Abstract
A gene encoding the mucin-desulfating sulfatase in Prevotella strain RS2 has been cloned, sequenced, and expressed in an active form. A 600-bp PCR product generated using primers designed from amino acid sequence data was used to isolate a 5,058-bp genomic DNA fragment containing the mucin-desulfating sulfatase gene. A 1,551-bp open reading frame encoding the sulfatase proprotein was identified, and the deduced 517-amino-acid protein minus its signal sequence corresponded well with the published mass of 58 kDa estimated by denaturing gel electrophoresis. The sulfatase sequence showed homology to aryl- and nonarylsulfatases with different substrate specificities from the sulfatases of other organisms. No sulfatase activity could be detected when the sulfatase gene was cloned into Escherichia coli expression vectors. However, cloning the gene into a Bacteroides expression vector did produce active sulfatase. This is the first mucin-desulfating sulfatase to be sequenced and expressed. A second open reading frame (1,257 bp) was identified immediately upstream from the sulfatase gene, coding in the opposite direction. Its sequence has close homology to iron-sulfur proteins that posttranslationally modify other sulfatases. By analogy, this protein is predicted to catalyze the modification of a serine group to a formylglycine group at the active center of the mucin-desulfating sulfatase, which is necessary for enzymatic activity.
Mucins are high-molecular-weight glycoproteins which form the structural component of the protective mucus gel layer at the surfaces of the gastrointestinal, respiratory, and female genital tracts. Colonic mucin, particularly in the proximal region, contains significant levels of sulfate covalently bound to the mucin oligosaccharide chains, and levels of 2.0 to 6.5 g of sulfate per 100 g of mucin have been found (9, 17). Heavily sulfated mucins (sulfomucins) have many of the general lubricating and barrier functions of mucins with lower sulfate levels. There is accumulating evidence that sulfomucins may, in addition, rate-limit mucin degradation by mucin-degrading bacterial enzymes (4, 7, 13, 15, 18, 24, 26, 27), and this role is thought to be particularly important in the colon, where approximately 1014 bacterial cells are located (10).
There have been reports of mucin-desulfating sulfatases that partially remove the sulfate from sulfomucin in a number of bacteria from the mouth, stomach, and colon and in feces. The effect of such sulfatases is to increase the susceptibility of the mucin to degradation by other mucin-degrading enzymes (4, 27). Elevated levels of bacterial mucin-desulfating sulfatases are found in feces of patients with ulcerative colitis (26). The sulfated sugar specificity of these fecal sulfatases, the number of different types present, the bacterial origin of the elevated levels, and the conditions which regulate bacterial production of such enzymes are unknown. The types of mucin-desulfating sulfatases that might be predicted include sulfatases specific for galactose-3-sulfate, galactose-6-sulfate, and N-acetylglucosamine-6-sulfate, and they could act on terminal or internal sulfated sugars (20) of the mucin oligosaccharide chain.
Two mucin-desulfating sulfatases have been purified to date. One was obtained from human fecal fluid and was produced by unknown bacteria (27). It has a molecular size of 15 kDa and a pH optimum of 4.5 and was isolated from extrcellular fluid. The other, from the periplasm of Prevotella strain RS2, has a pH optimum of 7.5, an estimated molecular size of 111 kDa as determined by gel exclusion chromatography, and a subunit size of 58 kDa as determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (19). It will desulfate [35S]mucin, free N-acetylglucosamine-6-sulfate, and glucose-6-sulfate (19), suggesting that N-acetylglucosamine-6-sulfate is its sulfated sugar specificity in mucin side chains.
We are interested in clarifying the nature of bacterial enzymes involved in desulfation of mucins during mucin degradation. As there is no information available on any of the bacterial mucin-desulfating sulfatases with respect to their gene sequences and relationships to other proteins, we have cloned and characterized the gene encoding the Prevotella sulfatase and expressed active protein in a Bacteroides recombinant system.
MATERIALS AND METHODS
Bacterial culture and sulfatase purification.
Prevotella strain RS2 is an isolate from pig colon, selected for its ability to grow anaerobically on colonic mucin as an energy source (24). The mucin-desulfating sulfatase protein from Prevotella strain RS2 was purified to homogeneity as previously described (19). The classification of Prevotella strain RS2 was further clarified by PCR amplification of the small subunit rRNA gene (16S), using genomic DNA (gDNA) as the template. The sequence was compared with ribosomal DNA sequences of the Bacteroides subgroup of the Cytophaga-Flavobacter-Bacteroides group (16) in the GenBank database. Strain RS2 fell within the Prevotella cluster, showing the greatest similarity to Prevotella oris (89.5% identity).
Reduction and S-carboxymethylation of the sulfatase and sequence analysis of peptides.
