Abstract
The emergence of the placenta is a revolutionary event in the evolution of therian mammals, to which some LTR retroelement–derived genes, such as PEG10, RTL1, and syncytin, are known to contribute. However, therian genomes contain many more LTR retroelement–derived genes that may also have contributed to placental evolution. We conducted large-scale evolutionary genomic and transcriptomic analyses to comprehensively search for LTR retroelement–derived genes whose origination coincided with therian placental emergence and that became consistently expressed in therian placentae. We identified NYNRIN as another Ty3/Gypsy LTR retroelement–derived gene likely to contribute to placental emergence in the therian stem lineage. NYNRIN knockdown inhibited the invasion of HTR8/SVneo invasive-type trophoblasts, whereas the knockdown of its nonretroelement-derived homolog KHNYN did not. Functional enrichment analyses suggested that NYNRIN modulates trophoblast invasion by regulating epithelial-mesenchymal transition and extracellular matrix remodeling and that the ubiquitin-proteasome system is responsible for the functional differences between NYNRIN and KHNYN. These findings extend our knowledge of the roles of LTR retroelement–derived genes in the evolution of therian mammals.
Keywords: evolutionary genomics, ancestral state reconstruction, placenta, LTR retrotransposon
Introduction
The emergence of the placenta is an important event in the evolutionary history of therian mammals. Although therian placentae vary in shape, form, and degree of invasiveness, placentotrophy is assumed to have originated only once in the evolutionary history of mammals (Blackburn 2015) and is believed to be a key factor behind the survival of mammals during the K-Pg extinction event. The process of placental attachment requires the invasion of fetal tissues into the maternal uterine endometrium for the subsequent establishment of fetomaternal adhesive forces. The invasion of therian fetal placentae is led by the collective invasion of trophoblast-lineage cells that are derived from trophectoderm cells residing in the external layer of therian embryos (fig. 1A). Trophoblast-lineage cells in therian mammals differentiate into specialized trophoblast cells that transform the uterine environment for maternal acceptance and embryonic support (Jones et al. 2014; Martinez et al. 2015; Kolahi et al. 2017; Pollheimer et al. 2018). The emergence of such a special system is an evolutionary innovation whose genomic bases are of general interest in evolutionary and developmental biology (Griffith and Wagner 2017).
Fig. 1.
Many LTR retroelement–derived genes could be involved in therian placental evolution. (A) Schematic differentiation of trophoblast-lineage cells in the tammar wallaby (nearly full-term) and human (third trimester) placentae from blastocyst trophectoderm cells, as examples of the differentiation in marsupials and that in placentals, respectively. (B) A simplified phylogeny and the numbers of tetrapod genomes or transcriptomes that were used in this study. (C and D) Schematic representations of transition events regarding (C) gene sequence and expression gain or loss or (D) integration of LTR retroelements. (E) A schematic diagram showing the reconstruction of ancestral states of genes and interpretation of transition events from character state changes. (F) Numbers of identified ortholog groups whose ancestral gene originated in the stem lineage of each clade but was not expressed in the placenta or chorioallantoic membrane of its MRCA (state 0 → 1) and those whose ancestral gene originated in the stem lineage of the MRCA and became expressed in the placenta or chorioallantoic membrane of that MRCA (state 0 → 2). (G) Numbers of identified ortholog groups whose ancestral gene in the stem lineage of each clade was newly integrated by at least an LTR retroelement, and all the integrated LTR retroelements became introns (state A → I) or at least an LTR retroelement became an exon (state A → E) in the gene of the MRCA of that clade. (H) Numbers of ortholog groups of LTR retroelement–derived genes that may have contributed to the placental evolution in the therian, marsupial, and placental lineages (states 0 → 2 and A → E).
LTR retroelements are repetitive genetic elements universally found in vertebrate genomes (Chalopin et al. 2015). In mammalian genomes, such as human and mouse genomes, LTR retroelements account for 8% and 10% of the total length, respectively (Lander et al. 2001; Waterston et al. 2002). Although the roles of most LTR retroelements in mammalian genomes remain elusive, several LTR retroelement–derived genes, such as PEG10, RTL1, and syncytin, have been shown to play crucial roles in the placental development of therian mammals (Ono et al. 2006; Sekita et al. 2008; Dupressoir et al. 2009; Cornelis et al. 2015) and Mabuya lizards (Cornelis et al. 2017). PEG10 functions in trophoblast stem-cell lineage specification (Abed et al. 2019) and trophoblast proliferation and invasion (Chen et al. 2015), whereas syncytin functions in trophoblast fusion (Mi et al. 2000; Cornelis et al. 2015; Cornelis et al. 2017). Altogether, their origination is considered to be a key evolutionary event that underlies the functional evolution of the placenta in therian mammals and placental reptiles.
The recent increase in the number of tetrapod genomes and transcriptomes in public databases provides us with an opportunity to comprehensively investigate the evolutionary history and expression pattern of LTR retroelement–derived genes across therian genomes. More than 250 curated tetrapod genomes are available in the NCBI RefSeq (O’Leary et al. 2016) and Ensembl (Howe et al. 2021) databases. Transcriptomes from the placenta of 13 therian mammals (Bernstein et al. 2010; Wang et al. 2013b; Armstrong et al. 2017; Guernsey et al. 2017) and the chorioallantoic membranes of a saurian species (Griffith et al. 2017) are also available. In addition, recent bioinformatics tools, such as PastML (Ishikawa et al. 2019), enabled the rapid reconstruction of ancestral states of genomes and transcriptomes from large-scale information of extant species. Because placental development requires a complex intermingling of cells from both the mother and fetus, which would have to be driven by a multitude of new factors, we expected that additional LTR retroelement–derived genes that have contributed to the evolution of the therian placenta would remain to be discovered.
In this study, we conducted comparative evolutionary genomic and transcriptomic analyses and revealed that NYN domain and retroviral integrase containing (NYNRIN, formerly called CGIN), an LTR retroelement–derived gene, originated in the therian stem lineage and became consistently expressed in therian placentae in parallel with the therian placental emergence. Furthermore, NYNRIN knockdown (KD) and RNA-seq experiments confirmed that NYNRIN functions in trophoblast invasion and would be another key factor that contributed to the emergence of the therian placenta.
Results
Nineteen LTR Retroelement–Derived Genes may have Contributed to the Placental Evolution in the Therian, Marsupial, or Placental Lineages
We first screened for LTR retroelement–derived genes that have potentially contributed to therian placental evolution. Screening started by using OrthoFinder (Emms and Kelly 2019) to reconstruct ortholog groups across 267 tetrapod species (fig. 1B). From the 41,884 ortholog groups that were identified, we assigned the states of genes in each ortholog group to each extant tetrapod species based on the presence/absence of gene sequences, their expression in the placenta or chorioallantoic membrane, and the presence/absence of LTR retroelements that became gene introns or exons (fig. 1C–E). Then, we inferred the states of the ancestral species using the maximum parsimony–based DOWNPASS and maximum likelihood–based marginal posterior probability approximation methods in PastML (Ishikawa et al. 2019). Both methods concordantly estimated 45, 85, and 311 ortholog groups whose ancestral genes originated in the therian, marsupial, and placental stem lineages, respectively, and became expressed in the placentae of the most recent common ancestor (MRCA) of therians, marsupials, and placentals (fig. 1F). Moreover, the ancestral gene of 1, 1, and 17 of these ortholog groups in the stem lineages of therian, marsupial, and placental mammals were, respectively, estimated to be newly integrated with (an) LTR retroelement(s) and at least an LTR retroelement became exon(s) in the gene of their MRCA (fig. 1GandH; supplementary table S1, Supplementary Material online).
NYNRIN is Another LTR Retroelement–Derived Gene that may have Contributed to the Therian Placental Emergence
Among the 19 LTR retroelement–derived genes, we focused on NYNRIN because its origination coincided with the emergence of the therian placenta (fig. 2AandB). NYNRIN originated from a duplication of a pre-existing gene, KH and NYN domain containing (KHNYN), and a subsequent fusion of the duplicate with the Ty3/Gypsy pol gene–derived segment that contained a reverse transcriptase ribonuclease H (RNase H) domain and an integrase (INT) domain (fig. 2C; Anantharaman and Aravind 2006; Marco and Marín 2009; Llorens et al. 2011). NYNRIN has been shown to be associated with a predisposition to Wilms tumor (Mahamdallie et al. 2019) and metastasis of melanoma (Leonard et al. 2021). Since cancer metastasis and placental invasion could be evolutionarily linked in the therian lineage (Kshitiz et al. 2019; Wagner et al. 2022), we hypothesized that NYNRIN, like PEG10, RTL1, and syncytin, may also play a role in placental development in therian mammals.
Fig. 2.
NYNRIN originated in the therian stem lineage and its expression was recruited into the placenta of the therian MRCA. (A and B) Reconstruction of the ancestral states of NYNRIN in terms of (A) the presence/absence of gene sequences and their expression in the placenta or chorioallantoic membrane or (B) the presence/absence of LTR retroelements that became exons of NYNRIN. Marginal posterior probability approximation method was used. Node color represents the estimated states. Blue arrow: species whose transcriptome data from the placenta or chorioallantoic membrane were available, crescent: mammalian MRCA, star: therian MRCA. (C) Domain architecture of the human NYNRIN, KHNYN, and other phylogenetically related NYN-containing N4BP1, ZC3H12C, and ZC3H12A proteins. KH-like, K homology-like domain; NYN, Nedd4-BP1 YacP nuclease domain; INT, integrase domain; CUBAN, cullin-binding domain associating with NEDD8 domain; CoCUN, cousin of CUBAN domain; UBA, ubiquitin-associated domain.
Reconstructions of the ancestral states of NYNRIN consistently showed that NYNRIN originated in the therian stem lineage and became expressed in placental tissues of the therian MRCA. NYNRIN was present in all extant therian species, except for the tammar wallaby (Notamacropus eugenii; supplementary table S2, Supplementary Material online). Comparative transcriptomic analysis also revealed that NYNRIN was actively expressed (zFPKM average > −3) in the placental tissues of all therian species whose data were available, except for the tammar wallaby. The absence of NYNRIN in the genome and transcriptome of the tammar wallaby could be due to the low data quality, where 22.2% of the conserved proteins were missing (supplementary fig. S1, Supplementary Material online). In addition, although a protein in a common garter snake (Thamnophis sirtalis) was annotated as NYNRIN (XP_013909250.1), this was likely a misannotation because its sequence similarity to other NYNRIN proteins was < 31.5%.
NYNRIN Proteins Localized in Trophoblast-Lineage Cells and its KD Inhibited Trophoblast Invasion
Next, we experimentally investigated whether NYNRIN functions in placental development and, hence, was likely to contribute to the therian placental emergence. First, the expression and localization pattern of NYNRIN in human placental tissues were identified by immunostaining of placental tissues from healthy individuals and patients with preeclampsia and intrauterine growth restriction (fig. 3A). NYNRIN localized in the cytoplasm of human trophoblast-lineage cells, including cytotrophoblasts, syncytiotrophoblasts, and extravillous trophoblasts, rather than in cells in the mesenchymal core of placental villi or decidual stromal cells. The localization pattern of NYNRIN was consistent with our hypothesis that NYNRIN contributes to placental development.
