SUMMARY
In December 2019, the severe acute respiratory syndrome 2 (SARS-CoV-2) coronavirus outbreak began in Wuhan, China, and quickly spread to practically every corner of the globe, killing millions of people. SARS-CoV-2 produced numerous variants, five of which have been identified as variants of concern (VOC) by the World Health Organization (WHO) (Alpha, Beta, Gamma, Delta, and Omicron). We conducted a comparative epidemiological analysis of SARS-CoV-2 and its VOC in this paper. We compared the effects of various spike (S) protein mutations in SARS-CoV-2 and its VOC on transmissibility, illness severity, hospitalization risk, fatality rate, immunological evasion, and vaccine efficacy in this review. We also looked into the clinical characteristics of patients infected with SARS-CoV-2 and its VOC.
Keywords: SARS-CoV-2, variants of concern, COVID-19, epidemiology, genetic mutation
INTRODUCTION
SARS-CoV-2 is a virus that engendered COVID-19, which evolved from the SARS-CoV wild-type virus. Several novel variations developed during the COVID-19 pandemic, some of which are known as Variants of Concern (VOC). VOCs are classified as Alpha (B.1.1.7), Beta (B.1.351), Gamma (P.1), Delta (B.1.617.2), and Omicron (B.1.1.529) by WHO [1]. These five SARS-CoV-2 VOCs have a substantial level of pathogenicity and transmissibility. New SARS-CoV-2 variants widely recognized as variations of interest (VOI) have been determined based on their transmissibility, severity of sickness, diagnostic escape, and immunological escape.
Over the outbreak of COVID-19, SARS-CoV-2 experiences genetic mutations in the virus’s S protein and RBD region, altering various characteristics of the virus, including transmission rate, pathogenicity, immune evasion, and testing failure, and vaccine efficacy. SARS-CoV-2, for example, which developed from the wild strain SARS-CoV by a single mutation (D614G), demonstrated enhanced transmission rate but decreased illness severity [2]. Furthermore, compared to SARS-CoV-2 and its VOC, the recent novel VOC Omicron contained many changes in the S and RBD area, resulting in a very high transmission rate and a relatively mild illness severity [3]. Fever, cough, myalgia (tiredness), diarrhea, sputum production, headache, dyspnea, rhinorrhea, nausea, sneezing, and sore throat are all symptoms of genetic alterations in the virus.
SARS-CoV-2 and its VOCs are the subjects of this narrative review. We found the information for this review by searching Google, Google Scholar, PubMed, and Scopus for phrases like SARS-CoV-2, Variants of Concern, COVID-19, Epidemiology of COVID-19, Genetic Mutation in SARS-CoV-2, COVID-19 vaccinations, and clinical characteristics of COVID-19.
We conducted a detailed comparative investigation of the epidemiology of SARS-CoV-2 and its VOC in this paper. We’ve also gone through the consequences of mutation on their biological roles in SARS-CoV-2 and VOC in great depth. Furthermore, we conducted a comparative investigation of clinical symptoms in SARS-CoV-2 and its VOC affected patients.
GENETIC MUTATIONS AND THEIR EFFECTS
SARS-CoV and SARS-CoV-2 are Beta-coronaviruses that comprise single-strand RNA. The analysis indicated that coronavirus has the biggest genome of all RNA viruses and has a high G and C concentration. Spike (S), membrane (M), envelope (E), and nucleocapsid (N) proteins are the four types of structural proteins found in them. The coronavirus produces various variants and sub-variants as a result of genetic mutation. We’ve covered the most crucial aspects of VOC in this section.
SARS-CoV-2
SARS-CoV-2 corresponds to the beta-coronavirus 2B lineage [4]. It differs from SARS-CoV in that it shares 79.5% of its sequence with this virus. Scientists ascertained the genetic sequences of multiple coronaviruses to SARS-CoV-2 and deduced that they are 96% identical to BatCov RaTG13. Based on these findings, several researchers hypothesized that SARS-CoV-2 is a bat-borne virus that spontaneously evolved from the coronavirus strain BatCov RaTG13. SARS-CoV-2 has a spherical form and a genomic size of 29.9 kb. It contains single-strand positive RNA (+ssRNA) [5]. The ORF1ab polyprotein encoding 16 non-structural protein (nsp1)16 is found at the 5 end of this genome, accounting for around 66.6% of the genome. The three end sides, on the other hand, encode all four critical proteins: the S protein, the nucleocapsid (N) protein, the membrane (M) protein, and the envelope (E) protein [6]. The genome is encircled by capsid, which is made up of N protein, and the entire thing is further coated by three structural proteins, M, S, and E (Figure 1) [6].