The purified protein was reduced and carboxymethylated prior to digestion with trypsin or cleavage with cyanogen bromide. Peptides were separated by reverse-phase-high-pressure liquid chromatography (HPLC), and their sequences were analyzed using a gas phase sequencer (PE Biosystems, Foster City, Calif.).
Molecular techniques.
After partial sequencing of selected peptides, degenerate PCR primers (22- to 23-mers) complementary to the forward (F) and reverse (R) strands of the cyanogen bromide peptides CB2 and CB11 were synthesized (Table 1). Primer sequences based on CB2 were TR4(F), 5′ TI CA(A/G) AA(A/G) CA(A/G) GCI TGG GA(C/T) GC 3′, and TR5(R), 5′ TA IGC (A/G)TC CCA IGC (C/T)TG (C/T)TT (C/T)TG 3′. Those based on CB11 were TR6(F), 5′ ATG CA(A/G) AA(C/T) GA(C/T) CCI AA(C/T) GA(A/G) (C/T)T 3′, and TR7(R), 5′ A(A/G) (C/T)TC (A/G)TT IGG (A/G)TC (A/G)TT (C/T)TG CAT 3′. PCR amplifications were conducted by standard methods with Prevotella gDNA as the template. Southern analysis, DNA restriction enzyme digestion, and cloning were performed according to procedures described by Sambrook et al. (22). Sequencing of plasmid inserts was performed by automated sequencing. All DNA sequences were constructed from overlapping fragments of DNA sequenced in both directions.
TABLE 1.
Amino acid sequences of mucin-desulfating sulfatase peptidesa
| Peptide | Sequence | Amino acids |
|---|---|---|
| T6a | FMYEE | 360–364 |
| T6b | QAWDAYY | 276–282 |
| T16 | EQGYATDIVTEHAVE | 166–180 |
| T17 | LI( )FY | 455–459 |
| T23 | YMHDYLSTIH | 307–316 |
| CB2 | (M)( )( )LQKQAWDAYY | 270–282 |
| CB11 | (M)QNDPNELN( )L | 478–488 |
| CB14a | (M)PNLKYLGLYDKVEFP | 205–220 |
| CB14b | (M)YEES(F,L)R(T,D)P | 361–369 |
Tryptic peptides are given a T prefix, and cyanogen bromide peptides have a CB prefix. The T6a and T6b and CB14a and CB14b sequences were determined by sequence analysis of samples which contained both peptides. Empty parentheses indicate an unidentified amino acid. (M), deduced methionine. Two amino acids within parentheses indicates that a decision between the two amino acids was equivocal. Amino acid numbering corresponds to the MdsA sequence shown in Fig. 2.
Expression of mucin-desulfating sulfatase protein in Escherichia coli.
An MseI 1,615-bp fragment (bases 3141 to 4755 from the 5,058-bp XhoI Prevotella fragment containing the entire sulfatase sequence) was subligated in the NdeI site of plasmid pET17b (Novagen Inc., Madison, Wis.) and used to transform E. coli BL21pLysS. Plasmids with the correct insert orientation were identified, and expression of the sulfatase protein was induced by isopropyl-β-d-thiogalactopyranoside (IPTG).
Expression of the Prevotella mucin-desulfating sulfatase gene in Bacteroides thetaiotaomicron.
A gDNA fragment containing all of the Prevotella sulfatase sequence was amplified by PCR with the 5,058-bp XhoI insert in pGEM-7 as the template. Primer sequences were TR22(F), 5′ CTT TGG ATC CAA TTA TGG GCA CCC GTG 3′ (bases 3144 to 3170 in Fig. 1, modified to contain a BamHI site), and SP3(R), 5′ TAG GTG ATA TCG TAG TGA GCG CCA ACA AG 3′ (bases 4811 to 4783 in Fig. 1 modified to contain an EcoRV site). PCR amplification was performed with ELONGase (Life Technologies, Gaithersburg, Md.), and the product was digested with BamHI and EcoRV.
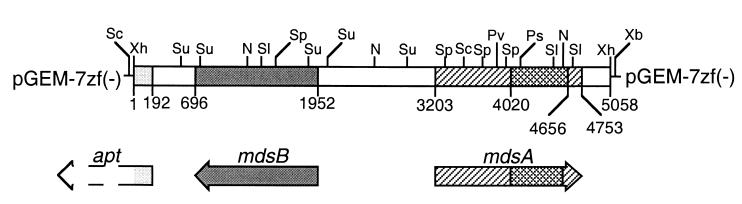
FIG. 1.
Restriction map of the 4.7-kb gDNA fragment from Prevotella strain RS2 cloned in pGEM-7zf(−). Arrows indicate the positions and directions of ORFs encoding the mucin-desulfating sulfatase (mdsA), the putative sulfatase posttranslational modification protein (mdsB), and a gene sequence tentatively identified as the N-terminal region of anthranilate phosphoribosyltransferase (apt). The cross-hatched region in mdsA shows the location of the 600-bp PCR product formed using primers TR4(F) and TR7(R). Nucleotide numbers refer to the 5,058 bp found after sequencing of the gDNA fragment. Restriction sites are indicated by the letters N (NcoI), Ps (PstI), Pv (PvuII), Sc (SacI), Sl (SalI), Sp (SphI), Su (Sau3AI), Xb (XbaI), and Xh (XhoI).
