Abstract
The Pseudomonas derived ς54-dependent regulators DmpR and XylR control the expression of genes involved in catabolism of aromatic compounds. Binding to distinct, nonoverlapping groups of aromatic effectors controls the activities of these transcriptional activators. Previous work has derived a common mechanistic model for these two regulators in which effector binding by the N-terminal 210 residues (the A-domain) of the protein relieves repression of an intrinsic ATPase activity essential for its transcription-promoting property and allows productive interaction with the transcriptional apparatus. Here we dissect the A-domains of DmpR and XylR by DNA shuffling to identify the region(s) that mediates the differences in the effector specificity profiles. Analysis of in vivo transcription in response to multiple aromatic effectors and the in vitro phenol-binding abilities of regulator derivatives with hybrid DmpR/XylR A-domains reveals that residues 110 to 186 are key determinants that distinguish the effector profiles of DmpR and XylR. Moreover, the properties of some mosaic DmpR/XylR derivatives reveal that high-affinity aromatic effector binding can be completely uncoupled from the ability to promote transcription. Hence, novel aromatic binding properties will only be translated into functional transcriptional activation if effector binding also triggers release of interdomain repression.
DmpR and XylR are highly homologous Pseudomonas-derived transcriptional regulators (12, 28) that share many mechanistic features. DmpR is the specific regulator of the Po promoter, which controls the (methyl)phenol catabolic dmp operon of pVI150 (reviewed in reference 26). XylR performs an analogous regulatory role at the Pu promoter controlling the upper pathway operon of the TOL plasmid pWW0, which encodes the enzymes for conversion of toluene and xylenes to carboxylic acids (reviewed in reference 23). Although the sequences of the Po and Pu promoter regions differ, the RNA polymerase and the regulator binding motifs are sufficiently similar to allow efficient cross regulation (7). DmpR and XylR belong to the prokaryotic family of enhancer binding proteins that regulate transcription via RNA polymerase utilizing the alternative sigma factor, ς54 (ςN), that recognizes −24 GG and −12 GC promoters (14). Like other family members, DmpR and XylR have a distinct domain structure consisting of an N-terminal signal reception A-domain linked to a central activation C-domain by a short B-domain, and a C-terminal D-domain mediating DNA binding (Fig. 1) (15).
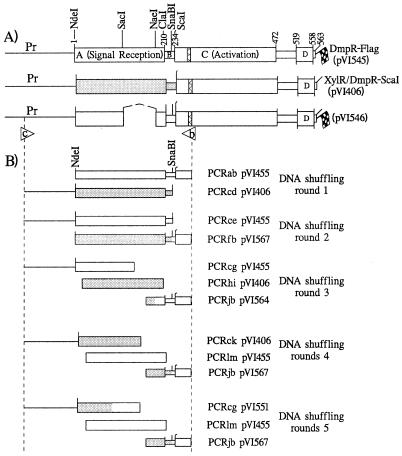
FIG. 1.
(A) Schematic representation of dmpR- (lines and open boxes) and xylR (shaded boxes)-derived DNAs in key constructs used in this study. The domain structure of DmpR and XylR, described in the text, is superimposed on the DNA restriction map. The hatched boxes represent the extent of the NTP Walker motif (32) found in this class of regulators (G--G-GKE--A---H--S [15]). The flags indicate the presence of an 8-amino-acid carboxy-terminal fusion with the Flag epitope tag. (B) Extents of PCR-generated fragments used in DNA-shuffling experiments described in the text. The lowercase letters refer to the primers used (see below), while the plasmid numbers refer to the template used in each case. The open arrowheads indicate the locations of the primers (b and c) used in the final step of the DNA-shuffling protocol and to identify derivatives of pVI546 harboring full-length hybrid A-domains, as described in Materials and Methods. The primer sequences, with NdeI and SnaBI sites underlined where applicable, are as follows: a, 5′-CGCCATATGCCGATCAAGTACAAG-3′; b, 5′-CGATCAGGTCCCCCGGGATCGCC-3′; c, 5′-CCGTCGATTGATCATTTGGTTG-3′; d, 5′-TCAGGCGGTTACGTAGGTTGGCAA-3′; e, 5′-GGTTGGTACGTAGCGACAACAGTTGCGAT-3′; f, 5′-GCCATATGTCGCTTACATACAAACC-3′; g, 5′-CAGATTTCCACCTCGAAGGAGTC-3′; h, 5′-CAGCCAGATCCGTTTCGTTGCCGCC-3′; i, 5′-CTACGATCGGGTCGCTTTTGAAGT-3′; j, 5′-CCGCAGAATCATTTTCCAGGAAACT-3′; k, 5′-CTCATTCATCAGGCCCAGCTCAGT-3′; l, 5′-GCCCATATGATCCACTTCCAGAGCATG-3′; m, 5′-CGCGGATCCGTTCTTGAAATACTGTTT-3′.
Most ς54-dependent activators are constitutively produced, but their activities are controlled in response to environmental signals (reviewed in reference 27). DmpR and XylR belong to a subgroup of ligand-responsive regulators. In each case, the activity of the regulator is controlled by interdomain repression where, in the absence of aromatic effectors, the regulatory A-domain inhibits the transcription-promoting activities of the C-domain (6, 16, 17, 21, 22, 30). DmpR and XylR are responsive to distinct, nonoverlapping profiles of aromatic compounds (1, 29, 30), which include some priority pollutants. In this respect, these transcriptional regulators serve as the “aromatic sensors” of the cell, which directly detect the presence of the substrates for the catabolic pathways they control. Utilization of these natural sensing systems and their mutant derivatives, which can respond to different aromatic effectors, provides a potentially inexpensive and simple way to monitor environmental contamination.
Effector specificity mutants and domain-swapping experiments with DmpR and XylR have shown that the effector specificity is mediated via the A-domain (4, 20, 30). However, the magnitude of the aromatic-stimulated transcriptional response varies depending on the position and nature of substituents on the aromatic ring. Recently, the A-domain of DmpR (amino acid residues 1 to 210) has been shown to be both necessary and sufficient to bind its ligand, phenol. Analysis of the binding data revealed a single phenol-binding site per monomer of DmpR (17). Furthermore, competition experiments using DmpR and a panel of aromatic compounds have shown that all the effectors tested act through the same binding site and that the affinity of DmpR for the different effectors determines the concentration at which a response can be detected, but not the magnitude of the response (18). Given the functional homology, it appears likely that all the multiple aromatic effectors of XylR/DmpR family members act through a single binding site on each protein. DNA shuffling between genes that share homology but encode proteins with distinctive properties is a powerful technique, pioneered by Stemmer and coworkers (3, 31), for directed evolution of desirable properties of individual enzymes and whole-enzyme systems (reviewed in reference 8). Here we use DNA shuffling between the A-domains of DmpR and XylR to identify a subregion of the A-domain that is primarily responsible for determining the distinct effector profiles of the two regulators.
