Abstract
The COVID-19 pandemic has made it necessary to create antivirals active against the SARS-CoV-2 coronavirus. One of the widely used strategies to fight off viral infections is the use of modified nucleoside analogues that inhibit viral replication by incorporating DNA or RNA into the growing chain, thus stopping its synthesis. The difficulty of using this method of treatment in the case of SARS-CoV-2 is that coronaviruses have an effective mechanism for maintaining genome stability. Its central element is the nsp14 protein, which is characterized by exonuclease activity, due to which incorrectly included and noncanonical nucleotides are removed from the 3' end of the growing RNA chain. Inhibitors of nsp14 exonuclease and nucleoside analogues resistant to its action are viewed as potential targets for anticoronavirus therapy.
Keywords: COVID-19, SARS-CoV-2, nucleoside inhibitors, replication, genome stability, RNA-dependent RNA polymerase, corrective exonuclease
Throughout its history, humanity has repeatedly encountered dangerous viruses causing serious diseases. The current COVID-19 pandemic, caused by the spread of the SARS-CoV-2 coronavirus, has led to a global health crisis. Despite the success of immunoprophylaxis, the number of cases continues to grow. Hence, there is an urgent need to create antiviral drugs that could facilitate the course of coronavirus infection and prevent the development of complications.
According to the latest classification of the International Committee on Taxonomy of Viruses [1], coronaviruses belong to the subfamily Orthocoronavirinae of the family Coronaviridae of the order Nidovirales. This subfamily consists of four genera—alpha-, beta-, gamma-, and deltacoronaviruses; representatives of the first two genera mainly affect mammals, while viruses of the other two infect birds. The highly pathogenic betacoronaviruses SARS-CoV, SARS-CoV-2, and MERS-CoV cause severe respiratory syndromes in humans, while the other four coronaviruses found in the human population (alphacoronaviruses HCoV-NL63 and HCoV-229E and betacoronaviruses HCoV-OC43 and HCoV- HKU1) usually cause only relatively mild upper respiratory disease; however, infants, young children, and the elderly can be infected much more severely [2].
Genomic sequence studies show that all human coronaviruses originally had wild mammalian hosts as natural ones: SARS-CoV, SARS-CoV-2, MERS-CoV, HCoV-NL63, and HCoV-229E most likely passed to humans from bats, while HCoV-OC43 and HKU1 came from rodents [2, 3]. In addition to human viruses, important and relatively well-studied representatives of coronaviruses include porcine transmissible gastroenteritis virus and some other viruses that infect farm animals and birds; feline enteritis/infectious peritonitis virus; and murine hepatitis virus (MHV), which is often used as a safe laboratory model of coronavirus biology. In contrast to the species-specific interaction of the virus with the surface receptors of infected cells and with the body’s immune system, the processes of actualizing genetic information inside the cell are almost identical for all coronaviruses, and the current state of knowledge about coronavirus replication mainly relies on the study of MHV, SARS-CoV, and SARS-CoV-2.
ORGANIZATION AND REPLICATION OF THE CORONAVIRUS GENOME
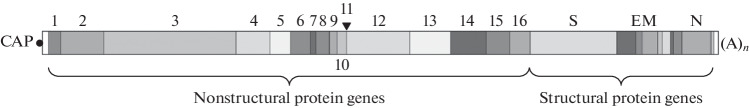
The genome of coronaviruses is a single-stranded polycistronic (+)-RNA 26–32 kb long, encoding 14 overlapping open reading frames [4, 5]. The genomic RNA contains a cap at the 5' end, a poly(A) tail at the 3' end, and short 5' and 3' untranslated regions forming hairpins with regulatory functions. Two large reading frames, ORF1a and ORF1b, occupying in total approximately two-thirds of the genome, are translated to form two polyproteins: pp1a and pp1ab, the latter being formed upon a shift in the reading frame by one nucleotide [6]. They are in turn cleaved into 16 nonstructural proteins (nsp) by ORF1a-encoded proteases: nsp5 chymotrypsin-like protease and nsp3 papain-like protease. In addition to protease activity, nonstructural proteins are involved in the modification of the internal environment of the host cell; fix the virus replication complexes on subcellular structures; and control the processes of RNA replication, transcription, and processing. The remaining third of the genome contains the genes for the viral spike, envelope, membrane, and nucleocapsid structural proteins and includes several ORFs encoding additional proteins (Fig. 1).
Fig. 1.
SARS-CoV-2 genome organization scheme.
As with other (+)-single-stranded RNA viruses, coronavirus genome replication begins with the synthesis of the (–)-strand RNA, which is then copied to form new (+)-RNA molecules. The main role in replication is played by the viral RNA-dependent RNA polymerase (RDRP), the nsp12 protein [7, 8]. Virus replication occurs in the cytoplasm of infected cells and begins when the RDRP complex with a number of other nonstructural proteins (the replicative-transcriptional complex, RTC) binds to the 3' end of the (+)-RNA. This process is stimulated by the secondary structures of the 3'-untranslated region of RNA [9]. RTC binds to modified membranes derived from the endoplasmic reticulum of the host cell, thereby forming viral replication factories [10]. Anchoring of RTCs to membranes is mediated by the nsp3 transmembrane domain together with the transmembrane proteins nsp4 and nsp6. In addition to RDRP, the central part of RTC includes nsp13 RNA helicase, processivity factors nsp7 and nsp8, and proteins nsp10 and nsp14 [9, 11–13]. The nsp13 protein can unwind double-stranded RNA in the 5' → 3' direction, resulting in the formation of a single-stranded template for RNA synthesis. The activity of nsp13 is increased by the direct protein–protein interaction with nsp12, suggesting that the efficiency of viral RNA synthesis is increased when the two proteins interact in a functional RTC.