The B. thetaiotaomicron shuttle plasmid pVGA-3 (6, 8) contains a promoter-operator region of the chondroitin sulfate-regulated operon csuCB controlling the expression of an E. coli reporter gene encoding β-glucuronidase (GUS). A polylinker region between the operator and the GUS gene has BamHI and SmaI sites. pVGA-3 was digested with these restriction enzymes, and the PCR product encoding the Prevotella sulfatase was ligated into the plasmid. E. coli S17-1, a strain that contains the transfer region of plasmid RP4 in its chromosome, was transformed by pVGA-3 containing the sulfatase insert and by control uncut pVGA-3. The transformed E. coli S17-1 was mated with B. thetaiotaomicron ATCC 29148, and the shuttle plasmid was transferred by conjugation (28). Bacteroides cells containing the plasmid were selected by anaerobic growth on supplemented brain-heart infusion-agar plates containing tetracycline (3 μg ml−1) and gentamicin (0.2 mg ml−1). For measurements of enzyme expression, B. thetaiotaomicron cells containing plasmids were grown in medium 10 broth with 0.3% (wt/vol) chondroitin sulfate, glucose, or maltose as the sole energy source in the presence of tetracycline (3 μg ml−1) to maintain the plasmid. Mucin-desulfating sulfatase, measured by the glucose-6-sulfatase assay (19), and GUS (6) activities were measured in cell extracts made by sonication followed by centrifugation or in the periplasm.
Antibody against Prevotella mucin-desulfating sulfatase and Western blotting.
A chimeric gene construct was made in plasmid pGEX-2T (Amrad Pharmacia Biotech, Uppsala, Sweden) which contains a glutathione S-transferase (GST) gene under the control of a lac promoter. A DNA fragment (bases 4431 to 4915 in Fig. 1) encoding the 3′ end of the Prevotella sulfatase gene was amplified by PCR with primers DPW10(F) (5′ C GGA TCC TCA TTC CTC GAC CTC TTC G 3′) and TR15(R) (5′ TCA GGG AAT TCC CAA GCG GCT TCA CGC TTT C 3′), which have been modified to incorporate BamHI and EcoRI sites. After restriction digestion, this sulfatase gene fragment was inserted in frame in the polycloning site at the 3′ end of the GST sequence. A chimeric protein of GST fused to 105 amino acids from the C terminus of the sulfatase was expressed and purified by glutathione–Sepharose CL-4B affinity chromatography (Amrad Pharmacia Biotech). Antibodies against the fusion protein were raised in New Zealand White rabbits. Antibody production was monitored by enzyme-linked immunosorbent assay (ELISA) using inactive recombinant Prevotella sulfatase produced in an E. coli expression vector as the bound antigen. Rabbit serum was collected, and antibody against GST epitopes was removed by chromatography through Sepharose 4B-GST resin. The remaining polyclonal antibody showed good reactivity with Prevotella sulfatase. Western blotting with this antibody was performed by standard procedures and with alkaline phosphatase-labeled second antibody (22) after proteins were electrophoresed on SDS–10% PAGE gels.
Nucleotide sequence accession number.
The 5,058-bp XhoI insert has been given GenBank accession number AF248951.
RESULTS
Sulfatase peptide sequences.
Initial attempts to sequence the N terminus of the intact 58-kDa mucin-desulfating sulfatase were unsuccessful, suggesting that it was blocked. The protein was subsequently cleaved with trypsin and cyanogen bromide, and five tryptic peptides and four cyanogen bromide peptides were isolated by reverse-phase HPLC and partially sequenced (Table 1).
Screening of a partial XhoI gDNA library for sulfatase gene sequence.
Forward and reverse primers based on sulfatase peptide sequences CB2 and CB11 and Prevotella strain RS2 gDNA template were used for PCR. TR5(R) plus TR6(F) gave no product, but TR4(F) plus TR7(R) gave an approximately 600-bp product. This was ligated into M13mp18 and sequenced. Sequences encoding peptides T6a, T23, T17, and CB14b and sequence 3′ to primer CB2 (Table 1) were present, confirming that the 600-bp product encoded part of the sulfatase protein.
Southern analysis of XhoI-digested Prevotella gDNA with the 600-bp PCR product as a probe revealed a 4.7-kb XhoI fragment. This was large enough to contain the sulfatase gene, predicted to be about 1.5 kb. A partial genomic library was constructed by ligating XhoI-digested gDNA (4.2- to 5.2-kb fragments) into pGEM-7zf(−) and transforming E. coli DH5αF′ to ampicillin resistance. Colonies were screened with the 600-bp probe, and a positive recombinant plasmid containing an approximately 4.7-kb gDNA insert was isolated.