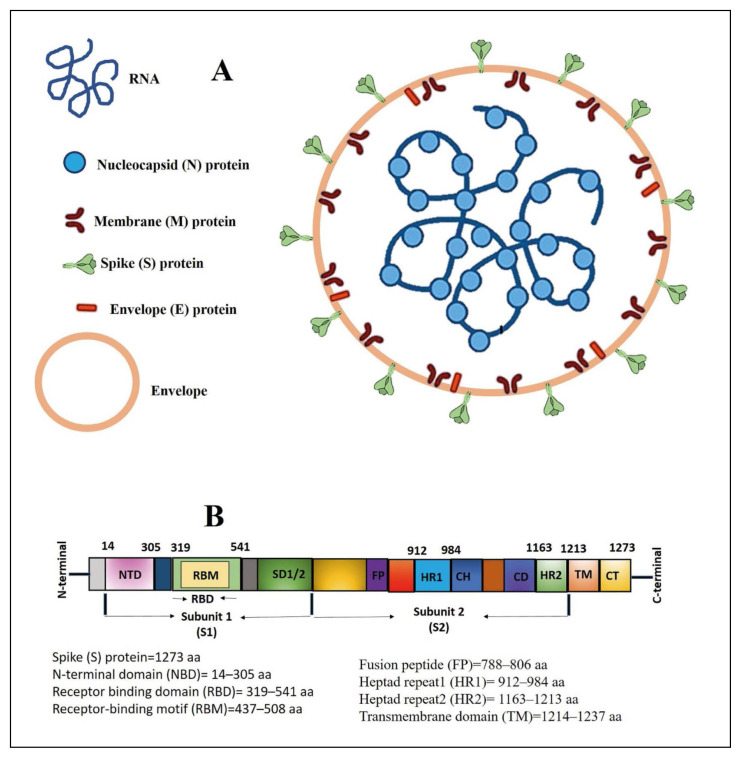
Figure 1.
An illustration showing different proteins, RNA and envelope of the SARS-CoV-2 (Panel A) and the various functional domains in the S protein (Panel B).
SARS-CoV-2 S is a transmembrane glycoprotein that has been leveraged in the development of vaccines and antiviral therapeutics. S glycoprotein is a homotrimer that aids in viral attachment and cellular internalization into host cells [7]. HR1 and HR2 combine to create a 6-helical bundle (6-HB) that aids membrane fusing. The RBD (Arg-319Phe541) binds to the human ACE2 receptor and facilitates entrance into host cells [8]. The RBD has a significant number of binding sites for human ACE2 receptors in a section known as the receptor-binding motif (RBM, (437–508 aa)) (Figure 1) [9]. The furin cleavage site, which is missing in SARS-CoV and is placed between S1 and S2, has four essential amino acids, the PRRA sequence (P681, R682, R683, and A684), which aid viral entrance into host cells and is one of the reasons for SARS-high CoV-2’s virulence [10]. They serve a decisive part in the replication and pathogenesis of SARS-CoV-2 [11].
Notably in the literature SARS-CoV-2 (Wuhan strain) has only one mutation, D614G, which is placed on the (Carboxy) C-end of the S1 protein (Table 1) [2]. The aspartic acid amino acid was substituted by glycine in this single mutation, which altered the behavior of SARS-CoV-2 in a variety of ways [2]. SARS-CoV-2 transmission, for example, has increased several times, and viruses have been observed to contain significant viral loads and infectivity in many COVID-19 patients, although the current evidence shows that it does not affect disease severity [12].
Table 1.
Genetic mutations in the S protein of SARS-CoV-2 and its variants of concern (VOC), as well as their consequences.
| SARS-CoV2 (Wuhan strain) | Alpha variant | Beta | Gamma | Delta | Omicron | References | |
|---|---|---|---|---|---|---|---|
| Lineage | B.1.1.7 | B.1.351 | P.1 | B.1.617.2 | B.1.1.529 | [1, 5, 6, 20, 28, 32, 38, 45, 61, 62, 64] | |
| Total mutations in spike protein | 1(D614G) | 9 (ΔH69-V70 deletion, ΔY144 deletion, N501Y, A570D, D614G, P681H, T716I, S982A, D1118H) | 10 (L18F, D80A, D215G, Δ242-244, R246I, K417N, E484K, N501Y, D614G, and A701V) | 12 (L18F, T20N, P26S, D138Y, R190S, K417T, E484K, N501Y, D614G, H655Y, T1027I, and V1176F) | 17 (T19R, V70F, G142D, 156del, 157del, R158G, Δ213-214, A222V, W258L, K417N, L452R, T478K, E484Q, D614G, P681R, D950N, T95I) | 39 (A67V, H69del, V70del, T95I, G142del, V143del, Y144del, Y145D, N211del, L212I, ins214E, ins215P, ins216E, G339D, S371L, S373P, S375F, K417N, N440K, G446S, S477N, T478K, E484A, Q493R, G496S, Q498R, N501Y, Y505H, T547K, D614G, H655Y, N679K, P681H, N764K, D796Y, N856K, Q954H, N969K, L981F | [2, 3, 8–10, 20–22, 32, 33, 37, 38, 45] |
| Mutations in NTD (14–305 aa) | 0 | 2 (Δ69-70 deletion, Δ144 deletion) | 5 (L18F, D80A, D215G, Δ242-244. R246I) | 5 (L18F, T20N, P26S, D138Y, R190S) | 9 (T19R, V700F, G142D, 156del, 157del, R158G, A222V, W258L, Δ213-214) | 13 (A67V, H69del, V70del, T95I, G142del, V143del, Y144del, Y145D, N211del, L212I, ins214E, ins215P, ins216E) | [2, 3, 5, 6, 8–10, 20–22, 32, 33, 37, 38, 45] |
| Mutation in RBD region (319–541 aa) | 0 | 1 (N501Y) | 3 (K417, E484K, N501Y) | 3 (K417T, E484K, N501Y) | 4 (K417N, L452R, T478K, E484K) | 15 (G339D, S371L, S373P, S375F, K417N, N440K, G446S, S477N, T478K, E484A, Q493R, G496S, Q498R, N501Y, Y505H,) | [2, 3, 8, 9, 22, 28, 37, 38, 45] |