ANTIVIRAL NUCLEOSIDE INHIBITORS
The action of a large number of therapeutic analogues of purine and pyrimidine nucleosides and nitrogenous bases relies on their ability to be metabolized in the cell to ribo- and deoxyribonucleoside triphosphate derivatives and to incorporate into DNA and RNA. Compounds of this class are widely used as anticancer and immunosuppressive drugs, as well as antiviral agents. When incorporated into genomic DNA or RNA, these modified nucleotides distort its structure or cause the termination of the chain to be synthesized. They also often inhibit replicative polymerases, competing with canonical dNTPs and NTPs for the active site of the enzyme. For example, the dG analogue acyclovir is used to combat infections caused by herpes viruses—simple and genital herpes, shingles, severe forms of chicken pox. Once in an infected cell, acyclovir is phosphorylated by viral thymidine kinase and then converted to triphosphate under the action of cellular nucleotide kinases. After the incorporation of the acyclovir residue into the growing DNA chain, the viral DNA polymerase binds the next dNTP but cannot synthesize the phosphodiester bond because of the absence of the 3′-OH group in acyclovir, and the growing chain is terminated. Moreover, viral DNA polymerase has a very high affinity for such a modified 3' end and forms a kind of dead-end complex with it, from which it is released very slowly. Many nucleoside inhibitors of RNA-dependent DNA polymerase (reverse transcriptase)—azidothymidine, lamivudine, adefovir, entecavir, etc.—have been developed for therapy aimed at human immunodeficiency virus and hepatitis B virus.
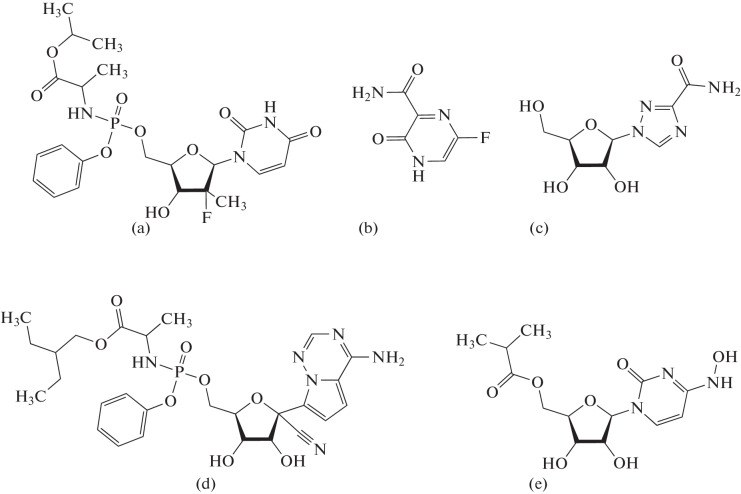
For viruses that contain an RNA genome and replicate it using RDRP, there are quite few nucleoside inhibitors (Fig. 2). Sofosbuvir, used in the treatment of hepatitis C, is an analogue of uridine. Favipiravir, which exhibits combined A and G matrix properties, is approved in some countries for the treatment of influenza and COVID-19 (in Russia, only for COVID-19). Ribavirin also acts on many RNA viruses by inhibiting RDRP and simultaneously suppressing the activity of enzymes capping the 5' end of the viral RNA.
Fig. 2.
Structure of some nucleoside RDRP inhibitors: (a) sofosbuvir, (b) favipiravir, (c) ribavirin, (d) remdesivir, and (e) molnupiravir.
The two known nucleoside inhibitors active against coronaviruses differ from the above-mentioned compounds in their mechanism of action. Remdesivir, an adenosine analogue, was developed as a broad-spectrum antiviral agent, was successfully used in the experimental treatment of Ebola and Marburg hemorrhagic fevers, and was approved in 2020 for COVID-19 therapy. After activation in cells, remdesivir is incorporated into the growing RNA chain, but synthesis does not stop immediately after that. After the remdesivir residue, RDRP incorporates another three nucleotides, moving one step down the template after each inclusion. After that, the nitrile group at C1' of the remdesivir molecule collides with the Ser861 RDRP residue, and the reaction stops [14–16]. The nucleoside analogue molnupiravir, approved for COVID-19 therapy in late 2021, does not cause chain termination at all and, after activation, is incorporated into viral RNA with high efficiency [17, 18]. However, since the molnupiravir molecule contains N 4-hydroxycytosine as a nitrogenous base, which easily pairs with A and G in various tautomeric forms, many inactivating mutations very quickly accumulate in the virus genome. The lethal level of mutagenesis is also partly responsible for the antiviral activity of favipiravir [19].
CORRECTION OF REPLICATION ERRORS IS A MECHANISM TO ENSURE GENOME STABILITY
Replication of viruses with RNA genomes is usually accompanied by numerous RDRP errors, which leads to their existence as populations of genomic mutants, or quasi-species [20, 21]. Low replication fidelity allows viruses with RNA genomes to adapt to different environmental conditions owing to selection pressure, but it also increases the likelihood of lethal mutations. This leads to the need for a balance between quasi-species diversity and replicative stability [22, 23]. In coronaviruses, which have the longest genome of all RNA viruses, replication fidelity is much more critical.
The concept of exonucleolytic correction of erroneously incorporated nucleotides (proofreading) as a general property of any replication complex was finally formulated in the 1980s on the basis of biochemical and genetic data for a number of bacterial and bacteriophage DNA polymerases [24, 25]. Many polypeptides that function as DNA polymerases also have 3' → 5'-exonuclease activity, which is more specific for unpaired 3'-end nucleotides, and the introduction of inactivating amino acid substitutions in the corresponding regions of the protein leads to a sharp increase in the overall level of mutagenesis. The most studied in this regard are still the Klenow fragment of E. coli DNA polymerase I and DNA polymerase of bacteriophages T7 and RB69, for which the existence of a separate 3′ → 5′-exonuclease domain in the polymerase structure has been shown. For them in addition, the kinetics of transfer of the primer end with incorrectly inserted dNMP into the exonuclease center and hydrolysis of its phosphodiester bond with incorporation for corresponding to the dNMP template and for not corresponding to it has been studied in detail. In general, polymerase error correction involving specific 3' → 5'-exonuclease activity is 5–6 orders of magnitude more efficient than pyrophosphorolysis of the same erroneously incorporated nucleotide.
