Abstract
Being a highly conserved catabolic process, autophagy is induced by various forms of cellular stress, and its modulation has considerable potential as a cancer therapeutic approach. In the present study, it was demonstrated that dicitrinone B (DB), a rare carbon-bridged citrinin dimer, may exert anticancer effects by blocking autophagy at a late stage, without disrupting lysosomal function in MCF7 breast cancer and MDA-MB-231 triple-negative breast cancer cells. Furthermore, it was discovered that DB significantly enhanced intracellular reactive oxygen species (ROS) production and that the removal of ROS was followed by the attenuation of autophagy inhibition. In addition, DB exerted notable inhibitory effects on the proliferation and promoting effects on the apoptosis of MCF7 and MDA-MB-231 cells. In combination with conventional chemotherapeutic drugs, DB exhibited a further enhanced synergistic effect than when used as a single agent. Overall, the data of the present study demonstrate that DB may prove to be a promising autophagy inhibitor with anticancer activity against breast cancer.
Keywords: dicitrinone B, autophagy inhibitor, breast cancer, reactive oxygen species, adriamycin
Introduction
Breast cancer is known as one of the leading causes of cancer-related mortality and morbidity among women worldwide (1-3). There is strong evidence for considering early-stage breast cancer potentially curable; however, concerning late-stage or metastatic breast cancer, currently available therapeutic approaches are only able to prolong the survival and maintain the quality of life of patients. Due to its highly drug-resistant and invasive nature, and its proclivity for recurrence and metastasis, triple-negative breast cancer [human epidermal growth factor receptor-2 (HER2)-, estrogen receptor (ER)- and prostaglandin receptor (PgR)-] is particularly lethal, with no effective treatments available (4-7).
Autophagy is an ubiquitous catabolic process in animal cells. It has been suggested that autophagy plays a crucial role in the growth and development of a variety of cells (8,9). As an evolutionary conserved adaptive process, autophagy can sequester long-lived, aggregated and misfolded proteins along with damaged organelles via the formation of autophagosomes, which then fuse with lysosomes, in which cargos are degraded and recycled (10,11). The biological and clinical significance of autophagy in cancer stems from its complex role in the tumor microenvironment (12). Autophagy has been revealed to suppress early cancer development, while facilitating advanced tumor progression (13-17). The pharmacological regulation of autophagy as a valid strategy in certain types of cancer, including breast cancer, and has been demonstrated to enhance the efficacy of therapeutics and to overcome resistance (18-21).
Marine microbes are a significant source of lead compounds in drugs that have been scientifically validated to exert marked anticancer, anti-bacterial and pro-apoptotic effects, and to regulate immunity in cell and animal models (22,23). There is accumulating evidence to indicate that purified natural therapeutics may remove impurities and toxic components, and enhance the curative effects in relation to fully synthetic therapeutics. In addition, various natural Chinese medicines such as Genkwadaphnin, dihydroartemisinin, etc. when combined with radiotherapy and chemotherapy, have been reported to not only reduce the side-effects of drugs, but to also significantly enhance their antitumor effects (24). Therefore, there is considerable interest in investigating marine natural products. Since 2010, dicitrinone A-F (DA-DF) has been successively discovered from the volcano ash-derived or marine-derived fungus, Penicillium citrinum. Those novel carbon-bridged citrinin dimers, as natural polyketones, have anti-tumor, anti-bacterial, anti-oxidant and microtubule targeting properties (25-28). Previous studies have demonstrated that DA-DD has a similar structure and can significantly inhibit the proliferation of HL-60, MOLT-4, A-549, BEL-7402 and SPC-A1 cells, of which dicitrinone B (DB) has the best anti-tumor activity (25-27). The structures of DE and DF are relatively different with those of DA-DD, and they have no antitumor activity (28). Previous research by the authors has revealed that DB induces the apoptosis of human malignant melanoma A375 cells by increasing reactive oxygen species (ROS) generation, and this process is related to the regulation of Bcl-2 family proteins (25). However, the anticancer effects of DB and its detailed mechanisms of action in breast cancer remain unclear. The novel findings of the present study (to the best of our knowledge) indicate that DB may be a potential autophagy inhibitor with anticancer activity; however, further preclinical research is required in order to develop effective treatments for breast cancer.
Materials and methods
Reagents and antibodies
The compound DB was separated from the fermentation product of Penicillium citrinum and purified by various separation and purification methods, such as extraction, column chromatography, and high-performance liquid chromatography, the purity of which was >95%, dissolved in dimethyl sulphoxide (DMSO, cat. no. 196055, MP Biomedicals, LLC) to yield a stock solution at 20 mM and stored at -20°C. Chloroquine (CQ; C129284) was purchased from Shanghai Aladdin Biochemical Technology Co., Ltd. and dissolved in phosphate-buffered saline (PBS). Adriamycin (ADM; cat. no. D807083) was purchased from Shanghai Macklin Biochemical Co., Ltd. and dissolved in PBS. Rapamycin (RAPA; cat. no. HY-10219) was purchased from MedChemExpress and dissolved in PBS. Acridine orange (AO; cat. no. cM07364) was purchased from Beijing Bai'aolaibo Technology Co., Ltd. LysoTracker Red (cat. no. C1046) and N-acetyl-L-cysteine (NAC; cat. no. S0077) were purchased from the Beyotime Institute of Biotechnology. Bicinchoninic acid (BCA; cat. no. MPK002) was purchased from MACGENE Biotechnology. Microtubule-associated protein 1 light chain 3 beta (LC3 B), microtubule-associated protein 1 light chain 3 alpha/beta (LC3A/B), p62, Bcl-2, Bax, poly(ADP-ribose) polymerase (PARP), mTOR, Akt and cleaved PARP antibodies (cat. nos. 3868S, 12741S, 16177S, 3498S, 5023S, 9532S, 2983S, 4691S and 5625S, respectively; rabbit) and HRP-linked goat anti-rabbit IgG, HRP-linked anti-mouse IgG (cat. nos. 7074P2 and 7076P2, respectively) were purchased from Cell Signaling Technology, Inc. Cathepsin D (CTSD; cat. no. BM1577; mouse), β-actin (cat. no. BM3873; rabbit), cofilin (cat. no. PB9033; rabbit) and cathepsin B (CTSB; cat. no. A01456-3, rabbit) antibodies were purchased from Wuhan Boster Biological Technology, Ltd. FITC-linked goat anti-rabbit IgG (cat. no. ZF-0311) was purchased from Beijing Zhongshan Jinqiao Biotechnology Co., Ltd. Puromycin was purchased from InvivoGen (cat. no. ant-pr-1).
Cells and cell culture
The human breast cancer cell line MCF7 (cat. no. TCHu 74) was purchased from the Shanghai Cell Resource Center and cultured in Dulbecco's modified Eagle's medium (HyClone; Cytiva) supplemented with 10% fetal bovine serum (Gemini Bio Products; cat. no. 900-108) and 1.0% penicillin/streptomycin (cat. no. MA0110; Dalian Meilun Biology Technology Co., Ltd.). The MDA-MB-231 cells were a gift from Dr. Liu (Fujian Cancer Hospital, Fujian, China) and were maintained in RPMI-1640 medium (HyClone; Cytiva) containing 10% fetal bovine serum (Gemini Bio Products; cat. no. 900-108) and 1% penicillin/streptomycin (cat. no. MA0110; Dalian Meilun Biotech, Co., Ltd.). All cells were cultured at 37°C in a humidified atmosphere with 5% CO2.
Cell viability assay
Cell growth inhibition activity was examined using a CellTiter 96® AQueous One Solution Cell Proliferation Assay (cat. no. G3581, Promega Corporation) following the manufacturer's instructions. Briefly, ~5,000 MCF7 or MDA-MB-231 cells were seeded per well in three replicates into 96-well plates and treated with DMSO or serial dilutions of DB (5, 10, 20, 40 and 60 µM), ADM (0.5, 2.5 and 10 µM) or PTX (0.05, 0.1, 0.2 and 0.4 µM) for 24 or 48 h, cells were incubated at 37°C in a humidified atmosphere with 5% CO2. In addition, ~5,000 MCF7 or MDA-MB-231 cells were seeded per well in three replicates into 96-well plates and treated with DB (10 µM), ADM (0.2 µM) and PTX (0.005 µM) alone or in combination with DB (10 µM), ADM (0.2 µM) or PTX (0.005 µM) for 24 and 48 h. Following treatment, MTS solution (provided with the kit) was used to evaluate the viability of cells. The absorbance was detected at 490 nm using a microplate reader (SH-1000; Corona Electric Co., Ltd.).