MATERIALS AND METHODS
DNA manipulations.
DNA isolation and manipulations were performed using standard techniques (25). The construction of T7 expression plasmids based on pET3a (24) encoding either the entire DmpR regulator or various regions of DmpR as NdeI-to-BamHI fragments, with the BamHI site in frame with the eight codons of a Flag tag (10), has been described previously (17, 30). Plasmids expressing Flag-tagged peptides spanning residues 1 to 190 (pVI540), 6 to 200 (pVI541), 11 to 200 (pVI542), 16 to 200 (pVI543), and 21 to 200 (pVI544) were constructed in an analogous manner by PCR amplification of the corresponding coding regions as NdeI-to-BamHI fragments. T7 expression plasmids for DNA-shuffling products (see below) were constructed by cloning NdeI-to-HindIII fragments, spanning the entire regulator and Flag tag, into pET3H (30). Plasmid pVI577 expresses DmpR/XylR-4 (derived from pVI571) and pVI578 expresses XDX-2 (derived from pVI574), while pVI579 expresses DXD-1 (derived from pVI575).
Plasmid pVI455 is an RSF1010-based broad-host-range plasmid carrying dmpR-Flag under control of its native promoter (30). A unique ClaI site (introducing a Glu-210-to-Asp substitution) and a silent SnaBI-site (overlapping codons 221 to 222) were introduced by PCR into the B-domain region of the dmpR gene of this plasmid, generating plasmid pVI545 (Fig. 1). An internal A-domain deletion between the SacI and NaeI sites of pVI545 was constructed with the aid of a linker to generate pVI546, illustrated in Fig. 1. Plasmids pVI550 to pVI572 (Table 1) carry hybrid A-domains of dmpR and xylR, generated as described below, which were cloned as NdeI (codon 1)-to-SnaBI (codon 221) fragments between these sites of pVI546. Note that the mutation introduced by the ClaI site is replaced in these derivatives. Plasmids pVI573 to pVI576 (see Fig. 6) were constructed from pVI546 by insertion of two PCR-generated fragments that introduced a diagnostic silent ClaI site overlapping residues I-129 and D-130, common to DmpR and XylR. NdeI (codon 1)-to-ClaI (codons 129 and 130) fragments were PCR amplified from the DNA-shuffling hybrids pVI550, pVI551, pVI570, and pVI571 and assembled with ClaI-to-SnaBI (residue 221) fragments amplified from pVI572 or pVI564 to generate full-length in-frame A-domain sequences. The DNA sequences of both strands of all PCR-generated DNAs were determined with Thermo Sequenase (Amersham Pharmacia Biotech) or custom sequenced by CyberGene AB (Stockholm, Sweden) using PRISM BigDye terminators and an ABI3377 DNA sequencer (PE Biosystems, Foster City, Calif.).
TABLE 1.
Characteristics of DNA-shuffling derivativesa
| Derivative | Hybrid junction | Mutation(s) | No. isolated | Plasmid no. |
|---|---|---|---|---|
| Round 1 | ||||
| XylR/DmpR-1 | XylR 1–26-EGKIW-DmpR 32–221 | 1 | pVI550* | |
| XylR/DmpR-2 | XylR 1–75-DA-DmpR 78–221 | Q138R; Y159H | 1 | pVI551* |
| XylR/DmpR-3 | XylR 1–130-S-DmpR 132–221 | 1 | pVI552* | |
| XylR/DmpR-4 | XylR 1–148-CW-DmpR 151–221 | 3 | pVI553* | |
| XylR 1–148-CW-DmpR 151–221 | R60R silent | 1 | pVI554 | |
| XylR 1–148-CW-DmpR 151–221 | I129I silent | 1 | pVI555 | |
| XylR 1–148-CW-DmpR 151–221 | N123D | 1 | pVI556 | |
| XylR/DmpR-5a | XylR 1–160-S-DmpR 162–221 | 1 | pVI557* | |
| XylR 1–160-S-DmpR 162–221 | E211E silent | 1 | pVI558 | |
| XylR 1–160-S-DmpR 162–221 | I208I silent | 1 | pVI559 | |
| XylR/DmpR-5b | XylR 1–162-FMG-DmpR 166–221 | 7 | pVI560 | |
| XylR 1–162-FMG-DmpR 166–221 | L143P | 1 | pVI561 | |
| XylR 1–162-FMG-DmpR 166–221 | A190V | 1 | pVI562 | |
| XylR 1–162-FMG-DmpR 166–221 | A190A silent | 1 | pVI563 | |
| XylR/DmpR-6 | XylR 1–186-GK-DmpR 189–221 | 1 | pVI564* | |
| XylR/DmpR-7 | XylR 1–212-YE-DmpR 215–221 | 1 | pVI565* | |
| XylR/DmpR-8 | XylR 1–215-DmpR 216–221 | S197G | 1 | pVI566* |
| XylR/DmpR-9 | XylR 1–221 | 1 | pVI567* | |
| Round 2 | ||||
| DmpR/XylR-1 | DmpR 1–48-XylR 49–221 | 1 | pVI568* | |
| DmpR/XylR-2 | DmpR 1–60-AK-XylR 63–221 | 1 | pVI569* | |
| DmpR/XylR-3 | DmpR 1–105-V-XylR 107–221 | 1 | pVI570* | |
| DmpR/XylR-4 | DmpR 1–162-FMG-XylR 166–221 | 1 | pVI571* | |
| DmpR-F132L-Flag | DmpR 1–221 | F132L | 1 | pVI572* |
Numbers refer to codons or residues of the coding sequences. Where the DNA sequences of DmpR and XylR are identical, and thus the exact location of the junction is not discernible, the amino acid residues common to both sequences are listed. Note that the XylR/DmpR-5a and -5b derivatives have the same amino acid sequence; only the junction point differs as detected by the different codon sequences. One derivative of each type was retained, and asterisks in the last column indicate the derivatives used for the quantitative studies shown in Fig. 3. All sequences of xylR-derived DNA harbored a G-C exchange (underlined below) relative to the database sequence (M101343, M15810, and M20635), giving LL-39 (CTGCTA) rather than the published LV-39 (CTCGTA). This G-C switch was also found in the xylR DNA sequence of pEZ6 (5). Thus, this variation in xylR appears in DNA derived from two different sources.
