathogen conflicts. Science 373(534):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lourenço M, Chaffringeon L, Lamy-Besnier Q, Pédron T, Campagne P, Eberl C, Bérard M, Stecher B, Debarbieux L, De Sordi L. 2020. The spatial heterogeneity of the gut limits predation and fosters coexistence of bacteria and bacteriophages. Cell Host Microbe 28:390–401.e5. [DOI] [PubMed] [Google Scholar]
- Mangalea MR, Duerkop BA. 2020. Fitness trade-offs resulting from bacteriophage resistance potentiate synergistic antibacterial strategies. Infect Immun. 88(7):e00926-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCandlish DM, Shah P, Plotkin JB. 2016. Epistasis and the dynamics of reversion in molecular evolution. Genetics 203:1335–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGlothlin JW, Kobiela ME, Feldman CR, Castoe TA, Geffeney SL, Hanifin CT, Toledo G, Vonk FJ, Richardson MK, Brodie ED, et al. . 2016. Historical contingency in a multigene family facilitates adaptive evolution of toxin resistance. Curr Biol. 26:1616–1621. [DOI] [PubMed] [Google Scholar]
- Meaden S, Paszkiewicz K, Koskella B. 2015. The cost of phage resistance in a plant pathogenic bacterium is context-dependent. Evolution 69:1321–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer JR, Agrawal AA, Quick RT, Dobias DT, Schneider D, Lenski RE. 2010. Parallel changes in host resistance to viral infection during 45,000 generations of relaxed selection. Evolution 64:3024–3034. [DOI] [PubMed] [Google Scholar]
- Newman M-A, Maxwell Dow J, Molinaro A, Parrilli M. 2007. Invited review: Priming, induction and modulation of plant defence responses by bacterial lipopolysaccharides. J Endotoxin Res. 13:69–84. [DOI] [PubMed] [Google Scholar]
- Pal C, Maciá MD, Oliver A, Schachar I, Buckling A. 2007. Coevolution with viruses drives the evolution of bacterial mutation rates. Nature 450:1079–1081. [DOI] [PubMed] [Google Scholar]
- Pennings PS, Ogbunugafor CB, Hershberg R. 2022. Reversion is most likely under high mutation supply when compensatory mutations do not fully restore fitness costs. G3 Genes Genom Genet. jkac190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picken RN, Beacham IR. 1977. Bacteriophage-resistant mutants of Escherichia coli K12. Location of receptors within the lipopolysaccharide. J Gen Microbiol. 102:305–318. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Brito B, Li L, Wegley L, Furlan M, Angly F, Breitbart M, Buchanan J, Desnues C, Dinsdale E, Edwards R, et al. . 2010. Viral and microbial community dynamics in four aquatic environments. ISME J. 4:739–751. [DOI] [PubMed] [Google Scholar]
- Rojas Echenique JI, Kryazhimskiy S, Nguyen Ba AN, Desai MM. 2019. Modular epistasis and the compensatory evolution of gene deletion mutants. PLoS Genet. 15:e1007958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Lopez A, Marshall CW, Haas AL, Turner C, Rasero J, Cooper VS. 2021. The roles of history, chance, and natural selection in the evolution of antibiotic resistance. eLife 10:e70676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scanlan PD, Buckling A, Hall AR. 2015. Experimental evolution and bacterial resistance: (co)evolutionary costs and trade-offs as opportunities in phage therapy research. Bacteriophage 5:e1050153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah P, McCandlish DM, Plotkin JB. 2015. Contingency and entrenchment in protein evolution under purifying selection. Proc Natl Acad Sci U S A. 112:E3226–E3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma RS, Mishra V, Mohmmed A, Babu CR. 2008. Phage specificity and lipopolysaccharides of stem- and root-nodulating bacteria (Azorhizobium caulinodans, Sinorhizobium spp., and Rhizobium spp.) of Sesbania spp. Arch Microbiol. 189:411–418. [DOI] [PubMed] [Google Scholar]