Western blot analysis
Western blot analysis was performed as previously described with minor modifications (25). In brief, the MCF7 or MDA-MB-231 cells were scraped in modified RIPA buffer (Wuhan Boster Biological Technology, Ltd.) containing 1 mM PMSF (cat. no. P0100, Beijing Solarbio Science & Technology Co., Ltd.) to prepare whole-cell lysates. BCA Protein Assay kit (cat. no. P0012, Beyotime Institute of Biotechnology) was used for protein quantification. Equal aliquots of protein (30 µg) were separated on 12 or 15% SDS-PAGE gels, transferred to a nitrocellulose membrane using the wet transfer method, blocked using blocking buffer (1X TBST with 5% w/v non-fat dry milk; purchased from Cell Signaling Technology, Inc.; cat. no. 9999) for 1 h at room temperature and incubated with antibodies specific for LC3B, p62, Bcl-2, Bax, PARP, cleaved PARP, mTOR, Akt, CTSD, CTSB, β-actin and cofilin (all antibody cat. nos. as described above; 1:1,000) overnight at 4°C and finally incubated with HRP-linked goat anti-rabbit IgG (1:3,000) or HRP-linked goat anti-mouse IgG (1:3,000) (all antibody cat. nos. as described above) for 1 h at room temperature. Bands were automatically visualized using a FluorChem E digital darkroom system (ProteinSimple; Bio-Techne). Band intensities were quantified using Image J 1.8.0.1 (National Institutes of Health), and densitometric analysis was performed using GraphPad Prism 7.0 software (GraphPad Software, Inc.).
Immunofluorescence assay
The MCF7 or MDA-MB-231 cells were grown on coverslips in 12-well plates and treated with 20 µM DB or 5 µM RAPA or 60 µM CQ for 6 h. Following treatment, the cells were fixed with 100% methanol for 5 min, blocked with BSA (5%) (cat. no 9048-46-8; Beijing bai'aolaibo Technology Co., Ltd.) and incubated with anti-LC3A/B (cat. no. D3U4C; 1:200) and sequestosome 1 (SQSTM1)/p62 (Abcam, cat. no. D6M5X; 1:200) primary antibodies at 4°C overnight. The coverslips were then incubated with FITC-linked goat anti-rabbit IgG (1:100) at room temperature for 1 h in the dark and examined under an Olympus inverted fluorescence microscope (Olympus Corporation). Images were randomly captured.
Analysis of autophagic flux
To analyze autophagic flux, ~10,000 MCF7 and MDA-MB-231 cells were plated in 96-well plates, incubated at 37°C in a humidified atmosphere with 5% CO2 for 24 h, then transfected with the lentivirus pGMLV-CMV-RFP-GFP-hLC3-Puro (cat. no. GM-3394LV, Genomeditech Co., Ltd.) at a MOI of 30 for 72 h at 37°C and then treated with 20 µM DB in the presence or absence of 5 µM RAPA (MedChemExpress, cat. no. HY-10219) or 60 µM CQ (Aladdin, cat. no. C129284) for a further 6 h. The MCF7 and MDA-MB-231 cells were pre-treated with 10 mM NAC (Beyotime Instittue of Biotechnology, cat. no. S0077) for 1 h and incubated with 20 µM DB for 6 h at 37°C. The stably transfected cells were constructed by puromycin selection (cells were cultured in culture medium with additional 10 µg/ml puromycin). Puromycin was purchased from InvivoGen (cat. no. ant-pr-1).
Brightfield microscopy
Cell monolayers were cultured for 24 h in 12-well glass-covered chamber slides and treated with 0, 5, 10 or 20 µM DB for 6 h. Micrographs were obtained using an Olympus inverted fluorescence microscope (Olympus Corporation).
Apoptosis assay
An apoptosis assay was performed as we previously described (29). In brief, ~1×106 MCF7 or MDA-MB-231 cells treated with 20 µM DB for 48 h (incubated at 37°C in a humidified atmosphere with 5% CO2) were collected and washed twice with 1X Annexin V binding buffer and resuspended in 100 µl of 1X Annexin V binding buffer. Subsequently, the cells were stained using an Annexin V-FITC/PI kit (BD Biosciences, cat. no. 556547). The staining conditions were as follows: Incubation for 15 min at room temperature in the dark. The samples were analyzed using a FACScan flow cytometer (BD Biosciences). Statistical analysis was performed using BD FlowJo™ V10 software.
AO staining
The MCF7 or MDA-MB-231 cells were cultured in 12-well glass-covered chamber slides and then treated with either DB or CQ for 6 h. Following treatment, the cells were incubated with AO for 15 min at 37°C in the dark and fixed with 100% methanol. Fluorescent images were obtained using an Olympus inverted fluorescence microscope (Olympus Corporation).
DCFH-DA and LysoTracker Red staining
The MCF7 or MDA-MB-231 cells (~1×106) were collected and incubated with pre-warmed LysoTracker Red or DCFH-DA (cat. no. C1046, Beyotime Institute of Biotechnology) for 30 min at 37°C in the dark. The cells were then washed twice in PBS and resuspended in PBS. Fluorescence was measured with a flow cytometer in the PI channel (emission=530 nm).
Transmission electron microscopy (TEM)
Untreated and DB-treated cells were carefully digested using trypsin (HyClone; Cytiva), washed with PBS, and fixed in 2.5% glutaraldehyde (Phygene) for 24 h. Following washing with PBS three times, the samples were dehydrated in graded acetone solutions (30-100%) and then embedded in low-viscosity resin. Subsequently, ultrathin sections were cut using the Leica Ultracut UCT ultramicrotome (Leica Microsystems, Inc.) and stained with uranyl acetate (Weill Corning Medicine) for 5 min at room temperature. Samples were observed by using a FEI Tecnai 12 BioTwin transmission electron microscope (FEI Company).
In vivo xenograft experiment
In this experiment, the mice used were female BALB/cJ nude mice (5 to 6 weeks old, weighting 15-18 g, n=20 in total) purchased from Gempharmatech Co., Ltd. All mice were kept in an environment at 24°C with a 12-h light/dark cycle, with water and food freely available. The human endpoints of the study were primarily determined by tumors that should not exceed 20 mm in any dimension, and also that mouse weight loss should be <20%. All procedures complied with the standards of euthanasia according to Laboratory Animal Guidelines (GB/T 39760-2021) issued by the National Standardization Management Committee. The animal experiments were reviewed and approved by The Animal Care and Use Committee of Fujian Medical University (approval no. 2020-CAARM015) and were carried out in accordance with the National Institutes of Health Guide for Care and Use of Laboratory Animals. Mice used in these studies were maintained in a clean, modified-barrier animal facility, fed regular commercial mouse diet (Gempharmatech) under a controlled light/dark cycle (12/12 h) and a controlled temperature (20-23°C). In order to relieve mouse pain, inhalation anesthesia was preferred; the mice inhaled 5% isoflurane (cat. no. 792632, MilliporeSigma) until death. The mice were observed for a lack of a heartbeat and respiration and for graying of mucous membranes for at least 10 min to confirm death. Subsequently, 5×106 cells were injected subcutaneously into the left hindlimbs of the nude mice. After 7 days of culture, the tumor volume was ~100 mm3. The mice were randomly divided into four groups [PBS control, DB, adriamycin (ADM) and DB + ADM; 5 mice per group]. During this experiment, mouse health and behavior were monitored daily. The injection dose of DB and ADM (Shanghai Macklin Biochemical Co., Ltd.) was 10 mg/kg. Intratumoral injection was used. The injection regimen was administered once every 2 days for 10 consecutive injections. The tumor length and width were measured every 3 days, and volume was calculated according to the formula (tumor volume=shortest diameter2 × longest diameter/2). The body weight of the mice was measured every 3 days. All experiments involving living mice followed Chinese experimental animal welfare and ethical guidelines and made every effort to minimize the pain of the animals. At the end of the experiment, the mice were euthanized by intravenous injection of 2% sodium pentobarbital (150 mg/kg). Death was verified by respiratory arrest and dilated pupils.
Histological examination
After 21 days, the tumors were removed, and the tumor tissue was fixed with 4% tumor tissue fixation solution. Finally, the tissue was sent to Wuhan Sevicebio Technology Co., Ltd. for the immunohistochemical detection of Ki-67, TUNEL and LC3.
Statistical analysis
Statistical analysis was performed using GraphPad Prism 7.0 software (GraphPad Software, Inc.). All data are expressed as the mean ± standard error of the mean (SEM) values, and each experiment was performed at least three times. One-way analysis of variance (ANOVA) followed by the Bonferroni's post hoc test and the unpaired Student t-test were used to assess statistical significance where appropriate. P<0.05 was considered to indicate a statistically significant difference.