The expansion of the list of studied DNA polymerases and replication systems showed that the corrective activity often does not belong to the DNA polymerase itself (more precisely, not to the polypeptide in which the polymerase activity is localized) but to a separate polypeptide. In some cases, for example, for the ε subunit of E. coli DNA polymerase III (DnaQ or MutD), it can be part of a strong replication complex. In other replication systems, corrective exonucleases exist as separate enzymes or as optional subunits of the replication complex and often perform additional functions in the cell. Thus, in humans, 3′ → 5′ exonucleases TREX1 and TREX2 [26] and exo/endonuclease APEX1 [27, 28] can perform corrective functions. In one replication system, DNA polymerases with corrective activity can coexist with individual 3' → 5' exonucleases, which is observed, in particular, in mammals since DNA polymerases δ and ε themselves can correct errors [29]. In addition to incorrectly inserted canonical nucleotides, corrective exonuclease activities also remove from DNA many 3'-terminal residues of nucleoside inhibitors [30‒34]; it has been recently discovered that DNA repair exonucleases CtIP and MRE11 from the pathway of nonhomologous reunion of the ends of double-strand breaks also participate in this process [35].
As opposed to the replication systems of bacteriophages, bacteria, and eukaryotes, we know very little about the possibility of error correction in viruses that infect cells of higher eukaryotes, including humans. In some DNA-containing viruses (adenoviruses, herpesviruses, orthopoxviruses), DNA polymerases have their own corrective activity [36–39], but there is no information about the participation of individual 3' → 5'-exonucleases in their replication. Regarding viruses with an RNA genome, the prevailing opinion was that they lack correction systems during replication, and replication itself must balance in a rather narrow range of accuracy between the generation of high genetic quasi-species diversity, which is important for adaptation to a changing environment, and lethal mutagenesis [20, 21, 40]. Extremely high mutation rates have been recorded for RNA viruses with fundamentally different life cycles—HIV, poliovirus, and influenza virus [41–45]. For coronaviruses, however, the observed level of mutagenesis is much lower than for other viruses with an RNA genome [46].
CORONAVIRUS EXONUCLEASE NSP14
Although the accuracy of nucleotide incorporation of coronavirus RDRP is higher than that of polymerases of other RNA viruses [47], it cannot ensure the observed low level of mutagenesis. In 2006, it was found that the SARS-CoV nonstructural protein nsp14 (also called ExoN in the literature), which is part of the RTC, is a 3' → 5' exonuclease, belonging to the same DnaQ superfamily as the ε subunit of DNA polymerase III and human TREX proteins and necessary for virus replication [48]. The activity of nsp14 depends on the presence of divalent metal ions and hydrolyzes single-stranded RNA and unpaired nucleotides of double-stranded RNA from the 3' end [48, 49]. In MHV, inactivation of the nsp14 exonuclease function reduces the viability of the virus in cell culture and pathogenicity in mice [50–52] and causes a sharp increase in the frequency of mutations during replication [53, 54]. Similar consequences are observed in the porcine transmissible gastroenteritis virus model [55]. The exonuclease activity of nsp14 is stimulated by the small viral nonstructural protein nsp10 [49, 56, 57], but in vitro nsp14 can also function as an isolated recombinant protein produced in E. coli [48].
In addition to its exonuclease function, the nsp14 protein has the activity of guanine-N7 methyltransferase and initiates the capping process of viral mRNAs [58, 59]. Interestingly, despite the physical separation of the centers of the two activities, the presence of an exonuclease domain (even a catalytically inactive one) is necessary for the manifestation of the methyltransferase function [58, 60]. Presumably, the methyltransferase function and capping are important for preventing the cellular interferon response to the appearance of coronavirus RNAs, but data on the need for exonuclease activity for this remain contradictory [55, 61, 62].
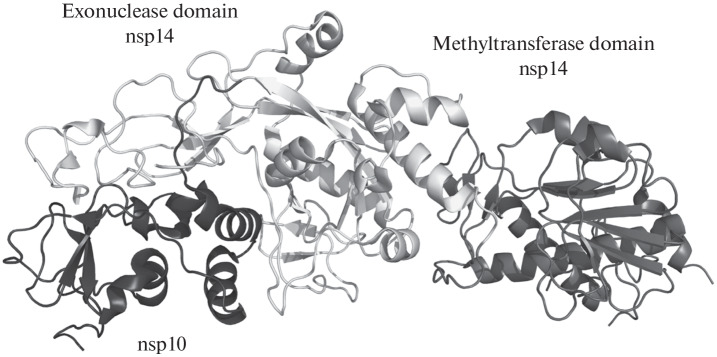
The spatial structure of the nsp14/nsp10 SARS-CoV complex, established in 2015 by X-ray diffraction analysis [63] (Fig. 3), revealed certain unique features of the exonuclease function of nsp14. Although the general structure of the exonuclease domain is similar to that of other exonucleases of the DnaQ family, including the DEEDh catalytic motif, the nsp14 protein additionally carries two Cys3His- and Cys2His2-type zinc fingers, important for its activity. The exonuclease and methyltransferase domains are connected by a linker without a pronounced secondary structure. The nsp10 protein is associated with the exonuclease domain of nsp14 and does not form contacts with the methyltransferase domain. In 2018, another structure of the same complex was published [64] with a significantly different mutual arrangement of the exonuclease and methyltransferase domains, which indicates the significant flexibility of the interdomain linker and conformational polymorphism of nsp14. In the absence of nsp10, the catalytic exonuclease center is partially disorganized, which explains the decrease in the activity of the isolated nsp14 protein.
