Abstract
The origin of nitrogen fixation is an important issue in evolutionary biology. While nitrogen is required by all living organisms, only a small fraction of bacteria and archaea can fix nitrogen. The prevailing view is that nitrogen fixation first evolved in archaea and was later transferred to bacteria. However, nitrogen-fixing (Nif) bacteria are far larger in number and far more diverse in ecological niches than Nif archaea. We, therefore, propose the bacteria-first hypothesis, which postulates that nitrogen fixation first evolved in bacteria and was later transferred to archaea. As >30,000 prokaryotic genomes have been sequenced, we conduct an in-depth comparison of the two hypotheses. We first identify the six genes involved in nitrogen fixation in all sequenced prokaryotic genomes and then reconstruct phylogenetic trees using the six Nif proteins individually or in combination. In each of these trees, the earliest lineages are bacterial Nif protein sequences and in the oldest clade (group) the archaeal sequences are all nested inside bacterial sequences, suggesting that the Nif proteins first evolved in bacteria. The bacteria-first hypothesis is further supported by the observation that the majority of Nif archaea carry the major bacterial Mo (molybdenum) transporter (ModABC) rather than the archaeal Mo transporter (WtpABC). Moreover, in our phylogeny of all available ModA and WtpA protein sequences, the earliest lineages are bacterial sequences while archaeal sequences are nested inside bacterial sequences. Furthermore, the bacteria-first hypothesis is supported by available isotopic data. In conclusion, our study strongly supports the bacteria-first hypothesis.
Keywords: nitrogen fixation, nitrogenase, molybdenum transporter, bacteria, archaea
Introduction
Nitrogen (N), which accounts for almost 80% of the earth’s atmosphere, is essential for all living creatures but no organism can directly utilize nitrogen (N2) (Erisman et al. 2008). Nitrogen fixation is the conversion of N2 to bio-available NH3 (Boyd and Peters 2013; Mus et al. 2019). Only a small fraction of bacteria and archaea can fix nitrogen (Boyd and Peters 2013). However, nitrogen-fixing (Nif) bacteria are physiologically and ecologically diverse. Most of them are free-living, including aerobic chemoorganotrophs (Setubal et al. 2009); anoxygenic and oxygenic phototrophs (Philippi et al. 2021; Watanabe and Horiike 2021); and anaerobic chemoorganotrophs (Li et al. 2019). Many of the remaining Nif bacteria are symbiotic with plants (Li et al. 2015; de Lajudie and Young 2017; Roy et al. 2020) or animals (Russell et al. 2009; König et al. 2016; Shukla et al. 2016). In contrast, most Nif archaea are exclusively free-living and belong to the chemolithotrophic methanogens (Mehta and Baross 2006; Boyd, Anbar, et al. 2011; Koirala and Brözel 2021). Free-living Nif species play an important role in nitrogen cycle in the modern biosphere as well as in the ancient environments.
Biological nitrogen fixation is catalyzed by nitrogenases, which have three variants, the Mo (molybdenum)-dependent nitrogenase (Nif), V (vanadium)-dependent nitrogenase (Vnf), and Fe-only nitrogenase (Anf) (Mus et al. 2018). Nitrogenases are composed of two distinct metalloproteins, namely dinitrogenase (the catalytic component) and dinitrogenase reductase (the electron transfer component) (Burén et al. 2020). Both components contain Fe–S clusters, and the dinitrogenase component contains a characteristic iron−molybdenum cofactor, FeMoco. Alternative nitrogenases contain either V + Fe or Fe-only in their dinitrogenase component (Mus et al. 2018). The nif (nitrogen-fixing) genes of conventional Mo-dependent nitrogenase, which is encoded by nifH (the dinitrogenase reductase subunit) and nifD and nifK (the dinitrogenase subunits) (Rubio and Ludden 2005; Hu and Ribbe 2016; López-Torrejón et al. 2016), have been well studied (Dos Santos et al. 2012; Mus et al. 2018, 2019; Zheng et al. 2018; Albright et al. 2019; Lee et al. 2019; Burén et al. 2020; Garcia et al. 2020). The nifD and nifK genes encode, respectively, the α- and β-subunit of dinitrogenase, which form a α2β2-tetramer (Rubio and Ludden 2005; Hu and Ribbe 2016; López-Torrejón et al. 2016). The nifE and nifN genes encode for another α2β2-tetramer, which is essential for the assembly of the metal cofactor (Rubio and Ludden 2005; Dos Santos et al. 2012; Hu and Ribbe 2016; López-Torrejón et al. 2016). The nifB gene product functions in the biosynthesis of the Fe and S donors for the metal cofactor (Rubio and Ludden 2005; Hu and Ribbe 2016; López-Torrejón et al. 2016). Although various sets of nif genes have been used as criteria to identify Nif species (Dos Santos et al. 2012; Poudel et al. 2018; Albright et al. 2019; Koirala and Brözel 2021), the entire set of the above six nif genes (i.e., nifHDKENB) has been proposed to be the definition of Nif species (Dos Santos et al. 2012; Mus et al. 2019). However, nifN or both nifE and nifN may not be essential because this gene is absent in the genomes of some Nif Chloroflexota (bacteria) (Ivanovsky et al. 2021), Firmicutes_A (bacteria) (Chen et al. 2021), Elusimicrobiota (bacteria) (Zheng, Dietrich, et al. 2016), and Nif methanogens (archaea) (Mehta and Baross 2006). We therefore will also consider the criterion of “the five-nif-gene set” (nifHDKEB), which includes no nifN, and that of “the four-nif-gene set” (nifHDKB), which includes no nifEN.
The molybdenum (Mo) is an essential element and participates in a variety of metalloenzymes in both bacteria and archaea (Zhang and Gladyshev 2008; Peng et al. 2018). Previous studies showed that the Mo-dependent enzymes (molybdoenzymes) evolved very early in prokaryotes, and it may have evolved their unique systems in bacteria and in archaea in very ancient times (Zhang and Gladyshev 2008; Peng et al. 2018). Moreover, in prokaryotes, nitrogenase is the only molybdoenzyme that contains the iron–Mo cofactor (FeMoco), while the remaining molybdoenzymes contain the molybdenum cofactor (Moco) (Zhang and Gladyshev 2008; Peng et al. 2018). The Mo transporter, which is the essential protein for prokaryotes to uptake Mo, may regulate nitrogenase (Demtröder et al. 2019). Therefore, the Mo transporter might have played an important role in the evolution of nitrogen fixation.
It has been proposed that nitrogen fixation first evolved in archaea and was later transferred to bacteria (Raymond et al. 2004; Boyd, Hamilton, et al. 2011; Boyd and Peters 2013; Mus et al. 2018, 2019; Koirala and Brözel 2021); we call this hypothesis “the archaea-first hypothesis”. This hypothesis was based on the observation that the earliest lineages in the reconstructed phylogeny of NifH, D, K, E, and N sequences and in that of the radical S-adenosyl-L-methionine (SAM) domains of NifB proteins were archaeal sequences (Raymond et al. 2004; Boyd, Anbar, et al. 2011). As an alternative to the archaea-first hypothesis, it has been proposed that nitrogen fixation evolved in the last universal common ancestor (LUCA) (Raymond et al. 2004; Weiss et al. 2016); we call it the LUCA-first hypothesis. Here, we propose “the bacteria-first hypothesis”, which postulates that nitrogen fixation first evolved in bacteria and was later transferred to archaea. These three hypotheses are mutually exclusive, so that only one of them can be true.
In this study, we applied the criterion of the six-nif-gene set (nifHDKENB) to identify Nif species in the available genomic sequence data (Parks et al. 2018, 2020; Mendler et al. 2019); we also identified those species that carry nifHDKEB and those that carry nifHDKB or nifHDKEB. To distinguish between the archaea-first and the bacteria-first hypothesis, we reconstructed the following gene trees: The first one was the phylogeny of NifH sequences because among the Nif proteins, NifH is the widest spread in prokaryotes. The second tree was the phylogeny of NifD, K, E, and N sequences because these proteins are paralogous. The third tree was the phylogeny of radical SAM domains of NifB proteins. The fourth tree was the phylogeny of the concatenated sequences of NifH, D, and K sequences that are tightly linked in the genome. In each of these trees, our aim was to find out whether a gene under study first appeared in bacteria or archaea. In addition, we identified modABC- and wtpABC-gene-carrying species. It is known that ModABC is the bacterial Mo transporter while WtpABC is the major Mo transporter in archaea (Peng et al. 2018). Thus, studying the association of these two Mo-transporters with Nif species (i.e., the six-nif-gene set) is helpful for distinguishing between the two hypotheses. For this purpose, we also reconstructed the phylogeny of ModA and WtpA sequences. Using these phylogenetic trees, we argued for the bacteria-first hypothesis.
Results
Identifying nif Genes
We analyzed 30,238 bacterial and 1,672 archaeal genomes from AnnoTree and GTDB (Parks et al. 2018, 2020; Mendler et al. 2019), and found 3,485 bacterial species and 373 archaeal species that contain at least one of the nifH, nifD, nifK, nifE, nifN, and nifB genes (table 1). The number of species that carry the six-nif-gene set is ∼18 times higher in bacteria (1,420) than in archaea (78) (table 1). Moreover, the number of species carrying the five-nif-gene set is ∼4 times higher in bacteria (53) than in archaea (13); and the number of species carrying the four-nif-gene set is ∼9 times higher in bacteria (171) than in archaea (18) (table 1) Thus, most known Nif species are bacteria. However, the proportion of bacterial species carrying the six-gene set (1,420/30,238 = 4.70%) is almost the same as that of archaeal species carrying the six-gene set (78/1,672 = 4.67%). The detailed information of nif-gene-harboring species in our collection and their genomic characteristics is given in supplementary data S1, Supplementary Material online.
Table 1.
Numbers of Bacterial and Archaeal Species With Different Gene Sets (Data from AnnoTree v1.2.0 GTDB Release R95).
| Nitrogenase-related genes | Number of species | |||||||
|---|---|---|---|---|---|---|---|---|
| Bacteria | Archaea | |||||||
| nif | H | 3,291 | 329 | |||||
| nif | D | 2,256 | 116 | |||||
| nif | K | 2,237 | 130 | |||||
| nif | E | 2,333 | 132 | |||||
| nif | N | 1,873 | 83 | |||||
| nif | B | 2,293 | 357 | |||||
| nif | H | D | K | B | 169 | 15 | ||
| nif | H | D | K | E | B | 53 | 13 | |
| nif | H | D | K | E | N | B | 1,420 | 78 |
| Containing at least one nif gene | 3,485 | 373 | ||||||
| vnf G | 25 | 8 | ||||||
| anf G | 101 | 6 | ||||||
Previous studies suggested that Mo-nitrogenase evolved earlier than V- and Fe-nitrogenase and most anf/vnf-gene-harboring species also contain nif genes (Mus et al. 2018, 2019; Garcia et al. 2020). Our collection identified conventional Mo-nitrogenase, so it included species that also contain the V- or Fe-nitrogenase. The additional γ-subunit (encoded by vnfG or anfG), of which the function has been unraveled only recently, is only present in V- and Fe-nitrogenases (Mus et al. 2018; Garcia et al. 2020), and the γ-subunit (vnf/anfG) is attached to the α-subunit (vnf/anfD). So, to identify V- and Fe-nitrogenases, we used the location of vnf/anfG genes in the genomes to identify vnf/anfH, D, K genes. We identified 25 bacteria and 8 archaea that contain vnfG, and 101 bacteria and 6 archaea that contain anfG (table 1 and supplementary data S1, Supplementary Material online). In total, our collection includes 114 bacteria and 5 archaea that contain Mo-nitrogenase and V- or Fe-nitrogenase, and 6 bacteria and 4 archaea that contain all three types of nitrogenases. In total, we identified 130 Nif species that contain V- or Fe-nitrogenase or both.