Results
DB is a novel autophagy modulator of MCF7 and MDA-MB-231 cells
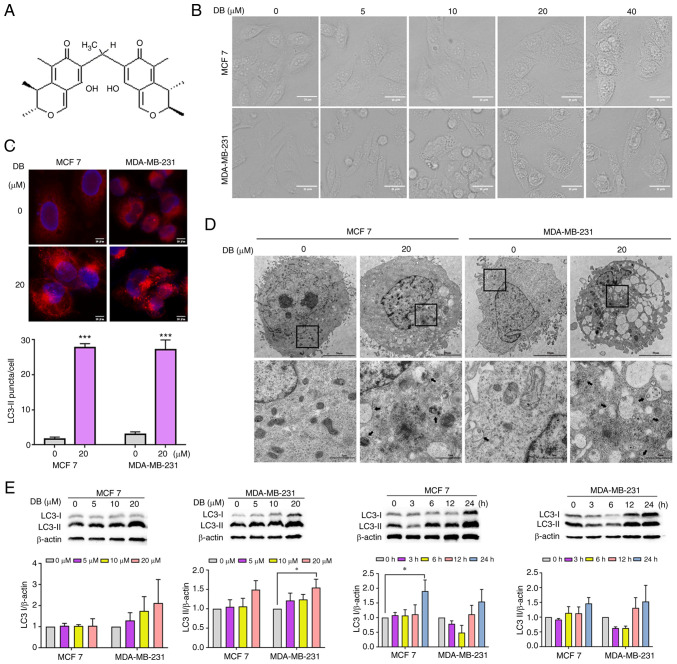
To discover novel autophagy modulators for human breast cancer, compounds isolated and purified from the marine fungus, Penicillium citrinum, were all screened and it was observed that treatment with one of the compounds, DB (Fig. 1A) for 6 h culminated in an increase in cellular perinuclear vacuoles in a concentration-dependent manner in MCF7 breast cancer cells and MDA-MB-231 triple-negative breast cancer cells (Fig. 1B). Thus, it was hypothesized that DB may be a potent autophagy modulator candidate and was thus used for further analysis.
Figure 1.
DB modulates autophagy in MCF7 and MDA-MB-231 cells. (A) Chemical structure of DB. The molecular weight of DB is 438 g/mol. (B) Representative microscopy images of cells treated with increasing concentrations (0-40 µM) of DB for 6 h. Scale bars, 20 µm. (C) Microscopy images of MCF7 and MDA-MB-231 cells following exposure to 20 µM DB for 6 h. Scale bar, 10 µm. Representative fluorescence images of MCF7 and MDA-MB-231 cells expressing RFP-LC3 following treatment with DB (10 µM) for 6 h. The numbers of RFP-LC3 puncta in each cell were quantified using Image J 1.8.0.1 software. Scale bars, 10 µm. n=five microscopic fields per group; ***P<0.001 vs. the control. (D) Transmission electron micrographs of MCF7 and MDA-MB-231 cells following incubation with 20 µM DB for 6 h; autophagic vacuoles are indicated by black arrows. The bottom panels (scale bars, 1 µm) represent a magnified image of the boxed region in the top panels (scale bars, 50 µm). (E) Representative western blots and corresponding protein quantification plots of LC3 I/II protein expression in MCF7 and MDA-MB-231 cells incubated with increasing concentrations of DB for 6 h or DB (20 µM) for various periods of time. β-actin was used as the loading control. *P<0.05 vs. the control. DB, dicitrinone B; RFP, red fluorescent protein; LC3, microtubule associated protein 1 light chain 3; LC3 I, cytoplasmic LC3; LC3 II, membrane-bound LC3.
To explore this hypothesis, the analyses of LC3 immunofluorescence were performed in the MCF7 and MDA-MB-231 cells. Presenting on the autophagosome during and after its formation, LC3 has been extensively studied as an autophagosome marker to monitor autophagy (30,31). The results of the present study demonstrated that treatment with DB for 6 h led to an increase in the cell punctate number of fluorescence as compared with diffuse fluorescence in the controls (Fig. 1C), confirming the influence of DB on the autophagic process. Representative TEM photomicrographs depicted that DB increased the number of double-membrane autophagosomes containing undigested cytoplasmic cargos, further highlighting the capacity of DB to modulate autophagy (Fig. 1D). For a comparative and more detailed evaluation of DB to modulate autophagy in MCF7 and MDA-MB-231 cells, western blot analysis of cytoplasmic LC3 (LC3-I) and membrane-bound LC3 (LC3-II) was performed. Notably, treatment with DB resulted in a marked increase in LC3-II levels in both MCF7 and MDA-MB-231 cells in a concentration- and time-dependent manner with respect to the control cells, indicating the accumulation of autophagosomes (Fig. 1E).
Inhibition of autophagic flux by DB in MCF7 and MDA-MB-231 cells
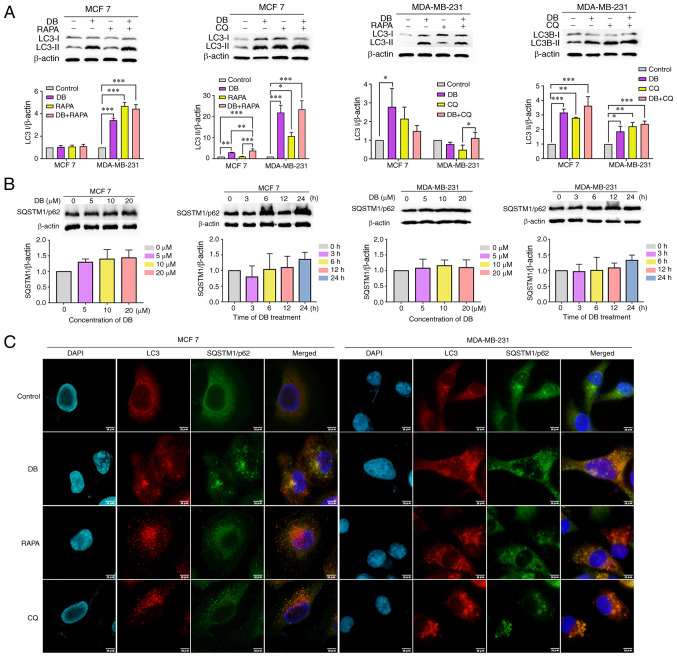
The accumulation of autophagosomes can be associated either with the induction of autophagy or the inhibition of late-stage autophagy (32). To distinguish the role of DB in autophagy in MCF7 and MDA-MB-231 cells, the levels of LC3-II in the presence and absence of the autophagy inducer, RAPA, or the autophagy inhibitor, CQ, were examined. As demonstrated in Fig. 2A, compared with cells treated with RAPA only, cells treated with both RAPA and DB exhibited elevated LC3-II levels. By contrast, co-incubation with DB and CQ did not result in a significant increase in LC3-II levels, as compared with those in cells treated with CQ only, indicating that DB is not an autophagy inducer.
Figure 2.
DB disrupts the autophagic flux. (A) Representative western blots and corresponding protein quantification plots of LC3-I/II protein expression in MCF7 and MDA-MB-231 cells following treatment with DB in the presence or absence of 5 µM RAPA or 60 µM CQ for 6 h. β-actin was used as the loading control. *P<0.05, **P<0.01 and ***P<0.001. (B) Representative western blots and corresponding protein quantification plots of SQSTM1/p62 protein expression in MCF7 and MDA-MB-231 cells following treatment with increasing concentrations of DB for 6 h or for different time periods (0-24 h) with 20 µM DB. β-actin was used as a loading control. (C) Representative images of MCF7 and MDA-MB-231 cells treated with 20 µM DB in the presence or absence of 5 µM RAPA or 60 µM CQ for 6 h and subjected to anti-LC3-II (red), anti-SQSTM1/p62 (green) and 4,6-DAPI (blue) staining. Scale bars, 10 µm. LC3, microtubule associated protein 1 light chain 3; RAPA, rapamycin; CQ, chloroquine; DB, dicitrinone B; SQSTM1, sequestosome 1; DAPI, diamidino-2-phenylindole.
To further explore whether DB is an autophagy inhibitor, the expression levels of SQSTM1/p62 were monitored using western blot analysis. SQSTM1/p62 is known as an autophagy inhibited marker, which is implicated in autophagic cargo recognition and was lost in autolysosome degradation (33). Correspondingly, a concentration- or time-dependent increase in the level of SQSTM1/p62 was observed in the MCF7 and MDA-MB-231 cells following treatment with DB, suggesting that DB probably blocked the autophagic flux (Fig. 2B). This phenomenon was further verified using immunofluorescence-based assays of intracellular accumulation of LC3-II and SQSTM1/p62 puncta. As depicted in Fig. 2C, LC3-II colocalized well with SQSTM1/p62 during DB treatment, similar to the finding in cells treated with CQ, a late-stage inhibitor. By contrast, the RAPA-treated cells exhibited a separated localization of LC3-II and SQSTM1/p62, suggesting the autophagy-inducing role of RAPA. Based on these findings, it was hypothesized that DB may modulate autophagy by blocking the fusion of autophagosomes with lysosomes. Taken together, these results demonstrated that DB interrupted the autophagic flux at the late stage in MCF7 and MDA-MB-231 cells, leading to autophagosome accumulation.