Fig. 3.
Structure of the SARS-CoV nsp14/nsp10 complex (no. 5C8U in the Protein Databank [63]).
In 2021, several structures of the SARS-CoV-2 nsp14/nsp10 heterodimer were published and deposited, as determined by X-ray diffraction analysis and cryoelectron microscopy [65–67]. In particular, the nsp14/nsp10 structure was obtained for the first time in a complex with substrate RNA, which made it possible to analyze the structural aspects of the mechanism of the phosphodiester bond hydrolysis reaction [66]. The enzyme inverts the unpaired 3′-terminal ribonucleotide into its active center and introduces the Phe146 residue in its place, which forms stacking interactions with the penultimate nitrogenous base of the RNA primer. Unlike many RNases, hydrolysis of the phosphodiester bond by the nsp14 enzyme proceeds without involvement of the hydroxyl group at position 2'. The active center contains two well-coordinated divalent cations, one of which activates the water molecule for nucleophilic attack on the phosphorus atom, and the other of which stabilizes the O3′ leaving group.
CORRECTIVE ACTIVITY OF NSP14 WITHIN THE REPLICATION‒TRANSCRIPTION COMPLEX
The structure of SARS-CoV-2 RNA-bound RTC, established in 2021 by cryoelectron microscopy [68, 69], provides the most detailed picture of coronavirus replication to date. The most complete of the complexes studied contains the template and primer chains of RNA and has a stoichiometry of protein subunits 1 molecule nsp12 (RDRP) : 1 nsp7 : 2 nsp8 (processivity factors) : 2 nsp13 (RNA helicase) : 1 nsp14 : 1 nsp10 : 1 nsp9 (single-stranded RNA binding protein). Note that the complete complex exists as a dimer formed due to the interaction of several subunits with each other. Removal of nsp14/nsp10 leads to the transition of RTC to a monomeric state, the organization of which is compatible with RNA polymerization and capping reactions.
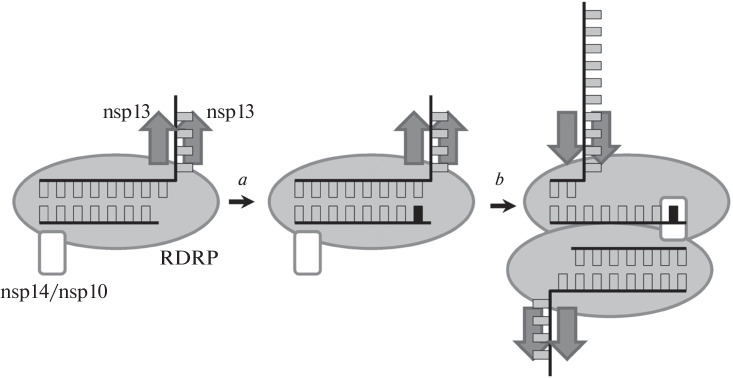
In the complete RTC, the active center of the nsp14 exonuclease domain is located at a significant distance from the end of the RNA primer (~80 Å), which makes it impossible for them to interact within one RTC monomer. Yan et al. [69] proposed a model (Fig. 4) in which the dimeric organization of RTC and the “return” of RDRP about 6 nucleotides back are necessary for the manifestation of exonuclease activity, which requires helicase activity of nsp13. In this case, the end of the RNA primer in one RTC monomer is within reach of the exonuclease subunit in the other monomer, which makes it possible to hydrolyze one or more 3'-terminal nucleotide(s). Such a mechanism could explain the need for nsp14 for the resistance of coronaviruses to remdesivir, which causes chain synthesis termination not immediately after incorporation but after completion of several RNA units [70, 71]. On the other hand, the above-described structure of the isolated nsp14/nsp10/RNA complex has a completely different character of interactions with RNA, and the dimeric organization of RTC in vivo has not yet been confirmed. It is possible that, in addition to the complexes studied, RTC may also exist in other architectures with more convenient access of nsp14 to the RNA primer.
Fig. 4.
Scheme of the actualization of the corrective activity of nsp14/nsp10 in the RTC dimer (according to [69]). When the wrong nucleotide (a) is incorporated, the polarity of the movement of the nsp13 helicase is reversed, RDRP returns 6 positions backward, and the 3'-terminal nucleotide enters the active site of nsp14 on another RTC complex.
CORONAVIRUS CORRECTIVE EXONUCLEASE AS A POTENTIAL PHARMACOLOGICAL TARGET
The importance of nsp14 in avoiding mutagenesis during coronavirus replication has raised the question of how much this protein can contribute to protecting the virus from nucleoside inhibitors. MHV and SARS-CoV lacking nsp14 exonuclease activity are hypersensitive to 5-fluorouracil, azacitidine, and ribavirin, and sensitivity to 5-fluorouracil is accompanied in such strains by a lethal level of mutagenesis [47, 57, 72]. Disruption of the interaction between nsp14 and the auxiliary protein nsp10 causes similar consequences [57]. Long-term passaging of exonuclease-deficient viruses causes the appearance of compensatory mutations in RDRP, which increase its accuracy and reduce sensitivity to nucleotide inhibitors; however, the spectrum of such mutations is very limited [73, 74]. Deficiency in the corrective activity of nsp14 also causes hypersensitivity to remdesivir [70]. These observations highlight the potential value of coronavirus exoribonuclease inhibitors and nucleoside terminators of RNA synthesis that are resistant to cleavage by this enzyme. At present, molnupiravir is the only, largely by chance, actualized example of such an approach [75, 76], but its use is complicated by the significant mutagenicity of this compound for human cells.