Phylogenetic Tree of Nif⁄Vnf⁄AnfH Proteins
The nifH gene encodes the dinitrogenase reductase and many studies used nifH to investigate the diversity of Nif species in environments (Zehr and McReynolds 1989; Affourtit et al. 2001; Hamelin et al. 2002; Langlois et al. 2005; Hamilton et al. 2011). Therefore, we first reconstruct the phylogeny of Nif/Vnf/AnfH protein sequences (fig. 1). Our study includes 3,535 Nif/Vnf/AnfH protein sequences and uses 376 Bch/ChlL and BchX sequences, which are NifH homologous proteins (Nomata et al. 2006; Tahon et al. 2016; Garcia et al. 2020), as the outgroup. We use the IQ-TREE method to reconstruct phylogenetic trees (Minh et al. 2020) and ModelFinder (Kalyaanamoorthy et al. 2017), which is implemented in IQ-TREE, to select the best-fit model of protein sequence evolution with ultrafast bootstrap (1,000 replicates) (Hoang et al. 2018). The model was selected according to the Bayesian information criterion (BIC) (Posada and Buckley 2004) is LG + R10.
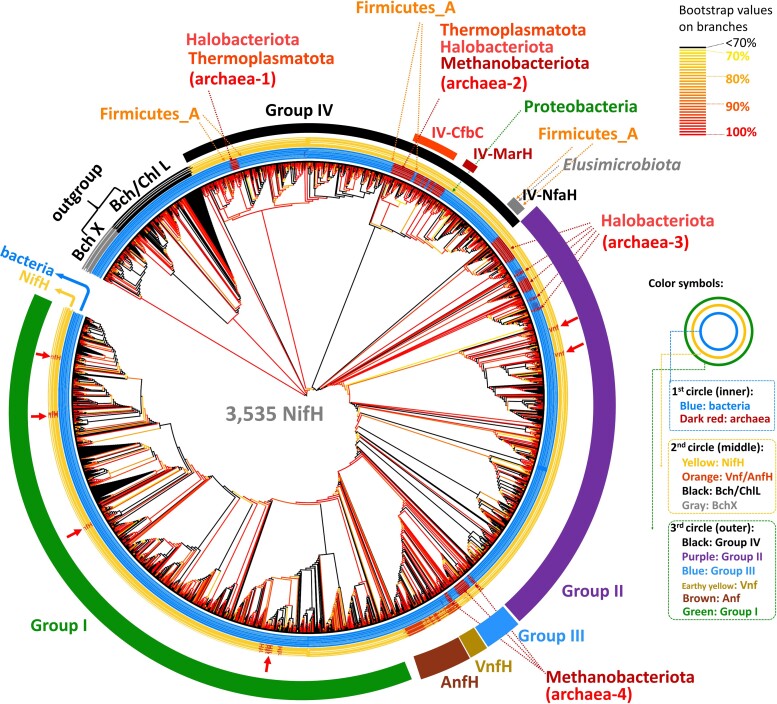
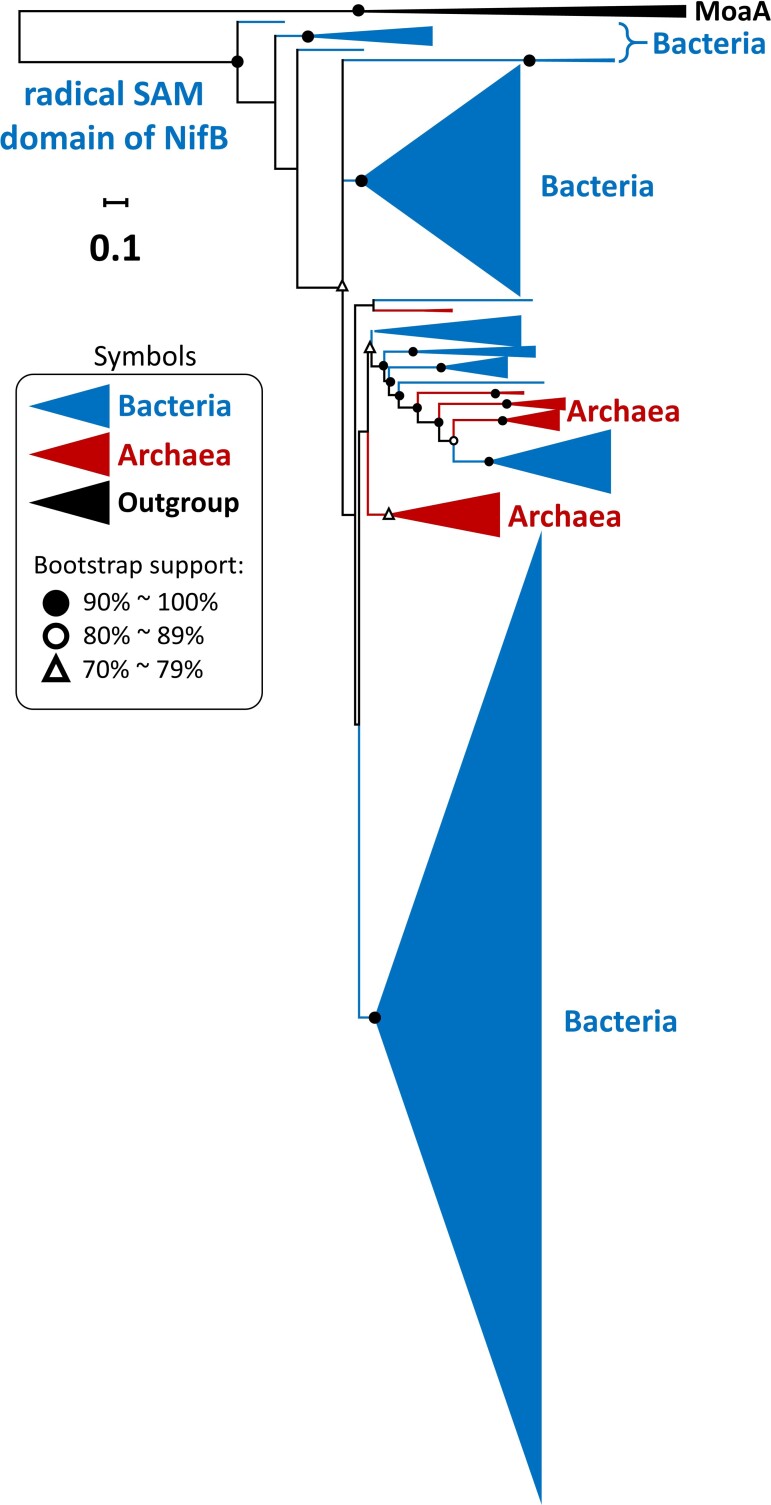
Fig. 1.
Simplified phylogeny of 3,535 Nif/Vnf/AnfH sequences. The scale bar denotes 0.1 amino acid substitutions per residue site. The solid and empty black circles denote the bootstrap supports of 90–100% and 80–89%, respectively, and the empty triangle denotes a bootstrap value of 70–79%. The outgroup is labeled by black color. The bacterial group is labeled by blue color and archaeal groups by dark red color. We use the light-independent protochlorophyllide reductase subunit L (Bch/ChlL) and chlorophyllide a reductase subunit X (BchX) sequences as the outgroup. We use the IQ-TREE method to reconstruct phylogenetic trees and ModelFinder to select the best-fit model of protein sequence evolution with ultrafast bootstrap (1,000 replicates). The model selected according to the BIC is LG + R10.
In figure 1, bacterial sequences (labeled by blue color) form well-supported earliest lineages while archaeal sequences (labeled by red color) are all nested inside bacterial sequences. This result suggests that NifH first evolved in bacteria. Moreover, the Vnf/AnfH sequences appear in the latest lineages, consistent with the previous inference that V- and Fe-nitrogenases evolved later than Mo-nitrogenase (Mus et al. 2018, 2019; Garcia et al. 2020).
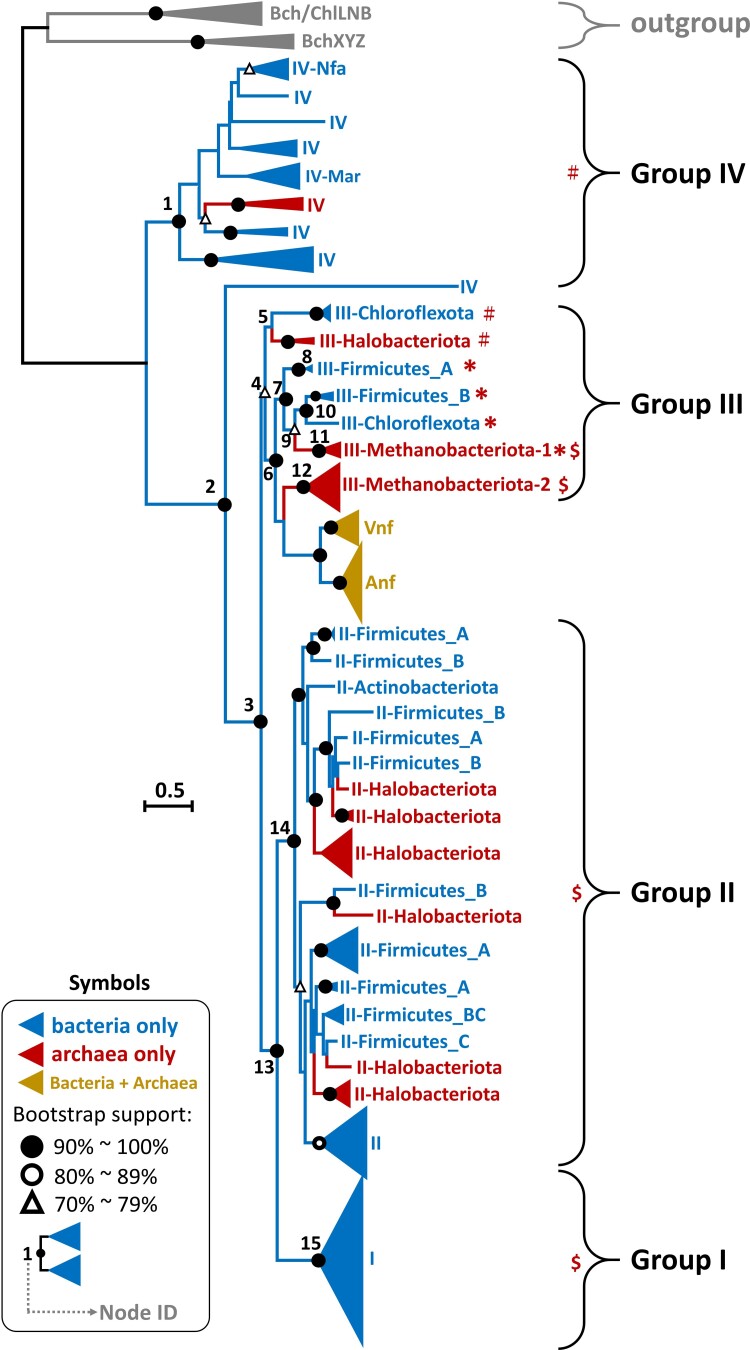
To show more details of the phylogeny of Nif/Vnf/AnfH proteins, we classify the protein sequences under study into Groups I, II, III, IV, Vnf, and Anf according to previous studies (Raymond et al. 2004; Howard et al. 2013; Zheng, Dietrich, et al. 2016; North et al. 2020) (fig. 2, supplementary fig. S1, Supplementary Material online). Figure 2 indicates that Group IV is the oldest group and Groups II, III/Vnf/Anf, and I are younger. As the sequences of archaea-1 and -2 are nested inside bacterial Groups IV sequences, none of them are likely the ancestral sequences. The archaeal sequences in archaea-3 are split into five subgroups but all belong to Halobacteriota. One of Halobacteriota subgroups is the early lineage in Group II, while the other four are nested inside Group II bacterial sequences, implying five different HGT events between bacteria and Halobacteriota because in each case the gene tree is drastically different from the species tree. Bacterial sequences form the early lineage in Group III, and the archaea-4 sequences, most of which belong to Methanobacteriota, are nested inside Group III bacterial sequences. Group I contains only bacteria, so its common ancestor should be a bacterium. In Group Vnf/AnfH, bacterial sequences form the earliest lineages and the archaeal Vnf/AnfH sequences are nested inside bacterial Vnf/AnfH sequences, implying that Vnf/AnfH first evolved in bacteria. Additionally, most Vnf/AnfH sequences are located next to Group III while some Vnf/AnfH sequences are nested inside Groups I and II NifH sequences (the red arrows in fig. 2) as in a previous study (Angel et al. 2018). This finding suggests that Vnf/AnfH evolved from the duplication of NifH as previous proposed (Mus et al. 2018; Garcia et al. 2020).