Fig. 3.
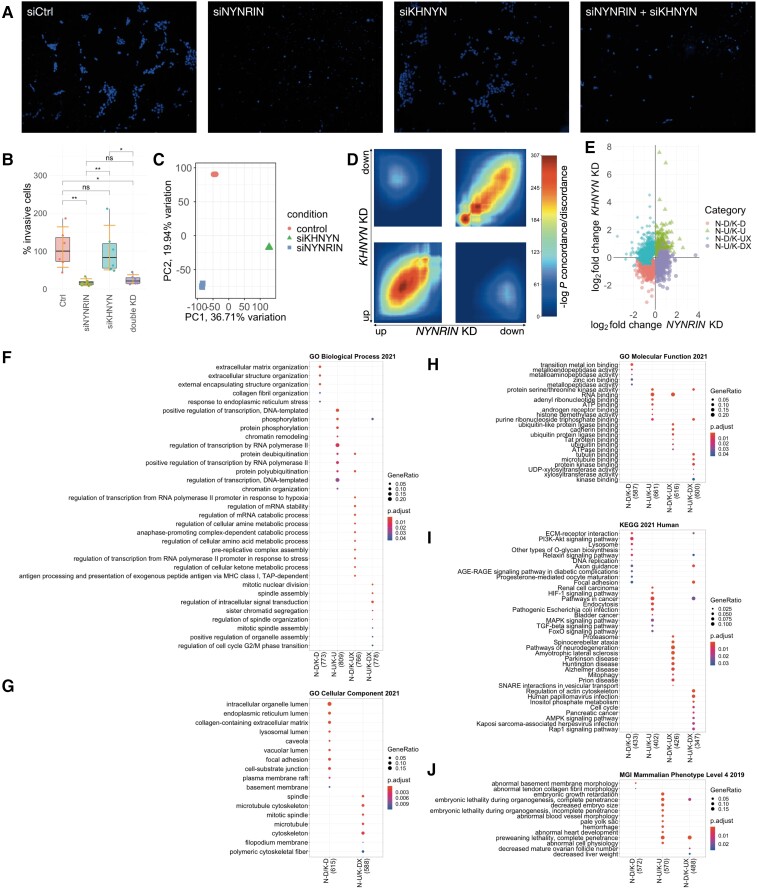
NYNRIN promotes trophoblast invasion rather than trophoblast fusion. (A) Human placental tissues from a healthy individual and those from a patient with preeclampsia and intrauterine growth restriction (IUGR) stained with anti-NYNRIN, anti-HLA-G (a marker of extravillous trophoblast), or normal rabbit IgG (negative control) (20X). ET (red arrow): extravillous trophoblast in decidual tissue, CT (yellow arrow): cytotrophoblast, ST (blue arrow): syncytiotrophoblast layer. (B) BeWo cells treated with siCtrl (control), siCtrl + Db, or siNYNRIN3 + Db (20×). Blue: nuclei, red: cell membranes, white arrow: fused syncytium. (C) Microscopic images (10×) and (D) proportions of HTR8/SVneo cells that invaded through Matrigel after treatment with siCtrl or each of the three siNYNRINs (siNYNRIN2 for the microscopic image). Blue: nuclei, *P < 0.05, **P < 0.01, ****P < 0.0001, orange lines: 95% confidence intervals for the median.
We also investigated the roles of NYNRIN in two important processes during placental development, trophoblast fusion, and trophoblast invasion, using siRNA KD. The role of NYNRIN in trophoblast fusion was explored by performing in vitro cell fusion assay in human BeWo trophoblastic choriocarcinoma cells. Three sets of NYNRIN siRNA (siNYNRIN) effectively reduced NYNRIN expression in BeWo cells compared with that in the control (n = 3; supplementary fig. S2A, Supplementary material online). However, there were no clear morphological differences in cell fusion, induced by dibutyryl cAMP (Db), between BeWo cells treated or untreated with siNYNRIN (fig. 3B). The expression of CGB, a molecular marker gene of trophoblast fusion, in Db-treated BeWo cells was also unchanged by NYNRIN KD (n = 4; supplementary fig. S2B, Supplementary material online).
We then investigated the role of NYNRIN in trophoblast invasion using HTR8/SVneo invasive-type trophoblast cells. Although HTR8/SVneo cells were often described as extravillous trophoblasts, we assume that their cytotrophoblast-like characteristics, including progenitor cell-like self-renewal properties (Weber et al. 2013), the ability to undergo cell-cell fusion by forskolin induction (Liu et al. 2015), and the ability to undergo epithelial-mesenchymal transition under normal culture conditions (Abou-Kheir et al. 2017; Msheik et al. 2020) make HTR8/SVneo cells good representatives of invasive trophoblast-lineage cells rather than only being representatives of extravillous trophoblasts. We confirmed that all the three sets of siNYNRIN reduced NYNRIN expression in HTR8/SVneo cells compared with that in the control (n = 3; supplementary fig S2C, Supplementary material online) and that NYNRIN KD significantly reduced the median proportion of invasive HTR8/SVneo cells compared with that in the control (n = 10; 11.7–44.7%; Dunn’s post hoc, adjusted P ≤ 0.0233; fig. 3CandD). These results suggest that NYNRIN may have contributed to the emergence of the therian placenta by promoting trophoblast invasion, rather than trophoblast fusion.
RNA-seq of NYNRIN-KD HTR8/SVneo Cells Suggested Roles of NYNRIN in Epithelial-Mesenchymal Transition and Extracellular Matrix Remodeling
We further examined the molecular and physiological functions of NYNRIN by comparative transcriptomic analysis of NYNRIN-KD HTR8/SVneo cells and the control. Principal component analysis showed that the gene expression profiles of the control and NYNRIN-KD groups clustered into two distinct groups, as expected (fig. 4A). Between the two groups, 6,507 differentially expressed genes (DEGs; adjusted P < 0.05, zFPKM averages in all groups > −3) were identified, including 3,174 downregulated and 3,333 upregulated genes in the NYNRIN-KD group (fig. 4B). The top 1,000 most significant DEGs (adjusted P < 1.89 × 10−16), including 672 and 328 downregulated and upregulated genes, respectively, were selected for functional enrichment analysis. The top ten Gene Ontology (GO; Gene Ontology Consortium, 2021) and Kyoto Encyclopedia of Genes and Genomes (KEGG; Kanehisa et al. 2021) terms (adjusted P < 0.05) enriched with the NYNRIN-KD downregulated genes were related to endoplasmic reticulum stress, extracellular matrix remodeling, cell motility, and Fcγ receptor signaling pathway (fig. 4C–F, supplementary table S3, Supplementary Material online), most of which are known to be related to trophoblast invasion (Kaloglu and Onarlioglu 2010; Zhu et al. 2012; Lee et al. 2019), except for the Fcγ receptor signaling pathway. The top ten GO and KEGG terms enriched with the NYNRIN-KD upregulated genes were related to glycolysis, HIF-1 signaling pathway, and cell cycle, all of which are known to be related to cell invasion (Dahl et al. 2005; Kohrman and Matus 2017; Zhu et al. 2017; Tsai et al. 2021). Notably, an enrichment analysis using the MGI Mammalian Phenotype terms (Smith and Eppig 2009) indicated that NYNRIN KD may affect abnormal embryonic growth and lethality, which are linked to placental development (fig 4G). We additionally applied the |log2 fold change| > 0.5 cutoff criterion to investigate the effect of highly dysregulated DEGs (supplementary fig. S3, Supplementary Material online). The top 1,000 most significant DEGs (adjusted P-value < 1.73×10−8) were enriched to a part of the top ten ontology terms related to trophoblast invasion previously found such as “cell-substrate junction,” “focal adhesion,” “HIF-1 signaling pathway,” and “decreased angiogenesis,” indicating that those functions are associated with the highly dysregulated DEGs.
Fig. 4.
NYNRIN promotes trophoblast invasion by regulating genes related to epithelial-mesenchymal transition and extracellular matrix remodeling. (A) Principal component plot of gene expression profiles of HTR8/SVneo cells treated with siCtrl or siNYNRIN. (B) Volcano plot showing negative log adjusted P-values from DESeq2 and log2 fold change of genes in NYNRIN-KD HTR8/SVneo cells. The top ten most significantly downregulated or upregulated DEGs are indicated. (C–G) Functional enrichment plots showing gene ratios and adjusted P-values, from clusterProfiler, of the top ten (C) GO Biological Process, (D) GO Molecular Function, (E) GO Cellular Component, (F) KEGG, or (G) MGI Mammalian Phenotype terms enriched with downregulated or upregulated genes among the top 1,000 most significant DEGs in NYNRIN-KD HTR8/SVneo cells. The number in parentheses indicates the number of DEGs that have annotations in each functional database. (H) Heatmap showing the differences in expression of selected genes in NYNRIN-KD (siNYNRYN) and KHNYN-KD (siKHNYN) HTR8/SVneo cells against the expression in the control cells. EpiM, epithelial marker genes; MesM, mesenchymal marker genes; MMP, matrix metalloproteinase; PLAU, plasminogen; IL, interleukin.
Additionally, we checked the changes in the expression of genes related to epithelial-mesenchymal transition and extracellular matrix remodeling because these processes play a key role in trophoblast invasion (Kaloglu and Onarlioglu 2010; Zhu et al. 2012; Oghbaei et al. 2022). The expression of mesenchymal marker genes N-cadherin (CDH2), VIM, and ZEB1 was significantly decreased by NYNRIN KD (fig. 4H), whereas the expression of epithelial marker genes E-cadherin (CDH1), DSP, and TJP1 was unchanged or increased. These findings suggest that NYNRIN KD inhibited the epithelial-mesenchymal transition of HTR8/SVneo cells. The decreased expression of several matrix metalloproteinase genes, plasminogen-related PLAU and PLAUR genes, and the pro-inflammatory cytokine IL6 gene in NYNRIN-KD HTR8/SVneo cells also suggested a role for NYNRIN in inhibiting extracellular matrix degradation. The top ten most significantly downregulated or upregulated genes also included HSP90AB1 (Voss et al. 2000), PRNP (Alfaidy et al. 2013), SPARC (Tossetta et al. 2019), COL5A (Anton et al. 2014), BNIP3 (Stepan et al. 2005; Zhou et al. 2021), AGO2 (Cheloufi et al. 2010), HIPK3 (Wang et al. 2022), and ARID3A (Rhee et al. 2017) which are associated with or functional in placental development or placenta diseases.