The literature describes several recent attempts at in silico and in vitro screening of low molecular weight nonnucleoside compounds inhibiting the exonuclease or methyltransferase function of the nsp14 protein [77–81]. However, there are no reports on the search for new nucleoside inhibitors that could not be removed by exonuclease. Knowledge of the mechanism of action of the nsp14 enzyme, based on structural data, may provide a basis for the rational design of such compounds.
FUNDING
This study was supported by the Russian Foundation for Basic Research, project no. 20-04-60433.
CONFLICT OF INTEREST
The authors declare that they have no conflicts of interest.
Footnotes
Sofiya Konstantinovna Yuyukina is an Engineer at the Laboratory of Genomic and Protein Engineering of the Institute of Chemical Biology and Fundamental Medicine of the RAS Siberian Branch (ICBFM SB RAS). RAS Corresponding Member Dmitrii Olegovich Zharkov is Head of the above laboratory at the same institute.
Translated by B. Alekseev
Contributor Information
S. K. Yuyukina, Email: s.iuiukina@g.nsu.ru
D. O. Zharkov, Email: dzharkov@niboch.nsc.ru
REFERENCES
- 1.Walker P. J., Siddell S. G., Lefkowitz E. J. Changes to virus taxonomy and to the International Code of Virus Classification and Nomenclature ratified by the International Committee on Taxonomy of Viruses (2021) Arch. Virol. 2021;166:2633–2648. doi: 10.1007/s00705-021-05156-1. [DOI] [PubMed] [Google Scholar]
- 2.Su S., Wong G., Shi W. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 2016;24:490–502. doi: 10.1016/j.tim.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miranda C., Silva V., Igrejas G., Poeta P. Genomic evolution of the human and animal coronavirus diseases. Mol. Biol. Rep. 2021;48:6645–6653. doi: 10.1007/s11033-021-06632-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brian D. A., Baric R. S. Coronavirus genome structure and replication. Curr. Top. Microbiol. Immunol. 2005;287:1–30. doi: 10.1007/3-540-26765-4_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fehr A. R., Perlman S. Coronaviruses: An overview of their replication and pathogenesis. Methods Mol. Biol. 2015;1282:1–23. doi: 10.1007/978-1-4939-2438-7_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dos Ramos F., Carrasco M., Doyle T., Brierley I. Programmed −1 ribosomal frameshifting in the SARS coronavirus. Biochem. Soc. Trans. 2004;32:1081–1083. doi: 10.1042/BST0321081. [DOI] [PubMed] [Google Scholar]
- 7.Hillen H. S. Structure and function of SARS-CoV-2 polymerase. Curr. Opin. Virol. 2021;48:82–90. doi: 10.1016/j.coviro.2021.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jiang Y., Yin W., Xu H. E. RNA-dependent RNA polymerase: Structure, mechanism, and drug discovery for COVID-19. Biochem. Biophys. Res. Commun. 2021;538:47–53. doi: 10.1016/j.bbrc.2020.08.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sola I., Almazán F., Zúñiga S., Enjuanes L. Continuous and discontinuous RNA synthesis in coronaviruses. Annu. Rev. Virol. 2015;2:265–288. doi: 10.1146/annurev-virology-100114-055218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.E. J. Snijder, R. W. A. L. Limpens, A. H. de Wilde, et al., “A unifying structural and functional model of the coronavirus replication organelle: Tracking down RNA synthesis,” PLoS Biol. 18 (6), e3000715 (2020). [DOI] [PMC free article] [PubMed]
- 11.A. von Brunn, C. Teepe, J. C. Simpson, et al., “Analysis of intraviral protein-protein interactions of the SARS coronavirus ORFeome,” PLoS ONE 2 (5), e459 (2007). [DOI] [PMC free article] [PubMed]
- 12.J. A. Pan, X. Peng, Y. Gao, et al., “Genome-wide analysis of protein-protein interactions and involvement of viral proteins in SARS-CoV replication,” PLoS ONE 3 (10), e3299 (2008). [DOI] [PMC free article] [PubMed]
- 13.Subissi L., Posthuma C. C., Collet A. One severe acute respiratory syndrome coronavirus protein complex integrates processive RNA polymerase and exonuclease activities. Proc. Natl. Acad. Sci. U. S. A. 2014;111:E3900–E3909. doi: 10.1073/pnas.1323705111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gordon C. J., Tchesnokov E. P., Feng J. Y. The antiviral compound remdesivir potently inhibits RNA-dependent RNA polymerase from Middle East respiratory syndrome coronavirus. J. Biol. Chem. 2020;295:4773–4779. doi: 10.1074/jbc.AC120.013056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gordon C. J., Tchesnokov E. P., Woolner E. Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute respiratory syndrome coronavirus 2 with high potency. J. Biol. Chem. 2020;295:6785–6797. doi: 10.1074/jbc.RA120.013679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kokic G., Hillen H. S., Tegunov D. Mechanism of SARS-CoV-2 polymerase stalling by remdesivir. Nat. Commun. 2021;12:279. doi: 10.1038/s41467-020-20542-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.C. J. Gordon, E. P. Tchesnokov, R. F. Schinazi, and M. Götte, “Molnupiravir promotes SARS-CoV-2 mutagenesis via the RNA template,” J. Biol. Chem. 297 (1), 100770 (2021). [DOI] [PMC free article] [PubMed]