Fig. 2.
Expanded phylogeny of Nif/Vnf/AnfH sequences. Figure 2 is an expanded version of figure 1. Branches are colored according to their bootstrap values; see the color bar on the top left. Bacteria and archaea are labeled in blue and dark red on the first (inner) circle. In the second circle, NifH is labeled by yellow while Vnf/AnfH by orange. The protein sequences are classified into Groups I, II, III, IV, Vnf, and Anf, which are labeled in different colors on the third (outer) circle. We use the IQ-TREE method to reconstruct the phylogenetic tree and ModelFinder to select the best-fit model of protein sequence evolution with ultrafast bootstrap (1,000 replicates). The model selected is LG + R10.
Phylogenetic Tree of Nif/Vnf/AnfD, K, E, and N Proteins
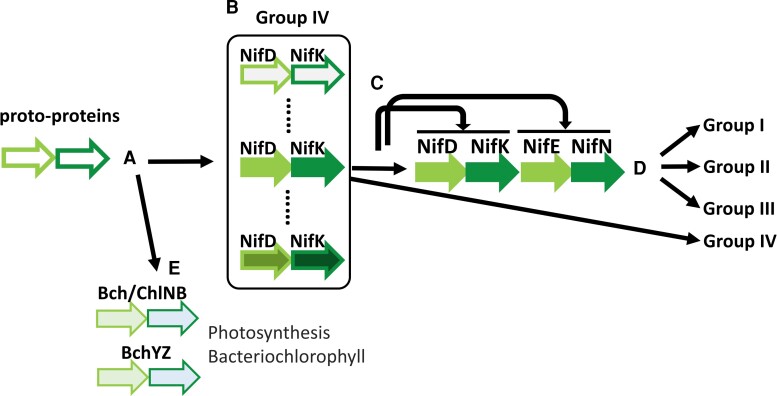
It has been proposed that NifDK were the ancestral sequences and that NifEN were derived from a duplication of NifDK (Fani et al. 2000; Raymond et al. 2004; Boyd, Anbar, et al. 2011; Dos Santos et al. 2012; Garcia et al. 2020). We call this hypothesis the NifDK-first hypothesis. This hypothesis can be depicted by figure 3, in which two linked proto-proteins evolved in one lineage into Bch/ChlNB and BchYZ (Muraki et al. 2010; Boyd, Anbar, et al. 2011; Garcia et al. 2020), and in another lineage into NifDK. NifDK then expanded into a group of NifDK sequences (Group IV), one of which was duplicated to become NifDKEN, which then evolved into Groups I, II, and III. Boyd, Anbar, et al. (2011) constructed a phylogeny of ∼150 Nif/Vnf/AnfD, K, E, and N protein sequences from 40 genomic sequences and proposed the archaea-first hypothesis. We reconstruct a phylogeny of 7,421 Nif/Vnf/AnfD, K, E, and N sequences from 1,489 genomes (fig. 4A), using the IQ-tree software (Minh et al. 2020) and using 20 Bch/ChlN and Bch/ChlB sequences as the outgroup as in a previous study (Garcia et al. 2020). The best-fit model of protein sequence evolution is LG + F + R10. Supplementary figure S2, Supplementary Material online is a detailed version of figure 4.
Fig. 3.
Proposed model for the origins and evolution of NifD, K, E, and N sequences. The NifDK-first hypothesis postulates: (A) Originally there were two linked proto-protein genes that evolved to NifDK in one lineage and to Bch/ChlNB and BchYZ in another lineage. (B) The NifDK was expanded to Group IV sequences. (C) One of the NifDK sequences in Group IV was tandemly duplicated to become NifDKEN. In this scheme, NifE was derived from NifD and NifN was derived from NifK. (D) NifDKEN was expanded into Groups I, II, and III, whereas Group IV, which contains only NifDK sequences but no NifEN, continue to evolve.
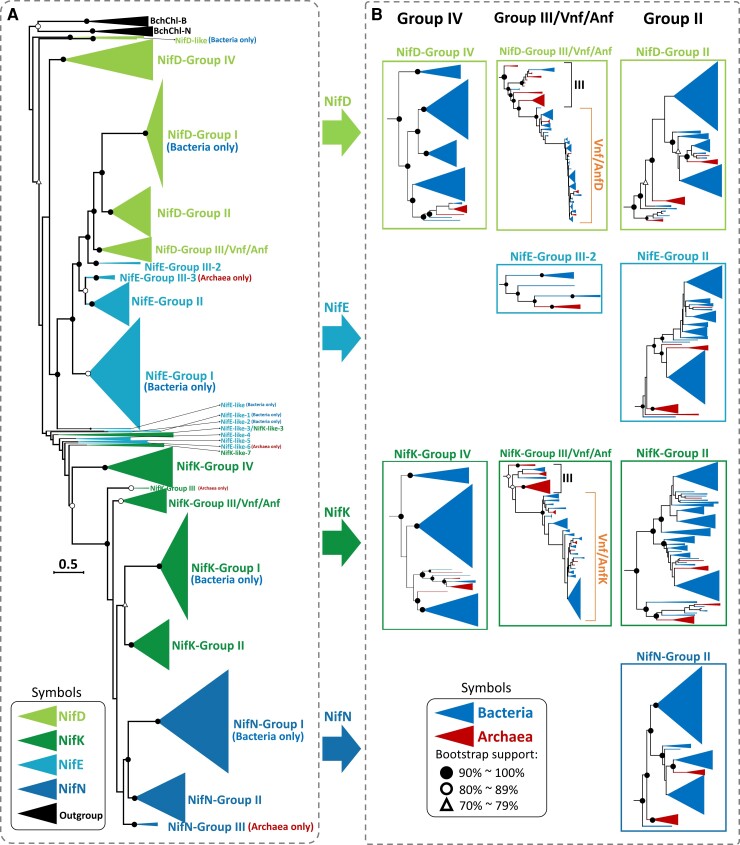
Fig. 4.
Phylogeny of 7,421 Nif/Vnf/AnfD, K, E, and N protein sequences. The scale bar denotes 0.5 amino acid substitutions per residue site. The solid and empty black circles denote the bootstrap values of 90–100% (solid circle) and 80–89% (empty circle) and the empty triangle denotes a bootstrap value of 70–79%. The light-independent protochlorophyllide reductase subunit N (Bch/ChlN) and light-independent protochlorophyllide reductase subunit B (Bch/ChlB) are used as the outgroup. The protein sequences are classified into Groups I, II, III, IV, Vnf, and Anf. (A) The outgroup is labeled by black color, NifD by light green, NifK by dark green, NifE by light blue, and NifN by dark blue. (B) is an expanded version of each subtree contained both bacteria and archaea in (A). The bacterial group is labeled by blue color and archaeal groups by dark red color. We use the IQ-TREE method to reconstruct the phylogenetic tree and ModelFinder to select the best-fit model of protein sequence evolution with ultrafast bootstrap (1,000 replicates). The model selected is LG + F + R10.
In figure 4A, we label NifD by light green, NifK by dark green, NifE by light blue and NifN by dark blue. To make it easier to understand, we divided the Nif protein sequences into Groups I, II, III, and IV as in previous studies (Raymond et al. 2004; Howard et al. 2013; Zheng, Dietrich, et al. 2016; Méheust et al. 2020; North et al. 2020; Pan et al. 2022). Inside each group, the bacterial sequences are labeled by dark blue while the archaeal sequences by dark red (fig. 4B). The earliest lineages in Group I (NifD, K, E, and N), Group II (NifD, K, E, and N), and Group III-2 (NifE) are bacterial sequences while archaeal sequences appear as the earliest lineages only in Group III (NifD and K), Group III-3 (NifE), and Group III (NifN). These observations suggest that NifD, K, E, and N all first evolved in bacteria.
It has been proposed that the Vnf/AnfDK were derived from NifDK by gene duplication (Mus et al. 2018; Garcia et al. 2020). This hypothesis is supported by our data, because NifDK and Vnf/AnfDK are found in the same genome (supplementary data S1, Supplementary Material online) and many bacterial species in Group I-DK and Group II-DK also carry Vnf/AnfDK. Moreover, in Group Vnf/Anf bacterial sequences form the earliest lineages and the Vnf/AnfDK sequences of archaea are nested inside Vnf/AnfDK sequences of bacteria (fig. 4B and supplementary fig. S2, Supplementary Material online). These observations suggest that Vnf/AnfDK first evolved in bacteria.
Figure 4 A largely supports the NifDK-first hypothesis because Group IV (NifD and K) sequences are the ancestral sequences, as suggested by many previous studies (Raymond et al. 2004; Staples et al. 2007; Howard et al. 2013; North et al. 2020) and because the NifD, K, E, N of Groups III, II, and I were derived from NifD, K of Group IV. Indeed, the lower half of figure 4A suggests that an ancestral K (in Group IV) was duplicated into NifK and NifN, which were then expanded into Groups I, II, and III. The only problem with figure 4A is that Groups I, II, and III NifD sequences are clustered with Groups II and III NifE sequences. Only this part of figure 4A supports the NifEN-first hypothesis, which postulates that NifEN were the ancestral sequences and that NifDK were a duplication of NifEN (GBE 2022; Garcia et al. 2022).
To see why Groups I, II, and III NifD sequences are clustered with Groups II and III NifE sequences in figure 4A, we construct a tree of NifD and NifE sequences (fig. 5A) and a tree of NifK and NifN sequences (fig. 5B), using the full-version maximum likelihood (ML) method (Mega X tool) (Le and Gascuel 2008; Kumar et al. 2018) instead of the IQ-tree method. To reduce the computational load, we select only 215 species (supplementary data S2, Supplementary Material online); we retain most of the archaea species in figure 4A that contain NifDK and eliminate mainly Group I sequences, which are all bacterial. The best-fit model of protein evolution is LG + G. Figure 5AandB shows that Group IV NifD and NifK sequences are ancestral to the other sequences in the tree, in agreement with the NifDK-first hypothesis and with previous studies (Raymond et al. 2004; Boyd, Hamilton, et al. 2011; Mus et al. 2019).
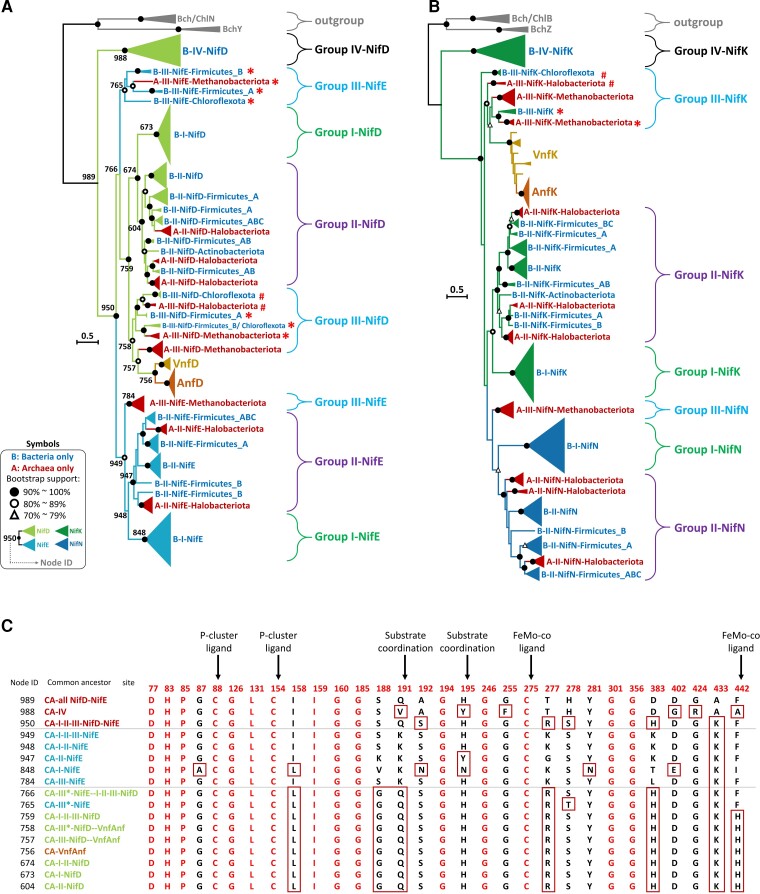
Fig. 5.