Sequence determination of the 4.7-kb XhoI insert.
The insert was characterized by restriction mapping (Fig. 1). The 600-bp probe bound to a region 1.0 to 0.4 kb from one end. The insert was sequenced in both directions and contained 5,058 bp.
Nucleotide and amino acid sequences of the sulfatase.
The 5,058-bp XhoI insert sequence revealed three open reading frames (ORFs). One of these, nucleotides 3203 to 4753, is 1,551 bp long and contains the 600-bp PCR product sequence (Fig. 1). There are two possible ATG codons for the initiation of translation, but subsequent expression of protein in an E. coli system suggests that translation may start at the first ATG (nucleotides 3203 to 3205) rather than the second (nucleotides 3218 to 3220) (see below). The ORF from the first ATG codon encodes a deduced protein of 517 amino acids, with a calculated molecular mass of 60 kDa. The deduced sequence is shown as the mucin-desulfating sulfatase A (MdsA-prev) in Fig. 2. The sequence contains a characteristic signal sequence (Met1 to Ala23) for targeting to the periplasm (29) and all nine sulfatase peptides found previously by microsequencing (Table 1).
FIG. 2.
Alignment of the deduced amino acid sequence of Prevotella strain RS2 mucin-desulfating sulfatase (MdsA-prev) with three other sulfatases: Yidj-esch, from the SWISS-PROT database, a putative E. coli sulfatase (f497 in reference 2); Gl6s-human, human N-acetylglucosamine-6-sulfatase (G6S in reference 21); and AtsA-kleb, K. pneumoniae arylsulfatase (14). Amino acids that occur in three of the four sequences are highlighted in reverse type, and a consensus sequence is included. Where the consensus sequence is highly conserved, the amino acids are underlined.
Comparisons with primary structures of sulfatases.
The deduced amino acid sequence of the mucin-desulfating sulfatase was compared with protein sequences in the SWISS-PROT database. Sequence homology was found with 26 aryl- and nonarylsulfatases. The deduced protein sizes of these sulfatases (including signal peptides, where present) were 464 to 649 amino acids. Figure 2 shows the alignment of the Prevotella mucin-desulfating sulfatase with two bacterial sulfatases, a putative E. coli sulfatase (Yidj-esch) and Klebsiella pneumoniae arylsulfatase, and a human lysosomal sulfatase specific for N-acetylglucosamine-6-sulfate. Regions of similarity extend throughout the sequences.
Expression of the sulfatase protein in E. coli. An attempt was made to express the Prevotella sulfatase gene in E. coli.
An MseI restriction fragment containing the entire sulfatase gene was inserted into pET17b, and the recombinant plasmid was used to transform E. coli BL21pLysS. A major protein band of the expected molecular size was induced by IPTG. However, the protein was concentrated in the cytoplasm rather than the periplasm, and no enzymatic activity was detected in extracts. N-terminal analysis of the band gave the sequence MKSDN, indicating that the first methionine (nucleotides 3203 to 3205) was the start of translation in this system and that the signal sequence had not been removed from the bulk of the protein. Attempts were made to express the sulfatase as an active enzyme in other E. coli expression systems, including plasmids pJLA602 and pET22b. In every case the engineered protein was the expected size on SDS-PAGE gels but precipitated out in inclusion bodies and had no enzymatic activity. Attempts to refold the engineered protein, with or without its signal sequence, were unsuccessful.
Expression of the Prevotella mucin-desulfating sulfatase gene in B. thetaiotaomicron.
One possible explanation for the results obtained using E. coli expression systems is that there is significant phylogenetic distance between Prevotella and Escherichia, and though protein translation has occurred, the subsequent processing of the sulfatase chain—chaperone interaction, posttranslational modification, translocation to the periplasm, or folding—has not functioned properly. Therefore, we attempted to express the Prevotella mucin-desulfating sulfatase in the closely related genus Bacteroides.
A 1,668-bp genomic DNA fragment (bases 3144 to 4811 of the 5,058-bp XhoI insert) containing the Prevotella sulfatase sequence was amplified by PCR. After restriction digestion to produce compatible ends, this fragment was ligated into the polylinker region of pVGA-3, downstream from the chondroitin sulfate promoter-operator region and upstream from the GUS reporter gene. This shuttle plasmid was then transferred from E. coli S17-1 into B. thetaiotaomicron. A control pVGA-3 plasmid without an insert was also transferred.