- 18.Kabinger F., Stiller C., Schmitzová J. Mechanism of molnupiravir-induced SARS-CoV-2 mutagenesis. Nat. Struct. Mol. Biol. 2021;28:740–746. doi: 10.1038/s41594-021-00651-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shannon A., Selisko B., Le N.-T.-T. Rapid incorporation of Favipiravir by the fast and permissive viral RNA polymerase complex results in SARS-CoV-2 lethal mutagenesis. Nat. Commun. 2020;11:4682. doi: 10.1038/s41467-020-18463-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Domingo E., Holland J. J. RNA virus mutations and fitness for survival. Annu. Rev. Microbiol. 1997;51:151–178. doi: 10.1146/annurev.micro.51.1.151. [DOI] [PubMed] [Google Scholar]
- 21.Bordería A. V., Rozen-Gagnon K., Vignuzzi M. Fidelity variants and RNA quasispecies. Curr. Top. Microbiol. Immunol. 2016;392:303–322. doi: 10.1007/82_2015_483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Denison M. R., Graham R. L., Donaldson E. F. Coronaviruses: An RNA proofreading machine regulates replication fidelity and diversity. RNA Biol. 2011;8:270–279. doi: 10.4161/rna.8.2.15013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith E. C., Denison M. R. Implications of altered replication fidelity on the evolution and pathogenesis of coronaviruses. Curr. Opin. Virol. 2012;2:519–524. doi: 10.1016/j.coviro.2012.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson K. A. Conformational coupling in DNA polymerase fidelity. Annu. Rev. Biochem. 1993;62:685–713. doi: 10.1146/annurev.bi.62.070193.003345. [DOI] [PubMed] [Google Scholar]
- 25.Kunkel T. A., Bebenek K. DNA replication fidelity. Annu. Rev. Biochem. 2000;69:497–529. doi: 10.1146/annurev.biochem.69.1.497. [DOI] [PubMed] [Google Scholar]
- 26.Mazur D. J., Perrino F. W. Identification and expression of the TREX1 and TREX2 cDNA sequences encoding mammalian 3′ → 5′ exonucleases. J. Biol. Chem. 1999;274:19655–19660. doi: 10.1074/jbc.274.28.19655. [DOI] [PubMed] [Google Scholar]
- 27.Chou K.-M., Cheng Y.-C. An exonucleolytic activity of human apurinic/apyrimidinic endonuclease on 3′-mispaired DNA. Nature. 2002;415:655–659. doi: 10.1038/415655a. [DOI] [PubMed] [Google Scholar]
- 28.Chou K.-M., Cheng Y.-C. The exonuclease activity of human apurinic/apyrimidinic endonuclease (APE1): Biochemical properties and inhibition by the natural dinucleotide Gp4G. J. Biol. Chem. 2003;278:18289–18296. doi: 10.1074/jbc.M212143200. [DOI] [PubMed] [Google Scholar]
- 29.Albertson Y. M., Ogawa M., Bugni J. M. DNA polymerase ε and δ proofreading suppress discrete mutator and cancer phenotypes in mice. Proc. Natl. Acad. Sci. U. S. A. 2009;106:17101–17104. doi: 10.1073/pnas.0907147106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chou K.-M., Kukhanova M., Cheng Y.-C. A novel action of human apurinic/apyrimidinic endonuclease: Excision of L-configuration deoxyribonucleoside analogs from the 3′ termini of DNA. J. Biol. Chem. 2000;275:31009–31015. doi: 10.1074/jbc.M004082200. [DOI] [PubMed] [Google Scholar]
- 31.Hanes J. W., Johnson K. A. Exonuclease removal of dideoxycytidine (zalcitabine) by the human mitochondrial DNA polymerase. Antimicrob. Agents Chemother. 2008;52:253–258. doi: 10.1128/AAC.00778-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lam W., Park S.-Y., Leung C.-H., Cheng Y.-C. Apurinic/apyrimidinic endonuclease-1 protein level is associated with the cytotoxicity of L-configuration deoxycytidine analogs (troxacitabine and β-L-2′,3′-dideoxy-2′,3′-didehydro-5-fluorocytidine) but not D-configuration deoxycytidine analogs (gemcitabine and β-D-arabinofuranosylcytosine) Mol. Pharmacol. 2006;69:1607–1614. doi: 10.1124/mol.105.021527. [DOI] [PubMed] [Google Scholar]
- 33.Chen H., Beardsley G. P., Coen D. M. Mechanism of ganciclovir-induced chain termination revealed by resistant viral polymerase mutants with reduced exonuclease activity. Proc. Natl Acad. Sci. U. S. A. 2014;111:17462–17467. doi: 10.1073/pnas.1405981111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsuda M., Terada K., Ooka M. The dominant role of proofreading exonuclease activity of replicative polymerase ε in cellular tolerance to cytarabine (Ara-C) Oncotarget. 2017;8:33457–33474. doi: 10.18632/oncotarget.16508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mohiuddin M., Rahman M. M., Sale J. E., Pearson C. E. CtIP-BRCA1 complex and MRE11 maintain replication forks in the presence of chain terminating nucleoside analogs. Nucleic Acids Res. 2019;47:2966–2980. doi: 10.1093/nar/gkz009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.King A. J., Teertstra W. R., van der Vliet P. C. Processive proofreading by the adenovirus DNA polymerase. Association with the priming protein reduces exonucleolytic degradation. Nucleic Acids Res. 1997;25:1745–1752. doi: 10.1093/nar/25.9.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gammon D. B., Evans D. H. The 3′-to-5′ exonuclease activity of vaccinia virus DNA polymerase is essential and plays a role in promoting virus genetic recombination. J. Virol. 2009;83:4236–4250. doi: 10.1128/JVI.02255-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tian W., Hwang Y. T., Lu Q., Hwang C. B. C. Finger domain mutation affects enzyme activity, DNA replication efficiency, and fidelity of an exonuclease-deficient DNA polymerase of herpes simplex virus type 1. J. Virol. 2009;83:7194–7201. doi: 10.1128/JVI.00632-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lawler J. L., Coen D. M. HSV-1 DNA polymerase 3′-5′ exonuclease-deficient mutant D368A exhibits severely reduced viral DNA synthesis and polymerase expression. J. Gen. Virol. 