Phylogenies of NifD and NifK sequences and of NifE and NifN sequences. (A) The phylogeny of 487 Nif/Vnf/AnfD and E sequences. (B) The phylogeny of 476 Nif/Vnf/AnfD and E sequences. The scale bar denotes 0.5 amino acid substitutions per residue site. The solid and empty black circles denote the bootstrap values of 90–100% and 80–89%, respectively, and the empty triangle denotes a bootstrap value of 70–79%. The protein sequences are classified into Groups I, II, III, IV, Vnf, and Anf. We label archaeal groups by dark red. The four-nif-gene set is labeled by # and the five-nif-gene set is labeled by *. We use 6 BchY, Z sequences and 10 Bch/Chl N, B sequences as the outgroup. (C) The ancestral sequences at the branch nodes numbered in (A). The residues on the sequences are numbered according to the Azotobacter vinelandii proteins. We use the full-version ML method to construct the tree and ModelFinder to select the best-fit model of protein sequence evolution with bootstrap (500 replicates). The model selected is LG + G.
Figure 5 A is consistent with the NifDK-first hypothesis, except that a small group of Group III NifE sequences (indicated by *) is clustered with the NifD sequences. This group includes Firmicutes_A, Firmicutes_B, Chloroflexota, and Methanobacteriota, which all lack NifN. A previous study found that the NifE* of Caldicellulosiruptor spp. (Firmicutes_A) showed low similarities with other known NifE’s (Chen et al. 2021). To see why this happened, we inferred the ancestral sequence at each branching node of the Nif/Vnf/AnfD, E tree (fig. 5A) and found that the common ancestor of NifE* (node 765) is more similar to NifD than to the other NifE sequences. To illustrate this point, figure 5C shows the alignment of ancestral sequences at 30 amino acid positions, which, according to previous studies (Raymond et al. 2004; Howard et al. 2013; Zheng, Dietrich, et al. 2016; North et al. 2020), are functionally important. Note that sites 188, 191, 277, and 383 of NifE* are the same as all NifD sequences but different from all other NifE sequences. This observation suggests that the substrate coordination and P-cluster ligand of NifE* are more similar to NifD than NifE. Because the NifE* sequences are more similar to NifD sequences than NifE sequences, they were clustered with the Groups III, II, and I NifD sequences in figure 5A.
Figure 5 B is completely consistent with the NifDK-first hypothesis as depicted in figure 3. It provides strong evidence against the NifEN-first hypothesis because it clearly indicates that the NifN sequences were derived from a NifK sequence.
The NifEN-first hypothesis is contrary to the observations that Group IV NifD and NifK were the ancestral sequences of Groups I, II, and III NifD, K, E, N sequences (fig. 5AandB) and that in both figure 5AandB Group IV includes no NifE and NifN sequences. These two observations were not made by Garcia et al. (2022), because their study included no Group IV NifD and NifK sequences.
Phylogenetic Tree of the Radical SAM Domains of NifB Proteins
Boyd, Anbar, et al. (2011) inferred that the earliest lineages in the phylogeny of the 30 radical SAM domains of NifB proteins were archaeal sequences (Boyd, Anbar, et al. 2011). Here, we reconstruct a phylogeny of 1,712 radical SAM domains of NifB proteins (fig. 6 and supplementary fig. S3, Supplementary Material online); we use 13 MoaA sequences as the outgroup as in Boyd, Anbar, et al. (2011); we use the IQ-tree software (Minh et al. 2020). The best-fit model of protein sequence evolution is LG + R10.
Fig. 6.
Phylogeny of the radical SAM domains of 1,712 NifB sequences. The scale bar denotes 0.1 amino acid substitutions per residue site. The two black circles denote the bootstrap values of 90–100% (solid circle) and 80–89% (empty circle) and the empty triangle denotes a bootstrap value of 70–79%. We use the radical SAM domains of molybdenum biosynthesis protein (MoaA) sequences as the outgroup. The outgroup is labeled by black color. The bacterial group is labeled by blue color and the archaeal groups by dark red color. We use the IQ-TREE method to reconstruct the phylogenetic tree and ModelFinder to select the best-fit model of protein sequence evolution with ultrafast bootstrap (1,000 replicates). The model selected is LG + R10.
In figure 6, many of the nodes have low bootstrap values. However, it is clear that the earliest lineages are bacterial sequences (blue color) and the archaeal sequences (red color) are all nested inside bacterial sequences. This result suggests that NifB first evolved in bacteria. Moreover, our result indicates that many NifB proteins of Firmicutes_B species, which contain only the SAM domain (supplementary fig. S3, Supplementary Material online), form the earliest lineages. A previous study inferred that Methanococcus spp. (Methanobacteriota phylum) formed the earliest lineages in the phylogeny of radical SAM domains of NifB proteins (Boyd, Anbar, et al. 2011) and the bacterial species, which contain the fused “SAM–NifX” proteins (the orange small box in supplementary fig. S3, Supplementary Material online), appeared in later lineages. In our species collection, almost every bacterial phylum contains both types of NifB (with only the SAM domain or with the fused “SAM–NifX” protein) as in a previous study (Arragain et al. 2017). Moreover, in our inferred phylogeny, the SAM domain sequences of Methanococcus spp. are nested inside the NifB sequences of bacteria, which contain only the SAM domain (supplementary fig. S3, Supplementary Material online).
Phylogenetic Tree of Concatenated Nif/Anf/VnfHDK Proteins
We concatenate the NifH, D, K sequences (structural components of nitrogenase) because concatenation increases the number of amino acid positions to infer the tree (Boyd and Peters 2013; Boyd et al. 2015; Garcia et al. 2020). We use the ML method with LG + G + I as the model of protein sequence evolution to construct the phylogeny of 326 concatenated Nif/Vnf/AnfHDK sequences (fig. 7). We also construct an IQ-tree using 1,487 concatenated Nif/Vnf/AnfHDK sequences (supplementary fig. S4, Supplementary Material online) and find that the two trees have similar topologies.
Fig. 7.
The phylogeny of 326 concatenated Nif/Vnf/AnfHDK sequences. The protein sequences are classified into Groups I, II, III, IV, Vnf, and Anf. We use the full-version ML method to construct the tree and ModelFinder to select the best-fit model of protein sequence evolution with bootstrap (500 replicates). The model selected is LG + G + I. The scale bar denotes 0.5 amino acid substitutions per residue site. The solid and empty black circles denote the bootstrap values of 90–100% and 80–89%, respectively, and the empty triangle denotes a bootstrap value of 70–79%. The bacteria are labeled by blue color and the archaea by dark red color. The Vnf/Anf groups contain both bacteria and archaea, labeled by earthy yellow. The four-nif-gene set is labeled by #, the five-nif-gene set by *, and the six-nif-gene set by $. We use 6 concatenated BchXYZ sequences and 10 concatenated Bch/Chl LNB sequences as the outgroup.
In figure 7, Group IV is the oldest group and Groups III, II, and I are younger, as in the NifH tree (figs. 1 and 2) and the NifD, K, E, N trees (figs. 4A, 5AandB). Note that in each group (I, II, III, IV, Vnf and Anf), bacterial sequences form well-supported earliest lineages while archaeal sequences are nested inside bacterial sequences. Thus, these three nif genes likely first evolved in bacteria. Most of the bacterial species that appear in the earliest lineages belong to Firmicutes_A or Firmicutes_B phyla (fig. 7 and supplementary fig. S4, Supplementary Material online). Moreover, Group IV species and the earliest lineages of Group III Chloroflexota (indicated by #) contain no NifE and NifN, implying that NifDK is older than NifEN. Thus, this tree also supports the NifDK-first hypothesis.
Group I contains only bacteria, so its common ancestor should be a bacterium (Firmicutes_B form the early lineages, supplementary fig. S4, Supplementary Material online). The majority of bacterial species in the earliest lineages of Group II belong to Firmicutes_A or Firmicutes_B (fig. 7 and supplementary fig. S4, Supplementary Material online).
Association of the Mo Transporter With Nitrogen Fixation
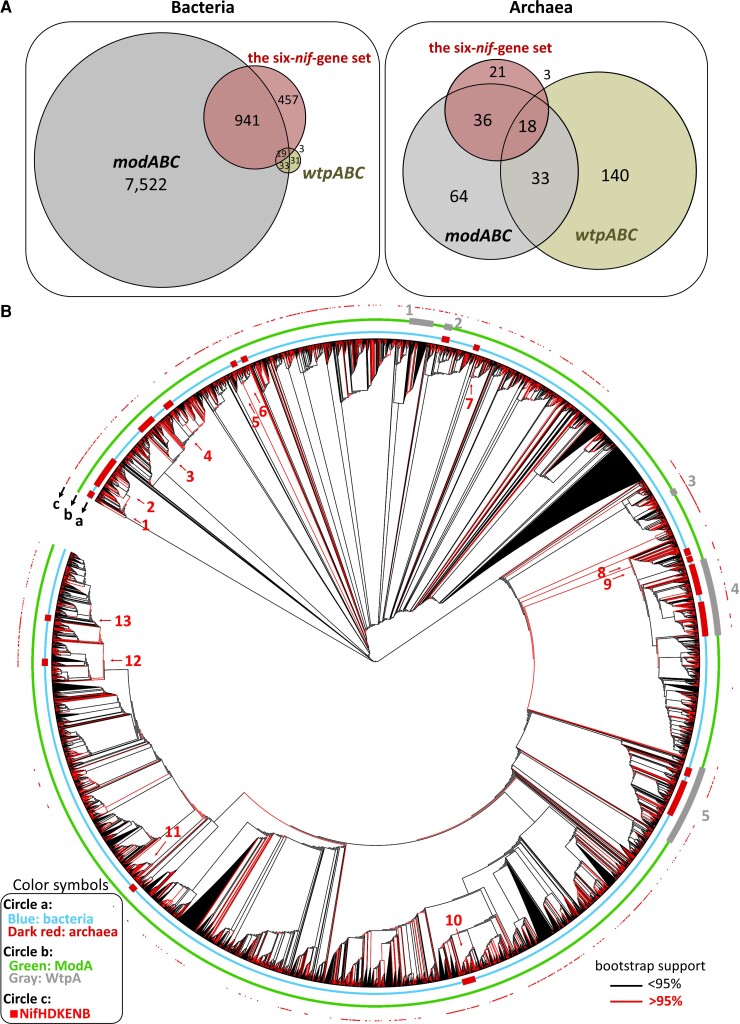
We identify the modABC- and wtpABC-gene-harboring species (supplementary data S3, Supplementary Material online) and study their association with nitrogen fixation (i.e., the six-nif-gene set). ModABC is the major Mo transporter in both Nif bacteria and Nif archaea (fig. 8A). Among the 1,420 bacterial species that carry the six-nif-gene set (table 1), 960 (941 + 19) are associated with ModABC whereas only 22 (19 + 3) are associated with WtpABC (see fig. 8A). Similarly, among the 78 archaeal species that carry the six-nif-gene set (table 1), 54 (36 + 18) are associated with ModABC whereas only 21 (3 + 18) are associated with WtpABC (fig. 8A). Moreover, most wtpABC-gene-harboring Nif archaea also harbor modABC genes (18/21). Under the archaea-first hypothesis, the six-nif-gene set first evolved in archaea, so it should be associated more often with wtpABC than with modABC, but the opposite is true in both archaea and bacteria. A recent study suggested that purple sulfur bacteria (Proteobacteria) could fix nitrogen and used ModABC transporter as early as 2.5–0.5 Ga (Philippi et al. 2021). As purple sulfur bacteria are not the oldest Nif species (Group I), there could be even older Nif bacteria that carried the modABC genes. The above observations strongly support the bacteria-first hypothesis.