DB has no effect on the pH of lysosomes
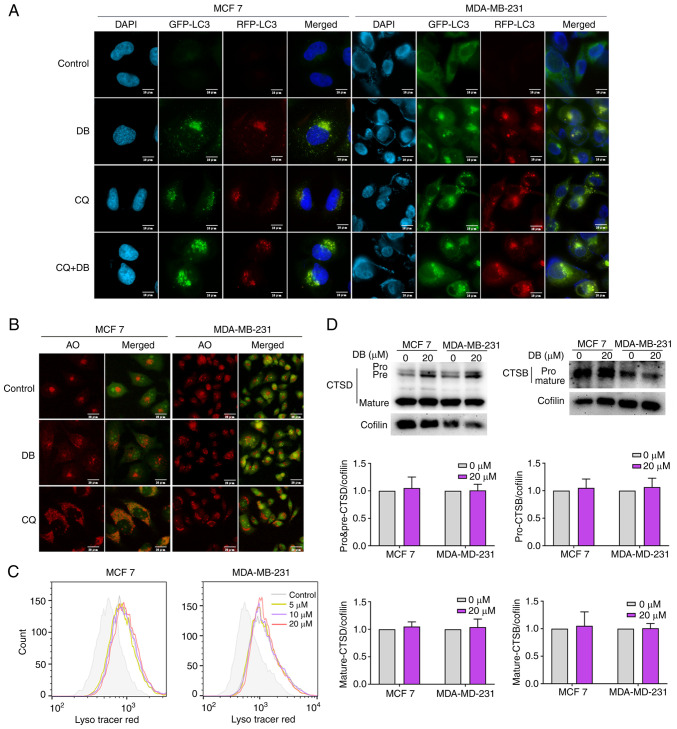
Considering that the blockade of autophagic flux may be caused by the inhibition of autophagosome-lysosome fusion or the impairment of lysosomal degradation, the effects of DB on the autophagic flux were further investigated using a tandem RFP-GFP-LC3 reporter. GFP is sensitive to the acidic environment of lysosomes or autophagolysosomes, whereas RFP fluorescence remains stable (34,35). Thus, autolysosomes can be observed using RFP-LC3 fluorescence. The results revealed that similar to treatment with the positive control CQ, DB treatment alone evoked the accumulation of both RFP and GFP puncta, visible as yellow fluorescence, in the MCF7 and MDA-MB-231 cells. Moreover, the autophagosome/autolysosome ratios were further increased when CQ was combined with DB treatment compared to DB treatment only, indicating that most autophagosomes and lysosomes cannot fuse (Fig. 3A).
Figure 3.
DB does not affect the pH or the hydrolytic function of lysosomes. (A) Fluorescence images of MCF7 and MDA-MB-231 cells transfected with double fluorescent mRFP-GFP-LC3 lentivirus and treated with DB, CQ alone or in combination for 6 h. Scale bars, 10 µm. (B) Fluorescence microscopy with AO staining. MCF7 and MDA-MB-231 cells were treated with 20 µM DB or 60 µM CQ for 6 h and then stained with AO. The red fluorescence represents the acidic vesicles. Scale bars, 20 µm. (C) Flow cytometry for LysoTracker Red in MCF7 and MDA-MB-231 cells treated with DB (0, 5, 10 and 20 µM) for 6 h. (D) Representative western blots and corresponding protein quantification plots of CTSD and CTSB protein expression in MCF7 and MDA-MB-231 cells treated with 20 µM DB for 6 h. Cofilin was used as a loading control. DB, dicitrinone B; RFP, red fluorescent protein; GFP, green fluorescent protein; LC3, microtubule associated protein 1 light chain 3; AO, acridine orange; CQ, chloroquine; DB, dicitrinone B; CTSD, cathepsin D; CTSB, cathepsin B.
To explore the detailed mechanisms of DB in the autophagic inhibition in MCF7 and MDA-MB-231 cells, the present study then examined whether DB can alter the lysosomal pH. The MCF7 and MDA-MB-231 cells were stained with AO following exposure to DB for 6 h. A large number of acidic vesicles with red fluorescence were observed to be significantly augmented in response to DB treatment, suggesting that DB could not inhibit lysosomal acidification (Fig. 3B). To further corroborate that the lysosomal pH was not altered by DB, flow cytometry was used to evaluate the LysoTracker Red fluorescence, which is used for labeling lysosomes and can be quenched by increasing the pH. As demonstrated in Fig. 3C, compared with CQ treatment, DB treatment resulted in a considerable increase in the acidic compartment, suggesting that lysosomal pH was unaltered.
CTSD is known to hydrolyze proteins in the acidic environment of lysosomes. Within the pH range of 2.8-5, CTSD can degrade hormones, polypeptide precursors, polypeptides, structural and functional proteins; however, it loses activity when the pH is greater than 5.5. CTSB is a cysteine proteolytic enzyme in lysosomes. It is active at pH 3.0~7.0 and is irreversibly inactivated under alkaline conditions (36,37). Herein, to investigate the effects of DB on lysosomal function, the levels of CTSD and CTSB in MCF7 and MDA-MB-231 cells were measured, and it was observed that the expression levels of the pro-form and the mature forms of CTSD and CTSB in cells treated with DB were not significantly different from those in control cells (Fig. 3D). Collectively, the findings of the present study suggested that the DB-mediated blockade of autophagosome-lysosome fusion was not due to impaired pH or lysosomal functionality.
DB inhibits autophagy via the overproduction of ROS
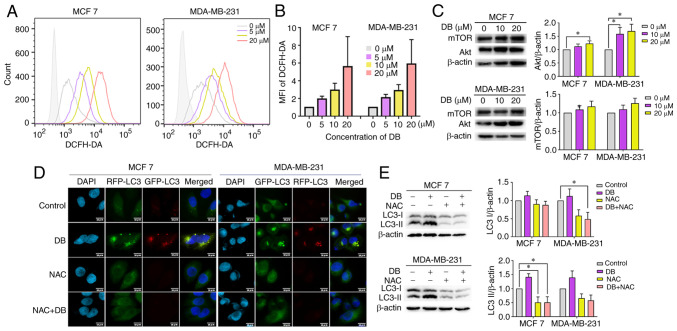
It is worth mentioning that the natural compound, DB, has previously been demonstrated to significantly augment cellular ROS in the human malignant melanoma cell line, A375 (25). Therefore, it was hypothesized that the role of DB in autophagy inhibition may be associated with an increase in intracellular ROS production. To verify this hypothesis, the ROS levels after DB treatment were examined in MCF7 and MDA-MB-231 cells using the DCFH-DA probe. Data from flow cytometric analysis revealed that cell exposure to DB led to a consistent increase in the level of intracellular ROS (Fig. 4A and B). In order to verify further whether ROS is related to the PI3K/Akt pathway, the expression levels of the PI3K/Akt pathway related proteins, Akt and mTOR, were evaluated in MCF7 and MDA-MB-231 cells following DB treatment. The results demonstrated that the expression of Akt and mTOR in both cell lines increased along with DB concentration, particularly in MDA-MB-231 cells (Fig. 4C). In addition, NAC, a scavenger of ROS, was administered to determine whether excess ROS is responsible for blocking autophagy by DB treatment. Flow cytometric and western blot analyses of LC3-II revealed that pre-treatment with NAC markedly reduced the conversion of LC3B-I to LC3B-II (Fig. 4D).
Figure 4.
Promotion of intracellular ROS generation is required for DB-induced autophagy inhibition. (A) Flow cytometry of ROS levels in DB (0, 5, 10 and 20 µM 6 h)-treated MCF7 and MDA-MB-231 cells. (B) The fluorescence intensity of each component was calculated according to the median value of Fig. 4A. (C) Representative western blots and corresponding protein quantification plots of Akt and mTOR protein expression in MCF7 and MDA-MB-231 cells, treated with 10, 20 µM DB for 6 h. β-actin was used as the loading control. *P<0.05. (D) Fluorescence images of mRFP-GFP-LC3 puncta in MCF7 and MDA-MB-231 cells treated with DB or NAC only or in combination. Scale bars, 10 µm. (E) The cells of MCF7 and MDA-MB-231 were pre-treated with NAC (10 mM) for 1 h and incubated with 20 µM DB for 6 h. Representative western blots and corresponding protein quantification plots of LC3-I and LC3-II expression levels were examined using western blot analysis. β-actin was used as a loading control. *P<0.05. ROS, reactive oxygen species; DB, dicitrinone B; RFP, red fluorescent protein; GFP, green fluorescent protein; NAC, N-acetyl-L-cysteine; LC3, microtubule associated protein 1 light chain 3; LC3 I, cytoplasmic LC3; LC3 II, membrane-bound LC3.