A Homologous Nonretroelement–Derived Gene KHNYN would not Function in Trophoblast Invasion
As described earlier, NYNRIN was derived from the cellular gene KHNYN which contains the K homology-like (KH-like) domain and Nedd4-BP1 YacP nuclease (NYN) domain, but not the LTR retroelement–derived RNase H nor INT domain (fig. 2C). Multiple sequence alignment of NYNRIN, KHNYN, and other NYN-containing proteins is provided in supplementary fig. S4A, Supplementary material online. To clarify whether the role of NYNRIN in trophoblast invasion was due to its nonretroelement-derived KH-like and NYN domains, we performed an invasion assay and RNA-seq analysis of KHNYN-KD HTR8/SVneo cells. If KHNYN-KD cells exhibit phenotypes and transcriptome profiles similar to those of NYNRIN-KD cells, the function of NYNRIN in trophoblast invasion could be attributed to the nonretroelement-derived segment, rather than the retroelement-derived segment of NYNRIN.
After confirming that all three sets of KHNYN siRNA (siKHNYN) effectively reduced KHNYN expression in HTR8/SVneo cells compared with that in the control (n = 3; supplementary fig. S2D, Supplementary material online), we performed an invasion assay. The results clearly showed that KHNYN KD did not inhibit trophoblast invasion (n = 6; Dunn’s post hoc, adjusted P = 0.903; fig. 5AandB). In addition, double KD of NYNRIN and KHNYN significantly inhibited trophoblast invasion compared with the control (adjusted P = 0.0117), without a significant difference from the effect of NYNRIN single KD (adjusted P = 0.820). This supports the hypothesis that the retroelement-derived segment of NYNRIN is responsible for its function in trophoblast invasion.
Fig. 5.
NYNRIN and KHNYN regulated trophoblast invasion differently. (A) Microscopic images (10×) and (B) proportions of HTR8/SVneo cells that invaded through Matrigel after treatment with siCtrl, siNYNRIN, siKHNYN, or both siNYNRIN and siKHNYN. Blue: nuclei, ns: nonsignificant, *P < 0.05, **P < 0.01, orange lines: 95% confidence intervals for the median. (C) Principal component plot of gene expression profiles of HTR8/SVneo cells treated with siCtrl, siNYNRIN, or siKHNYN. (D) Rank-rank hypergeometric overlap plot showing the degrees of concordance/discordance between upregulated and downregulated genes in NYNRIN-KD and KHNYN-KD HTR8/SVneo cells (negative log P-values). (E) Scatter plot showing log2 fold change of the top 1,000 genes with the highest disco scores (Domaszewska et al. 2017) whose expression was upregulated or downregulated by either NYNRIN KD or KHNYN KD compared with the control. N-D/K-D, N-U/K-U, N-D/K-UX, and N-U/K-DX denote gene groups with different expression regulation patterns (e.g., N-D/K-UX indicates the group of genes whose expression was downregulated in NYNRIN-KD but upregulated or unchanged in KHNYN-KD). (F–J) Functional enrichment plots showing gene ratios and adjusted P-values, from clusterProfiler, of the top ten (F) GO Biological Process, (G) GO Molecular Function, (H) GO Cellular Component, (I) KEGG, or (J) MGI Mammalian Phenotype terms enriched with the top 1,000 genes with the highest disco scores shown in (E). The number in parentheses represents the number of DEGs that have annotations in each functional database.
Principal component analysis of RNA-seq of KHNYN KD showed that the gene expression profiles of the control, NYNRIN-KD, and KHNYN-KD groups were clearly distinct (fig. 5C), indicating that NYNRIN and KHNYN have distinct functions. There were 8,813 DEGs (adjusted P < 0.05, zFPKM averages in all groups > −3) between the KHNYN-KD and control groups, comprising 4,325 downregulated and 4,488 upregulated genes, respectively. Interestingly, KHNYN KD decreased the expression of NYNRIN and vice versa (fig. 4B, supplementary fig. S5, Supplementary Material online) despite the distinct gene expression profiles of NYNRIN KD and KHNYN KD. It should be noted that off-target silencing effects were unlikely because all siRNAs had been designed to contain at least two mismatches against the transcript of each nontarget gene (supplementary fig. S6, Supplementary Material online).
Thereafter, we investigated the discordance in expression changes of genes related to epithelial-mesenchymal transition and extracellular matrix remodeling between the two KD experiments. The expression of mesenchymal marker genes CDH2, VIM, and ZEB1 was significantly increased by KHNYN KD but decreased by NYNRIN KD (fig. 4H). KHNYN KD also increased the expression of other mesenchymal marker genes, ZEB2, SNAI1 (SNAIL), and SNAI2 (SLUG), whose expression was unchanged by NYNRIN KD. These results indicate that KHNYN KD did not inhibit the epithelial-mesenchymal transition of HTR8/SVneo cells. In contrast, the expression of the epithelial marker genes CDH1, DSP, and TJP1 was upregulated by both KHNYN KD and NYNRIN KD. The expression of PLAU and several matrix metalloproteinase genes was similarly decreased in both the KD experiments.
Rank-rank hypergeometric overlap analysis revealed sets of genes whose expression was concordantly regulated by NYNRIN KD and KHNYN KD, possibly because of the shared nonretroelement-derived domains as well as those that were independently affected (fig. 5D). We identified 1,201 genes whose expression was downregulated by both NYNRIN KD and KHNYN KD (N-D/K-D); 1,336 genes whose expression was upregulated by both NYNRIN KD and KHNYN KD (N-U/K-U); 1,973 genes whose expression was downregulated by NYNRIN KD but not by KHNYN KD (N-D/K-UX); and 1,997 genes whose expression was upregulated by NYNRIN KD but not by KHNYN KD (N-U/K-DX). We conducted a functional enrichment analysis by comparing the top 1,000 genes with the highest absolute values of disco scores (Domaszewska et al. 2017) from each set (fig. 5E) to focus on genes whose expression was specifically affected by NYNRIN (fig. 5F–H, supplementary table S4, Supplementary Material online). The ontology terms enriched with the top 1,000 N-D/K-UX genes were related to embryonic or preweaning lethality (MGI Mammalian Phenotype), proteasome (KEGG), and ubiquitin or ubiquitin-like protein ligase binding (GO Molecular Function), which indicated a link between NYNRIN-specific function and the ubiquitin-proteasome system.
Losing the Ubiquitin-Binding Domain may have Contributed to the Trophoblast Invasion Function of NYNRIN
Furthermore, we investigated the molecular-level differences between the KH-like and NYN domains of NYNRIN and those of KHNYN to search for nonretroelement-derived factors that may contribute to the unique function of NYNRIN. The KH-like domains have putative RNA-binding activity (Nicastro et al. 2015), whereas the NYN domains have widely conserved nuclease activity (Anantharaman and Aravind 2006). Both domains have been reported to possess antiviral activity (Ficarelli et al. 2019).
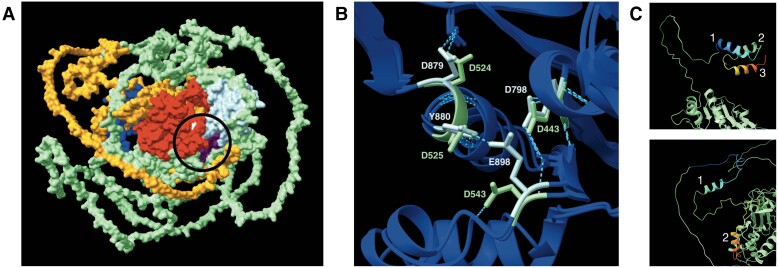
The predicted 3D structures of NYNRIN and KHNYN generated by AlphaFold (Jumper et al. 2021) suggest that their KH-like and NYN domains have similar overall structures (fig. 6A, supplementary fig. S7, Supplementary Material online). Notably, both AlphaFold and IUPred3 predicted that NYNRIN and KHNYN would have substantial intrinsically disordered regions with > 100 amino acids (supplementary fig. S8, Supplementary Material online). We also noted that NYNRIN has more hydrogen bonds among the KH-like, NYN, and INT domains than among the KH-like and NYN domains of KHNYN does (supplementary fig. S9, Supplementary Material online), although this might be due to structural estimation biases. Subcellular localization prediction indicated that the integration of the LTR retroelement did not change the localization pattern of NYNRIN from that of KHNYN. Both were expected to localize in the cytoplasm (supplementary fig. S10, Supplementary material online).
Fig. 6.
Roles of NYNRIN in promoting trophoblast invasion could be derived from its putative ubiquitin-binding domain, but not its NYN domain. (A) A predicted 3D structure of human NYNRIN (see Materials and Methods). The circle indicates where the INT domain connects the KH-like and NYN domains with hydrogen bonds. Purple: KH-like domain, light blue: NYN domain, blue: RNase H domain, red: INT domain, orange: hypothetical LTR retroelement–derived segment excluding the RNase H and INT domains, light green: hypothetical nonretroelement-derived segment excluding the KH-like and NYN domains. (B) 3D superimposition of NYN domains of NYNRIN and KHNYN showing mutations in DDDD motifs. Light blue: NYNRIN, light green: KHNYN, blue dashed lines: hydrogen bonds. (C) Predicted 3D structures of KHNYN CUBAN (top) and the region of NYNRIN aligned with KHNYN CUBAN (bottom). The aligned regions were colored by the rainbow rule, from blue on the N-terminus to red on the C-terminus. Numbers represent the order of ɑ-helices along the proteins.
Our primary structure analysis found that whereas NYNRIN of marsupials, KHNYN, and other NYN-containing proteins, including NEDD4-binding protein 1 (N4BP1), Zinc Finger CCCH-Type Containing 12C (ZC3H12C), and Zinc Finger CCCH-Type Containing 12A (ZC3H12A), have strictly conserved DDDD motifs in the active site in the NYN domain, NYNRIN of placentals has two distal substitutions (fig. 6B, supplementary fig. S4B, Supplementary material online), indicating the loss of nuclease activity (Habacher and Ciosk 2017) during the metatherian-eutherian split. In addition, NYNRIN lost a ubiquitin-binding domain homologous to the cullin-binding domain associating with NEDD8 (CUBAN) domain of KHNYN (fig. 2C). Although there is a region on NYNRIN that is somewhat similar to the CUBAN domain of KHNYN and cousin of CUBAN (CoCUN) domain of N4BP1, their sequences were poorly aligned, and their secondary structures were also different (fig. 6C, supplementary fig. S4C, Supplementary material online). In combination with our previous observation from RNA-seq analyses that ubiquitination-proteasome system–related genes were differentially affected by NYNRIN KD and KHNYN KD, the loss of the ubiquitin-binding domain may also have contributed to the functions of NYNRIN in trophoblast invasion.