Fig. 8.
The distribution of ModABC and WtpABC in bacteria and archaea, and the phylogeny of ModA and WtpA sequences. (A) The Venn diagrams of Nif species (red color), modABC-gene-harboring species (gray color) and wtpABC-gene-harboring species (olive green). (B) The phylogeny of 10,119 ModA and 829 WtpA sequences. In Circle a, the bacterial groups are labeled by blue and the archaeal groups by dark red. In Circle b, ModA groups are labeled by green and WtpA groups by gray. In Circle c, the Nif species are labeled by small red squares. The tree is unrooted, but for simplicity of presentation we took one group of ModA sequences as the outgroup. The number of ultrafast bootstrap replicates was 1,000. A red line (branch) means a bootstrap value of ≧95%, whereas a black line means a bootstrap value of <95%.
We reconstruct a phylogeny of 10,119 ModA and 829 WtpA sequences, which represent all available ModA and WtpA sequences in our collection (fig. 8B). ModA (WtpA) is the molybdate-binding protein component of the ModABC (WtpABC) transporter and is the first key protein involved in the absorption of molybdate by prokaryotes. We use the IQ-tree software (Minh et al. 2020) and the best-fit model (Q.pfam + R7) of protein sequence evolution to construct the tree. Our tree topology is similar to that of a previous tree of 4,623 ModA/WtpA sequences (Ge et al. 2020). In our tree (fig. 8B), the branches are represented by red and black lines, which indicate bootstrap support of ≧95% and <95%, respectively. Thus, the many clades that are preceded by a red branch are supported by a bootstrap value of ≧95% and so are likely reliable. In Circle b, each gray segment represents a group of WtpA sequences. As all of the five gray segments are deeply nested inside the green segments (ModA sequences), a simple explanation is that the WtpA sequences were derived from ModA sequences. A previous study also suggested that WtpA evolved later than ModA (Aguilar-Barajas et al. 2011). In Circle a, each dark red segment represents an archaeal clade. As all of the 13 dark red (archaeal) clades that carry ModA are nested inside the bacterial ModA sequences (blue segments), the archaeal ModA sequences were likely derived from bacterial ModA sequences, probably via horizontal gene transfers (HGTs). Thus, figure 8 supports the bacteria-first hypothesis.
The dispersal of the gray segments on Circle b might have three possible non-exclusive explanations. First, the dispersal might be partly due to tree reconstruction errors. This is, however, unlikely the case for those segments that are far separated on the phylogeny because such separations are supported by very high bootstrap values and so are unlikely to have occurred by chance. Second, some gray segments had evolved independently from green segments. Third, gene conversion had occurred between gray segments and green segments and distorted the phylogenetic relationships. The relative likelihoods of the second and the third scenario are difficult to tell. The observation that some gray segments are carried by archaea while the others by bacteria suggests that HGTs had occurred between archaea and bacteria. For example, the fourth gray segment contains a subsegment of bacterial WtpA sequences (blue color in Circle a) that lies in between two archaeal gray subsegments (dark red in Circle a), so it was likely derived from archaeal sequences via a HTG.
Discussion
In this study, we have used a large number of bacterial and archaeal genomes to conduct an in-depth study of the origin and evolution of nitrogen fixation. We identified many more Nif prokaryotes than did previous studies (Poudel et al. 2018; Albright et al. 2019; Mus et al. 2019; Koirala and Brözel 2021). Our data suggest that many HGT events have occurred between bacteria and archaea but the majority of them were apparently from bacteria to archaea because most archaeal nif genes are phylogenetically nested inside bacterial nif genes (figs. 1, 2 and 4–7). Moreover, we found that while the majority of archaea use WtpABC as the Mo transporter, the majority of Nif archaeal species, like bacteria, use the ModABC as the Mo transporter (fig. 8A). This finding strongly supports the bacteria-first hypothesis. Furthermore, our phylogeny of ModA and WtpA protein sequences shows that the archaeal sequences are nested inside bacterial sequences (fig. 8B), providing further support for the bacteria-first hypothesis.
In constructing the concatenated NifHDK tree, we used only linked HDK sequences, so that the three genes have the same evolutionary history. The NifHDK tree (fig. 7) is congruent with the NifH, NifD and NifK trees (figs. 1, 5AandB) in that Group IV is older than Groups I, II, and III. It is also congruent with the NifD and NifK trees (fig. 5AandB) in that Groups I and II are joined together before they are jointed to Group III, that is ([Group I, Group II], Group III), which is the same as in previous studies (Enkh-Amgalan et al. 2006; Boyd and Peters 2013; Garcia et al. 2020; Chen et al. 2021). It is incongruent with the NifH tree in that the NifH tree shows ([Group I, Group III], Group II), which is the same as in previous studies (Angel et al. 2018; Nishihara, Thiel, et al. 2018; Pan et al. 2022). The observation that concatenation of NifH, NifD, and NifK leads to ([Group I, Group II], Group III) suggests that NifD and NifK together provide stronger phylogenetic signals for the branching order of Groups I, II, and III than does NifH. Note that in all of these trees, Group IV is the oldest and that in Group IV the archaeal sequences are all nested inside bacterial sequences, so that no archaeal sequence was likely the ancestral sequence of Groups III, II, and I. As most nodes in the NifHDK tree (fig. 7) are supported by a higher than 90% bootstrap value, the tree appears to be largely reliable. We therefore will include this tree in the following inferences.
The NifH, NifDE, and NifKN trees (figs. 1, 5AandB) suggest that NifH, NifD, and NifK first evolved in bacteria (Group IV). Moreover, the NifB tree (fig. 6) suggests that NifB first evolved in bacteria because bacterial NifB sequences form the earliest lineages. Thus, the four-nif-gene set (nifHDKB) should have first evolved in bacteria. This view is supported by the NifHDK tree (fig. 7) because all Group IV genomes contain NifB as well as NifHDK. Note that Endomicrobium proavitum, which is a Group IV-Nfa bacterium, has been experimentally shown to be able to fix nitrogen (Zheng, Dietrich, et al. 2016). Moreover, several studies found that only bacteria can use Group IV-Nfa NifHDKB to fix nitrogen (Zheng, Dietrich, et al. 2016; Méheust et al. 2020; North et al. 2020; Koirala and Brözel 2021). Note further that Oscillocholris trichoides has also been shown experimentally to fix nitrogen (Ivanovsky et al. 2021). As O. trichoides belongs to Group III (Chloroflexota) while E. proavitum belongs to Group IV, there might be other species with the four-nif-gene set (nifHDKB) that can fix nitrogen. However, there seems to be no experimental evidence to show that an archaeon with nifHDKB can fix nitrogen. In our phylogenies of NifH, NifD, K, E, N, and NifHDK (supplementary figs. S1, S2 and S4, Supplementary Material online), Firmicutes_A species or Firmicutes_B species form the earliest lineages in Group IV-Nfa and E. proavitum is nested inside Group IV-Nfa sequences of Firmicutes_A (e.g., genus Clostridium). Thus, Firmicutes_A might be the first species to use Group IV-Nfa proteins (NifHDKB) for nitrogen fixation.
We now argue that the six-nif-gene set (nifHDKENB) first evolved in bacteria. In the above, we have provided evidence for the NifDK-first hypothesis. According to this hypothesis, NifEN was derived from the duplication of a Group IV NifDK (fig. 3). This NifDK should be in a bacterium because Group IV archaeal NifDK sequences were all nested inside bacterial NifDK sequences, so that no archaeal NifDK could be the ancestral sequence of Groups I, II, and III; that is, there is no branch connecting an archaeon in Group IV to the common ancestor of Groups III, II, and I. Note further that when NifDK was duplicated in a bacterium, that bacterium immediately possessed the six-nif-gene set because before the duplication it already possessed the four-nif-gene set. Thus, we propose that the six-nif-gene set first arose in bacteria.
How the five-nif-gene set (nifHDKEB) arose is less certain. It could have arisen from a duplication of NifD without the duplication of NifK, but then the gene order would have been DEK instead of DKE. A simpler scheme is that the five-nif-gene set was derived from the six-nif-gene set by losing NifN. This view is supported by the following inference. As most Groups I genomes carry the six-nif-gene set, their common ancestor (node 15 in fig. 7) would carry the six-nif-gene set. The same comment applies to the common ancestor of Group II genomes (node 14). Therefore, the common ancestor of Groups I and II (node 13) should also carry the six-nif-gene set. The nif-gene set at the common ancestor (node 4) of Group III is less certain because Group III contains species that carry 4 (denoted by #), 5 (denoted by *) or 6 (denoted by $) nif genes. In particular, the subgroup denoted as Methanobacteriota-1 contains a clade of 4 species carrying nifHDKEB and a clade of 2 species carrying nifHDKENB; we denote these two clades by Methanobacteriota-1-1 and Methanobacteriota-1-2. First, assume that the common ancestor (node 4) of Group III carried nifHDKB. Then this gene set gained nifEN to become nifHDKENB at node 6 (the assumption of nifHDKEB at node 6 can be shown to be less parsimonious). The nifHDKENB gene set then lost nifN at node 8, at node 10 and in the common ancestor of Methanobacteriota-1-1 to become nifHDKEB at these three nodes. Moreover, this scheme implies that node 3 carried nifHDKB, so it requires the gain of nifEN to explain the nifHDKENB at node 13. Thus, this scheme requires two gains of nifEN and three losses of nifN. Second, assume that node 4 carried nifHDKEB (the five gene set). As this scheme requires that nifHDKEB was derived from nifHDKB in Group IV, it requires the gain of nifE at node 4 and also a gain of nifEN at node 13 to explain nifHDKENB at node 13. As mentioned above, the gain of nifE from nifD without the simultaneous duplication of nifK would produce the gene order of DEK instead of DKE. Moreover, it requires a gain of nifN in the common ancestor of Methanobacteriota-1-2 and another gain of nifN at node 12 and a loss of nifEN at node 5. Thus, this scheme requires one gain of nifE, one gain of nifEN, two gains of nifN and one loss of nifEN. Therefore, this scheme is complex and not necessarily more parsimonious than the first scheme. Third, assume that node 4 carried nifHDKENB. This scheme requires the loss of nifEN at node 5, and the loss of nifN at node 8, node 10 and in the common ancestor of Methanobacteriota-1-1. Thus, this scheme requires one loss of nifEN, three losses of nifN and also one gain of nifEN (at node 3). Compared to the first scheme, it requires one additional loss of nifEN, but one fewer gain of nifEN. As losing nifEN is simpler than gaining nifEN at a specific location (Albalat and Cañestro 2016), the third scheme is more plausible than the first. In conclusion, it seems that nifHDKEB was derived from nifHDKENB due to a loss of nifN. Note that Caldicellulosiruptor spp, which possesses the five-nif-gene set (nifHDKEB) and has been experimentally shown to fix nitrogen (Chen et al. 2021), is a Firmicutes_A species in Group III.
In conclusion, regardless of whether the four-nif-gene set, the five-nif-gene set or the six-nif-gene is used as the criterion for nitrogen fixation, nitrogen fixation appears to have first evolved in bacteria. Thus, our study strongly favors the bacteria-first hypothesis over the archaea-first hypothesis and the LUCA-first hypothesis. We note that a previous phylogenetic analysis of protein families including nitrogenase did not support the existence of Nif LUCA (Berkemer and McGlynn 2020).
Garcia et al. (2020) proposed the following sequential evolutionary events of the nitrogenase complex formation: 1) NifHDKB, 2) NifHDKEB, 3) NifHDKENB, and 4) Vnf/AnfHDK. Step 1 is supported by our observation that Group IV is the oldest group and most of its members possess the four-nif-gene set (node 1 of fig. 7). However, Step 2 is less certain because as discussed above the five-nif-gene set could have been derived from the six-nif-gene set; in this case, Steps 2 and 3 are reversed. The NifHDK tree suggests that Vnf/AnfHDK (Step 4) was derived from a duplication of NifHDK (fig. 7).