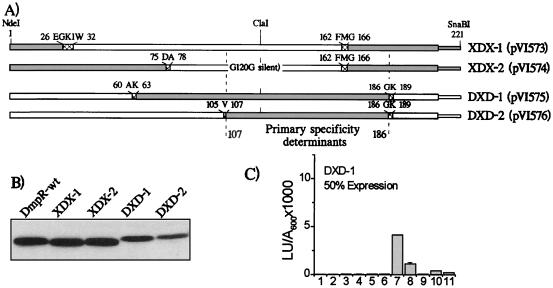
FIG. 6.
Analysis of mosaic DmpR/XylR regulators. (A) Schematic illustration of the relative contributions of DmpR (open boxes) and XylR (shaded boxes) amino acid sequences to the A-domains of the indicated derivatives. Western analysis (B) and aromatic response profile (C) of DXD-1, as described in the legend to Fig. 3. DXD-1 is expressed at about 50% of the level of DmpR-wt (wild type). LU, luciferase units.
Generation of hybrid dmpR/xylR A-domain-encoding DNA was performed essentially as described by Stemmer (31). In brief, PCR-generated fragments were subjected to DNase I treatment at 25°C, and random fragments of approximately 30 to 300 bp were recovered from 2% agarose gels using DEAE membranes (Schleicher & Schuell). The purified fragments were then mixed in a self-priming PCR (without primers) to generate extended hybrid products. The resulting products were diluted 10- to 50-fold in a new PCR containing specific primers. To ensure the production of full-length hybrid products, the most distal primer used to generate the starting PCR products was used in each case (primers c and b [Fig. 1]). The products from these reactions were purified, digested with NdeI and SnaBI, and used for cloning into the shuffling cassette of pVI546. Derivatives containing full-length hybrid A-domains were identified by colony PCR using the same primers.
Luciferase assays.
For plate test screening, colonies of Pseudomonas putida KT2440::Po-luxAB (30) harboring various plasmids were replica plated and grown overnight on Luria agar plates containing antibiotics selective for the resident plasmid and supplemented with 2 mM effector or exposed to effector vapor (toluene or m-xylene). Inverted plates were exposed to decanal vapor, and the light emission was recorded by placing film over the plates. For quantification of luciferase activity, cultures were grown to late exponential phase (A650, 2.5); at that time, aliquots of the cells were left unsupplemented or were supplemented with 2 mM effector or vapor and were further incubated with rigorous shaking for 3 h. Light emission due to luciferase expression was assayed as previously described (20).
Expression, purification, and [14C]phenol binding.
Flag-tagged proteins were expressed, affinity purified, and tested for the ability to bind universally labeled [14C]phenol (5.22 GBq/mmol; Amersham Pharmacia Biotech) as previously described (18). Experiments were performed at a final concentration of 16 μM radiolabeled phenol (the Kd of wild-type DmpR). Background levels bound to a ΔA-NifA-Flag derivative were subtracted to determine the specific binding of phenol as previously described (17). Values are expressed as percentages of phenol binding mediated by wild-type DmpR-Flag.
Protein analysis.
Crude extracts of cytosolic proteins, sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), transfer to nitrocellulose filters, and Western blot analysis with polyclonal rabbit anti-ΔA1-DmpR serum were as previously described (30). Antibody-decorated bands were revealed by using chemiluminescence reagents as directed by the supplier (Amersham Pharmacia Biotech). Differences in expression levels were assessed by comparison of dilution series of the test samples with those of wild-type DmpR-Flag. For N-terminal sequence analysis, samples were transferred to a polyvinylidene difluoride nitrocellulose filter (Bio-Rad), briefly stained, destained, and extensively washed prior to excision of a strip of filter from the center of the band corresponding to the unknown 60,000-Da protein.
RESULTS AND DISCUSSION
Deletions in the A-domain cause association with GroEL.
Previous analysis of the [14C]phenol-binding capacity of Flag epitope-tagged peptides of DmpR has shown that amino acid residues 1 to 200 are sufficient for full phenol binding (compared to that of the wild-type protein) (17). As a first strategy to further define the region of DmpR responsible for aromatic-ligand binding, we constructed a series of derivatives that would express truncated derivatives of the A-domain as Flag epitope-tagged peptides. However, further deletions resulting in peptides spanning residues 1 to 190, 6 to 200, 11 to 200, 16 to 200, and 21 to 200 all gave rise to preparations in which the peptide copurified with an unknown protein with an Mr of 60,000 and were incapable of binding [14C]phenol (data not shown). N-terminal sequencing of the copurified protein revealed the amino acid sequence AAKDVKFGNDARV, which is identical to that of the protein-folding chaperone GroEL of Escherichia coli. These results suggests that the A-domain of DmpR, defined as residues 1 to 210 by sequence alignment with other members of the family (28), is a discrete domain and that truncation of the domain at either the N or C terminus results in unfolded peptides. This finding prompted us to use DNA shuffling to generate hybrid A-domains in the wild-type context of the remainder of DmpR as a strategy to identify the key region(s) involved in the specificity of ligand binding.
Construction of a dmpR-shuffling template.
The domain structures of DmpR and XylR are schematically illustrated in Fig. 1. Previous manipulation to introduce a silent NdeI site overlapping the ATG initiation codons of dmpR and xylR, and introduction of a silent mutation generating a ScaI site in the coding region of xylR analogous to that in dmpR, allowed construction of a hybrid gene in which codons 1 to 233 of dmpR were exchanged for those of xylR (Fig. 1A, pVI406). The encoded protein, XylR/DmpR-ScaI, was tightly regulated in response to aromatic effectors of XylR but was unable to respond to effectors of DmpR, demonstrating that the A-domain of XylR is fully functional and capable of exerting its repressive property and effector response in the context of DmpR (see below) (29). Because of the presence of two other ScaI sites in the vector, this site was not useful for DNA-shuffling purposes. Therefore, a unique silent SnaBI was introduced into the B linker of dmpR so that replacement of the NdeI (codon 1)-to-SnaBI (overlapping codons 221 to 222) region would result in replacement of the entire A-domain (Fig. 1A, pVI545). To facilitate the identification of clones encoding full-length hybrid A-domains, two other manipulations were made. First, a diagnostic unique ClaI site was also introduced, and second, an internal deletion between the SacI site (overlapping codons 112 to 113) and the NdeI site (overlapping codons 188 to 190) was made to give the final template into which hybrid NdeI-to-SnaBI cassettes would be cloned (Fig. 1A, pVI546).