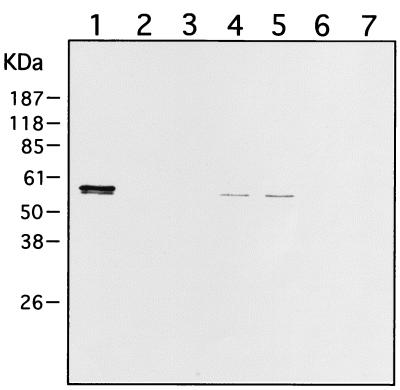
Table 2 shows that the strain containing pVGA-3 with the sulfatase insert expresses both active sulfatase (as measured by the glucose-6-sulfatase assay) and reporter enzyme GUS when cells are grown on chondroitin sulfate but not on glucose or maltose. The control strain containing pVGA-3 with no sulfatase insert expresses only GUS when grown on chondroitin sulfate but not the other substrates. Clearly the Bacteroides expression system is able to synthesize and fold the Prevotella sulfatase. The results of Western blotting of the Bacteroides recombinant protein were consistent with these enzyme activity results (Fig. 3). Only cells containing the plasmid with the mdsA insert and grown on chondroitin sulfate produced a band at approximately 58 kDa detected by antibody against Prevotella sulfatase (lane 1). The enzyme was concentrated in the periplasm rather than in spheroplasts, as expected (compare lanes 1 and 5), confirming that the mdsA signal sequence is recognized. Neither the above cells grown on glucose (lane 3) nor cells containing a plasmid without the mdsA insert grown on chondroitin sulfate (lane 2) showed bands. A positive control containing periplasm from Prevotella cells grown on mucin is shown in lane 4. When the Prevotella strain was grown on galactose, no sulfatase band was seen (result not shown).
TABLE 2.
Specific activities of GUS and mucin-desulfating sulfatase (MDS) in cell extracts from B. thetaiotaomicron containing plasmid pVGA-3 with or without the mdsA insert after the chondroitin sulfate promoter-operator region grown on medium 10 with various sugar substrates
| Plasmid | Sugar substrate | GUS sp acta | MDS sp actb |
|---|---|---|---|
| pVGA-3 | Chondroitin sulfate | 4.4 | 0.5 |
| Glucose | 0.02 | 0 | |
| Maltose | 0.02 | 0.5 | |
| pVGA-3 + mdsA | Chondroitin sulfate | 8.5 | 49 |
| Glucose | 0.03 | 0 | |
| Maltose | 0 | 1.3 |
GUS specific activity in nanomoles of p-nitrophenol per minute per milligram of cell extract protein.
MDS specific activity in nanomoles of glucose per minute per milligram of cell extract protein.
FIG. 3.
Western blot analysis of recombinant and normal Prevotella strain RS2 mucin-desulfating sulfatases after SDS-PAGE. Samples analyzed included the periplasm of chondroitin sulfate-grown B. thetaiotaomicron containing plasmid pVGA-3 with the mdsA insert (7 μg of protein) (lane 1) and without the mdsA insert (7 μg of protein) (lane 2), periplasm of glucose-grown B. thetaiotaomicron containing plasmid pVGA-3 with the mdsA insert (7 μg of protein) (lane 3), periplasm of mucin-grown Prevotella strain RS2 (7 μg of protein) (lane 4), lysed spheroplasts of chondroitin sulfate-grown B. thetaiotaomicron containing plasmid pVGA-3 with the mdsA insert (16 μg of protein) (lane 5) and without the mdsA insert (23 μg of protein) (lane 6), and lysed spheroplasts of glucose-grown B. thetaiotaomicron containing plasmid pVGA-3 with the mdsA insert (9 μg of protein) (lane 7). Molecular size standards are shown on the left.
The specificity of the sulfatase made using the Bacteroides expression system was compared with that of authentic enzyme from Prevotella strain RS2 (Table 3). Glucose-6-sulfate was a substrate for both enzymes. The activity of the periplasmic enzyme from the recombinant source was higher than that of the periplasmic enzyme from mucin-grown Prevotella cells. Two competitive inhibitors, N-acetylglucosamine and N-acetylglucosamine-6-phosphate, inhibited both enzymes.
TABLE 3.
Comparison of mucin-desulfating sulfatase (MDS) activities in the periplasm of mucin-grown Prevotella strain RS2 and the periplasm of chondroitin sulfate-grown B. thetaiotaomicron containing plasmid pVGA-3 with an mdsA insert
| Conditions | Periplasmic MDS activity, nmol/min/mg of protein (% inhibition)
|
|
|---|---|---|
| Prevotella strain RS2 | B. thetaiotaomicron | |
| Glucose-6-sulfate substrate | 24 | 82 |
| Glucose-6-sulfate substrate + 2 mM N-acetylglucosamine-6-phosphate | 3 (87) | 12 (85) |
| Glucose-6-sulfate substrate + 3 mM N-acetylglucosamine | 10 (58) | 51 (38) |
Identification of a second ORF within the 5,058-bp XhoI insert.