Previous studies suggested that the Group IV-Cfb protein, which is a methane biogenesis nickel-containing tetrapyrrole (known as coenzyme F430) (Zheng, Ngo, et al. 2016; Moore et al. 2017), is the common ancestor of both Nif proteins and Bch/Chl proteins (Raymond et al. 2004; Boyd, Hamilton, et al. 2011). This hypothesis was proposed because Group IV-Cfb separated the Nif and Bch/Chl lineages in the phylogeny (Raymond et al. 2004; Boyd, Hamilton, et al. 2011). The “Cfb” is reclassified here as a subgroup of Group IV-CfbC (fig. 2) according to previous studies (Zheng, Ngo, et al. 2016; Moore et al. 2017; North et al. 2020). Because only archaea were found to contain Cfb, the archaea-first hypothesis was proposed (Boyd, Hamilton, et al. 2011; Mus et al. 2019). However, a recent study (North et al. 2020) found that another subgroup of Group IV-Mar (fig. 2) can biosynthesize methionine, ethylene, and methane, which contribute to sulfur and carbon recycling. Moreover, MAR has been found only in bacteria. With the new homologous Group IV protein sequences, our figure 2 suggests that bacterial sequences form the early lineages and archaeal sequences are nested inside bacterial sequences. Moreover, there are many bacteria that contain multiple types of these homologous proteins in their genomes; for example, Rhodospeudomonas palustris CGA009 contains Bch/Chl, IV-Mar, and Nif/Vnf/Anf proteins in its genome (North et al. 2020), implying that these homologous proteins originally evolved in bacteria. As only bacterial species contain Group IV-Nfa and -Mar sequences, this observation implies that the ancient bacteria play an important role not only in the nitrogen cycle but probably also in the carbon or even sulfur cycle.
In our phylogenetic trees, anaerobic firmicutes form the earliest lineages (supplementary figs. S1–S4, Supplementary Material online), and many of them can fix CO2 through the Wood–Ljungdahl pathway (also termed the acetyl CoA pathway) and possess the sulfate-reducing ability (supplementary data S4, Supplementary Material online). The acetyl CoA pathway has been considered the earliest metabolic pathway on earth (Martin 2020). Moreover, carbon isotope evidence consistent with the presence of the acetyl CoA pathway was found in rocks dated ∼3.95 billion years old (Tashiro et al. 2017). Therefore, we suggest that the earliest Nif species is an anaerobic firmicutes. Consistently, anaerobic firmicutes were found to play a major role in nitrogen fixation in environments analogous to the early earth. For example, thermophilic Caldicellulosiruptor spp. carrying nifH were highly abundant in alkaline sulfidic hot springs in Japan (Nishihara, Thiel, et al. 2018). Furthermore, the nitrogenase activity in an alkaline sulfidic hot spring was not affected by the addition of 2-bromo-ethane sulfonate, a specific inhibitor of methanogenesis, but was apparently inhibited by the addition of molybdate, a specific inhibitor of assimilatory sulfate reduction and sulfur disproportionation (biotransformation of elemental sulfur or thiosulfate into sulfide and sulfate), suggesting that sulfate-reducing/sulfur-metabolizing bacteria, rather than methanogenic archaea, were responsible for nitrogen fixation in the early-earth-like environments (Nishihara, Haruta, et al. 2018).
According to isotopic records, ancient anaerobic Fe-oxidizing bacteria might have evolved around 3.77–4.28 Ga (Dodd et al. 2017; Lepot 2020; Papineau et al. 2022). As Fe-oxidizing bacteria are phylogenetically nested inside Firmicutes (and Nitrospirota, Proteobacteria, Bacteroidota) (Hedrich et al. 2011; Quaiser et al. 2014; Scott et al. 2015), the common ancestor of firmicutes should be older than the Fe-oxidizing bacteria; that is, it is at least 3.77 Ga. It is thus possible that firmicutes had evolved nitrogen fixation very early during the evolution of bacteria.
The isotopic data obtained from pyrite suggested the occurrence of bacterial sulfate reduction around 3.47 billion years ago (Ga) (Reitner and Thiel 2011). From a thermodynamic perspective, the use of sulfate as an electron acceptor allows chemolithotrophic bacteria to utilize H2 at a threshold level much lower than that in chemolithotrophic methanogenic archaea (Lovley 1985) and to acquire more free energy for ATP generation and nitrogen fixation. Although Archaeoglobus spp. are sulfate-reducing archaea, they are mostly thermophilic aerotolerant anaerobes (Abreu et al. 2000) and are not considered an ancient lineage. Moreover, we did not find any Nif Archaeoglobus spp. as in two previous studies (Dahl et al. 1994; Steinsbu et al. 2010).
Recent studies reported cases of aerobic bacterial methane synthesis from methyl-phosphonate, methylamine, and betaine by cyanobacteria and aerobic proteobacteria (Bižić et al. 2020; Lepot 2020; Wang et al. 2021). Therefore, the 3.48 Ga carbon isotopic data might be from aerobic bacteria rather than archaea (Lepot 2020). Aerobic nitrogen fixation evolved only after Nif bacteria had evolved the defense against oxygen (Boyd et al. 2015; Li et al. 2019). A previous study hypothesized that members of Cyanobacteria (aerobes) evolved nitrogen fixation after archaea (Mus et al. 2019). However, recent findings indicate that the ancestor of Cyanobacteria already existed >3.63 Ga (Garcia-Pichel et al. 2019; Bižić et al. 2020; Ohmoto 2020), which is older than the record of microbial methanogenesis (3.48 Ga). Moreover, cyanobacterial nitrogen fixation can be traced back to 2.7–3.2 Ga (Thomazo et al. 2018; Yang et al. 2019). Since Cyanobacteria is only a phylum in bacteria, aerobic nitrogen fixation in bacteria might have evolved before 2.7–3.2 Ga, which is as old as the estimated minimum age of 2.77 Ga for archaea (Javaux 2019; Lepot 2020). As Nif archaea are a small fraction of archaea, nitrogen fixation could have evolved considerably later than the common ancestor of archaea. From these arguments and those above, we propose that nitrogen fixation evolved first in anaerobic bacteria and later at similar times in aerobic bacteria and archaea.
Materials and Methods
Data Collection
We downloaded the protein sequences from AnnoTree (v1.2.0) (Mendler et al. 2019) and the GTDB (Release R95) database (Parks et al. 2018, 2020). The genomic characteristics and the taxonomic classification of each selected species were downloaded from GTDB (Parks et al. 2020) (supplementary data S1, Supplementary Material online). The nitrogenase genes and other genes were downloaded from the AnnoTree database based on the functional annotation of TIGRFAM (v15.0) and the homology information of KEGG (UniRef100) (supplementary data S5, Supplementary Material online). The genes of the Wood–Ljungdahl pathway were identified according to previous studies (Adam et al. 2018, 2019; Jiao et al. 2021) (supplementary data S4, Supplementary Material online).
Identification of nif Genes
In our BLASTp search, we set the e-value threshold = 1e−5, and cutoffs of 30% percent amino acid sequence identity, 70% subject alignment sequence coverage and 70% query sequence alignment, which are more stringent than a previous study (Poudel et al. 2018). The genes collected are listed in supplementary data S1, Supplementary Material online. However, to more accurately identify nifH, nifD, nifK, nifE, nifN, and nifB genes in a genome, we increased the amino acid sequence identity from 30% to 60%. The role of each nif gene in nitrogen fixation has been described in previous studies (Rubio and Ludden 2005; Burén et al. 2020).
Because it is difficult to distinguish among the three types of nitrogenase (nif, vnf, and anf) by structural protein sequences (Hu and Ribbe 2015), we selected the major difference between conventional and alternative nitrogenase as follows. We used the γ-subunit genes (vnfG and anfG) to detect alternative nitrogenases. We also identified the vnfHDK and anfHDK genes by comparing the KEGG and TIGRFAMS databases of AnnoTree v1.2.0. In some cases, it was difficult to distinguish between vnfHDK and anfHDK genes, and we distinguished them by their clustering with vnfG or anfG in the genome.
Boyd, Anbar, et al. (2011) identified the VnfE and VnfN genes and included them in the phylogeny of NifD, NifK, NifE, and NifN sequences. These genes are named “Nitrogenase_VnfE/N_like”, “nitrogenase associated protein E/N”, or “Nitrogenase molybdenum-iron protein like” in the NCBI database. Therefore, we label these genes as “-like” in figure 4.
Identification of Mo Transporter
To collect the Mo transporter ModABC and WtpABC, we set the E-value threshold = 1e−5, and cutoffs of 60% amino acid sequence identity, 70% subject alignment sequence coverage and 70% query sequence alignment. The 60% identity cutoffs strength is same as a previous study (Ge et al. 2020). These data are also collected from AnnoTree (v1.2.0) (Mendler et al. 2019) and the GTDB (Release R95) database (Parks et al. 2018, 2020).
Phylogenetic Tree Reconstruction
Amino acid sequences were aligned using MAFFT (v7.487) (Katoh and Standley 2013). Pairwise alignments of NifH, NifD, NifK, NifE, and NifN sequences and the radical SAM domains of NifB protein sequences are listed in supplementary data S6, Supplementary Material online. The pairwise alignments were imported into a MS Excel® spreadsheet (supplementary data S6, Supplementary Material online); the sequences were numbered to correspond to the Azotobacter vinelandii proteins; and the key conserved residues of each protein sequence are according to previous studies (Howard et al. 2013; Fay et al. 2015; North et al. 2020).
We use the IQ-TREE tool (Minh et al. 2020) to reconstruct phylogenetic trees and ModelFinder (Kalyaanamoorthy et al. 2017), which is implemented in IQ-TREE, to select the best-fit model of protein sequence evolution with ultrafast bootstrap (1,000 replicates) (Hoang et al. 2018). We also use the MEGA X tool (Kumar et al. 2018) to reconstruct full-version ML (Le and Gascuel 2008) trees and ModelFinder, which is implemented in MEGA X (Kumar et al. 2018), to select the best-fit model of protein sequence evolution with bootstrap (500 replicates). The model is selected according to the BIC (Posada and Buckley 2004). Each phylogeny was annotated by the tool “Interactive tree of life (iTOL) v4” (Letunic and Bork 2019). We classify the nitrogenase genes into different groups by the pairwise sequence alignments and the phylogenetic relationships as in previous studies (Raymond et al. 2004; Howard et al. 2013; North et al. 2020).
Venn Diagrams
The area-proportional Venn diagrams in figure 8A are created by the online tool “BioVenn” (Hulsen et al. 2008). We used the GTDB IDs and names of species from supplementary data S1 and S3, Supplementary Material online to create figure 8A. The data collection is mentioned above.
Supplementary Material
Acknowledgments
We thank Dr Chun-Ping Yu for configurating the server and Ciao-Jyun Pi for configurating the Microsoft azure for bioinformatics analysis. We thank Prof. William B Whitman, Prof. William Martin, Dr Sen-Lin Tang, and Dr Chase W. Nelson for valuable suggestions. We thank the National Center for High-performance Computing (NCHC) of Taiwan for providing computational resources. The work was supported by Academia Sinica, Taiwan (AS-KPQ-109-ITAR-11) and by the Ministry of Science and Technology (MOST) in Taiwan (MOST 110-2311-B-001-035).
Contributor Information
Hong-Wei Pi, Ph.D. Program in Microbial Genomics, National Chung Hsing University and Academia Sinica, Taiwan; Biodiversity Research Center, Academia Sinica, Taipei 11529, Taiwan.
Jinn-Jy Lin, Biodiversity Research Center, Academia Sinica, Taipei 11529, Taiwan.
Chi-An Chen, Biodiversity Research Center, Academia Sinica, Taipei 11529, Taiwan; Genome and Systems Biology Degree Program, Academia Sinica and National Taiwan University, Taipei 10617, Taiwan.
Po-Hsiang Wang, Graduate Institute of Environmental Engineering, National Central University, Taoyuan 32001, Taiwan; Earth-Life Science Institute, Tokyo Institute of Technology, Tokyo 145-0061, Japan.