To corroborate the findings from western blot analysis, the present study then investigated the effects on autophagic flux using tandem monomeric RFP-GFP-tagged LC3 in response to NAC. Consistently, immunofluorescence micros-copy revealed that pre-treatment with NAC significantly delayed the formation of yellow fluorescence induced by DB in both MCF7 and MDA-MB-231 cells (Fig. 4E), revealing the significant involvement of ROS regulatory mechanisms. Taken together, the results of the present study suggested that DB-induced autophagy inhibition in breast cancer cells was in fact modulated via the overproduction of ROS.
DB induces MCF7 and MDA-MB-231 cell apoptosis via autophagy inhibition
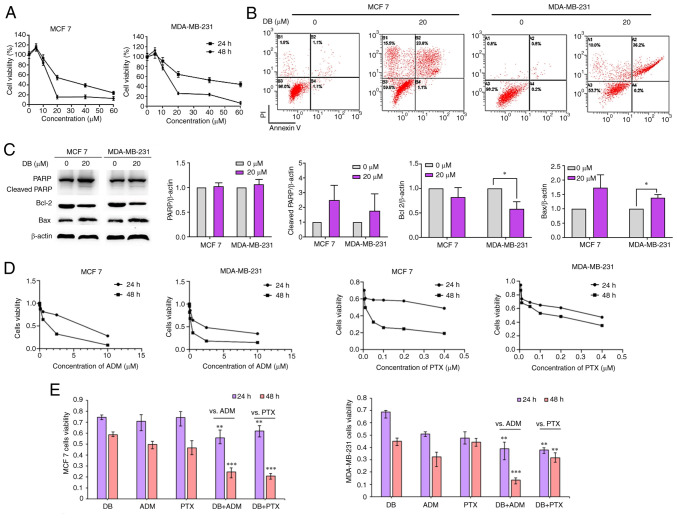
To reveal the cell fate of MCF7 and MDA-MB-231 after autophagy inhibition and ROS explosion evoked by DB treatment, MTS assays were then performed to evaluate the cytotoxicity of DB in MCF7 and MDA-MB-231 cells. The data demonstrated that the number of MCF7 and MDA-MB-231 cells and their metabolic activity markedly decreased with the increasing concentrations and treatment times (Fig. 5A). The IC50 values of DB for MCF7 and MDA-MB-231 cells were 14.29 and 15.70 µM, respectively, for 48 h.
Figure 5.
Effect of DB on apoptosis when singly used or in combination with conventional chemotherapeutic drugs in MDA-MB-231 and MCF7 cells. (A) CellTiter 96® AQueous One Solution Cell Proliferation assay was used to detect the cell survival rate of MDA-MB-231 and MCF7 cells treated with DB (20 µM) for 24 h. (B) Representative images of Annexin V/PI staining assay using flow cytometry in control cells and cells treated with DB (0 and 20 µM, 48 h). (C) Representative western blots and corresponding protein quantification plots of PARP, cleaved PARP, Bcl-2 and Bax protein expression in MCF-7 and MDA-MB-231 cells treated with DB (20 µM) for 6 h. *P<0.05. (D) The viability of MCF7 and MDA-MB-231 cells treated with ADM and PTX. (E) CellTiter 96® AQueous One Solution Cell Proliferation Assay was used to determine the survival rate of MDA-MB-231 cells and MCF7 cells treated with DB, ADM and PTX alone or in combination with DB, ADM or PTX (10 µM DB, 0.2 µM ADM, 0.005 µM PTX, 24 and 48 h). **P<0.01 vs. ADM or PTX, ***P<0.001 vs. ADM or PTX. DB, dicitrinone B; PARP, poly (ADP-ribose) polymerase; ADM, adriamycin; PTX, paclitaxel.
Considering that apoptosis is the major pathway of cell death mediated by chemotherapeutics, it was further attempted to identify the association between apoptosis and DB-induced autophagy inhibition in MCF7 and MDA-MB-231 cells. The results of the flow cytometric Annexin V-FITC/propidium iodide staining assay results demonstrated that apoptotic cell numbers increased with DB concentration in MCF7 and MDA-MB-231 cells (Fig. 5B). Apoptotic rates were markedly increased up to ~24.9 and ~36.4% in the MCF7 and MDA-MB-231 cells, respectively, after the cells were exposed to DB for 48 h, indicating that DB-induced autophagy inhibition may eventually lead to apoptosis.
The expression of the apoptosis-related proteins, PARP, Bax and Bcl-2, were then evaluated in DB-treated cells using western blot analysis. As demonstrated in Fig. 5C, DB treatment resulted in an upregulation of cleaved PARP and Bax and a downregulation of Bcl-2 expression, confirming that DB could promote apoptosis by blocking autophagy in MCF7 and MDA-MB-231 cells.
It is known that autophagy inhibition can enhance the sensitivity of tumor cells to chemotherapeutic drugs or targeted drugs (38). The change in the sensitivity of MCF7 and MDA-MB-231 cells to the conventional chemotherapeutics, ADM and paclitaxel (PTX) was also investigated (Fig. 5D). The results revealed that the inhibitory effects observed following combined treatment with DB and ADM or PTX were significantly greater than those observed with DB, ADM or PTX treatment alone in MDA-MB-231 and MCF7 cells (Fig. 5E), indicating that DB-induced autophagy inhibition may enhance the sensitivity of MCF7 and MDA-MB-231 cells to ADM and PTX.
Antitumor efficacy of combined treatment with DB and the chemotherapeutic drug, ADM in vivo
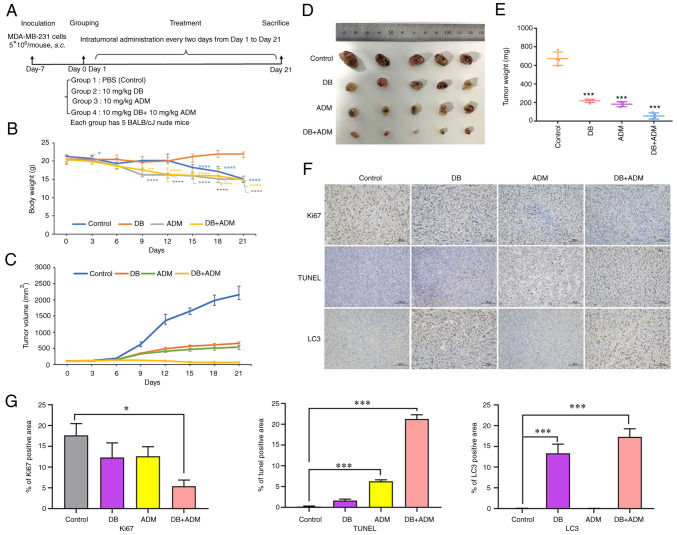
To explore the antitumor efficacy of DB in vivo, a nude mouse subcutaneous planting tumor model was constructed using MDA-MB-231 cells. In the experiment, 5×106 cells were injected subcutaneously into the left hindlimb of nude mice. After 7 days, the tumor volumes reached ~100 mm3, and the mice were randomly divided into four groups with 5 mice in each group (PBS, DB, ADM and DB + ADM). The mice were treated with DB and/or ADM via an intratumoral injection every 2 days at a dose of 10 mg/kg (Fig. 6A). Following 12 days of treatment, the body weight of mice in the control group began to decrease significantly, while that of the mice in the DB group instead slightly increased, indicating that the use of DB in vivo could be considered safe (Fig. 6B). After 21 days of administration (10 consecutive injections), the mice were sacrificed, and the tumors were dissected. It was found that the tumors in the blank control group were the largest, while the tumors in the combined treatment group were the smallest in size. Moreover, the efficacy of DB treatment and ADM treatment demonstrated almost no difference, indicating that when used as a single therapeutic, DB can be comparable to ADM, and there is a good synergistic effect when used in combination (Fig. 6C-E). The results of immunohistochemical analysis revealed that compared with the control group, the number of TUNEL-positive cells increased significantly in the combined group and increased slightly in the DB or ADM treatment group (Fig. 6F and G). Simultaneously, the number of Ki-67-positive cells decreased significantly in the combined group and decreased slightly in the DB or ADM treatment group (Fig. 6F and G). These data suggested that DB or ADM treatment can promote tumor apoptosis and inhibit tumor proliferation, and the combination of DB and ADM may enhance the antitumor efficacy. Of note, the results of the immunohistochemical analysis of both the DB and combined administration groups revealed more LC3-positive cells, indicating the blockade of autophagy after DB treatment in tumor cells (Fig. 6F and G), which was consistent with the experimental results obtained in vitro. Taken together, the aforementioned results revealed that DB was safe and effective as an antitumor drug by regulating autophagy in vivo.
Figure 6.