In the region upstream from the sulfatase gene, another ORF 1,257 nucleotides in length was identified, encompassing nucleotides 1952 to 696 (Fig. 1) and coding in the reverse direction from the sulfatase gene. The predicted peptide of 419 amino acids corresponds to a 49.1-kDa protein. Upstream from this ORF are two sets of putative RNA polymerase-binding sequences and a putative ribosome-binding site, while a putative transcription termination sequence is downstream from the stop codon.
The deduced protein sequence from this ORF (mdsB in Fig. 1) was compared with sequences in the database. Striking similarities were found to three proteins formerly thought to be involved in transcriptional regulation of sulfatases but recently shown to be involved instead in sulfatase posttranslational modification (25). The Prevotella protein showed 65% identity to B. thetaiotaomicron chondro-6-sulfatase “regulatory protein” ChuR (3), 44% identity to E. coli putative arylsulfatase “regulatory protein” AslB (5), and 37% identity to K. pneumoniae putative arylsulfatase “regulatory protein” AtsB (14). Alignment showed that homology with the putative Prevotella protein MdsB occurred over the length of the proteins. This homology suggests that the putative Prevotella protein MdsB, like the Klebsiella protein AtsB, is likely to encode an iron-sulfur protein involved in posttranslational modification of the sulfatase, rather than a sulfatase regulatory protein (25).
Identification of a third partial ORF within the 5,058-bp XhoI insert.
Nucleotides 192 to 1 in the 5,058-bp gDNA fragment contain another partial ORF (Fig. 1). Amino acid homology suggests that the sequence codes for the N terminus of an anthranilate phosphoribosyltransferase. This protein is not relevant to mucin degradation or sulfatases, and investigation of the DNA in this direction was not continued.
DISCUSSION
The Prevotella sulfatase gene mdsA encodes the first mucin-desulfating sulfatase to be fully characterized. The gene encodes a 517-amino-acid proprotein, and the deduced size of 57.6 kDa is consistent with the subunit size of 58 kDa estimated from SDS-PAGE (19). Sequence homologies occur throughout the length between this mucin-desulfating sulfatase and other aryl- and nonarylsulfatases, which are proteins of 464 to 649 amino acids. Only one other mucin-desulfating sulfatase has been purified, a 15-kDa protein (27), which is much smaller than any sequenced sulfatase. Given the significant sequence homology between sulfatases, it may be difficult to fit this fecal mucin-desulfating sulfatase into the existing sulfatase family, and information on its protein sequence would be very helpful.
The crystal structures of two human sulfatases have been solved (1, 11), and the critical amino acids participating in sulfatase substrate and metal binding are highly conserved (30). In the Prevotella mucin-desulfating sulfatase, 9 of 10 of these highly conserved active-site residues (30) are found, the exception being a conservative replacement of glutamine for asparagine. One of the conserved active-site residues is a formylglycine, made by posttranslational modification (23) of a cysteine or serine residue (Ser79 in the Prevotella protein).
A prime objective of the present work was to prepare large amounts of the Prevotella mucin-desulfating sulfatase by recombinant expression, to confirm the identity of the cloned gene, and to simplify preparation of the enzyme for future research. However, all attempts to make this sulfatase as an active enzyme in E. coli expression systems were unsuccessful. This result was puzzling, as protein synthesis occurred and the signal sequence should have been usable. Our initial conclusion was that E. coli is probably too distant phylogenetically from Prevotella for recognition by the host chaperones or translocation machinery. We therefore made use of a Bacteriodes expression system, developed in A. A. Salyers' laboratory, and were successful in expressing active sulfatase.
However, a recent paper on expression of the Klebsiella sulfatase gene (atsA) in an E. coli expression system has provided a different explanation (25). E. coli contains three sulfatase gene homologs, but no sulfatase activity has been detected in this bacterium. When the Klebsiella sulfatase gene (atsA) was cloned into E. coli, the expressed protein was inactive unless an adjacent Klebsiella gene (atsB) was cloned simultaneously (25). The researchers have provided evidence that atsB is not involved in regulation of sulfatase transcription, as previously thought, but encodes an iron-sulfur enzyme catalyzing the critical serine-to-formylglycine posttranslational modification necessary for AtsA active-center function. The E. coli homolog of atsB is inactive (25). Our work provides independent support for this explanation as well as an alternative way of producing recombinant sulfatase. The Prevotella sulfatase mdsA is not expressed in E. coli but is expressed in the alternative Bacteroides expression system, which is known to contain chuR, a homolog of atsB, known to be necessary for chondroitin sulfate desulfation in the host organism. The mdsB and mdsA genes are adjacent in Prevotella strain RS2, as are their homologs atsB and atsA in K. pneumoniae. Thus, we speculate that lack of posttranslational serine modification activity in E. coli expression systems accounts for the absence of activity of the Prevotella sulfatase.