Yin-Ru Chiang, Biodiversity Research Center, Academia Sinica, Taipei 11529, Taiwan.
Chieh-Chen Huang, Department of Life Sciences, National Chung Hsing University, Taichung 402, Taiwan.
Chiu-Chung Young, Department of Soil and Environmental Sciences, College of Agriculture and Natural Resources, National Chung Hsing University, Taichung 402, Taiwan.
Wen-Hsiung Li, Biodiversity Research Center, Academia Sinica, Taipei 11529, Taiwan; Department of Ecology and Evolution, University of Chicago, Chicago 60637, IL, USA.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Author Contributions
H.-W.P., J.-J.L., and W.-H.L. designed the study. H.-W.P. and J.-J.L. collected the data. H.-W.P. and C.-A.C., and J.-J.L. performed the bioinformatic analyses. H.-W.P. wrote the first manuscript draft with the help of W.-H.L. W.-H.L., P.-H.W., and Y.-R.C. advised the study. W-H.L. supervised the study. All authors participated in revising the manuscript.
Data Availability
All our relevant data are provided in Supplementary files of our paper.
References
- Abreu IA, Saraiva LM, Carita J, Huber H, Stetter K, Cabelli D, Teixeira M. 2000. Oxygen detoxification in the strict anaerobic archaeon Archaeoglobus fulgidus: superoxide scavenging by neelaredoxin. Mol Microbiol. 38:322–334. [DOI] [PubMed] [Google Scholar]
- Adam PS, Borrel G, Gribaldo S. 2018. Evolutionary history of carbon monoxide dehydrogenase/acetyl-CoA synthase, one of the oldest enzymatic complexes. Proc Natl Acad Sci U S A. 115:E1166–E1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adam PS, Borrel G, Gribaldo S. 2019. An archaeal origin of the Wood–Ljungdahl H 4 MPT branch and the emergence of bacterial methylotrophy. Nat Microbiol. 4:2155–2163. [DOI] [PubMed] [Google Scholar]
- Affourtit J, Zehr J, Paerl H. 2001. Distribution of nitrogen-fixing microorganisms along the Neuse River Estuary, North Carolina. Microb Ecol. 41:114–123. [DOI] [PubMed] [Google Scholar]
- Aguilar-Barajas E, Díaz-Pérez C, Ramírez-Díaz MI, Riveros-Rosas H, Cervantes C. 2011. Bacterial transport of sulfate, molybdate, and related oxyanions. Biometals 24:687–707. [DOI] [PubMed] [Google Scholar]
- Albalat R, Cañestro C. 2016. Evolution by gene loss. Nat Rev Genet. 17:379. [DOI] [PubMed] [Google Scholar]
- Albright MB, Timalsina B, Martiny JB, Dunbar J. 2019. Comparative genomics of nitrogen cycling pathways in bacteria and archaea. Microb Ecol. 77:597–606. [DOI] [PubMed] [Google Scholar]
- Angel R, Nepel M, Panhölzl C, Schmidt H, Herbold CW, Eichorst SA, Woebken D. 2018. Evaluation of primers targeting the diazotroph functional gene and development of NifMAP–a bioinformatics pipeline for analyzing nifH amplicon data. Front Microbiol. 9:703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arragain S, Jiménez-Vicente E, Scandurra AA, Burén S, Rubio LM, Echavarri-Erasun C. 2017. Diversity and functional analysis of the FeMo-cofactor maturase NifB. Front Plant Sci. 8:1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkemer SJ, McGlynn SE. 2020. A new analysis of archaea-bacteria domain separation: variable phylogenetic distance and the tempo of early evolution. Mol Biol Evol. 37(8):2332–2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bižić M, Klintzsch T, Ionescu D, Hindiyeh M, Günthel M, Muro-Pastor AM, Eckert W, Urich T, Keppler F, Grossart H-P. 2020. Aquatic and terrestrial cyanobacteria produce methane. Sci Adv. 6:eaax5343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd E, Anbar A, Miller S, Hamilton T, Lavin M, Peters J. 2011. A late methanogen origin for molybdenum-dependent nitrogenase. Geobiology 9:221–232. [DOI] [PubMed] [Google Scholar]
- Boyd ES, Costas AMG, Hamilton TL, Mus F, Peters JW. 2015. Evolution of molybdenum nitrogenase during the transition from anaerobic to aerobic metabolism. J Bacteriol. 197:1690–1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd ES, Hamilton TL, Peters JW. 2011. An alternative path for the evolution of biological nitrogen fixation. Front Microbiol. 2:205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd E, Peters JW. 2013. New insights into the evolutionary history of biological nitrogen fixation. Front Microbiol. 4:201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burén S, Jiménez-Vicente E, Echavarri-Erasun C, Rubio LM. 2020. Biosynthesis of nitrogenase cofactors. Chem Rev. 120(12):4921–4968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Nishihara A, Haruta S. 2021. Nitrogen-fixing ability and nitrogen fixation-related genes of thermophilic fermentative bacteria in the genus Caldicellulosiruptor. Microbes Environ. 36:ME21018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl C, Speich N, Trüper HG. 1994. Enzymology and molecular biology of sulfate reduction in extremely thermophilic archaeon Archaeoglobus fulgidus. Methods Enzymol. 243:331–349. [DOI] [PubMed] [Google Scholar]
- de Lajudie PM, Young JPW. 2017. International committee on systematics of prokaryotes subcommittee for the taxonomy of rhizobium and agrobacterium minutes of the meeting, Budapest, 25 August 2016. Int J Syst Evol Microbiol. 67:2485–2494. [DOI] [PubMed] [Google Scholar]
- Demtröder L, Narberhaus F, Masepohl B. 2019. Coordinated regulation of nitrogen fixation and molybdate transport by molybdenum. Mol Microbiol. 111:17–30. [DOI] [PubMed] [Google Scholar]
- Dodd MS, Papineau D, Grenne T, Slack JF, Rittner M, Pirajno F, O’Neil J, Little CT. 2017. Evidence for early life in Earth’s oldest hydrothermal vent precipitates. Nature 543:60–64. [DOI] [PubMed] [Google Scholar]
- Dos Santos PC, Fang Z, Mason SW, Setubal JC, Dixon R. 2012. Distribution of nitrogen fixation and nitrogenase-like sequences amongst microbial genomes. BMC Genomics 13:162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enkh-Amgalan J, Kawasaki H, Seki T. 2006. Molecular evolution of the nif gene cluster carrying nifI1 and nifI2 genes in the Gram-positive phototrophic bacterium Heliobacterium chlorum. Int J Syst Evol Microbiol. 56:65–74. [DOI] [PubMed] [Google Scholar]
- Erisman JW, Sutton MA, Galloway J, Klimont Z, Winiwarter W. 2008. How a century of ammonia synthesis changed the world. Nat Geosci. 1:636. [Google Scholar]
- Fani R, Gallo R, Lio P. 2000. Molecular evolution of nitrogen fixation: the evolutionary history of the nifD, nifK, nifE, and nifN genes. J Mol Evol. 51:1–11. [DOI] [PubMed] [Google Scholar]
- Fay AW, Wiig JA, Lee CC, Hu Y. 2015. Identification and characterization of functional homologs of nitrogenase cofactor biosynthesis protein NifB from methanogens. Proc Natl Acad Sci U S A. 112:14829–14833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Pichel F, Lombard J, Soule T, Dunaj S, Wu SH, Wojciechowski MF. 2019. Timing the evolutionary advent of cyanobacteria and the later Great Oxidation Event using gene phylogenies of a sunscreen. MBio 10(3):e00561-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia AK, Kolaczkowski B, Kaçar B. 2022. Reconstruction of nitrogenase predecessors suggests origin from maturase-like proteins. Genome Biol Evol. 14:evac031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia AK, McShea H, Kolaczkowski B, Kaçar B. 2020. Reconstructing the evolutionary history of nitrogenases: evidence for ancestral molybdenum-cofactor utilization. Geobiology 18:394–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge X, Thorgersen MP, Poole FL, Deutschbauer AM, Chandonia J-M, Novichkov PS, Gushgari-Doyle S, Lui LM, Nielsen T, Chakraborty R. 2020. Characterization of a metal-resistant Bacillus strain with a high molybdate affinity ModA from contaminated sediments at the Oak Ridge reservation. Front Microbiol. 11:587127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamelin J, Fromin N, Tarnawski S, Teyssier-Cuvelle S, Aragno M. 2002. Nifh gene diversity in the bacterial community associated with the rhizosphere of Molinia coerulea, an oligonitrophilic perennial grass. Environ Microbiol. 4:477–481. [DOI] [PubMed] [Google Scholar]
- Hamilton TL, Boyd ES, Peters JW. 2011. Environmental constraints underpin the distribution and phylogenetic diversity of nifH in the Yellowstone geothermal complex. Microb Ecol. 61:860–870. [DOI] [PubMed] [Google Scholar]
- Hedrich S, Schlömann M, Johnson DB. 2011. The iron-oxidizing proteobacteria. Microbiology 157:1551–1564. [DOI] [PubMed] [Google Scholar]
- Hoang DT, Chernomor O, Von Haeseler A, Minh BQ, Vinh LS. 2018. UFBoot2: improving the ultrafast bootstrap approximation. Mol Biol Evol. 35:518–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard JB, Kechris KJ, Rees DC, Glazer AN. 2013. Multiple amino acid sequence alignment nitrogenase component 1: insights into phylogenetics and structure-function relationships. PLoS One 8:e72751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Ribbe MW. 2015. Nitrogenase and homologs. J Biol Inorg Chem. 20:435–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Ribbe MW. 2016. Biosynthesis of the metalloclusters of nitrogenases. Annu Rev Biochem. 85:455–483. [DOI] [PubMed] [Google Scholar]
- Hulsen T, de Vlieg J, Alkema W. 2008. BioVenn–a web application for the comparison and visualization of biological lists using area-proportional Venn diagrams. BMC Genomics 9:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanovsky R, Lebedeva N, Keppen O, Tourova T. 2021. Nitrogen metabolism of an anoxygenic filamentous phototrophic bacterium Oscillocholris trichoides strain DG-6. Microbiology 90:428–434. [Google Scholar]
- Javaux EJ. 2019. Challenges in evidencing the earliest traces of life. Nature 572:451–460. [DOI] [PubMed] [Google Scholar]
- Jiao J-Y, Fu L, Hua Z-S, Liu L, Salam N, Liu P-F, Lv A-P, Wu G, Xian W-D, Zhu Q. 2021. Insight into the function and evolution of the Wood–Ljungdahl pathway in actinobacteria. ISME J. 15(10):3005–3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalyaanamoorthy S, Minh BQ, Wong TK, Von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods 14:587–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koirala A, Brözel VS. 2021. Phylogeny of nitrogenase structural and assembly components reveals new insights into the origin and distribution of nitrogen fixation across bacteria and archaea. Microorganisms 9:1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- König S, Gros O, Heiden SE, Hinzke T, Thuermer A, Poehlein A, Meyer S, Vatin M, Mbéguié-A-Mbéguié D, Tocny J. 2016. Nitrogen fixation in a chemoautotrophic lucinid symbiosis. Nat Microbiol. 2:1–10. [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35:1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langlois RJ, LaRoche J, Raab PA. 2005. Diazotrophic diversity and distribution in the tropical and subtropical Atlantic Ocean. Appl Environ Microbiol. 71:7910–7919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le SQ, Gascuel O. 2008. An improved general amino acid replacement matrix. Mol Biol Evol. 25:1307–1320. [DOI] [PubMed] [Google Scholar]
- Lee TK, Han I, Kim MS, Seong HJ, Kim J-S, Sul WJ. 2019. Characterization of a nifH-harboring bacterial community in the soil-limited Gotjawal forest. Front Microbiol. 10:1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepot K. 2020. Signatures of early microbial life from the Archean (4 to 2.5 Ga) eon. Earth-Sci Rev. 209:103296. [Google Scholar]