DNA shuffling round 1: generation of XylR/DmpR hybrids.
The general strategy for all DNA-shuffling experiments followed the same basic steps, as detailed in Materials and Methods. First, PCR-generated fragments spanning the A-domains of dmpR and xylR (depicted in Fig. 1B) were subject to limited DNase I treatment, and the resulting random fragments of 30 to 300 bp were used to generate extended hybrid DNA fragments in a self-priming PCR. The resulting products were then used to generate DNA spanning from upstream of the NdeI site to downstream of the SnaBI site in primer-directed PCRs. Finally, purified NdeI-to-SnaBI fragments were cloned into the DNA-shuffling template pVI546 (Fig. 1A) to regenerate an entire regulator under control of the native promoter of dmpR.
For screening of the aromatic responsiveness of hybrid regulators expressed from DNA-shuffling derivatives, we used a previously constructed luciferase reporter system consisting of a single copy of the DmpR-regulated promoter Po fused to the luxAB genes on the chromosome of a P. putida host (KT2440::Po-luxAB [30]). Ninety derivatives harboring full-length A-domains generated from DNA shuffling round 1 (Fig. 1B) were introduced into the reporter strain and screened for the ability to respond to the DmpR effector 2-methylphenol and the XylR effectors toluene and 3-methylbenzyl alcohol. Twenty-six derivatives were found by the plate test described in Materials and Methods to respond to one or more of these compounds and were subjected to DNA sequence analysis. The results, summarized in Table 1, showed that this round of DNA shuffling generated a series of hybrid genes, all starting with xylR sequences but with a gradient of junction points with dmpR sequence. The shuffling derivatives could be classified on the basis of the DNA junction point into nine classes, although some derivatives in the different classes also harbored PCR-generated mutations (Table 1). Alignment of the DNA and amino acid sequences of the A-domains of DmpR and XylR are shown in Fig. 2. As expected, all junction points corresponded to regions of DNA homology of dmpR and xylR (Fig. 2). However, not all the possible junction points were found, and a “hot spot” between codons 160 and 167 was observed (Table 1), suggesting that some junction points do not produce functional proteins.
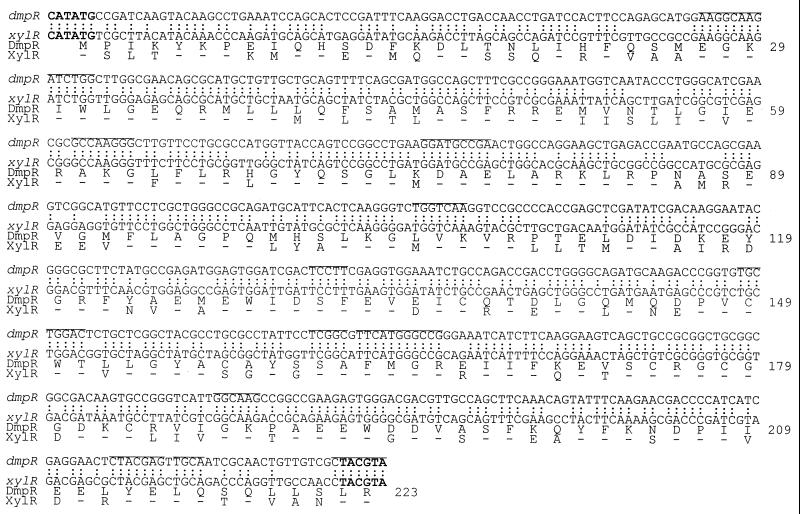
FIG. 2.
A-domain DNA and amino acid sequence alignments of DmpR and XylR. The NdeI and SnaBI sites used in the DNA-shuffling protocol are shown in boldface. The 12 different junction points found in the hybrid regulators are marked by lines. Colons indicate identical bases, and dashes indicate identical amino acid residues.
DNA shuffling round 2: generation of DmpR/XylR hybrid A-domains.
The results discussed above suggested that the strategy used in round 1 introduced a bias in the shuffling in favor of hybrids with 5′ ends derived from xylR DNA. Therefore, to generate derivatives with 5′ ends derived from dmpR DNA, we performed a second round of DNA shuffling in which the locations of the primers used to generate the starting DNA for the shuffling were reversed (Fig. 1B). Of 25 full-length derivatives tested as described above, 5 were found to be responsive to one or more of the test aromatic compounds. DNA sequence analysis of these five derivatives showed that this round of DNA shuffling, as expected, resulted in genes starting with dmpR sequences. On the basis of the junction point, the shuffling derivatives could be grouped into five classes (Table 1). The derivative with the most distal junction point at codon 221 regenerated full-length dmpR sequence. However, since it harbored a mutation that results in a single-amino-acid substitution (F132L) close to a previously defined effector specificity mutant (E135K [20]), this derivative was further analyzed as described below.
Effector responses of hybrid DmpR/XylR regulators.