Mucin is thought to provide one of several endogenous energy sources for colon bacteria (12), and there is accumulating evidence that heavy sulfation of mucin rate-limits its breakdown. Clearly, it is important to identify specific sulfatases from bacteria that remove sulfate from these sulfated mucin sugars and how the levels of these sulfatases are regulated. In the context of large-bowel inflammatory disease, fecal mucin-desulfating sulfatase levels are elevated in ulcerative colitis (26), but it is uncertain which bacteria are involved, which sulfated sugars are being desulfated, and what has caused the change in activities. The present research characterizes for the first time two adjacent genes, encoding a sulfatase and its putative posttranslational modifying enzyme, which may well be similar to genes encoding the proteins involved in the mucin-desulfating sulfatase changes recorded in this disease. Production of the Prevotella mucin-specific N-acetylglucosamine-6-sulfatase in a recombinant system should allow us to develop simple but specific assays for its measurement in colon contents.
ACKNOWLEDGMENTS
This work was supported by the Auckland Medical Research Foundation, the Auckland University Staff Research Fund, and the New Zealand Lottery Health Scientific Committee.
We thank A. A. Salyers and N. B. Shoemaker, University of Illinois, for their advice and the gift of the Bacteroides shuttle vector, and D. J. Saul for advice on the DNA analyses. We are grateful to Christine Partridge for her technical assistance during part of this work. D.P.W. was the recipient of an Auckland University Maori and Pacific Islander Doctoral Scholarship.
REFERENCES
- 1.Bond C S, Clements P R, Ashby S J, Collyer C A, Harrop S J, Hopwood J J, Guss J M. Structure of a human lysosomal sulfatase. Structure. 1997;5:277–289. doi: 10.1016/s0969-2126(97)00185-8. [DOI] [PubMed] [Google Scholar]
- 2.Burland V, Plunkett G, Daniels D L, Blattner F R. DNA sequence and analysis of 136 kilobases of the Escherichia coli genome: organisational symmetry around the origin of replication. Genomics. 1993;16:551–561. doi: 10.1006/geno.1993.1230. [DOI] [PubMed] [Google Scholar]
- 3.Cheng Q, Hwa V, Salyers A A. A locus that contributes to colonization of the intestinal tract by Bacteroides thetaiotaomicron contains a single regulatory gene (chuR) that links two polysaccharide utilization pathways. J Bacteriol. 1992;174:7185–7193. doi: 10.1128/jb.174.22.7185-7193.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Corfield A P, Wagner S A, O'Donnell L J D, Durdey P, Mountford R A, Clamp J R. The roles of enteric bacterial sialidase, sialate O-acetyl esterase, and glycosulfatase in the degradation of human colonic mucin. Glycoconjugate J. 1993;10:72–81. doi: 10.1007/BF00731190. [DOI] [PubMed] [Google Scholar]
- 5.Daniels D L, Plunkett G, Burland V, Blattner F R. Analysis of the Escherichia coli genome: DNA sequence of the region from 84.5 to 86.5 minutes. Science. 1992;257:771–778. doi: 10.1126/science.1379743. [DOI] [PubMed] [Google Scholar]
- 6.Feldhaus M J, Hwa V, Salyers A A. Use of an Escherichia coli β-glucuronidase gene as a reporter gene for investigation of Bacteroides promoters. J Bacteriol. 1991;173:4540–4543. doi: 10.1128/jb.173.14.4540-4543.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Houdret N, Ramphal R, Scharfman A, Perini J-M, Filliat M, Lamblin G, Roussel P. Evidence for the in vivo degradation of human respiratory mucins during Pseudomonas aeruginosa infection. Biochim Biophys Acta. 1989;992:96–105. doi: 10.1016/0304-4165(89)90055-x. [DOI] [PubMed] [Google Scholar]
- 8.Hwa V, Salyers A A. Evidence for differential regulation of genes in the chondroitin sulfate utilization pathway of Bacteroides thetaiotaomicron. J Bacteriol. 1992;174:342–344. doi: 10.1128/jb.174.1.342-344.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Irimura T, Wynn D M, Hager L G, Cleary K R, Ota D M. Human colonic sulphomucin identified by a specific colonic monoclonal antibody. Cancer Res. 1991;51:5728–5735. [PubMed] [Google Scholar]
- 10.Luckey T D. Introduction to intestinal microecology. Am J Clin Nutr. 1972;25:1292–1294. doi: 10.1093/ajcn/25.12.1292. [DOI] [PubMed] [Google Scholar]
- 11.Lukatala G, Krauss N, Theis K, Selmer T, Gieselmann V, von Figura K, Saenger W. Crystal structure of human arylsulfatase A: the aldehyde function and the metal ion at the active site suggest a novel mechanism for sulfate ester hydrolysis. Biochemistry. 1998;37:3654–3664. doi: 10.1021/bi9714924. [DOI] [PubMed] [Google Scholar]