Efficacy of DB combined with the conventional chemotherapeutic drug, ADM, in a MDA-MB-231 cell line-derived tumor mouse model. (A) Mouse experimental scheme map. (B) Changes in tumor volume in different groups of mice during treatment. Calculation formula: tumor volume=shortest diameter2 × longest diameter/2. *P<0.05, **P<0.01 and ****P<0.0001 vs. the control. (C) Changes in body weight in different groups of mice during treatment. (D) Images of tumors removed from nude mice after 21 days of treatment. (E) Tumor tissue weight of mice in different groups (control, DB, ADM and DB + ADM) after 21 days of treatment. (F) On day 21, all mice were sacrificed, and tumors were isolated for histopathological examination with TUNEL, Ki-67 and LC3 B staining assays (scale bar, 100 µm). The dose of DB and ADM was 10 mg/kg. DB, dicitrinone B; ADM, Adriamycin; LC3 B, microtubule associated protein 1 light chain 3 beta. ***P<0.001 vs. the control. (G) Relative expression quantification of histopathological examination with TUNEL, Ki-67 and LC3 B staining assays. *P<0.05 and ***P<0.001.
Discussion
As a highly heritable heterogeneous disease, breast cancer has diverse histological and molecular subtypes. The prognosis of triple-negative breast cancer patients is particularly poor due to the lack of effective targeted therapy, and in a large number of patients, advanced breast cancer eventually becomes refractory or relapses due to invasion and metastasis (39). In this context, targeting autophagy inhibition may represent a new therapeutic strategy for human breast cancer cells with apoptosis resistance or highly frequent metastasis. Previous studies have suggested that autophagy is a survival-promoting mechanism in cancer, particularly in advanced tumors (38,40,41). It can help cancer cells evade various environmental stressors by removing damaged organelles and recycling nutrients. The autophagy inhibitors, CQ and hydroxychloroquine (HCQ), have shown promising efficacy in breast cancer and triple-negative breast cancer pre-clinical models or in clinical trials when combined with other conventional chemotherapies (42,43). However, the retinal toxicity of CQ/HCQ has also attracted increased attention (44-47). Therefore, the research and development of novel autophagy inhibitors is of utmost clinical significance in cancer treatment. In the present study, the novel natural product, DB, was examined, which may impair autophagic flux and lead to a therapeutic benefit for breast cancer.
Similar to CQ/HCQ, in the present study, DB induced the accumulation of autophagosomes by inhibiting autophagosome-lysosome fusion. It was noted that the results of the immunofluorescence assay of LC3 appeared to be more notable in the MDA-MB-231 cells; however, the western blot results looked not so obvious. It was hypothesized that this may be attributed to the different morphological characteristics of the autophagosomes in MDA-MB-231 cells and MCF7 cells. The autophagosomes formed in MDA-MB-231 cells were usually of larger size than that those in MCF7 cells, leading to the potential confusion that the level of LC3 and p62 was higher in MDA-MB-231 cells. Actually, the effect of DB on autophagosome accumulation in MDA-MB-231 and MCF7 cells was relatively similar, based on the results of western blot analysis. Various studies have revealed that alkalization of lysosomes, defects in lysosomal proteolytic activity, and delayed trafficking of autophagosomes to lysosomes easily lead to the limitation of autophagosomes-lysosome fusion (48,49). However, the data from the AO and LysoTracker Red staining assays in the present study demonstrated that the pH was not altered in response to DB treatment. It is believed that lysosomal hydrolases, known as cathepsins, are the main tool for autolysosomes to decompose their contents (50-52). Western blot analysis of the hydrolases CTSD and CTSB indicated that although DB treatment blocked the autophagic flux and increased autophagosomes, it did not alter the activity and function of lysosomal cathepsins. To date, most compounds that affect autophagy in cancer cells perform this by locking the fusion of autophagosomes and lysosomes (53). Among these compounds, bafilomycin A1 can raise the pH of lysosomes, which leads to the inhibition of the activity of resident hydrolases and further blocks the fusion of autophagosomes and lysosomes (54). Liensinine, as an autophagy inhibitor, is able to affect the recruitment of the small GTP binding protein RAB7A to lysosomes but does not affect lysosomal pH, which in turn blocks the fusion of autophagosomes and lysosomes in breast cancer cells (55). CQ/HCQ decreases autophagosome lysosome fusion by interfering with autophagosomal SNARE protein SNAP29 recruitment and the Golgi complex without substantially changing lysosomal acidity (56,57). It cannot be excluded that DB may preferentially destabilize auxilin, which is involved in the fusion of autophagosomes and lysosomes, and consequently lead to autophagosome aggregation and disruption of autophagy (58).
As a type of highly reactive oxygen-free radical or non-radical molecules, ROS play an essential role in deciding cell fate. A previous study by the authors have revealed that the levels of ROS in A375 human malignant melanoma cells was obviously increased after DB treatment, due to the significant decrease in mitochondrial membrane potential induced by DB (25). In the present study, DB also caused an increase in ROS generation in MCF7 breast cancer and MDA-MB-231 TNBC cells. ROS perform important functions and are associated with numerous signaling pathways in cells (59). To determine the association between ROS and autophagy, the ROS scavenger, NAC, was used to treat the cells prior to DB-inhibited autophagy. Of note, the DB-induced autophagic flux inhibition was abolished by NAC. Therefore, the effect of DB on ROS generation may be related to the blocking of the autophagic flow in MCF7 and MDA-MB-231 cells. To date, the association linking autolysosomal accumulation and ROS accumulation remains largely unclear. It has been reported that high levels of intracellular ROS may stem from damaged mitochondria in the cytoplasm or undegraded mitochondria in autophagosomes (60,61). The accumulation of autophagosomes containing defective mitochondria and dysfunctional mitochondria after DB treatment may promote ROS release. Simultaneously, high levels of ROS trigger the formation of new autophagosomes and cellular damage, potentially initiating a vicious cycle that eventually leads to apoptosis. In addition, it has been reported that Akt, a downstream protein of the PI3K pathway, inhibits autophagy by activating rapamycin complex 1 (mTORC1) in response to the increase in ROS levels, and mTORC1 and rapamycin complex 2 (mTORC2) inhibit autophagy at medium ROS levels; however, mTORC2 can promote cell aging through autophagy at high ROS levels (62). The present study revealed that DB upregulated Akt and mTOR protein levels in breast cancer cells, particularly in MDA-MB-231 cells. This indicated that the blocking effect of DB on the autophagy of MCF7 and MDA-MB-231 cells may be related to ROS and PI3K pathways.
The present study only detected the anti-tumor and autophagy inhibitory activity of DB in two human breast cancer cell lines (MCF7 and MDA-MB-231) without making comparisons with a normal control cell line. Therefore, it was not possible to evaluate the toxicity of DB to normal cells at the cellular level. However, the mouse experiments revealed that the body weight of the mice in the doxorubicin group decreased significantly, while that of the mice in the DB group was not markedly altered, indicating that DB is safe to use in vivo.
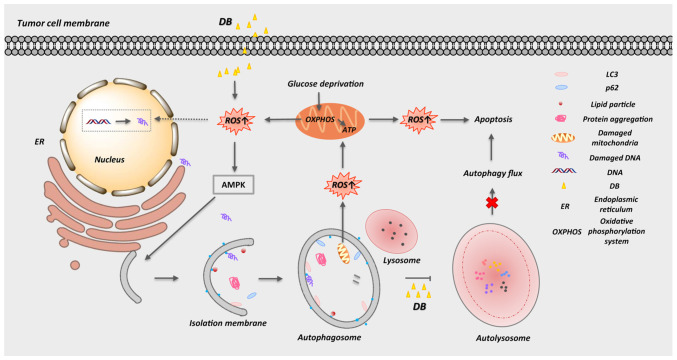
In conclusion, the present study provides biochemical evidence of a novel (to the best of our knowledge) autophagy modulator, DB, that can inhibit autophagy and induce apoptosis via the accumulation of autophagosomes and the promotion of ROS production in MCF7 breast cancer and MDA-MB-231 TNBC cells (Fig. 7). In view of its safety and efficacy in vivo and the necessity to enhance the sensitivity of tumors to chemotherapeutic drugs, DB is expected to become a new-generation antitumor drug.
Figure 7.
Schematic representation of DB-induced autophagy inhibition and apoptosis activation. DB, dicitrinone B.
Acknowledgments
The authors would like to thank Dr Liu Shijia from Fujian Cancer Hospital (Fuzhou, China) for the generous gift of the MDA-MB-231 cells.