Discussion
This study, which combines large-scale evolutionary genomic and transcriptomic analyses with in vitro experiments, revealed that another LTR retroelement–derived gene, NYNRIN, was likely to contribute to placental emergence in the therian stem lineage. Invasion assays and RNA-seq experiments suggested that the origination of NYNRIN may have contributed to the therian placental emergence at the cellular level by promoting trophoblast invasion. Comparison with KHNYN, a closely related homolog of NYNRIN, suggested that the unique function of NYNRIN may be attributed to the LTR retroelement–derived RNase H and INT domains, as well as the loss of the ubiquitin-binding domain. Our results shed light on another critical role for exogenous sequences in the evolution of therian mammals, which is one of the most fundamental questions in biology.
Apart from NYNRIN, our analyses also identified 18 LTR retroelement–derived genes that originated in the stem lineage of marsupials or placentals and became expressed in the placenta of their MRCA. These genes include eight zinc finger genes and four retrotransposon gag-like genes. Whereas the Human Protein Atlas database shows that the expression of some of these genes, including NYNRIN, is neither limited to placental tissues nor trophoblast-lineage cells (Uhlén et al. 2015; Karlsson et al. 2021), the identified LTR retroelement–derived genes actually included genes that have been known for the association with or crucial function in placental development or diseases, which are RTL1 (Sekita et al. 2008), PEG3 (Kim et al. 2013), LDOC1 (Naruse et al. 2014), CCDC8 (Wang et al. 2019b), and IFT88 (Ritter et al. 2020). Nevertheless, we would like to note that our analyses do not preclude the possible contribution of the identified LTR retroelement–derived genes to the evolution of other organs in the therian lineage.
The absence of NYNRIN in the tammar wallaby genome may be interesting to further investigate the association of NYNRIN expression with the degree of placental invasiveness because the tammar wallaby placenta is vaguely invasive (Jones et al. 2014). In marsupials that have invasive trophoblasts such as gray short-tailed opossums, NYNRIN is weakly expressed in their trophectoderm cells at the pre-invasive epiblast stage (Mahadevaiah et al. 2020), whereas the expression data during the invasive period is still unavailable. In placentals that have epitheliochorial placentae such as cattle and pigs, NYNRIN is expressed in bovine trophoblasts at the early blastocyst stage (Pillai et al. 2019) but absent at the early gastrulation stage (Pfeffer et al. 2017). The expression of NYNRIN was very low in porcine trophectoderm cells at the late blastocyst, peri-elongating, and elongating stages (Liu et al. 2021). Compared with humans that have more-invasive hemochorial placentae, the expression of NYNRIN was found in cytotrophoblasts, syncytiotrophoblasts (Pavličev et al. 2017; West et al. 2019), and extravillous trophoblasts (West et al. 2019) at peri-implantation or term periods. Although NYNRIN could have a role in trophoblast invasion of both marsupials and placentals, further investigation is required to indicate whether the expression level of NYNRIN in trophoblasts of therian mammals is associated with the degree of invasiveness. It would be noted that the absence of gene expression or the low gene expression in the single-cell RNA-Seq experiments could be attributed to technical limitations (Bacher and Kendziorski, 2016; Hicks et al. 2018).
Comparing NYNRIN to PEG10 would be of particular interest, as PEG10 also originated in the therian stem lineage (Kaneko-Ishino and Ishino 2012), derived from a Ty3/Gypsy, and is known to promote trophoblast invasion (Chen et al. 2015). Our analyses did not identify PEG10 because non-PEG10 LTR retroelement gag–derived proteins in two lissamphibian and eight saurian species were erroneously clustered into the PEG10 ortholog group (supplementary table S2, Supplementary Material online). In terms of domain architecture, NYNRIN lacks a gag-derived major homology domain, a gag-derived zinc knuckle, and a pol-derived protease domain, all of which are present in PEG10 of the representative therian species (Clark et al. 2007). On the other hand, NYNRIN contains RNase H and INT domains, which are mostly known for their functions in the integration of retroelements into host genomes (Finnegan 2012). The roles of RNase H and INT domains in therian hosts remain unclear. Given that the reverse transcriptase and INT proteins of HIV-1 retrovirus can interact with 37 and 136 human proteins, respectively (Fahey et al. 2011; Jäger et al. 2012), the RNase H domain, which is required for reverse transcriptase to be functional (Julias et al. 2002), and the INT domain of NYNRIN may have functions in therian hosts. Even though the ubiquitin-binding domain of NYNRIN was lost during the origination of NYNRIN, the INT domain of NYNRIN may still be able to interact with host proteins related to ubiquitination and proteasome degradation because the INT domains of HIV-1 and avian leukosis virus are targetable by both lysine and nonlysine ubiquitination (Zheng and Yao 2013; Wang et al. 2017). Notably, the first cysteine in the HHCC motifs of INT domains is conserved in all therian species and can be targeted by nonlysine ubiquitination (Wang et al. 2017; supplementary fig. S4D, Supplementary material online).
The collective invasion of trophoblast-lineage cells involves mechanisms similar to tumor migration and metastasis (Kshitiz et al. 2019). Our results showed that NYNRIN KD suppressed the invasion of HTR8/SVneo cells through downregulation of endoplasmic reticulum stress-related HSP90AB1 and PRNP, both of which have been shown to promote cancer cell invasion and metastasis (Wang et al. 2019a; Ding et al. 2021) and placental development (Voss et al. 2000; Alfaidy et al. 2013). However, NYNRIN KD was also shown to promote the invasion of human metastatic melanoma cells (Leonard et al. 2021). These observations suggest that NYNRIN has a complex role in trophoblast and cancer cell invasion. In addition, it remains unclear how NYNRIN mechanistically promotes trophoblast invasion. A previous study showed that mutating the proximal therian-conserved aspartic acid in the DDDD motif of the NYN domain of ZC3H12C did not affect the invasion of ZC3H12C-overexpressing human colorectal cancer cells (Suk et al. 2018). This indicates that the nuclease activity of the NYN domain would not contribute to trophoblast invasion in therian mammals. Further investigation, such as conducting in vivo experiments and deletion of the LTR retroelement–derived domains, is required to reveal the role of NYNRIN and its LTR retroelement–derived domains in the therian placental emergence at the organ level and to gain a broader understanding of viral-host interactions and mammalian evolution.
Methods
Data Collection and Preprocessing
Genomic and protein sequences of 267 tetrapod species (supplementary table S5, Supplementary Material online) were retrieved from the NCBI RefSeq (O’Leary et al. 2016) and Ensembl (Howe et al. 2021) databases (April 20, 2021). For human and mouse genomes, alternative loci were excluded to remove duplicated genes, and patch annotation was applied to substitute the original one. RNA-seq reads from placental tissues of 13 therian species and chorioallantoic membranes of a saurian species were retrieved from the NCBI SRA database (supplementary table S6, Supplementary Material online). Reads from the placentae of humans (GSE66622), whose transcriptomic expression may be affected by drinking, infection, diabetes, medication, and cesarean delivery were excluded. Adapters and low-quality leading and trailing edges (Q-score < 20) were trimmed using Trimmomatic v.0.36 (Bolger et al. 2014). Trimmed reads with lengths of < 36 base pairs were removed. The quality of the trimmed reads was first assessed by FastQC v.0.11.7 (Andrew et al. 2018) and then by aligning them to their reference genomic sequences using HISAT2 v.2.1.0 (Kim et al. 2015) in default mode. Only read samples with overall alignment rates exceeding 70% were retained for downstream analyses. We utilized the –merge mode of StringTie v.2.0.3 (Kovaka et al. 2019) to generate a set of nonredundant transcripts for each species. Gene expression levels were then quantified using Salmon v.1.4.0 (Patro et al. 2017) in default mode.
LTR Retroelement Searching
We integrated three search methods (supplementary fig. S11, Supplementary Material online) to identify LTR retroelements in 267 collected genomes. First, we utilized RepeatMasker v.4.1.1 (Smit et al. 2020) in the sensitive mode to search for LTR retroelements using the RMBlast search engine and tetrapod repetitive element search library. Other repetitive elements found using RepeatMasker were discarded. Second, we used LtrDetector (Valencia and Girgis 2019) to perform a de novo search for full-length LTR retroelements in the collected genomes. To identify and classify LTR retroelements found from LtrDetector, we collected additional data sets including 420 profile hidden Markov models of LTR retroelement and other related proteins from the Pfam database v.33.1 (Mistry et al. 2021) and Gypsy database v.2.0 (Llorens et al. 2011; supplementary table S7, Supplementary Material online), 477,258 RefSeq viral proteins from the NCBI Virus database (Brister et al. 2015), taxonomic data of orterviruses (Order Ortervirales; viral sources of LTR retroelements) from the NCBI Taxonomy database (Schoch et al. 2020), and 2,636 retroelement and other related proteins from the Gypsy core protein database (supplementary table S8, Supplementary Material online). Open reading frames (ORFs) in the putative full-length LTR retroelements, whose translated proteins are at least 80 amino acids in length, were predicted using EMBOSS getorf v.6.6.0.0 (Rice et al. 2000). Putative LTR retroelements were then retrieved from the predicted ORFs by performing a profile hidden Markov model search against the 420 collected models using HMMER v.3.3 (Mistry et al. 2013) with an E-value ≤ 1 × 10−5. LTR retroelements were also identified by aligning the predicted ORFs with the RefSeq viral proteins and GyDB proteins using Diamond v.2.0.9 (Buchfink et al. 2021) with the –more-sensitive mode and E-value ≤ 1 × 10−5. Putative LTR retroelements found by HMMER and Diamond were classified, as described in supplementary fig. S11, Supplementary Material online. Unidentified ORFs and non-ORFs were excluded from the downstream analysis. Third, we searched for LTR retroelements in RefSeq-translated coding sequences (CDSs) or Ensembl proteins using the same methods as those used for putative LTR retroelement ORFs found in putative full-length LTR retroelements.
Species Tree Reconstruction
We gathered CDSs of 66 genes (supplementary table S9, Supplementary Material online) that were present in all 267 tetrapod species for species tree reconstruction. The most optimal CDS isoform of each gene in each species for multiple sequence alignment was selected using IsoSel v.1.0 (Philippon et al. 2017) with the -auto mode. The selected CDSs of each gene were subjected to multiple sequence alignment using MAFFT v.7.427 (Katoh and Standley 2013) with the E-INS-i method and 1,000 maximum iterative refinements. The aligned CDSs were then concatenated without gap treatment to form the representative sequences of each species. The total length of each concatenated alignment was 324,405 base pairs, with 137,604 parsimony-informative sites. Subsequently, we used the maximum likelihood–based IQ-TREE v.2.0.7 (Minh et al. 2020) to reconstruct the species tree. The best-fit nucleotide substitution models and partitioning schemes were automatically selected by ModelFinder (Kalyaanamoorthy et al. 2017) and PartitionFinder (Chernomor et al. 2016) in the IQ-TREE suite, respectively. Branch support was assessed using ultrafast bootstrap approximation (Hoang et al. 2018) with 1,000 bootstrap replicates. There were 90.15% internal nodes with branch support values ≥ 95% (supplementary fig. S12, Supplementary Material online). The reconstructed tree was visualized using iTOL v6 (Letunic and Bork 2021).