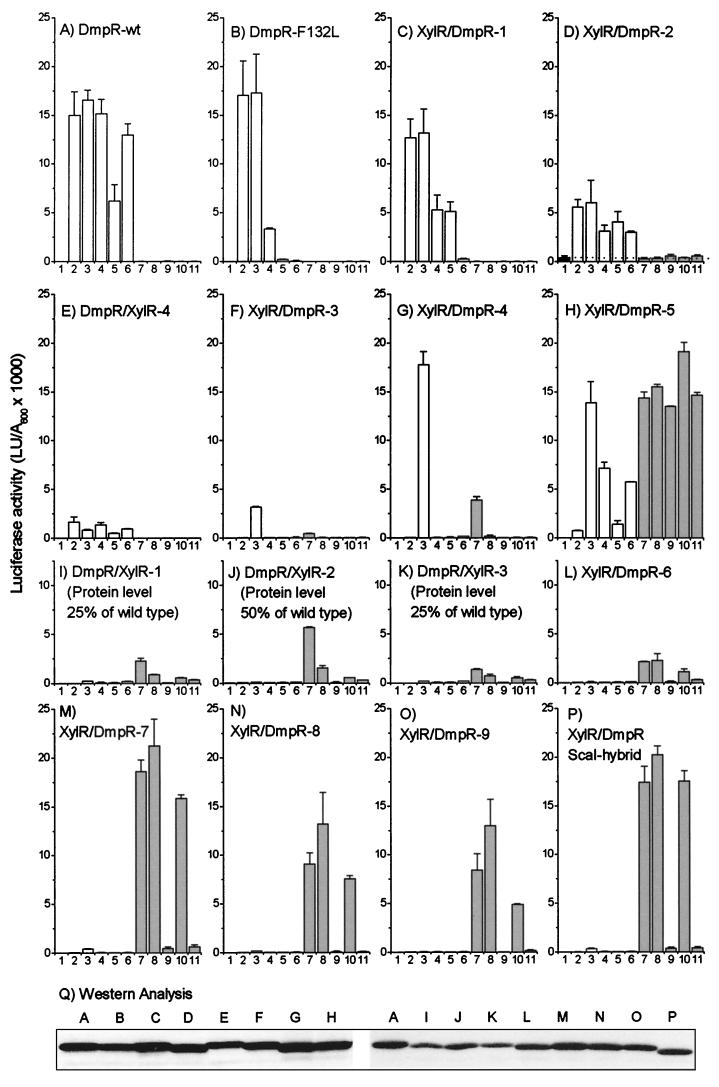
One representative from each class of hybrid regulators (Table 1) was quantitatively assayed in the luciferase reporter strain described above for the ability to respond to two sets of aromatic compounds. The responses are compared to those of wild-type DmpR and the XylR/DmpR-ScaI hybrid in Fig. 3. The first sets of aromatic compounds chosen (Fig. 3) are all effectors of DmpR and include phenol; 2-, 3-, and 4-methylphenol; and 3,4-dimethylphenol. The second set of aromatic compounds (Fig. 3) includes three effectors of XylR (toluene, m-xylene, and 3-methylbenzyl alcohol) and two structurally closely related compounds (2- and 4-methylbenzyl alcohol). The expression of the regulator in each case was also monitored by Western analysis using polyclonal rabbit antiserum directed against the C- to D-domains of DmpR, i.e., the region common to all the regulators tested (Fig. 3Q).
FIG. 3.
In vivo transcriptional response of Po mediated by DmpR and XylR hybrids. (A to P) Luciferase transcriptional response of P. putida KT2440::Po-luxAB harboring plasmids expressing the indicated derivatives (for plasmid designations, see Table 1) was measured in the absence of any effector (bars 1) or in the presence of two sets of aromatic effectors. The DmpR effectors (open bars) were phenol (bars 2), 2-methylphenol (bars 3), 3-methylphenol (bars 4), 4-methylphenol (bars 5), and 3,4-dimethylphenol (bars 6). The XylR and XylR-like effectors (shaded bars) were toluene (bars 7), m-xylene (bars 8), 2-methylbenzyl alcohol (bars 9), 3-methylbenzyl alcohol (bars 10), and 4-methylbenzyl alcohol (bars 11). The data are the averages of triplicate determinations in each of two independent experiments. The level of protein expression, estimated from panel Q (as described in Materials and Methods), is expressed as a percentage of that of DmpR-wt (wild type) for derivatives with significantly decreased expression levels (I, J, and K). (Q) Expression levels of the regulators. Western analysis of 30 μg of crude extract derived from cells exposed to 2-methylphenol shown in panels A to P, separated by SDS–11% PAGE, and probed with anti-DmpR serum as described in Materials and Methods is shown. LU, luciferase units. The error bars indicate standard deviations.
On the basis of responsiveness to aromatic compounds, the derivatives could be classified into different response groups. The members of the first group (Fig. 3A to E) are DmpR-like in that they are capable of responding, to different degrees, to some or all of the DmpR effectors but not to XylR effectors. Two of the derivatives in this group have narrower response profiles than wild-type DmpR. DmpR-F132L (Fig. 3B), due to a single-amino-acid substitution, has lost the ability to respond to 4-methylphenol and 3,4-dimethylphenol and has reduced ability to respond to 3-methyphenol. XylR/DmpR-1 (Fig. 3C) has lost the ability to respond to 3,4-dimethylphenol and has reduced ability to respond to 3-methylphenol. The members of a second group of derivatives (Fig. 3I to P) are XylR-like, responding to XylR effectors but not DmpR effectors. The members of a final group are hybrid in the type of effectors they respond to. Hybrid type 1 responders (Fig. 3F and G) have a narrow response profile, only responding to 2-methylphenol (a DmpR effector) and toluene (a XylR effector). Conversely, hybrid type 2 derivatives (Fig. 3H) have a broadened response profile, responding to the presence of all the XylR effectors and showing a good response to three of the DmpR effectors. In addition, these derivatives have gained the novel ability to respond to 2-methylbenzyl alcohol (Fig. 3, lane 9) and 4-methylbenzyl alcohol (lane 11), to which neither parent regulator could respond (Fig. 3A and P). Thus, this strategy has allowed the production of regulators with novel response abilities (Fig. 3H) and both broadened (Fig. 3H) and narrowed (Fig. 3B, C, F, and G) response profiles.
The linear array of XylR and DmpR contributions to the composition of the A-domains is illustrated in the four different response groups in Fig. 4. From the alignment, it becomes obvious that the nature of the residues between 107 and 186 is critical for determining the type of response observed. All derivatives that are composed of XylR residues in this region give XylR-like profiles, while all derivatives which are composed of DmpR residues in this region give DmpR-like profiles. With one exception, all the regulators that are hybrid in the composition of this region are also hybrid in the their response profiles. The exception, DmpR/XylR-4, has very low transcriptional response to its effectors. The nature of the defect in this derivative and other derivatives with low-level response is discussed below. The data strongly suggest that residues 107 to 186 are intimately involved in determining the specificity of ligand binding. Consistent with this conclusion is the observation that single-amino-acid substitutions in DmpR (E135K, D140K, and R184W [20, 30]) and XylR (E172K [4]) that confer novel effector response capabilities and the F132L substitution of DmpR that mediates a more limited response profile (Fig. 3B) all map in this region. However, since the A-domains of DmpR and XylR share 64.8% identity overall, it is perfectly possible, and even probable, that residues outside this region are also involved in forming the aromatic-effector binding site. A prediction from this is that alterations of residues outside this region that play a common role in the formation of the effector binding sites in DmpR and XylR might also alter the effector response profile. Hence, we limit the definition of the region between residues 107 and 186 to containing the primary specificity determinants that distinguish the effector response profile of DmpR from that of XylR. Comparison of the amino acid sequences between residues 107 and 186 of DmpR and XylR (Fig. 2) shows that the first residue that differs is 110. The overall identity of residues 110 to 186 of DmpR and XylR is 63.4%. This region is only slightly less conserved than the A-domains of the two regulators overall (64.8%) and could not have been predicted by visual comparison to be involved in determining effector specificity. Residues 110 to 186 define the outer boundaries that can be discerned from the data. The actual region involved could be smaller, but it presumably spans residues 130 to 160 (the left and right boundaries of the hybrid responders XylR/DmpR-3 and XylR/DmpR-5a [Fig. 4]).