- Letunic I, Bork P. 2019. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 47:W256–W259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Liu X, Zhang H, Chen S. 2019. Evolution and functional analysis of orf1 within nif gene cluster from Paenibacillus graminis RSA19. Int J Mol Sci. 20:1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H-L, Wang W, Mortimer PE, Li R-Q, Li D-Z, Hyde KD, Xu J-C, Soltis DE, Chen Z-D. 2015. Large-scale phylogenetic analyses reveal multiple gains of actinorhizal nitrogen-fixing symbioses in angiosperms associated with climate change. Sci Rep. 5:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Torrejón G, Jiménez-Vicente E, Buesa JM, Hernandez JA, Verma HK, Rubio LM. 2016. Expression of a functional oxygen-labile nitrogenase component in the mitochondrial matrix of aerobically grown yeast. Nat Commun. 7:11426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovley DR. 1985. Minimum threshold for hydrogen metabolism in methanogenic bacteria. Appl Environ Microbiol. 49:1530–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin WF. 2020. Older than genes: the acetyl CoA pathway and origins. Front Microbiol. 11:817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Méheust R, Castelle CJ, Matheus Carnevali PB, Farag IF, He C, Chen L-X, Amano Y, Hug LA, Banfield JF. 2020. Groundwater Elusimicrobia are metabolically diverse compared to gut microbiome Elusimicrobia and some have a novel nitrogenase paralog. ISME J. 14:2907–2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta MP, Baross JA. 2006. Nitrogen fixation at 92 C by a hydrothermal vent archaeon. Science 314:1783–1786. [DOI] [PubMed] [Google Scholar]
- Mendler K, Chen H, Parks DH, Lobb B, Hug LA, Doxey AC. 2019. AnnoTree: visualization and exploration of a functionally annotated microbial tree of life. Nucleic Acids Res. 47:4442–4448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, Von Haeseler A, Lanfear R. 2020. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 37:1530–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore SJ, Sowa ST, Schuchardt C, Deery E, Lawrence AD, Ramos JV, Billig S, Birkemeyer C, Chivers PT, Howard MJ. 2017. Elucidation of the biosynthesis of the methane catalyst coenzyme F430. Nature 543:78–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muraki N, Nomata J, Ebata K, Mizoguchi T, Shiba T, Tamiaki H, Kurisu G, Fujita Y. 2010. X-ray crystal structure of the light-independent protochlorophyllide reductase. Nature 465:110–114. [DOI] [PubMed] [Google Scholar]
- Mus F, Alleman AB, Pence N, Seefeldt LC, Peters JW. 2018. Exploring the alternatives of biological nitrogen fixation. Metallomics 10:523–538. [DOI] [PubMed] [Google Scholar]
- Mus F, Colman DR, Peters JW, Boyd ES. 2019. Geobiological feedbacks, oxygen, and the evolution of nitrogenase. Free Radic Biol Med. 140:250–259. [DOI] [PubMed] [Google Scholar]
- Nishihara A, Haruta S, McGlynn SE, Thiel V, Matsuura K. 2018. Nitrogen fixation in thermophilic chemosynthetic microbial communities depending on hydrogen, sulfate, and carbon dioxide. Microbes Environ. 33:10–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishihara A, Thiel V, Matsuura K, McGlynn SE, Haruta S. 2018. Phylogenetic diversity of nitrogenase reductase genes and possible nitrogen-fixing bacteria in thermophilic chemosynthetic microbial communities in Nakabusa hot springs. Microbes Environ. 33(4):357–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomata J, Mizoguchi T, Tamiaki H, Fujita Y. 2006. A second nitrogenase-like enzyme for bacteriochlorophyll biosynthesis: reconstitution of chlorophyllide a reductase with purified X-protein (BchX) and YZ-protein (BchY-BchZ) from rhodobacter capsulatus. J Biol Chem. 281:15021–15028. [DOI] [PubMed] [Google Scholar]
- North JA, Narrowe AB, Xiong W, Byerly KM, Zhao G, Young SJ, Murali S, Wildenthal JA, Cannon WR, Wrighton KC. 2020. A nitrogenase-like enzyme system catalyzes methionine, ethylene, and methane biogenesis. Science 369:1094–1098. [DOI] [PubMed] [Google Scholar]
- Ohmoto H. 2020. A seawater-sulfate origin for early Earth’s volcanic sulfur. Nat Geosci. 13:576–583. [Google Scholar]
- Pan J, Xu W, Zhou Z, Shao Z, Dong C, Liu L, Luo Z, Li M. 2022. Genome-resolved evidence for functionally redundant communities and novel nitrogen fixers in the deyin-1 hydrothermal field, Mid-Atlantic Ridge. Microbiome 10:1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papineau D, She Z, Dodd MS, Iacoviello F, Slack JF, Hauri E, Shearing P, Little CT. 2022. Metabolically diverse primordial microbial communities in Earth’s oldest seafloor-hydrothermal jasper. Sci Adv. 8:eabm2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks DH, Chuvochina M, Chaumeil P-A, Rinke C, Mussig AJ, Hugenholtz P. 2020. A complete domain-to-species taxonomy for bacteria and archaea. Nat Biotechnol. 38(9):1079–1086. [DOI] [PubMed] [Google Scholar]
- Parks DH, Chuvochina M, Waite DW, Rinke C, Skarshewski A, Chaumeil P-A, Hugenholtz P. 2018. A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat Biotechnol. 36(10):996–1004. [DOI] [PubMed] [Google Scholar]
- Peng T, Xu Y, Zhang Y. 2018. Comparative genomics of molybdenum utilization in prokaryotes and eukaryotes. BMC Genomics 19:691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippi M, Kitzinger K, Berg JS, Tschitschko B, Kidane AT, Littmann S, Marchant HK, Storelli N, Winkel LH, Schubert CJ. 2021. Purple sulfur bacteria fix N2 via molybdenum-nitrogenase in a low molybdenum Proterozoic ocean analogue. Nat Commun. 12:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posada D, Buckley TR. 2004. Model selection and model averaging in phylogenetics: advantages of Akaike information criterion and Bayesian approaches over likelihood ratio tests. Syst Biol. 53:793–808. [DOI] [PubMed] [Google Scholar]
- Poudel S, Colman DR, Fixen KR, Ledbetter RN, Zheng Y, Pence N, Seefeldt LC, Peters JW, Harwood CS, Boyd ES. 2018. Electron transfer to nitrogenase in different genomic and metabolic backgrounds. J Bacteriol. 200:e00757-00717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quaiser A, Bodi X, Dufresne A, Naquin D, Francez A-J, Dheilly A, Coudouel S, Pedrot M, Vandenkoornhuyse P. 2014. Unraveling the stratification of an iron-oxidizing microbial mat by metatranscriptomics. PLoS One 9:e102561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond J, Siefert JL, Staples CR, Blankenship RE. 2004. The natural history of nitrogen fixation. Mol Biol Evol. 21:541–554. [DOI] [PubMed] [Google Scholar]
- Reitner J, Thiel V. 2011. Sulfate-reducing bacteria. In: Encyclopedia of Geobiology. Amsterdam: Springer. p. 853–855. [Google Scholar]
- Roy S, Liu W, Nandety RS, Crook A, Mysore KS, Pislariu CI, Frugoli J, Dickstein R, Udvardi MK. 2020. Celebrating 20 years of genetic discoveries in legume nodulation and symbiotic nitrogen fixation. Plant Cell 32:15–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio LM, Ludden PW. 2005. Maturation of nitrogenase: a biochemical puzzle. J Bacteriol. 187:405–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell JA, Moreau CS, Goldman-Huertas B, Fujiwara M, Lohman DJ, Pierce NE. 2009. Bacterial gut symbionts are tightly linked with the evolution of herbivory in ants. Proc Natl Acad Sci U S A. 106:21236–21241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott JJ, Breier JA, Luther GW III, Emerson D. 2015. Microbial iron mats at the Mid-Atlantic Ridge and evidence that Zetaproteobacteria may be restricted to iron-oxidizing marine systems. PLoS One 10:e0119284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setubal JC, Dos Santos P, Goldman BS, Ertesvåg H, Espin G, Rubio LM, Valla S, Almeida NF, Balasubramanian D, Cromes L. 2009. Genome sequence of Azotobacter vinelandii, an obligate aerobe specialized to support diverse anaerobic metabolic processes. J Bacteriol. 191:4534–4545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla SP, Sanders JG, Byrne MJ, Pierce NE. 2016. Gut microbiota of dung beetles correspond to dietary specializations of adults and larvae. Mol Ecol. 25:6092–6106. [DOI] [PubMed] [Google Scholar]
- Staples CR, Lahiri S, Raymond J, Von Herbulis L, Mukhophadhyay B, Blankenship RE. 2007. Expression and association of group IV nitrogenase NifD and NifH homologs in the non-nitrogen-fixing archaeon Methanocaldococcus jannaschii. J Bacteriol. 189:7392–7398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinsbu BO, Thorseth IH, Nakagawa S, Inagaki F, Lever MA, Engelen B, Øvreås L, Pedersen RB. 2010. Archaeoglobus sulfaticallidus sp. nov., a thermophilic and facultatively lithoautotrophic sulfate-reducer isolated from black rust exposed to hot ridge flank crustal fluids. Int J Syst Evol Microbiol. 60:2745–2752. [DOI] [PubMed] [Google Scholar]
- Tahon G, Tytgat B, Willems A. 2016. Diversity of phototrophic genes suggests multiple bacteria may be able to exploit sunlight in exposed soils from the Sør Rondane Mountains, East Antarctica. Front Microbiol. 7:2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tashiro T, Ishida A, Hori M, Igisu M, Koike M, Méjean P, Takahata N, Sano Y, Komiya T. 2017. Early trace of life from 3.95 Ga sedimentary rocks in Labrador, Canada. Nature 549:516–518. [DOI] [PubMed] [Google Scholar]
- Thomazo C, Couradeau E, Garcia-Pichel F. 2018. Possible nitrogen fertilization of the early Earth Ocean by microbial continental ecosystems. Nat Commun. 9:2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Alowaifeer A, Kerner P, Balasubramanian N, Patterson A, Christian W, Tarver A, Dore JE, Hatzenpichler R, Bothner B. 2021. Aerobic bacterial methane synthesis. Proc Natl Acad Sci U S A. 118(27):e2019229118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Horiike T. 2021. The evolution of molybdenum dependent nitrogenase in cyanobacteria. Biology 10:329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss MC, Sousa FL, Mrnjavac N, Neukirchen S, Roettger M, Nelson-Sathi S, Martin WF. 2016. The physiology and habitat of the last universal common ancestor. Nat Microbiol. 1:1–8. [DOI] [PubMed] [Google Scholar]
- Yang J, Junium CK, Grassineau N, Nisbet E, Izon G, Mettam C, Martin A, Zerkle AL. 2019. Ammonium availability in the Late Archaean nitrogen cycle. Nat Geosci. 12:553–557. [Google Scholar]
- Zehr JP, McReynolds LA. 1989. Use of degenerate oligonucleotides for amplification of the nifH gene from the marine cyanobacterium Trichodesmium thiebautii. Appl Environ Microbiol. 55:2522–2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Gladyshev VN. 2008. Molybdoproteomes and evolution of molybdenum utilization. J Mol Biol. 379:881–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H, Dietrich C, Radek R, Brune A. 2016. Endomicrobium proavitum, the first isolate of Endomicrobia class. nov. (phylum Elusimicrobia)–an ultramicrobacterium with an unusual cell cycle that fixes nitrogen with a Group IV nitrogenase. Environ Microbiol. 18:191–204. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Harris DF, Yu Z, Fu Y, Poudel S, Ledbetter RN, Fixen KR, Yang Z-Y, Boyd ES, Lidstrom ME. 2018. A pathway for biological methane production using bacterial iron-only nitrogenase. Nat Microbiol. 3:281. [DOI] [PubMed] [Google Scholar]
- Zheng K, Ngo PD, Owens VL, Yang X-p, Mansoorabadi SO. 2016. The biosynthetic pathway of coenzyme F430 in methanogenic and methanotrophic archaea. Science 354:339–342. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All our relevant data are provided in Supplementary files of our paper.