Ortholog Group Inference
The completeness of the proteome data sets was assessed using BUSCO v.4.1.3 (Manni et al. 2021) with the OrthoDB tetrapod reference protein set. The longest protein isoform for each gene in the 267 species was retrieved to infer hierarchical ortholog groups (HOGs; N0.tsv) using OrthoFinder v.2.5.2 (Emms and Kelly 2019) with default parameters. The reconstructed species tree was input into OrthoFinder. To ensure the accuracy of the presence/absence of homologous proteins in any species, additional nonredundant proteins (supplementary table S5, Supplementary Material online) were retrieved from the NCBI Protein (May 10, 2021) and Ensembl Release 102 databases and aligned to the proteins that OrthoFinder already assigned to graph-based ortholog groups (GOGs; orthogroups.tsv). Alignment was performed using Diamond with the –more-sensitive mode and E-value ≤ 0.001, which were the same as those used by OrthoFinder to infer ortholog groups. Each additional protein was assigned to the inferred HOG of the best-match assigned protein (the protein with the highest bit score) if (1) the best-match assigned protein did not belong to the same species, as this implies that the two proteins are likely isoforms of each other; (2) the additional protein had alignment hits with at least 95% of all assigned proteins in the best-match GOG; and (3) each average of identity, query coverage, subject coverage, bit score, and alignment length of all alignment hits between the additional protein and assigned proteins in the best-match GOG fell within ± 2SD of the alignment statistics obtained when aligning assigned proteins within the best-match GOG.
Reconstruction of the Gene Ancestral States
We first assigned the states of the genes in each HOG to each extant tetrapod species. The states for the presence/absence of genes and their expression were categorized as “lack gene sequence and expression in placenta/chorioallantoic membrane” (state 0), “have gene sequence(s) but lack expression in placenta/chorioallantoic membrane” (state 1), and “have gene sequence(s) and expression in placenta/chorioallantoic membrane” (state 2). The presence/absence of gene sequences was directly derived from the presence/absence of homologous proteins in each HOG. The presence/absence of gene expression was determined by the zFPKM method using the zFPKM R package v.1.12.0 (Hart et al. 2013; Ammar and Thompson 2020). Genes with zFPKM averages > −3 were considered actively expressed (have expression), and those with zFPKM averages ≤ −3 were considered inactive (lack expression). For species that lacked expression information, an ambiguous state 1/2 was assigned. If there was more than one protein in each species, we used the rightmost possible state from the order [0 → 1 → 1/2 → 2] to represent the state of the protein group. Similarly, the states for the presence/absence of LTR retroelements that became gene introns or exons were categorized as “lack LTR retroelement” (state A), “all LTR retroelements are introns” (state I), and “at least an LTR retroelement is an exon” (state E). The order of the states was [A/I/E → A/I → A → I → E]. The locations of gene introns and exons were based on the NCBI RefSeq or Ensembl annotations. Ancestral states were then estimated using the DOWNPASS and marginal posterior probability approximation methods of PastML v.1.9.33 (Ishikawa et al. 2019) with default parameters. The gain and loss events of gene sequence, gene expression, and integration of LTR retroelements (fig. 1CandD) were then inferred from the state changes from each ancestral node to each of its immediate descendant nodes. The results were visualized using the phytools R package v.0.7.80 (Revell 2012).
Cell Culture
The human choriocarcinoma BeWo cell line was purchased from the JCRB Cell Bank (Osaka, Japan), whereas the human HTR8/SVneo invasive-type trophoblast cell line was kindly provided by Dr Charles Graham (Queen’s University Kingston, ON, Canada). BeWo cells were cultured in Ham’s F-12/DMEM (1:1; Fujifilm Wako Pure Chemical Corp., Osaka, Japan). HTR8/SVneo cells were cultured in Roswell Park Memorial Institute 1640 medium (Fujifilm Wako Pure Chemical Corp.). The media were supplemented with 10% fetal bovine serum (FBS; Nichirei Biosciences, Tokyo, Japan) and 1% penicillin-streptomycin-neomycin antibiotic mixture (Thermo Fisher Scientific, Waltham, MA, USA). Cells were maintained at 37 °C under 5% CO2 in a humidified incubator.
siRNA Transfection and Trial Experiments
All siRNAs for NYNRIN (siNYNRIN) and KHNYN (siKHNYN) were commercially predesigned (Sigma-Aldrich, Tokyo, Japan; supplementary table S10, Supplementary material online). BeWo or HTR8/SVneo cells that had been grown in 24-well plates (3 × 104 cells/well) for 24 h were transfected with nontargeting control siRNA (siCtrl; Thermo Fisher Scientific), siNYNRIN, or siKHNYN using 3 µl of Lipofectamine RNAiMAX reagent (Thermo Fisher Scientific) according to the manufacturer’s instructions. The final concentration of each siRNA was 50 nM. The cells were subsequently collected to quantify the expression levels of NYNRIN or KHNYN using quantitative real-time polymerase chain reaction (PCR). The off-target effects of siNYNRIN on KHNYN mRNA (XM_011536590.2) and siKHNYN on NYNRIN mRNA (NM_025081.3) were assessed by performing global alignment between the siRNA and the nontargeting gene using the EMBOSS Needle web services of EMBL-EBI (Madeira et al. 2019) with default parameters.
Quantification of Gene Expression Level by Quantitative Real-Time PCR
RNA was extracted from the cultured cells using the RNeasy Mini Kit (Qiagen, Tokyo, Japan) according to the manufacturer’s protocol. mRNA was reverse transcribed using the ReverTra Ace qPCR RT Kit (Toyobo, Osaka, Japan), and the cDNA produced was subjected to quantitative real-time PCR amplification in a PowerUP SYBR Green Master Mix (Thermo Fisher Scientific; supplementary table S11, Supplementary material online). GAPDH was used as the reference gene. The cycle threshold of each gene was determined using Sequence Detection System software v2.3 (Thermo Fisher Scientific). The relative expression level of each target gene was quantified with regard to the amplification efficiencies of the target and reference genes.
Cell Fusion Assay
BeWo cells that had been grown in 24-well plates (3 × 104 cells/well) for 24 h were transfected with siCtrl or each of the three siNYNRINs as described earlier. Fusion of BeWo cells in noncontrol groups was induced by treatment with dibutyryl cAMP (Db; 500 μM; Tokyo Chemical Industry Co., Tokyo, Japan) for 72 h. The BeWo cells were then fixed with methanol and stained with an antidesmoglein-1 antibody (clone 32-2B; 1:200; Merck Millipore, Burlington, MA, USA) together with an Alexa Fluor 594 goat antimouse antibody (Thermo Fisher Scientific) to distinguish cell surfaces. Nuclei were counterstained with 4′,6-diamino-2-phenylindole·2HCl (DAPI). The stained cells were examined under a digital microscope (BZ-X810; Keyence, Osaka, Japan). The cells for measuring CGB expression were collected separately without fixation and staining.
Cell Invasion Assay
HTR8/SVneo cells that had been grown in 24-well plates (3 × 104 cells/well) for 24 h were transfected with siCtrl or each of the three siNYNRINs or siKHNYNs as described earlier. An invasion assay of HTR8/SVneo cells was then performed using the transwell system (Chemotaxicell; Kurabo, Osaka, Japan) equipped with 8-µm-pore polycarbonate filters. The HTR8/SVneo cells were resuspended in the basal medium containing 2% FBS and loaded into the upper compartment of the filter, which had been coated with Matrigel (Corning, Inc., Corning, NY, USA). The filters were then placed into the 24-well plate containing the basal medium supplemented with 2% FBS for 48 h. Cells that invaded beyond the lower surface of the filters were fixed with cold methanol, stained with DAPI, and examined under the digital microscope. In each experiment, the number of cells in five randomly chosen microscopic fields per filter was counted.
Statistical Tests for Cell Invasion Assay and Quantitative Real-Time PCR
All statistical tests were performed using R v.4.0.3 (R Core Team 2020). Normality and homogeneity of variances of data were initially tested using the Shapiro–Wilk test and Levene’s test, respectively. The ɑ-value for both tests was set to 0.05. Based on the test results, the Kruskal–Wallis test was selected to compare differences between medians of samples in each experimental group, followed by Dunn’s post hoc test. Benjamini–Hochberg correction was applied for multiple comparisons. The 95% confidence interval for the median was calculated by the groupwiseMedian function of the rcompanion R package v.2.4.1 (Mangiafico 2021) using 5,000 bootstrap replicates.
Immunohistochemistry
Placental tissues of normal patients and those of patients with preeclampsia and intrauterine growth restriction were obtained in their third trimester (32 weeks of gestation; n = 3) during cesarean sections. The use of the tissues was approved by the Clinical Research Ethics Committee of Kyushu University and Tokyo University of Pharmacy and Life Sciences (#1512), and informed consent was obtained from the participants. The tissues were directly fixed in 0.2 M phosphate buffer (pH 7.45, room temperature) containing 4% paraformaldehyde for 8 h before being embedded in paraffin and sectioned into 6-μm sections. Before the tissues were immunostained for NYNRIN or major histocompatibility complex class I, G (HLA-G), the sections had been deparaffinized, rehydrated, and boiled in 10 mM citrate buffer (pH 6.0) for 20 min. The sections were then incubated overnight at 4 °C with a rabbit polyclonal anti-NYNRIN antibody (HPA018945; 1:100; Atlas Antibodies, Stockholm, Sweden), a mouse monoclonal anti-HLA-G antibody (ab52455; 1:100; Abcam, Tokyo, Japan), or a normal rabbit IgG (sc-2027; 1:100; Santa Cruz Biotechnology, Dallas, TX, USA). The sections were subsequently incubated with Histofine Simple Stain MAX-PO (Nichirei Biosciences) and DAB (Fujifilm Wako Pure Chemical Corp.), counterstained with hematoxylin, and examined under the digital microscope.