- Shaver AC, Dombrowski PG, Sweeney JY, Treis T, Zappala RM, Sniegowski PD. 2002. Fitness evolution and the rise of mutator alleles in experimental Escherichia coli populations. Genetics 162:557–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon BC, Verhulst S. 1996. Ecological immunology: costly parasite defences and trade-offs in evolutionary ecology. Trends Ecol Evol. 11:317–321. [DOI] [PubMed] [Google Scholar]
- Simmons EL, Bond MC, Koskella B, Drescher K, Bucci V, Nadell CD. 2020. Biofilm structure promotes coexistence of phage-resistant and phage-susceptible bacteria. mSystems 5(3):e00877-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl EA, Dwyer G, Mauricio R, Kreitman M, Bergelson J. 1999. Dynamics of disease resistance polymorphism at the Rpm1 locus of Arabidopsis. Nature 400:667–671. [DOI] [PubMed] [Google Scholar]
- Stenseth NC, Maynard Smith J. 1984. Coevolution in ecosystems: red queen evolution or stasis? Evolution 38:870. [DOI] [PubMed] [Google Scholar]
- Teotónio H, Rose MR. 2007. Perspective: reverse evolution. Evolution 55:653–660. [DOI] [PubMed] [Google Scholar]
- Testa S, Berger S, Piccardi P, Oechslin F, Resch G, Mitri S. 2019. Spatial structure affects phage efficacy in infecting dual-strain biofilms of Pseudomonas aeruginosa. Commun Biol. 2:405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurber RV. 2009. Current insights into phage biodiversity and biogeography. Curr Opin Microbiol. 12:582–587. [DOI] [PubMed] [Google Scholar]
- Toledo G, Hanifin C, Geffeney S, Brodie ED 3rd. 2016. Chapter four: Convergent evolution of tetrodotoxin-resistant sodium channels in predators and prey. Curr Top Membr. 78:87–113. [DOI] [PubMed] [Google Scholar]
- Travisano M, Mongold JA, Bennett AF, Lensk RE. 1995. Experimental tests of the roles of adaptation, chance, and history in evolution. Science 267(5194):87–90. [DOI] [PubMed] [Google Scholar]
- Triantafilou M, Triantafilou K. 2005. Invited review: The dynamics of LPS recognition: complex orchestration of multiple receptors. J Endotoxin Res. 11:5–11. [DOI] [PubMed] [Google Scholar]
- Wahida A, Tang F, Barr JJ. 2021. Rethinking phage-bacteria-eukaryotic relationships and their influence on human health. Cell Host Microbe 29:681–688. [DOI] [PubMed] [Google Scholar]
- Waterbury JB, Valois FW. 1993. Resistance to co-occurring phages enables marine Synechococcus communities to coexist with cyanophages abundant in seawater. Appl Environ Microbiol. 59:3393–3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinbauer MG, Rassoulzadegan F. 2004. Are viruses driving microbial diversification and diversity? Environ Microbiol. 6:1–11. [DOI] [PubMed] [Google Scholar]
- Weinreich DM, Delaney NF, Depristo MA, Hartl DL. 2006. Darwinian evolution can follow only very few mutational paths to fitter proteins. Science 312(5770):111–114. [DOI] [PubMed] [Google Scholar]
- Wielgoss S, Bergmiller T, Bischofberger AM, Hall AR. 2016. Adaptation to parasites and costs of parasite resistance in mutator and nonmutator bacteria. Mol Biol Evol. 33:770–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods RJ, Barrick JE, Cooper TF, Shrestha U, Kauth MR, Lenski RE. 2011. Second-order selection for evolvability in a large Escherichia coli population. Science 331:1433–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen P, Papin JA. 2017. History of antibiotic adaptation influences microbial evolutionary dynamics during subsequent treatment. PLoS Biol. 15(8):e2001586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JM, Luketina RC, Marshall AM. 1977. The effects on temperature on growth in vitro of Pseudomonas syringae and Xanthomonas pruni. J Appl Bacteriol. 42:345–354. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data and code supporting the figures in this paper can be found in the online repository https://github.com/reenadebray/loss-of-resistance.