FIG. 4.
Schematic illustration of the relative contributions of DmpR (open boxes) and XylR (shaded boxes) amino acid sequences to the A-domains of the derivatives tested for aromatic responsiveness (Fig. 3). The derivatives are classified into four response profile groups as described in the text. The arrowheads mark the locations of single-amino-acid substitutions that give rise to novel effector response abilities of DmpR (open arrowheads) and XylR (shaded arrowheads) as described in the text. wt, wild type. Narrow ends indicate the extent of β-domain linker sequences (Fig. 1), while stippled boxes indicate the junction points defined in Table 1.
Many hybrid DmpR/XylR regulators give low-level responses to aromatic effectors.
The scale for the magnitude of the transcriptional response in the presence of different effectors in Fig. 3 is the same for all the derivatives. It is notable that some of the derivatives are poor at promoting transcription in the presence of their effectors (Fig. 3E to F and I to L). All derivatives are expressed from the same cis-acting regulatory element, but it is possible that the different hybrid proteins exhibit different stabilities. From the Western blot analysis (Fig. 3Q), it appears that the protein level of the regulator can account for the poor responses observed in some but not all cases. Reduced protein levels could partially or completely account for the reduced response levels of the derivatives shown in Fig. 3I to K. However, no major difference in protein levels could be observed for the derivatives shown in Fig. 3E, F, or L. This suggests that these derivatives are defective in their binding or response to aromatic effectors. The A-domain represses the essential activities of the C-domain required for transcriptional activation, namely, ATPase activity and interaction with the transcriptional apparatus, until aromatic effectors are bound. Mechanistically, the A-domain-mediated control of these two processes can be uncoupled, since some aromatic compounds that can be bound by DmpR elicit an ATPase activity but not a transcriptional response (18). Therefore, the defect in the abilities of the hybrid regulators to promote a transcriptional response can be predicted to lie on any of three levels: (i) at the level of correct folding of the hybrid A-domain, (ii) at the level of effector binding affinity, or (iii) at the level of the ability of the effector-bound form to allow interaction with the transcriptional apparatus. To distinguish the basis for the low levels of response observed, we analyzed the [14C]phenol-binding ability of DmpR/XylR-4, which exhibits a low-level response to phenol despite approximately wild-type protein levels. The ability of this derivative (Fig. 3E) to bind [14C]phenol is compared to that of wild-type DmpR in Fig. 5. The results show that DmpR/XylR-4 has approximately twofold-reduced ability to bind 16 μM phenol. In terms of phenol binding, this decrease is reminiscent of the effector specificity mutant DmpR-E135K, which has two- to fourfold-reduced affinity for phenol. However, DmpR-E135K-Flag is capable of a full in vivo transcriptional response at the 2 mM effector concentration used in the transcriptional reporter assays here (18). Thus, it appears likely that, despite the reduced ability of DmpR/XylR-4 to bind phenol, the main defect in this derivative lies at the level of transmission of the binding signal to generate an effector-bound form that allows productive interaction with the transcriptional apparatus.
FIG. 5.
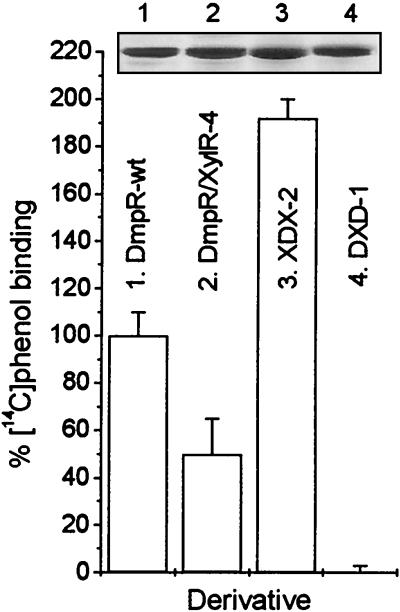
Comparison of [14C]phenol binding of DmpR-Flag and DNA-shuffling derivatives. The data are the averages (+ standard deviations) of triplicate determinations performed at the Kd for wild-type DmpR-Flag (16 μM) as described in Materials and Methods. Binding by wild-type DmpR-Flag was set at 100%. The inset shows a Coomassie blue stain of 3 μg of proteins released from 3 μl of the beads and separated by SDS–11% PAGE.
Mosaic DmpR/XylR A-domains are predominantly nonresponsive to aromatic effectors.
An initially surprising finding from the DNA shuffling rounds 1 and 2, was the complete absence of isolation of derivatives with mosaic A-domains in which more than two blocks of dmpR and xylR DNA had been shuffled. In these experiments, a total of 12 different junction points were found (Fig. 2), but no more than one junction was found per hybrid. This suggests that more complex shuffling of A-domain sequences may result in predominantly nonresponsive derivatives. To test this idea, we did three more rounds of shuffling with DNA, depicted in Fig. 1B (rounds 3 to 5). The design of the different DNA fragments used in these shuffling experiments served two purposes. First, it forced at least two junctions to generate a full-length A-domain. Second, it did not allow the possibility of a junction at the hybrid hot spot between codons 160 and 167 found in round 1, which results in the multiple effector responder XylR/DmpR-5 (Fig. 3H). From a total of 213 derivatives that had full-length A-domains derived from DNA shuffling rounds 3 to 5, no derivative capable of significant response to the three test effectors (2-methylphenol, toluene, and 3-methylbenzyl alcohol) was found. Therefore, to address the question of the aromatic responsiveness of mosaic A-domains, and as an independent test of whether residues 107 to 186 determine the specificity of activation, we artificially constructed four mosaic A-domains. These constructs were designed on the basis of the productive dmpR/xylR junction points found in rounds 1 and 2 and were generated as described in Materials and Methods. The extents of the contributions of DmpR and XylR in these derivatives are illustrated in Fig. 6A. In the XDX-1 and -2 derivatives, DmpR A-domain residues are flanked by XylR residues, while in the DXD-1 and -2 derivatives, XylR A-domain residues are flanked by DmpR residues. Of these four derivatives, only one, DXD-1, gave detectable responses to any of the aromatic effectors, again supporting the idea that a high proportion of mosaic A-domains are non-aromatic responsive, even when junction points that are productive in formation of hybrid A-domains are used (Fig. 6C and data not shown). Furthermore, the DXD-1 derivative had a XylR-like aromatic effector response profile, as would be predicted from the results of rounds 1 and 2.