Funding Statement
The present study was funded by grants from the National Natural Science Foundation of China (no. 81873045), The Natural Science Foundation of Fujian Province (nos. 2020J011118, 2020J011115 and 2021J01608), the Fujian Province Health Care Young and Middle-aged Backbone Talents Training Project (no. 2020GGA015), the Startup Fund for Scientific Research, Fujian Medical University (no. 2019QH1197), the Fujian Provincial Clinical Research Center for Cancer Radiotherapy and Immunotherapy (no. 2020Y2012) and the Fuzhou University Testing Fund of Precious Apparatus (no. 2022T041).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
QL, YY and QZ contributed to the experimental design. QL, YY, FC, MC and SC contributed to the experiments. QL, YY, YS and LC contributed to data analysis. QL, YY and LC contributed to the original manuscript preparation. YS and LC contributed to the supervision of the present study. YS and LC contributed to the manuscript review and editing. YS and LC confirm the authenticity of all the raw data. All authors contributed to drafting the manuscript. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
The animal experiments were reviewed and approved by The Animal Care and Use Committee of Fujian Medical University (approval no. 2020-CAARM015) and were carried out in accordance with the National Institutes of Health Guide for Care and Use of Laboratory Animals.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Ghoncheh M, Pournamdar Z, Salehiniya H. Incidence and mortality and epidemiology of breast cancer in the world. Asian Pac J Cancer Prev. 2016;17:43–56. doi: 10.7314/APJCP.2016.17.S3.43. [DOI] [PubMed] [Google Scholar]
- 2.Kocarnik JM, Compton K, Dean FE, Fu W, Gaw BL, Harvey JD, Henrikson HJ, Lu D, Pennini A, Xu R, et al. Cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life years for 29 cancer groups from 2010 to 2019: A systematic analysis for the global burden of disease study 2019. JAMA Oncol. 2021;8:420–444. doi: 10.1001/jamaoncol.2021.6987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 4.Gadi VK, Davidson NE. Practical approach to triple-negative breast cancer. J Oncol Pract. 2017;13:293–300. doi: 10.1200/JOP.2017.022632. [DOI] [PubMed] [Google Scholar]
- 5.Harbeck N, Gnant M. Breast cancer. Lancet. 2017;389:1134–1150. doi: 10.1016/S0140-6736(16)31891-8. [DOI] [PubMed] [Google Scholar]
- 6.Harbeck N, Penault-Llorca F, Cortes J, Gnant M, Houssami N, Poortmans P, Ruddy K, Tsang J, Cardoso F. Breast cancer. Nat Rev Dis Primers. 2019;5:66. doi: 10.1038/s41572-019-0111-2. [DOI] [PubMed] [Google Scholar]
- 7.Kwapisz D. Pembrolizumab and atezolizumab in triple-negative breast cancer. Cancer Immunol Immunother. 2021;70:607–617. doi: 10.1007/s00262-020-02736-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Glick D, Barth S, Macleod KF. Autophagy: Cellular and molecular mechanisms. J Pathol. 2010;221:3–12. doi: 10.1002/path.2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Onorati AV, Dyczynski M, Ojha R, Amaravadi RK. Targeting autophagy in cancer. Cancer. 2018;124:3307–3318. doi: 10.1002/cncr.31335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mizushima N, Klionsky DJ. Protein turnover via autophagy: Implications for metabolism. Annu Rev Nutr. 2007;27:19–40. doi: 10.1146/annurev.nutr.27.061406.093749. [DOI] [PubMed] [Google Scholar]
- 11.Morishita H, Mizushima N. Diverse cellular roles of autophagy. Annu Rev Cell Dev Biol. 2019;35:453–475. doi: 10.1146/annurev-cellbio-100818-125300. [DOI] [PubMed] [Google Scholar]
- 12.Camuzard O, Santucci-Darmanin S, Carle GF, Pierrefite-Carle V. Autophagy in the crosstalk between tumor and microenvironment. Cancer Lett. 2020;490:143–153. doi: 10.1016/j.canlet.2020.06.015. [DOI] [PubMed] [Google Scholar]
- 13.Li YJ, Lei YH, Yao N, Wang CR, Hu N, Ye WC, Zhang DM, Chen ZS. Autophagy and multidrug resistance in cancer. Chin J Cancer. 2017;36:52. doi: 10.1186/s40880-017-0219-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang B, Liu L. Autophagy is a double-edged sword in the therapy of colorectal cancer. Oncol Lett. 2021;21:378. doi: 10.3892/ol.2021.12639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parzych KR, Klionsky DJ. An overview of autophagy: Morphology, mechanism, and regulation. Antioxid Redox Signal. 2014;20:460–473. doi: 10.1089/ars.2013.5371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saha S, Panigrahi DP, Patil S, Bhutia SK. Autophagy in health and disease: A comprehensive review. Biomed Pharmacother. 2018;104:485–495. doi: 10.1016/j.biopha.2018.05.007. [DOI] [PubMed] [Google Scholar]
- 17.Singh SS, Vats S, Chia AY, Tan TZ, Deng S, Ong MS, Arfuso F, Yap CT, Goh BC, Sethi G, et al. Dual role of autophagy in hallmarks of cancer. Oncogene. 2018;37:1142–1158. doi: 10.1038/s41388-017-0046-6. [DOI] [PubMed] [Google Scholar]
- 18.Mowers EE, Sharifi MN, Macleod KF. Autophagy in cancer metastasis. Oncogene. 2017;36:1619–1630. doi: 10.1038/onc.2016.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mowers EE, Sharifi MN, Macleod KF. Functions of autophagy in the tumor microenvironment and cancer metastasis. FEBS J. 2018;285:1751–1766. doi: 10.1111/febs.14388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith AG, Macleod KF. Autophagy, cancer stem cells and drug resistance. J Pathol. 2019;247:708–718. doi: 10.1002/path.5222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vempati RK, Malla RR. Autophagy-induced drug resistance in liver cancer. Crit Rev Oncog. 2020;25:21–30. doi: 10.1615/CritRevOncog.2020034969. [DOI] [PubMed] [Google Scholar]
- 22.Demain AL, Vaishnav P. Natural products for cancer chemotherapy. Microb Biotechnol. 2011;4:687–699. doi: 10.1111/j.1751-7915.2010.00221.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Talmadge JE. Natural product derived immune-regulatory agents. Int Immunopharmacol. 2016;37:5–15. doi: 10.1016/j.intimp.2016.02.025. [DOI] [PubMed] [Google Scholar]
- 24.Yang Y, Liu Q, Shi X, Zheng Q, Chen L, Sun Y. Advances in plant-derived natural products for antitumor immunotherapy. Arch Pharm Res. 2021;44:987–1011. doi: 10.1007/s12272-021-01355-1. [DOI] [PubMed] [Google Scholar]
- 25.Chen L, Gong MW, Peng ZF, Zhou T, Ying MG, Zheng QH, Liu QY, Zhang QQ. The marine fungal metabolite, dicitrinone B, induces A375 cell apoptosis through the ROS-related caspase pathway. Mar Drugs. 2014;12:1939–1958. doi: 10.3390/md12041939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen L, Zhao YY, Lan RF, Du L, Wang BS, Zhou T, Li YP, Zhang QQ, Ying MG, Zheng QH, et al. Dicitrinone D, an antimitotic polyketide isolated from the marine-derived fungus Penicillium citrinum. Tetrahedron. 2017;73:5900–5911. doi: 10.1016/j.tet.2017.08.032. [DOI] [Google Scholar]
- 27.Du L, Li D, Zhang G, Zhu T, Ai J, Gu Q. Novel carbon-bridged citrinin dimers from a volcano ash-derived fungus Penicillium citrinum and their cytotoxic and cell cycle arrest activities. Tetrahedron. 2010;66:9286–9290. doi: 10.1016/j.tet.2010.09.036. [DOI] [Google Scholar]
- 28.Wang L, Li C, Yu G, Sun Z, Zhang G, Gu Q, Zhu T, Che Q, Guan H, Li D. Dicitrinones E and F, citrinin dimers from the marine derived fungus Penicillium citrinum HDN-152-088. Tetrahedron Letters. 2019;60:151182–151189. doi: 10.1016/j.tetlet.2019.151182. [DOI] [Google Scholar]
- 29.Zhao H, Zhang X, Wang M, Lin Y, Zhou S. Stigmasterol simultaneously induces apoptosis and protective autophagy by inhibiting Akt/mTOR pathway in gastric cancer cells. Front Oncol. 2021;11:629008. doi: 10.3389/fonc.2021.629008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee YK, Lee JA. Role of the mammalian ATG8/LC3 family in autophagy: Differential and compensatory roles in the spatiotemporal regulation of autophagy. BMB Rep. 2016;49:424–430. doi: 10.5483/BMBRep.2016.49.8.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tanida I, Ueno T, Kominami E. LC3 conjugation system in mammalian autophagy. Int J Biochem Cell Biol. 2004;36:2503–2518. doi: 10.1016/j.biocel.2004.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harris H, Rubinsztein DC. Control of autophagy as a therapy for neurodegenerative disease. Nat Rev Neurol. 2012;8:108–117. doi: 10.1038/nrneurol.2011.200. [DOI] [PubMed] [Google Scholar]