Gene Functional Comparisons
RNA samples from HTR8/SVneo cells transfected with siCtrl, siNYNRIN2, or siKHNYN1 (n = 3) were submitted to Macrogen Japan (Tokyo, Japan) for sequencing. The integrity of the RNA samples was assessed using Agilent Technologies 2200 TapeStation. All RNA samples passed the minimum criterion for RNA integrity. A cDNA library was then generated from each RNA sample using TruSeq-stranded mRNA and subjected to paired-end read sequencing using the Illumina NovaSeq 6000 platform. Transcriptomic reads of HTR8/SVneo cells were preprocessed, assessed for quality, and quantified in the same manner as those of placental tissue samples (see Data Collection and Preprocessing). Differential expression analyses were performed using DEseq2 (Love et al. 2014) with the apeglm method (Zhu et al. 2019) to estimate the fold change in gene expression. Genes were considered differentially expressed if their Benjamini–Hochberg adjusted P-values were < 0.05. Principal component analysis of gene expression profiles was performed using the PCAtools R package v.2.3.13 (Blighe and Lun 2020). Volcano plots were generated using the EnhancedVolcano R package v.1.9.13 (Blighe et al. 2018). Concordant and discordant gene expression signatures were identified and visualized using the RRHO2 R package v.1.0 (Cahill et al. 2018). The levels of concordance/discordance in gene regulation across the two RNA-seq experiments were ranked by absolute values of disco scores (Domaszewska et al. 2017) which were calculated from the equation |disco score| = |log2FCNYNRIN · log2FCKHNYN · (log PNYNRIN + log PKHNYN)|, where FC is the gene expression fold change and P is the significant value of the differential expression from DESeq2. The libraries of 2021 GO and KEGG terms and 2019 MGI Mammalian Phenotype Level 4 terms were retrieved from the Enrichr website (https://maayanlab.cloud/Enrichr/#libraries). Gene set enrichment analyses were performed and visualized using the clusterProfiler R package v.3.18.1 (Yu et al. 2012). Ontology terms were considered significantly enriched if their Benjamini–Hochberg adjusted P-values were < 0.05.
Protein Structural and Functional Comparisons
Protein sequences of NYNRIN, KHNYN, N4BP1, ZC3H12C, and ZC3H12A in 267 tetrapod species were subjected to optimal protein isoform selection using IsoSel with the -auto mode (supplementary table S12, Supplementary material online). Multiple sequence alignment of the selected proteins was performed using MAFFT with the L-INS-i method and 1,000 maximum iterative refinements. Multiple sequence alignment results were analyzed and visualized using AliView v.1.27 (Larsson 2014) or Jalview v.2.11.1.14 (Waterhouse et al. 2009). The residue positions of the KH-like, NYN, RT, and INT domains were defined based on the annotations of the human NYNRIN (UniProt ID: Q9P2P1) and KHNYN (UniProt ID: O15037) collected from the Pfam and InterPro databases. The residue positions of the human KHNYN CUBAN and N4BP1 CoCUN domains have been previously identified (Castagnoli et al. 2019). The residue positions of DDDD motifs in the NYN domains and HHCC motifs in the INT domains of NYNRIN were determined from the residue positions of the DDDD motif in the human ZC3H12A NYN domain (Habacher and Ciosk 2017) and the HHCC motifs in human and short-tailed opossum NYNRIN INT domains (Marco and Marín 2009), respectively. The 3D structures of human NYNRIN and KHNYN were retrieved from the AlphaFold protein structure database (Jumper et al. 2021). Intrinsically disordered regions of both proteins were predicted using the IUPred3 web server (Erdős et al. 2021) with long disorder and medium smoothing options. Structural alignments of specific domains were performed using the RaptorX Structure Alignment web server (Wang et al. 2013a). All 3D protein structures were visualized using Chimera-X v.1.2.5 (Pettersen et al. 2021). Protein subcellular localization was predicted using the DeepLoc-1.0 web server with the protein profiles option (Armenteros et al. 2017).
Supplementary Material
Acknowledgments
The authors thank Motomu Matsui, Salvatore Cosentino, and other members of Iwasaki Laboratory at the University of Tokyo for their helpful comments and suggestions regarding this research. This work was financially supported by the Japan Science and Technology Agency (CREST JPMJCR19S2) and Japan Society for the Promotion of Science (KAKENHI 16H06279 and 22H04925).
Contributor Information
Arnon Plianchaisuk, Department of Computational Biology and Medical Sciences, Graduate School of Frontier Sciences, The University of Tokyo, Kashiwa, Chiba 277-0882, Japan.
Kazuya Kusama, Department of Endocrine Pharmacology, Tokyo University of Pharmacy and Life Sciences, Hachioji, Tokyo 192-0392, Japan.
Kiyoko Kato, Department of Obstetrics and Gynecology, Graduate School of Medical Sciences, Kyushu University, Fukuoka, Fukuoka, Japan.
Sira Sriswasdi, Center of Excellence in Computational Molecular Biology, Research Affairs, Faculty of Medicine, Chulalongkorn University, Pathum Wan, Bangkok 10330, Thailand.
Kazuhiro Tamura, Department of Endocrine Pharmacology, Tokyo University of Pharmacy and Life Sciences, Hachioji, Tokyo 192-0392, Japan.
Wataru Iwasaki, Department of Computational Biology and Medical Sciences, Graduate School of Frontier Sciences, The University of Tokyo, Kashiwa, Chiba 277-0882, Japan; Department of Integrated Biosciences, Graduate School of Frontier Sciences, The University of Tokyo, Kashiwa, Chiba 277-0882, Japan; Department of Biological Sciences, Graduate School of Science, The University of Tokyo, Bunkyo-ku, Tokyo 113-0032, Japan; Atmosphere and Ocean Research Institute, The University of Tokyo, Kashiwa, Chiba 277-0882, Japan; Institute for Quantitative Biosciences, The University of Tokyo. Bunkyo-ku, Tokyo 113-0032, Japan; Collaborative Research Institute for Innovative Microbiology, The University of Tokyo, Bunkyo-ku, Tokyo 113-0032, Japan.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Author Contributions
A.P. handled the in silico data collection and analysis. K. Kazuya, K.T., and K. Kiyoko conducted laboratory experiments. A.P. and K. Kazuya analyzed the experimental laboratory data. A.P. and W.I. wrote the manuscript with contributions from the co-authors. W.I. directed and supervised the research, with contributions from S.S.
Data Availability
All new transcriptomic data from this study were submitted to the NCBI SRA database which can be accessed through the BioProject accession number PRJNA830988.
Conflict of interest. The authors declare no competing financial interests.
References
- Gene Ontology Consortium . 2021. The Gene Ontology resource: enriching a GOld mine. Nucleic Acids Res. 49:D325–D334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abed M, Verschueren E, Budayeva H, Liu P, Kirkpatrick DS, Reja R, Kummerfeld SK, Webster JD, Gierke S, Reichelt M, et al. 2019. The Gag protein PEG10 binds to RNA and regulates trophoblast stem cell lineage specification. PLoS One 14:e0214110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abou-Kheir W, Barrak J, Hadadeh O, Daoud G. 2017. HTR-8/SVneo cell line contains a mixed population of cells. Placenta 50:1–7. [DOI] [PubMed] [Google Scholar]
- Alfaidy N, Chauvet S, Donadio-Andrei S, Salomon A, Saoudi Y, Richaud P, Aude-Garcia C, Hoffman P, Andrieux A, Moulis J, et al. 2013. Prion protein expression and functional importance in developmental angiogenesis: role in oxidative stress and copper homeostasis. Antioxid Redox Signal. 18:400–411. [DOI] [PubMed] [Google Scholar]
- Ammar R, Thompson J.. 2020. zFPKM: a suite of functions to facilitate zFPKM transformations. R package version 1.12.0. [accessed 2020 Oct 29]. Available from: https://github.com/ronammar/zFPKM/.
- Anantharaman V, Aravind L. 2006. The NYN domains: novel predicted RNAses with a PIN domain-like fold. RNA Biol. 3:18–27. [DOI] [PubMed] [Google Scholar]
- Andrew S, Lindenbaum P, Howard B, Ewels P.. 2018. FastQC high throughput sequence QC report. Version 0.11.7. [accessed 2018 Jan 10]. Available from: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- Anton L, Brown AG, Bartolomei MS, Elovitz MA. 2014. Differential methylation of genes associated with cell adhesion in preeclamptic placentas. PLoS One. 9:e100148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armenteros JJA, Sønderby CK, Sønderby SK, Nielsen H, Winther O. 2017. DeepLoc: prediction of protein subcellular localization using deep learning. Bioinformatics 33:3387–3395. [DOI] [PubMed] [Google Scholar]
- Armstrong DL, McGowen MR, Weckle A, Pantham P, Caravas J, Agnew D, Benirchke K, Savage-Rumbaugh S, Nevo E, Kim CJ, et al. 2017. The core transcriptome of mammalian placentas and the divergence of expression with placental shape. Placenta 57:71–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacher R, Kendziorski C. 2016. Design and computational analysis of single-cell RNA-sequencing experiments. Genome Biol. 17:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Stamatoyannopoulos JA, Costello JF, Ren B, Milosavljevic A, Meissner A, Kellis M, Marra MA, Beaudet AL, Ecker JR, et al. 2010. The NIH Roadmap Epigenomics Mapping Consortium. Nat Biotechnol. 28:1045–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn DG. 2015. Evolution of vertebrate viviparity and specializations for fetal nutrition: a quantitative and qualitative analysis. J Morphol. 276:961–990. [DOI] [PubMed] [Google Scholar]
- Blighe K, Lun A. 2020. PCAtools: everything principal components analysis. R package version 2.3.13.
- Blighe K, Rana S, Lewis M. 2018. EnhancedVolcano: publication-ready volcano plots with enhanced colouring and labeling. R package version. 2.3.13.