Given the finding that the response-defective derivative DmpR/XylR-4 could still bind phenol, we reasoned that all the XDX and DXD derivatives may similarly still be able to bind effectors. If this is the case, then a second prediction can be derived from the conclusion that residues 110 to 186 distinguish the effector profiles of DmpR and XylR, namely, that XDX derivatives, despite a response defect, should bind phenol while DXD derivatives should not. To test this idea we employed XDX-2 as the derivative with the smallest span of DmpR-derived residues and DXD-1 as a functional derivative that does not respond to phenol. The [14C]phenol-binding analysis (Fig. 5) shows that the prediction holds true, with XDX-2 exhibiting approximately twofold-enhanced affinity for phenol compared to that of the wild-type DmpR and with DXD-1 being unable to bind phenol at all. In addition to demonstrating that a specific region of the regulator determines the aromatic-effector binding profile, the results for XDX-2 underscore the importance of the A-domain as a complete functional domain for a correct response to the effector binding signal. Previously it had been shown that wild-type DmpR could bind some noneffector aromatic compounds (e.g., 4-ethylphenol and 2,4-dimethylphenol) and that these compounds could elicit an ATPase response in vitro but did not serve as effectors for in vivo transcriptional activation (18). These results suggested that a major regulatory consequence of the binding of aromatic effectors by the A-domain is to allow productive interaction of the C-domain of the regulator with the transcriptional apparatus. XDX-2 provides a clear-cut demonstration of a mutant derivative in which binding of the potent natural effector phenol is completely uncoupled from the ability to respond to the binding signal.
Concluding remarks.
In this report we have demonstrated, using hybrid A-domains of DmpR and XylR, that the amino acid residues 110 to 186 are intimately involved in mediating the distinct effector profiles of these two regulators. The aromatic-sensing properties of natural regulators of catabolic pathways have been used in biosensor applications (2, 9, 11, 13, 19, 33). The identification of a subregion of the A-domain that is primarily responsible for determining effector specificity provides a target for directed mutagenesis to modify sensitivity and expand the effector specificity of this class of regulators for greater utility in such biosensor systems. The DNA-shuffling approach used here was designed to be unbiased, and identification of functional derivatives by screening did not involve any positive selection for a desired property. Nevertheless, derivatives with novel response abilities and broadened or narrowed response profiles were identified, demonstrating the power of the approach to generate variants with different response properties. Previous work has shown that the affinity of DmpR for four of its natural effectors tested determines the concentration at which a response can be detected but not the magnitude of the response (18). The finding that a hybrid derivative can have enhanced binding affinity for phenol but be completely deficient in transcriptional activation has important mechanistic implications and suggests that the conformation of the A-domain upon binding of the aromatic effector is crucial for productive interaction with the transcriptional apparatus. Hence, novel or enhanced binding affinities will only lead to productive in vivo transcriptional responses if the alterations made are also compatible with the release of interdomain repression and subsequent interaction of the C-domain with RNA polymerase.
ACKNOWLEDGMENTS
We thank Bo Ek, Uppsala Biomedical Center, for N-terminal sequencing and Martin Gullberg, Umeå University, for critical reading of the manuscript.
This work was supported by grants from the Swedish Research Councils for Natural and Engineering Sciences, the Swedish Foundation for Strategic Research, and the J. C. Kempe Foundation.
REFERENCES
- 1.Abril M-A, Michán C, Timmis K N, Ramos J L. Regulator and enzyme specificities of the TOL plasmid-encoded upper pathway for the degradation of aromatic hydrocarbons and expansion of the substrate range of the pathway. J Bacteriol. 1989;171:6782–6790. doi: 10.1128/jb.171.12.6782-6790.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Applegate B M, Kehrmeyer S R, Sayler G S. A chromosomally based tod-luxCDABE whole-cell reporter for benzene, toluene, ethylbenzene, and xylene (BTEX) sensing. Appl Environ Microbiol. 1998;64:2730–2735. doi: 10.1128/aem.64.7.2730-2735.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Crameri A, Dawes G, Rodríguez E, Jr, Silver S, Stemmer W P C. Molecular evolution of an arsenate detoxification pathway by DNA shuffling. Nat Biotechnol. 1997;15:436–438. doi: 10.1038/nbt0597-436. [DOI] [PubMed] [Google Scholar]
- 4.Delgado A, Ramos J-L. Genetic evidence for activation of the positive transcriptional regulator XylR, a member of the NtrC family of regulators, by effector binding. J Biol Chem. 1994;269:8059–8062. [PubMed] [Google Scholar]
- 5.de Lorenzo V, Herrero M, Metzke M, Timmis K N. An upstream XylR- and IHF-induced nucleoprotein complex regulates the sigma54-dependent Pu promoter of TOL plasmid. EMBO J. 1991;10:1159–1167. doi: 10.1002/j.1460-2075.1991.tb08056.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fernández S, Pérez-Martín J, de Lorenzo V. Activation of the transcriptional regulator XylR of Pseudomonas putida by release of intramolecular repression between functional domains. Mol Microbiol. 1995;16:205–213. doi: 10.1111/j.1365-2958.1995.tb02293.x. [DOI] [PubMed] [Google Scholar]
- 7.Fernández S, Shingler V, de Lorenzo V. Cross-regulation by XylR and DmpR activators of Pseudomonas putida suggest that transcriptional control of biodegradative operons evolves independently of catabolic genes. J Bacteriol. 1994;176:5052–5058. doi: 10.1128/jb.176.16.5052-5058.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harayama S. Artificial evolution by DNA shuffling. Trends Biotechnol. 1998;16:76–82. doi: 10.1016/s0167-7799(97)01158-x. [DOI] [PubMed] [Google Scholar]