- 12.Macfarlane G T, Gibson G R. Metabolic activities of the normal colonic flora. In: Gibson S A W, editor. Human health: the contribution of microorganisms. New York, N.Y: Springer-Verlag; 1994. pp. 18–52. [Google Scholar]
- 13.Mian N, Anderson C E, Kent P W. Neuraminidase inhibition by chemically sulphated glycopeptides. Biochem J. 1979;181:377–385. doi: 10.1042/bj1810377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Murooka Y, Ishibashi K, Yasumoto M, Sasaki M, Sugino H, Azakami H, Yamashita M. A sulfur- and tyramine-regulated Klebsiella aerogenes operon containing the arylsulfatase (atsA) gene and the atsB gene. J Bacteriol. 1990;172:2131–2140. doi: 10.1128/jb.172.4.2131-2140.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nieuw Amerongen A V, Bolscher J G M, Bloemena E, Veerman E C I. Sulfomucins in the human body. Biol Chem. 1998;379:1–18. doi: 10.1515/bchm.1998.379.1.1. [DOI] [PubMed] [Google Scholar]
- 16.Paster B J, Dewhirst F E, Olsen I, Fraser G J. Phylogeny of Bacteroides, Prevotella, and Porphyromonas and related bacteria. J Bacteriol. 1994;176:725–732. doi: 10.1128/jb.176.3.725-732.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Podolsky D K, Isselbacher K J. Composition of human colonic mucin: selective alteration in inflammatory bowel disease. J Clin Investig. 1983;72:142–153. doi: 10.1172/JCI110952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roberton A M, Corfield A P. Mucin degradation and its significance in inflammatory conditions of the gastrointestinal tract. In: Tannock G W, editor. Medical importance of normal microflora. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1999. pp. 222–261. [Google Scholar]
- 19.Roberton A M, McKenzie C G, Scharfe N, Stubbs L B. A glycosulphatase that removes sulphate from mucus glycoprotein. Biochem J. 1993;293:683–689. doi: 10.1042/bj2930683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roberton A M, Rosendale D I, Wright D P. Enzymic degradation of mucins—assays for bacterial mucin-desulfating sulfatases. Methods Mol Biol. 1999;125:417–426. doi: 10.1385/1-59259-048-9:417. [DOI] [PubMed] [Google Scholar]
- 21.Robertson D A, Freeman C, Morris C P, Hopwood J J. A cDNA clone for human glucosamine-6-sulphatase reveals differences between arylsulphatases and nonarylsulphatases. Biochem J. 1992;288:539–544. doi: 10.1042/bj2880539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 23.Schmidt B, Selmer T, Ingendoh A, von Figura K. A novel amino acid modification in sulfatases that is defective in multiple sulfatase deficiency. Cell. 1995;82:271–278. doi: 10.1016/0092-8674(95)90314-3. [DOI] [PubMed] [Google Scholar]
- 24.Stanley R A, Ram S P, Wilkinson R K, Roberton A M. Degradation of pig gastric and colonic mucins by bacteria isolated from the pig colon. Appl Environ Microbiol. 1986;51:1104–1109. doi: 10.1128/aem.51.5.1104-1109.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Szameit C, Miech C, Balleininger M, Schmidt B, von Figura K, Dierks T. The iron sulfur protein AtsB is required for posttranslational formation of formylglycine in Klebsiella sulfatase. J Biol Chem. 1999;274:15375–15381. doi: 10.1074/jbc.274.22.15375. [DOI] [PubMed] [Google Scholar]
- 26.Tsai H H, Dwarakanath A D, Hart C A, Milton J D, Rhodes J M. Increased faecal mucin sulfatase activity in ulcerative colitis: a potential target for treatment. Gut. 1995;36:570–576. doi: 10.1136/gut.36.4.570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsai H H, Sunderland D, Gibson G R, Hart C A, Rhodes J M. A novel mucin sulphatase from human faeces: its identification, purification and characterisation. Clin Sci. 1992;82:447–454. doi: 10.1042/cs0820447. [DOI] [PubMed] [Google Scholar]
- 28.Valentine P J, Shoemaker N B, Salyers A A. Mobilization of Bacteroides plasmids by Bacteroides conjugal elements. J Bacteriol. 1988;170:1319–1324. doi: 10.1128/jb.170.3.1319-1324.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.von Heijne G. Membrane proteins: the amino acid composition of membrane penetrating segments. Eur J Biochem. 1981;120:275–278. doi: 10.1111/j.1432-1033.1981.tb05700.x. [DOI] [PubMed] [Google Scholar]
- 30.Waldow A, Schmidt B, Dierks T, von Bülow R, von Figura K. Amino acid residues forming the active site of arylsulfatase A. Role in the catalytic activity and substrate binding. J Biol Chem. 1999;274:12284–12288. doi: 10.1074/jbc.274.18.12284. [DOI] [PubMed] [Google Scholar]