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brister JR, Ako-Adjei D, Bao Y, Blinkova O. 2015. NCBI viral genomes resource. Nucleic Acids Res. 43:D571–D577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchfink B, Reuter K, Drost HG. 2021. Sensitive protein alignments at tree-of-life scale using DIAMOND. Nat Methods 18:366–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahill KM, Huo Z, Tseng GC, Logan RW, Seney ML. 2018. Improved identification of concordant and discordant gene expression signatures using an updated rank-rank hypergeometric overlap approach. Sci Rep. 8:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castagnoli L, Mandaliti W, Nepravishta R, Valentini E, Mattioni A, Procopio R, Iannuccelli M, Polo S, Paci M, Cesareni G, et al. 2019. Selectivity of the CUBAN domain in the recognition of ubiquitin and NEDD8. FEBS J. 284:653–677. [DOI] [PubMed] [Google Scholar]
- Chalopin D, Naville M, Plard F, Galiana D, Volff J. 2015. Comparative analysis of transposable elements highlights mobilome diversity and evolution in vertebrates. Genome Biol Evol. 7:567–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheloufi S, Dos Santos CO, Chong MMW, Hannon GJ. 2010. A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature 465:584–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Sun M, Liu J, Tong C, Meng T. 2015. Silencing of paternally expressed gene 10 inhibits trophoblast proliferation and invasion. PLoS One 10:e0144845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernomor O, von Haeseler A, Minh BQ.. 2016. Terrace aware data structure for phylogenomic inference from supermatrices. Syst Biol. 65:997–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark MB, Jänicke M, Gottesbühren U, Kleffmann T, Legge M, Poole ES, Tate WP. 2007. Mammalian gene PEG10 expresses two reading frames by high efficiency -1 frameshifting in embryonic-associated tissues. J Biol Chem. 282:37359–37369. [DOI] [PubMed] [Google Scholar]
- Cornelis G, Funk M, Vernochet C, Leal F, Tarazona OA, Meurice G, Heidmann O, Dupressoir A, Miralles A, Ramirez-Pinilla MP, et al. 2017. An endogenous retroviral envelope syncytin and its cognate receptor identified in the viviparous placental Mabuya lizard. Proc Natl Acad Sci U S A. 114:E10991–E11000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelis G, Vernochet C, Carradec Q, Souquere S, Mulot B, Catzeflis F, Nilsson MA, Menzies BR, Renfree MB, Pierron G, et al. 2015. Retroviral envelope gene captures and syncytin exaptation for placentation in marsupials. Proc Natl Acad Sci U S A. 112:E487–E496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl KDC, Robertson SE, Weaver VM, Simon MC. 2005. Hypoxia-inducible factor regulates alphavbeta3 integrin cell surface expression. Mol Biol Cell. 16:1901–1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding M, Chen Y, Lang Y, Cui L. 2021. The role of cellular prion protein in cancer biology: a potential therapeutic target. Front Oncol. 11:742949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domaszewska T, Scheuermann L, Hahnke K, Mollenkopf H, Dorhoi A, Kaufmann SHE, Weiner J 3rd. 2017. Concordant and discordant gene expression patterns in mouse strains identify best-fit animal model for human tuberculosis. Sci Rep. 7:12094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupressoir A, Vernochet C, Bawa O, Harper F, Pierron G, Opolon P, Heidmann T. 2009. Syncytin-A knockout mice demonstrate the critical role in placentation of a fusogenic, endogenous retrovirus-derived, envelope gene. Proc Natl Acad Sci U S A. 106:12127–12132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emms DM, Kelly S. 2019. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 20:238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdős G, Pajkos M, Dosztányi Z. 2021. IUPred3: prediction of protein disorder enhanced with unambiguous experimental annotation and visualization of evolutionary conservation. Nucleic Acids Res. W1:W297–W303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahey ME, Bennett MJ, Mahon C, Jäger S, Pache L, Kumar D, Shapiro A, Rao K, Chanda SK, Craik CS, et al. 2011. GPS-Prot: a web-based visualization platform for integrating host-pathogen interaction data. BMC Bioinformatics 12:298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ficarelli M, Wilson H, Galão RP, Mazzon M, Antzin-Anduetza I, Marsh M, Neil SJ, Swanson CM. 2019. KHNYN is essential for the zinc finger antiviral protein (ZAP) to restrict HIV-1 containing clustered CpG dinucleotides. Elife 8:e46767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnegan DJ. 2012. Retrotransposons. Curr Biol. 22:R432–R437. [DOI] [PubMed] [Google Scholar]
- Griffith OW, Bradley MC, Whittington CM, Belov K, Thompson MB. 2017. Comparative genomics of hormonal signaling in the chorioallantoic membrane of oviparous and viviparous amniotes. Gen Comp Endocrinol. 244:19–29. [DOI] [PubMed] [Google Scholar]
- Griffith OW, Wagner GP. 2017. The placenta as a model for understanding the origin and evolution of vertebrate organs. Nat Ecol Evol. 1:72. [DOI] [PubMed] [Google Scholar]
- Guernsey MW, Chuong EB, Cornelis G, Renfree MB, Baker JC. 2017. Molecular conservation of marsupial and eutherian placentation and lactation. eLife 6:e27450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habacher C, Ciosk R. 2017. ZC3H12A/MCPIP1/Ragnase-1-related endonucleases: an evolutionary perspective on molecular mechanisms and biological functions. Bioessays 39:1700051. [DOI] [PubMed] [Google Scholar]
- Hart T, Komori HK, LaMere S, Podshivalora K, Salomon DR. 2013. Finding the active genes in deep RNA-seq gene expression studies. BMC Genomics 14:778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks SC, Townes FW, Teng M, Irizarry RA. 2018. Missing data and technical variability in single-cell RNA-sequencing experiments. Biostatistics 19:562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS.. 2018. UFBoot2: improving the ultrafast bootstrap approximation. Mol Biol Evol. 35:518–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe KL, Achuthan P, Allen J, Allen J, Alvarez-Jarreta J, Amode MR, Armean IM, Azov AG, Bennett R, Bhai J, et al. 2021. Ensembl 2021. Nucleic Acids Res. 49:D884–D891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa SA, Zhukova A, Iwasaki W, Gascuel O. 2019. A fast likelihood method to reconstruct and visualize ancestral scenarios. Mol Biol Evol. 36:2069–2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jäger S, Cimermancic P, Gulbahce N, Johnson JR, McGovern KE, Clarke SC, Shales M, Mercenne G, Pache L, Li K, et al. 2012. Global landscape of HIV-human protein complexes. Nature 481:365–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones CJP, Skipper J, Renfree MB, Aplin JD. 2014. Trophoblast specialisations during pregnancy in the tammar wallaby, Macropus eugenii: a morphological and lectin histochemical study. Placenta 35:467–475. [DOI] [PubMed] [Google Scholar]
- Julias JG, McWilliams MJ, Sarafianos SG, Arnold E, Hughes SH. 2002. Mutations in the RNase H domain of HIV-1 reverse transcriptase affect the initiation of DNA synthesis and the specificity of RNase H cleavage in vivo. Proc Natl Acad Sci U S A. 99:9515–9520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Židek A, Potapenko A, et al. 2021. Highly accurate protein structure prediction with AlphaFold. Nature 596:583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaloglu C, Onarlioglu B. 2010. Extracellular matrix remodelling in rat endometrium during early pregnancy: the role of fibronectin and laminin. Tissue Cell 42:301–306. [DOI] [PubMed] [Google Scholar]
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14:587–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Furumichi M, Sato Y, Ishiguro-Watanabe M, Tanabe M. 2021. KEGG: integrating viruses and cellular organisms. Nucleic Acids Res. 49:D545–D551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko-Ishino T, Ishino F. 2012. Evolution of viviparity and genomic imprinting in mammals by retrotransposons. In: Pontarotti P, editor. Evolutionary biology: mechanisms and trends. Heidelberg, New York, Dordrecht, London: Springer. p. 265–281. [Google Scholar]
- Karlsson M, Zhang C, Méar L, Zhong W, Digre A, Katona B, Sjöstedt E, Butler L, Odeberg J, Dusart P, et al. 2021. A single-cell transcriptomics map of human tissues. Sci Adv. 7:eabh2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Frey WD, He H, Kim H, Ekram MB, Bakshi A, Faisal M, Perera BPU, Ye A, Teruyama R. 2013. Peg3 mutational effects on reproduction and placenta-specific gene families. PLoS One 8:e3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Langmead B, Salzberg SL. 2015. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 12:357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohrman AQ, Matus DQ. 2017. Divide or conquer: cell cycle regulation of invasive behavior. Trends Cell Biol. 27:12–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolahi KS, Valent AM, Thornburg KL. 2017. Cytotrophoblast, not syncytiotrophoblast, dominates glycolysis and oxidative phosphorylation in human term placenta. Sci Rep. 7:42941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovaka S, Zimin AV, Pertea GM, Razaghi R, Salzberg SL, Pertea M. 2019. Transcriptome assembly from long-read RNA-seq alignments with StringTie2. Genome Biol. 20:278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kshitiz AJ, Maziarz JD, Hamidzadeh A, Liang C, Erkenbrack EM, Kim HN, Haeger J, Pfarrer C, Hoang T, et al. 2019. Evolution of placental invasion and cancer metastasis are causally linked. Nat Ecol Evol. 3:1743–1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. 2001. Initial sequencing and analysis of the human genome. Nature 409:860–921. [DOI] [PubMed] [Google Scholar]
- Larsson A. 2014. AliView: a fast and lightweight alignment viewer and editor for large datasets. Bioinformatics 30:3276–3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Veerbeek JHW, Rana TK, van Rijn BB, Burton GJ, Yung HW. 2019. Role of endoplasmic reticulum stress in proinflammatory cytokine-mediated inhibition of trophoblast invasion in placenta-related complications of pregnancy. Am J Pathol. 189:467–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard MK, Puts GM, Pamidimukkala N, Adhikary G, Xu Y, Kwok E, Jin Y, Snyder D, Matsangos N, Novak M, et al. 2021. Comprehensive molecular profiling of UV-induced metastatic melanoma in Nme1/Nme2-deficient mice reveals novel markers of survival in human patients. Oncogene 40:6329–6342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Bork P. 2021. Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49:W293–W296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T, Li J, Yu L, Sun H, Li J, Dong G, Hu Y, Li Y, Shen Y, Wu J, et al. 2021. Cross-species single-cell transcriptomic analysis reveals pre-gastrulation developmental differences among pigs, monkeys, and humans. Cell Discov. 7:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Zheng Q, Cui X, Dai K, Yang X, Li F, Yan Q. 2015. Expression of uPAR in human trophoblast and its role in trophoblast invasion. Int J Clin Exp Pathol. 8:14325–14334. [PMC free article] [PubMed] [Google Scholar]
- Llorens C, Futami R, Covelli L, Dominguez-Escriba L, Viu JM, Tamarit D, Aguilar-Rodríguez J, Vicente-Ripolles M, Fuster G, Bernet GP, et al. 2011. The Gypsy database (GyDB) of mobile genetic elements: release 2.0. Nucleic Acids Res. 39:D70–D74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Ander S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madeira F, Park YM, Lee J, Buso N, Gur T, Madhusoodanan N, Basutkar P, Tivey ARN, Potter SC, Finn RD, et al. 2019. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 47:W636–W641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahadevaiah SK, Sangrithi MN, Hirota T, Turner JMA. 2020. A single-cell transcriptome atlas of marsupial embryogenesis and X inactivation. Nature 586:612–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahamdallie S, Yost S, Poyastro-Pearson E, Holt E, Zachariou A, Seal S, Elliott A, Clarke M, Warren-Perry M, Hanks S, et al. 2019. Identification of new Wilms tumour predisposition genes: an exome sequencing study. Lancet Child Adolesc Health. 3:322–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangiafico S. 2021. rcompanion: functions to support extension education program evaluation. R package version 2.4.1.