- 9.Heitzer A, Webb O, Thonnard J, Sayler G. Specific and quantitative assessment of naphthalene and salicylate bioavailability by using a bioluminescent catabolic reporter bacterium. Appl Environ Microbiol. 1992;58:1839–1845. doi: 10.1128/aem.58.6.1839-1846.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hopp T P, Prickett K S, Price V L, Libby R T, March C J, Cerretti D P, Urdal D L, Conlon P J. A short polypeptide marker sequence useful for recombinant protein identification and purification. Bio/Technology. 1988;6:1204–1210. [Google Scholar]
- 11.Ikariyama Y, Nishiguchi S, Koyama T, Kobatake E, Azawa M. Fiber-optic based biomonitoring of benzene derivatives by recombinant E. coli bearing luciferase gene-fused TOL-plasmid immobilized on the fiber-optic end. Anal Chem. 1997;69:2600–2605. doi: 10.1021/ac961311o. [DOI] [PubMed] [Google Scholar]
- 12.Inouye S, Nakazawa A, Nakazawa T. Nucleotide sequence of the regulatory gene xylR of the TOL plasmid from Pseudomonas putida. Gene. 1988;66:301–306. doi: 10.1016/0378-1119(88)90366-6. [DOI] [PubMed] [Google Scholar]
- 13.Layton A C, Muccini M, Ghosh M M, Sayler G S. Construction of a bioluminescent reporter strain to detect polychlorinated biphenyls. Appl Environ Microbiol. 1998;64:5023–5026. doi: 10.1128/aem.64.12.5023-5026.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Merrick M J. In a class of its own—the RNA polymerase factor ς54 (ςN) Mol Microbiol. 1993;10:903–909. doi: 10.1111/j.1365-2958.1993.tb00961.x. [DOI] [PubMed] [Google Scholar]
- 15.Morett E, Segovia L. The ς54 bacterial enhancer-binding protein family: mechanism of action and phylogenetic relationship of their functional domains. J Bacteriol. 1993;178:6067–6074. doi: 10.1128/jb.175.19.6067-6074.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ng L C, O'Neill E, Shingler V. Genetic evidence for inter-domain regulation of the phenol responsive ς54-dependent activator DmpR. J Biol Chem. 1996;271:17281–17286. doi: 10.1074/jbc.271.29.17281. [DOI] [PubMed] [Google Scholar]
- 17.O'Neill E, Ng L C, Sze C C, Shingler V. Aromatic ligand binding and intra-molecular signalling of the phenol-responsive ς54-dependent regulator DmpR. Mol Microbiol. 1998;28:131–141. doi: 10.1046/j.1365-2958.1998.00780.x. [DOI] [PubMed] [Google Scholar]
- 18.O'Neill E, Sze C C, Shingler V. Novel effector control through modulation of a pre-existing binding site of the aromatic responsive ς54-dependent regulator DmpR. J Biol Chem. 1999;274:32425–32432. doi: 10.1074/jbc.274.45.32425. [DOI] [PubMed] [Google Scholar]
- 19.Palmer G, McFadzean R, Kilham K, Sinclair A, Paton G. Use of lux-based biosensors for rapid diagnosis of pollution in arable soils. Chemosphere. 1998;36:2683–2696. [Google Scholar]
- 20.Pavel H, Forsman M, Shingler V. An aromatic effector specificity mutant of the transcriptional regulator DmpR overcomes the growth constraints of Pseudomonas sp. strain CF600 on para-substituted methylphenols. J Bacteriol. 1994;176:7550–7557. doi: 10.1128/jb.176.24.7550-7557.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pérez-Martín J, de Lorenzo V. The amino-terminal domain of the prokaryotic enhancer-binding protein XylR is a specific intramolecular repressor. Proc Natl Acad Sci USA. 1995;92:9392–9396. doi: 10.1073/pnas.92.20.9392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pérez-Martín J, de Lorenzo V. Identification of the repressor subdomain within the signal reception module of the prokaryotic enhancer-binding protein XylR of Pseudomonas putida. J Biol Chem. 1996;271:7899–7902. doi: 10.1074/jbc.271.14.7899. [DOI] [PubMed] [Google Scholar]
- 23.Ramos J L, Marqués S, Timmis K N. Transcriptional control of the Pseudomonas TOL plasmid catabolic operands is achieved through an interplay of host factors and plasmid-encoded regulators. Annu Rev Microbiol. 1997;51:341–373. doi: 10.1146/annurev.micro.51.1.341. [DOI] [PubMed] [Google Scholar]
- 24.Rosenberg A H, Lade B N, Chui D-S, Lin S-W, Dunn J J, Studier F W. Vectors for selective expression of cloned DNAs by T7 RNA polymerase. Gene. 1987;56:125–135. doi: 10.1016/0378-1119(87)90165-x. [DOI] [PubMed] [Google Scholar]
- 25.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 26.Shingler V. Metabolic and regulatory check points in phenol degradation by Pseudomonas CF600. In: Nakazawa T, editor. Pseudomonas: molecular biology and biotechnology. Washington, D.C.: ASM Press; 1996. pp. 153–164. [Google Scholar]
- 27.Shingler V. Signal sensing by ς54-dependent regulators: derepression as a control mechanism. Mol Microbiol. 1996;19:409–416. doi: 10.1046/j.1365-2958.1996.388920.x. [DOI] [PubMed] [Google Scholar]
- 28.Shingler V, Bartilson M, Moore T. Cloning and nucleotide sequence of the positive regulator, DmpR, of the phenol catabolic pathway of pVI150: a member of the NtrC family of transcriptional activators. J Bacteriol. 1993;175:1596–1604. doi: 10.1128/jb.175.6.1596-1604.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shingler V, Moore T. Sensing of aromatic compounds by the DmpR transcriptional activator of phenol-catabolizing Pseudomonas sp. strain CF600. J Bacteriol. 1994;176:1555–1560. doi: 10.1128/jb.176.6.1555-1560.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shingler V, Pavel H. Direct activation of the ATPase activity of the transcriptional activator DmpR by aromatic compounds. Mol Microbiol. 1995;17:505–513. doi: 10.1111/j.1365-2958.1995.mmi_17030505.x. [DOI] [PubMed] [Google Scholar]
- 31.Stemmer W P C. Rapid evolution of a protein in vitro by DNA shuffling. Nature. 1994;370:389–391. doi: 10.1038/370389a0. [DOI] [PubMed] [Google Scholar]
- 32.Walker J E, Saraste M, Runswick M J, Gay N J. Distantly related sequences in the α- and β-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Willardson B, Wilkins J, Rand T, Schupp J, Hill K, Keim P, Jackson P. Development and testing of a bacterial biosensor for toluene-based environmental contaminants. Appl Environ Microbiol. 1998;64:1006–1012. doi: 10.1128/aem.64.3.1006-1012.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]