2018;99:1432–1437. doi: 10.1099/jgv.0.001138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tsukiyama-Kohara K., Kohara M. Hepatitis C virus: Viral quasispecies and genotypes. Int. J. Mol. Sci. 2018;19:23. doi: 10.3390/ijms19010023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jorba J., Campagnoli R., De L., Kew O. Calibration of multiple poliovirus molecular clocks covering an extended evolutionary range. J. Virol. 2008;82:4429–4440. doi: 10.1128/JVI.02354-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Abram M. E., Ferris A. L., Shao W. Nature, position, and frequency of mutations made in a single cycle of HIV-1 replication. J. Virol. 2010;84:9864–9878. doi: 10.1128/JVI.00915-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dapp M. J., Heineman R. H., Mansky L. M. Interrelationship between HIV-1 fitness and mutation rate. J. Mol. Biol. 2013;425:41–53. doi: 10.1016/j.jmb.2012.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cheung P. P.-H., Rogozin I. B., Choy K.-T. Comparative mutational analyses of influenza A viruses. RNA. 2015;21:36–47. doi: 10.1261/rna.045369.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.M. D. Pauly, M. C. Procario, and A. S. Lauring, “A novel twelve class fluctuation test reveals higher than expected mutation rates for influenza A viruses,” Elife 6, e26437 (2017). [DOI] [PMC free article] [PubMed]
- 46.Smith E. C., Sexton N. R., Denison M. R. Thinking outside the triangle: Replication fidelity of the largest RNA viruses. Annu. Rev. Virol. 2014;1:111–132. doi: 10.1146/annurev-virology-031413-085507. [DOI] [PubMed] [Google Scholar]
- 47.Sexton N. R., Smith E. C., Blanc H. Homology-based identification of a mutation in the coronavirus RNA-dependent RNA polymerase that confers resistance to multiple mutagens. J. Virol. 2016;90:7415–7428. doi: 10.1128/JVI.00080-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Minskaia E., Hertzig T., Gorbalenya A. E. Discovery of an RNA virus 3′ → 5′ exoribonuclease that is critically involved in coronavirus RNA synthesis. Proc. Natl Acad. Sci. U. S. A. 2006;103:5108–5113. doi: 10.1073/pnas.0508200103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bouvet M., Imbert I., Subissi L. RNA 3′-end mismatch excision by the severe acute respiratory syndrome coronavirus nonstructural protein nsp10/nsp14 exoribonuclease complex. Proc. Natl Acad. Sci. U. S. A. 2012;109:9372–9377. doi: 10.1073/pnas.1201130109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.S. G. Sawicki, D. L. Sawicki, D. Younker, et al., “Functional and genetic analysis of coronavirus replicase-transcriptase proteins,” PLoS Pathog. 1 (4), e39 (2005). [DOI] [PMC free article] [PubMed]
- 51.Sperry S. M., Kazi L., Graham R. L. Single-amino-acid substitutions in open reading frame (ORF) 1b-nsp14 and ORF 2a proteins of the coronavirus mouse hepatitis virus are attenuating in mice. J. Virol. 2005;79:3391–3400. doi: 10.1128/JVI.79.6.3391-3400.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Eckerle L. D., Brockway S. M., Sperry S. M. Effects of mutagenesis of murine hepatitis virus nsp1 and nsp14 on replication in culture. Adv. Exp. Med. Biol. 2006;581:55–60. doi: 10.1007/978-0-387-33012-9_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Eckerle L. D., Lu X., Sperry S. M. High fidelity of murine hepatitis virus replication is decreased in nsp14 exoribonuclease mutants. J. Virol. 2007;81:12135–12144. doi: 10.1128/JVI.01296-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.L. D. Eckerle, M. M. Becker, R. A. Halpin, et al., “Infidelity of SARS-CoV Nsp14-exonuclease mutant virus replication is revealed by complete genome sequencing,” PLoS Pathog. 6 (5), e1000896 (2010). [DOI] [PMC free article] [PubMed]
- 55.Becares M., Pascual-Iglesias A., Nogales A. Mutagenesis of coronavirus nsp14 reveals its potential role in modulation of the innate immune response. J. Virol. 2016;90:5399–5414. doi: 10.1128/JVI.03259-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bouvet M., Lugari A., Posthuma C. C. Coronavirus Nsp10, a critical co-factor for activation of multiple replicative enzymes. J. Biol. Chem. 2014;289:25783–25796. doi: 10.1074/jbc.M114.577353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Smith E. C., Case J. B., Blanc H. Mutations in coronavirus nonstructural protein 10 decrease virus replication fidelity. J. Virol. 2015;89:6418–6426. doi: 10.1128/JVI.00110-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen Y., Cai H., Pan J. A. Functional screen reveals SARS coronavirus nonstructural protein nsp14 as a novel cap N7 methyltransferase. Proc. Natl Acad. Sci. U. S. A. 2009;106:3484–3489. doi: 10.1073/pnas.0808790106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.M. Bouvet, C. Debarnot, I. Imbert, et al., “In vitro reconstitution of SARS-coronavirus mRNA cap methylation,” PLoS Pathog. 6 (4), e1000863 (2010). [DOI] [PMC free article] [PubMed]