- Manni M, Berkeley MR, Seppey M, Simão FA, Zdobnov EM. 2021. BUSCO update: novel and streamlined workflows along with broader and deeper phylogenetic coverage for scoring of eukaryotic, prokaryotic, viral genomes. Mol Biol Evol. 38:4647–4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marco A, Marín I. 2009. CGIN1: a retroviral contribution to mammalian genomes. Mol Biol Evol. 26:2167–2170. [DOI] [PubMed] [Google Scholar]
- Martinez F, Olvera-Sanchez S, Esparza-Perusquia M, Gomez-Chang E, Flores-Herrera O. 2015. Multiple functions of syncytiotrophoblast mitochondria. Steroids 103:11–22. [DOI] [PubMed] [Google Scholar]
- Mi S, Lee X, Li X, Veldman GM, Finnerty H, Racie L, LaVallie E, Tang X, Edouard P, Howes S, et al. 2000. Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature 403:785–789. [DOI] [PubMed] [Google Scholar]
- Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, Lanfear R.. 2020. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 37:1530–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mistry J, Chuguransky S, Williams L, Qureshi M, Salazar GA, Sonnhammer ELL, Tosatto SCE, Paladi L, Raj S, Richardson LJ, et al. 2021. Pfam: the protein families database in 2021. Nucleic Acids Res. 49:D412–D419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mistry J, Finn RD, Eddy SR, Bateman A, Punta M. 2013. Challenges in homology search: HMMER3 and convergent evolution of coiled-coil regions. Nucleic Acids Res. 41:e121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Msheik H, Azar J, El Sabeh M, Abou-Kheir W, Daoud G. 2020. HTR-8/SVneo: a model for epithelial to mesenchymal transition in the human placenta. Placenta 90:90–97. [DOI] [PubMed] [Google Scholar]
- Naruse M, Ono R, Irie M, Nakamura K, Furuse T, Hino T, Oda K, Kashimura M, Yamada I, Wakana S, et al. 2014. Sirh/Ldoc1 knockout mice exhibit placental P4 overproduction and delayed parturition. Development 141:4763–4771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicastro G, Taylor IA, Ramos A. 2015. KH-RNA interactions: back in the groove. Curr Opin Struct Biol. 30:63–70. [DOI] [PubMed] [Google Scholar]
- Oghbaei F, Zarezadeh R, Jafari-Gharabaghlou D, Ranjbar M, Nouri M, Fattahi A, Imakawa K. 2022. Epithelial-mesenchymal transition process during embryo implantation. Cell Tissue Res 388:1–17. [DOI] [PubMed] [Google Scholar]
- O’Leary NA, Wright MW, Brister JR, Ciufo S, Haddad D, McVeigh R, Rajput B, Robbertse B, Smith-White B, Ako-Adjei D, et al. 2016. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 44:D733–D745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono R, Nakamura K, Inoue K, Naruse M, Usami T, Wakisaka-Saito N, Hino T, Suzuki-Migishima R, Ogonuki N, Miki H, et al. 2006. Deletion of Peg10, an imprinted gene acquired from a retrotransposon, causes early embryonic lethality. Nat Genet. 38:101–106. [DOI] [PubMed] [Google Scholar]
- Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. 2017. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods. 14:417–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavličev M, Wagner GP, Chavan AR, Owens K, Maziarz J, Dunn-Fletcher C, Kallapur SG, Muglia L, Jones H. 2017. Single-cell transcriptomics of the human placenta: inferring the cell communication network of the maternal-fetal interface. Genome Res. 27:349–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Meng EC, Couch GS, Croll TI, Ferrin TE. 2021. UCSF ChimeraX: meeting modern challenges in visualization and analysis. Protein Sci. 30:70–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer PL, Smith CS, Maclean P, Berg DK. 2017. Gene expression analysis of bovine embryonic disc, trophoblast and parietal hypoblast at the start of gastrulation. Zygote 25:265–278. [DOI] [PubMed] [Google Scholar]
- Philippon H, Souvane A, Brochier-Armanet C, Perrière G. 2017. IsoSel: protein isoform selector for phylogenetic reconstructions. PLoS One 12:e0174250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai VV, Siqueira LG, Das M, Kei TG, Tu LN, Herren AW, Phinney BS, Cheong SH, Hansen PJ, Selvaraj V. 2019. Physiological profile of undifferentiated bovine blastocyst-derived trophoblasts. Biol Open. 8:bio037937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollheimer J, Vondra S, Baltayeva J, Beristain AG, Knöfler M. 2018. Regulation of placental extravillous trophoblasts by the maternal uterine environment. Front Immunol. 9:2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team . 2020. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. [accessed 2020 Dec 2]. Available from: https://www.R-project.org/. [Google Scholar]
- Revell LJ. 2012. phytools: an R package for phylogenetic comparative biology (and other things). Methods Ecol Evol. 3:217–223. [Google Scholar]
- Rhee C, Edwards M, Dang C, Harris J, Brown M, Kim J, Tucker HO. 2017. ARID3A is required for mammalian placenta development. Dev Biol. 422:83–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A. 2000. EMBOSS: the European molecular biology open software suite. Trends Genet. 16:276–277. [DOI] [PubMed] [Google Scholar]
- Ritter A, Roth S, Kreis N, Friemel A, Hoock SC, Souto AS, Eichbaum C, Neuhoff A, Chen Q, Solbach C, et al. 2020. Primary cilia in trophoblastic cells: potential involvement in preeclampsia. Hypertension 76:1491–1505. [DOI] [PubMed] [Google Scholar]
- Schoch CL, Ciufo S, Domrachev M, Hotton CL, Kannan S, Khovanskaya R, Leipe D, Mcveigh R, O’Neill K, Robbertse B, et al. 2020. NCBI Taxonomy: a comprehensive update on curation, resources and tools. Database (Oxford). 2020:baaa062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekita Y, Wagatsuma H, Nakamura K, Ono R, Kagami M, Wakisaka N, Hino T, Suzuki-Migishima R, Kohda T, Ogura A, et al. 2008. Role of retrotransposon-derived imprinted gene, Rtl1, in the feto-maternal interface of mouse placenta. Nat Genet. 40:243–248. [DOI] [PubMed] [Google Scholar]
- Smit AFA, Hubley R, Green P.. 2020. RepeatMasker Open-4.0. Version 4.1.1. [accessed 2020 Sep 22]. Available from: http://www.repeatmasker.org.
- Smith CL, Eppig JT. 2009. The mammalian phenotype ontology: enabling robust annotation and comparative analysis. Wiley Interdiscip Rev Syst Biol Med. 1:390–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepan H, Leo C, Purz S, Höckel M, Horn L-C. 2005. Placental localization and expression of the cell death factors BNip3 and Nix in preeclampsia, intrauterine growth retardation and HELLP syndrome. Eur J Obstet Gynecol Reprod Biol. 122:172–176. [DOI] [PubMed] [Google Scholar]
- Suk F, Chang C, Lin R, Lin S, Chen Y, Liang Y. 2018. MCPIP3 as a potential metastasis suppressor gene in human colorectal cancer. Int J Mol Sci. 19:1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tossetta G, Fantone S, Giannubilo SR, Busilacchi EM, Ciavattini A, Castellucci M, Di Simone N, Mattioli-Belmonte M, Marzioni D. 2019. Pre-eclampsia onset and SPARC: a possible involvement in placenta development. J Cell Physiol. 234:6091–6098. [DOI] [PubMed] [Google Scholar]
- Tsai KYF, Tullis B, Mejia J, Reynolds PR, Arroyo JA. 2021. Regulation of trophoblast cell invasion by pyruvate kinase isozyme M2 (PKM2). Placenta 103:24–32. [DOI] [PubMed] [Google Scholar]
- Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C, Sjöstedt E, Asplund A, et al. 2015. Tissue-based map of the human proteome. Science 347:1260419. [DOI] [PubMed] [Google Scholar]
- Valencia JD, Girgis HZ. 2019. LtrDetector: a tool-suite for detecting long terminal repeat retrotransposons de-novo. Software 20:450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss AK, Thomas T, Gruss P. 2000. Mice lacking HSP90beta fail to develop a placental labyrinth. Development 127:1–11. [DOI] [PubMed] [Google Scholar]
- Wagner GP K, Dighe A, Levchenko A. 2022. The coevolution of placentation and cancer. Annu Rev Anim Biosci. 10:259–279. [DOI] [PubMed] [Google Scholar]
- Wang H, Deng G, Ai M, Xu Z, Mou T, Yu J, Liu H, Wang S, Li G. 2019a. Hsp90ab1 stabilizes LRP5 to promote epithelial-mesenchymal transition via activating of AKT and Wnt/β-catenin signaling pathways in gastric cancer progression. Oncogene 38:1489–1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Hou X, Wang Y, Xu A, Cao W, Liao M, Zhang R, Tang J. 2017. Ubiquitination of non-lysine residues in the retroviral integrase. Biochem Biophys Res Commun. 494:57–62. [DOI] [PubMed] [Google Scholar]
- Wang W, Liu J, Pan E. 2022. CircHIPK3 contributes to human villous trophoblast growth, migration and invasion via modulating the pathway of miR-346/KCMF1. Placenta 118:46–54. [DOI] [PubMed] [Google Scholar]
- Wang S, Ma J, Peng J, Xu J. 2013a. Protein structure alignment beyond spatial proximity. Sci Rep. 3:1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Miller DC, Harman R, Antczak DF, Clark AG. 2013b. Paternally expressed genes predominate in the placenta. Proc Natl Acad Sci U S A. 110:10705–10710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Yan F, Li Z, Yu Y, Parnell SE, Xiong Y. 2019b. Impaired plasma membrane localization of ubiquitin ligase complex underlies 3-M syndrome development. J Clin Invest. 129:4393–4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ.. 2009. Jalview version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25:1189–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P, et al. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420:520–562. [DOI] [PubMed] [Google Scholar]
- Weber M, Knoefler I, Schleussner E, Markert UR, Fitzgerald JS. 2013. HTR8/SVneo cells display trophoblast progenitor cell-like characteristics indicative of self-renewal repopulation activity, and expression of “stemness-” associated transcription factors. Biomed Res Int. 2013:243649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West RC, Ming H, Logsdon DM, Sun J, Rajput SK, Kile RA, Schoolcraft WB, Roberts RM, Krisher RL, Jiang Z, et al. 2019. Dynamics of trophoblast differentiation in peri-implantation-stage human embryos. Proc Natl Acad Sci U S A. 116:22635–22644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G, Wang L, Han Y, He Q. 2012. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16:284–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Yao X. 2013. Posttranslational modifications of HIV-1 integrase by various cellular proteins during viral replication. Viruses 5:1787–1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Zhao X, Zhou W, Qi H, Zhang H, Han T, Baker P. 2021. Impaired placental mitophagy and oxidative stress are associated with dysregulated BNIP3 in preeclampsia. Sci Rep. 11:20469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu A, Ibrahim JG, Love MI. 2019. Heavy-tailed prior distrubutions for sequence count data: removing the noise and preserving large differences. Bioinformatics 35:2084–2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Pang Z, Yu Y. 2012. Regulation of trophoblast invasion: the role of matrix metalloproteinases. Rev Obstet Gynecol. 5:e137–e143. [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Wang K, Li T, Chen J, Xie D, Chang X, Yao J, Wu J, Zhou Q, Jia Y, et al. 2017. Hypoxia-induced TET1 facilitates trophoblast cell migration and invasion through HIF1α signaling pathway. Sci Rep. 7:8077. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All new transcriptomic data from this study were submitted to the NCBI SRA database which can be accessed through the BioProject accession number PRJNA830988.
Conflict of interest. The authors declare no competing financial interests.