- 33.Rogov V, Dotsch V, Johansen T, Kirkin V. Interactions between autophagy receptors and ubiquitin-like proteins form the molecular basis for selective autophagy. Mol Cell. 2014;53:167–178. doi: 10.1016/j.molcel.2013.12.014. [DOI] [PubMed] [Google Scholar]
- 34.Yang K, Tang M, Chang HH, Kanamala M, Davidson AJ, Wu Z. Mannosylation of pH-sensitive liposomes promoted cytoplasmic delivery of protein to macrophages: Green fluorescent protein (GFP) performed as an endosomal escape tracer. Pharm Dev Technol. 2021;26:1000–1009. doi: 10.1080/10837450.2021.1969406. [DOI] [PubMed] [Google Scholar]
- 35.Mizuno H, Sawano A, Eli P, Hama H, Miyawaki A. Red fluorescent protein from Discosoma as a fusion tag and a partner for fluorescence resonance energy transfer. Biochemistry. 2001;40:2502–2510. doi: 10.1021/bi002263b. [DOI] [PubMed] [Google Scholar]
- 36.Qiao S, Tao S, de la Vega M, Park SL, Vonderfecht AA, Jacobs SL, Zhang DD, Wondrak GT. The antimalarial amodiaquine causes autophagic-lysosomal and proliferative blockade sensitizing human melanoma cells to starvation- and chemotherapy-induced cell death. Autophagy. 2013;9:2087–2102. doi: 10.4161/auto.26506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jakoš T, Pišlar A, Jewett A, Kos J. Cysteine cathepsins in tumor-associated immune cells. Front Immunol. 2019;10:2037. doi: 10.3389/fimmu.2019.02037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dikic I, Johansen T, Kirkin V. Selective autophagy in cancer development and therapy. Cancer Res. 2010;70:3431–3434. doi: 10.1158/0008-5472.CAN-09-4027. [DOI] [PubMed] [Google Scholar]
- 39.Abramson VG, Lehmann BD, Ballinger TJ, Pietenpol JA. Subtyping of triple-negative breast cancer: Implications for therapy. Cancer. 2015;121:8–16. doi: 10.1002/cncr.28914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu B, Wen X, Cheng Y. Survival or death: Disequilibrating the oncogenic and tumor suppressive autophagy in cancer. Cell Death Dis. 2013;4:e892. doi: 10.1038/cddis.2013.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dower CM, Wills CA, Frisch SM, Wang HG. Mechanisms and context underlying the role of autophagy in cancer metastasis. Autophagy. 2018;14:1110–1128. doi: 10.1080/15548627.2018.1450020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rojas-Sanchez G, Garcia-Miranda A, Montes-Alvarado JB, Cotzomi-Ortega I, Sarmiento-Salinas FL, Jimenez-Ignacio EE, Ramirez-Ramirez D, Romo-Rodriguez RE, Reyes-Leyva J, Vallejo-Ruiz V, et al. Chloroquine induces ROS-mediated macrophage migration inhibitory factor secretion and epithelial to mesenchymal transition in ER-positive breast cancer cell lines. J Mammary Gland Biol Neoplasia. 2021;26:341–355. doi: 10.1007/s10911-021-09503-5. [DOI] [PubMed] [Google Scholar]
- 43.Dong J, Zhu C, Zhang F, Zhou Z, Sun M. 'Attractive/adhesion force' dual-regulatory nanogels capable of CXCR4 antagonism and autophagy inhibition for the treatment of metastatic breast cancer. J Control Release. 2022;341:892–903. doi: 10.1016/j.jconrel.2021.12.026. [DOI] [PubMed] [Google Scholar]
- 44.Ruamviboonsuk P, Lai TYY, Chang A, Lai CC, Mieler WF, Lam DSC and for Asia-Pacific Vitreo-Retina Society Chloroquine and hydroxychloroquine retinal toxicity consideration in the treatment of COVID-19. Asia Pac J Ophthalmol (Phila) 2020;9:85–87. doi: 10.1097/APO.0000000000000289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Muller R. Systemic toxicity of chloroquine and hydroxychloroquine: Prevalence, mechanisms, risk factors, prognostic and screening possibilities. Rheumatol Int. 2021;41:1189–1202. doi: 10.1007/s00296-021-04868-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Doyno C, Sobieraj DM, Baker WL. Toxicity of chloroquine and hydroxychloroquine following therapeutic use or overdose. Clin Toxicol (Phila) 2021;59:12–23. doi: 10.1080/15563650.2020.1817479. [DOI] [PubMed] [Google Scholar]
- 47.Askarian F, Firoozi Z, Ebadollahi-Natanzi A, Bahrami S, Rahimi HR. A review on the pharmacokinetic properties and toxicity considerations for chloroquine and hydroxychloroquine to potentially treat coronavirus patients. Toxicol Res. 2021;38:137–148. doi: 10.1007/s43188-021-00101-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Abokyi S, Shan SW, Lam CHI, Catral KP, Pan F, Chan HHL, To CH, Tse DYY. Targeting lysosomes to reverse hydroquinone-induced autophagy defects and oxidative damage in human retinal pigment epithelial cells. Int J Mol Sci. 2021;22:9042. doi: 10.3390/ijms22169042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen R, Jaattela M, Liu B. Lysosome as a central hub for rewiring PH homeostasis in tumors. Cancers. 2020;12:2437. doi: 10.3390/cancers12092437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hossain MI, Marcus JM, Lee JH, Garcia PL, Singh V, Shacka JJ, Zhang J, Gropen TI, Falany CN, Andrabi SA. Restoration of CTSD (cathepsin D) and lysosomal function in stroke is neuroprotective. Autophagy. 2021;17:1330–1348. doi: 10.1080/15548627.2020.1761219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Di YQ, Han XL, Kang XL, Wang D, Chen CH, Wang JX, Zhao XF. Autophagy triggers CTSD (cathepsin D) maturation and localization inside cells to promote apoptosis. Autophagy. 2021;17:1170–1192. doi: 10.1080/15548627.2020.1752497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cao M, Luo X, Wu K, He X. Targeting lysosomes in human disease: From basic research to clinical applications. Signal Transduct Target Ther. 2021;6:379. doi: 10.1038/s41392-021-00778-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li Z, Si W, Jin W, Yuan Z, Chen Y, Fu L. Targeting autophagy in colorectal cancer: An update on pharmacological small-molecule compounds. Drug Discov Today. 2022;27:2373–2385. doi: 10.1016/j.drudis.2022.05.011. [DOI] [PubMed] [Google Scholar]
- 54.Feng X, Zhang H, Meng L, Song H, Zhou Q, Qu C, Zhao P, Li Q, Zou C, Liu X, Zhang Z. Hypoxia-induced acetylation of PAK1 enhances autophagy and promotes brain tumorigenesis via phosphorylating ATG5. Autophagy. 2021;17:723–742. doi: 10.1080/15548627.2020.1731266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhou J, Li G, Zheng Y, Shen HM, Hu X, Ming QL, Huang C, Li P, Gao N. A novel autophagy/mitophagy inhibitor liensinine sensitizes breast cancer cells to chemotherapy through DNM1L-mediated mitochondrial fission. Autophagy. 2015;11:1259–1279. doi: 10.1080/15548627.2015.1056970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tian X, Teng J, Chen J. New insights regarding SNARE proteins in autophagosome-lysosome fusion. Autophagy. 2021;17:2680–2688. doi: 10.1080/15548627.2020.1823124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ganley IG, Wong PM, Gammoh N, Jiang X. Distinct autophagosomal-lysosomal fusion mechanism revealed by thapsigargin-induced autophagy arrest. Mol Cell. 2011;42:731–743. doi: 10.1016/j.molcel.2011.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vidyadhara DJ, Lee JE, Chandra SS. Role of the endolysosomal system in Parkinson's disease. J Neurochem. 2019;150:487–506. doi: 10.1111/jnc.14820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gibson SB. A matter of balance between life and death: Targeting reactive oxygen species (ROS)-induced autophagy for cancer therapy. Autophagy. 2010;6:835–837. doi: 10.4161/auto.6.7.13335. [DOI] [PubMed] [Google Scholar]
- 60.Kubota C, Torii S, Hou N, Saito N, Yoshimoto Y, Imai H, Takeuchi T. Constitutive reactive oxygen species generation from autophagosome/lysosome in neuronal oxidative toxicity. J Biol Chem. 2010;285:667–674. doi: 10.1074/jbc.M109.053058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Scherz-Shouval R, Elazar Z. ROS, mitochondria and the regulation of autophagy. Trends Cell Biol. 2007;17:422–427. doi: 10.1016/j.tcb.2007.07.009. [DOI] [PubMed] [Google Scholar]
- 62.Kma L, Baruah TJ. The interplay of ROS and the PI3K/Akt pathway in autophagy regulation. Biotechnol Appl Biochem. 2022;69:248–264. doi: 10.1002/bab.2104. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.