- 60.Chen Y., Tao J., Sun Y. Structure-function analysis of severe acute respiratory syndrome coronavirus RNA cap guanine-N7-methyltransferase. J. Virol. 2013;87:6296–6305. doi: 10.1128/JVI.00061-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Case J. B., Ashbrook A. W., Dermody T. S., Denison M. R. Mutagenesis of S-adenosyl-L-methionine-binding residues in coronavirus nsp14 N7-methyltransferase demonstrates differing requirements for genome translation and resistance to innate immunity. J. Virol. 2016;90:7248–7256. doi: 10.1128/JVI.00542-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Case J. B., Li Y., Elliott R. Murine hepatitis virus nsp14 exoribonuclease activity is required for resistance to innate immunity. J. Virol. 2018;92:e01531–17. doi: 10.1128/JVI.01531-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ma Y., Wu L., Shaw N. Structural basis and functional analysis of the SARS coronavirus nsp14–nsp10 complex. Proc. Natl Acad. Sci. U. S. A. 2015;112:9436–9441. doi: 10.1073/pnas.1508686112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ferron F., Subissi L., Silveira De Morais A. T. Structural and molecular basis of mismatch correction and ribavirin excision from coronavirus RNA. Proc. Natl Acad. Sci. U. S. A. 2018;115:E162–E171. doi: 10.1073/pnas.1718806115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lin S., Chen H., Chen Z. Crystal structure of SARS-CoV-2 nsp10 bound to nsp14-ExoN domain reveals an exoribonuclease with both structural and functional integrity. Nucleic Acids Res. 2021;49:5382–5392. doi: 10.1093/nar/gkab320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu C., Shi W., Becker S. T. Structural basis of mismatch recognition by a SARS-CoV-2 proofreading enzyme. Science. 2021;373:1142–1146. doi: 10.1126/science.abi9310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.N. H. Moeller, K. Shi, Ö. Demir, et al., “Structure and dynamics of SARS-CoV-2 proofreading exoribonuclease ExoN,” Proc. Natl. Acad. Sci. U. S. A. 119 (9), e2106379119 (2022). [DOI] [PMC free article] [PubMed]
- 68.Yan L., Ge J., Zheng L. Cryo-EM structure of an extended SARS-CoV-2 replication and transcription complex reveals an intermediate state in cap synthesis. Cell. 2021;184:184–193.e10. doi: 10.1016/j.cell.2020.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yan L., Yang Y., Li M. Coupling of N7-methyltransferase and 3′–5′ exoribonuclease with SARS-CoV-2 polymerase reveals mechanisms for capping and proofreading. Cell. 2021;184:3474–3485.e11. doi: 10.1016/j.cell.2021.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Agostini M. L., Andres E. L., Sims A. C. Coronavirus susceptibility to the antiviral remdesivir (GS-5734) is mediated by the viral polymerase and the proofreading exoribonuclease. MBio. 2018;9:e00221–18. doi: 10.1128/mBio.00221-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.A. Shannon, N. T.-T. Le, B. Selisko, et al., “Remdesivir and SARS-CoV-2: Structural requirements at both nsp12 RdRp and nsp14 Exonuclease active-sites,” Antiviral Res. 178, 104793 (2020). [DOI] [PMC free article] [PubMed]
- 72.E. C. Smith, H. Blanc, M. C. Surdel, et al., “Coronaviruses lacking exoribonuclease activity are susceptible to lethal mutagenesis: Evidence for proofreading and potential therapeutics,” PLoS Pathog. 9 (8), e1003565 (2013). [DOI] [PMC free article] [PubMed]
- 73.Graepel K. W., Lu X., Case J. B. Proofreading-deficient coronaviruses adapt for increased fitness over long-term passage without reversion of exoribonuclease-inactivating mutations. MBio. 2017;8:e01503–17. doi: 10.1128/mBio.01503-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Graepel K. W., Agostini M. L., Lu X. Fitness barriers limit reversion of a proofreading-deficient coronavirus. J. Virol. 2019;93:e00711–19. doi: 10.1128/JVI.00711-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Agostini M. L., Pruijssers A. J., Chappell J. D. Small-molecule antiviral β-D-N4-hydroxycytidine inhibits a proofreading-intact coronavirus with a high genetic barrier to resistance. J. Virol. 2019;93:e01348–19. doi: 10.1128/JVI.01348-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.T. P. Sheahan, A. C. Sims, S. Zhou, et al., “An orally bioavailable broad-spectrum antiviral inhibits SARS-CoV-2 in human airway epithelial cell cultures and multiple coronaviruses in mice,” Sci. Transl. Med. 12 (541), eabb5883 (2020). [DOI] [PMC free article] [PubMed]
- 77.Canal B., McClure A. W., Curran J. F. Identifying SARS-CoV-2 antiviral compounds by screening for small molecule inhibitors of nsp14/nsp10 exoribonuclease. Biochem. J. 2021;478:2445–2464. doi: 10.1042/BCJ20210198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.S. Khater, P. Kumar, N. Dasgupta, et al., “Combining SARS-CoV-2 proofreading exonuclease and RNA-dependent RNA polymerase inhibitors as a strategy to combat COVID-19: A high-throughput in silico screening,” Front. Microbiol. 12, 647693 (2021). [DOI] [PMC free article] [PubMed]
- 79.Narayanan N., Nair D. T. Ritonavir may inhibit exoribonuclease activity of nsp14 from the SARS-CoV-2 virus and potentiate the activity of chain terminating drugs. Int. J. Biol. Macromol. 2021;168:272–278. doi: 10.1016/j.ijbiomac.2020.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kozielski F., Sele C., Talibov V. O. Identification of fragments binding to SARS-CoV-2 nsp10 reveals ligand-binding sites in conserved interfaces between nsp10 and nsp14/nsp16. RSC Chem. Biol. 2022;3:44–55. doi: 10.1039/D1CB00135C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rona G., Zeke A., Miwatani-Minter B., Vries M. D. The NSP14/NSP10 RNA repair complex as a pan-coronavirus therapeutic target. Cell Death Differ. 2022;29:285–292. doi: 10.1038/s41418-021-00900-1. [DOI] [PMC free article] [PubMed] [Google Scholar]