| Common mutations in NTD (14–305aa) | – | Δ69–70 deletion, Δ144 deletion | L18F | L18F | – | H69del, V70del, Y144del | [2, 3, 5, 6, 8–10, 20–22, 32, 33, 37, 38, 45] |
| Common Mutation in RBD region (319–541 aa) | – | N501Y | N501Y, E484K | K417N, E484K N501Y, | K417N, T478K, E484K | K417N. T478K, E484K, N501Y | [2, 3, 8, 9, 22, 28, 37, 38, 45] |
| Mutations in FCS (681PRRAR 685) | – | P681H | – | – | P681H | P681H | [10, 11, 33, 37, 38, 45] |
| Binding affinity with human ACE2 | Increased | Increased | Increased | Increased | Increased | Decreased | [8, 22, 32, 33, 34, 39, 40] |
| Impact on transmissibility | Increased | Increased | Increased | Increased | Increased | Increased | [24, 25, 30, 32, 33, 62, 63,69] |
| Disease severity | Not increased compared to SARS-CoV | Increased | Increased | Increased | Increased | Reduced | [12, 34, 38, 46, 47, 66, 69] |
| Impact on infectivity | Increased compared to SARS-CoV | Increased | Increased | Increased | Increased | Increased | [34, 38, 46, 47, 66, 69] |
| Hospitalization Risk | Increased | Increased | Increased | Increased | Increased | Reduced | [28, 29, 35, 61, 64, 66, 69] |
| Mortality rate | Increased | Increased | Increased | Increased | Increased | Reduced | [28, 29, 61, 64, 66, 69] |
| Testing kit failure | – | Yes | No | – | Yes | Yes | [2, 23, 29, 60] |
| Immune escape | Yes | Yes | Yes | Yes | Yes | [1, 23, 34, 41, 51] | |
| Impact on effectiveness of vaccine | 96% (Pfize- BioNTech) | No significant changes | No significant changes | No significant changes | No significant changes | Little change | [13, 14, 15, 16, 17, 18, 19, 26, 27, 36, 38, 42, 43, 44, 48–50] |
Gram et al. in Denmark recently found that after 14 days, vaccination efficiency was 88% after taking ChAdOx1 nCov-19 (AstraZeneca) as the first dose and mRNA vaccine as the subsequent dose [13]. According to research conducted by Chadeau-Hyam et al. in England, vaccination effectiveness against COVID-19 infection after the second dosage was 44.8%, 71.3%, and 75.1% for AstraZeneca, Pfizer–BioNTech vaccine, and Moderna vaccine, respectively [14]. After the second dosage, according to a study conducted in Qatar, the Pfizer-BioNTech vaccine had 96% effectiveness against any severe, fatal, or critical instances of COVID-19 infection [15]. The vaccine efficacy of the Pfizer-BioNTech vaccine was also tested in children aged 5 to 11, and it was observed that two doses of vaccination had a 90.7% vaccine efficacy [16]. In a US health-care trial, Pfizer-BioNTech vaccine and Moderna vaccine exhibited 77.6% and 88.9% vaccine efficacy, respectively, after partial vaccination. Furthermore, following full vaccination, vaccine effectiveness for the Pfizer-BioNTech vaccine and the Moderna vaccine was 88.8% and 96.3%, respectively [17]. The Pfizer-BioNTech vaccine’s booster dosage (third dose) is critical for protecting against COVID-19 illness. This vaccine has a 93% vaccination effectiveness for hospital admission, 92% for severe illness, and 81% for mortality due to COVID-19 after a booster dose [18]. Furthermore, another study showed that people with COVID-19 benefit from the third dosage. The vaccination effectiveness was 92.9% crude and 89.1% adjusted, according to the data [19].
Alpha variant (B.1.1.7)
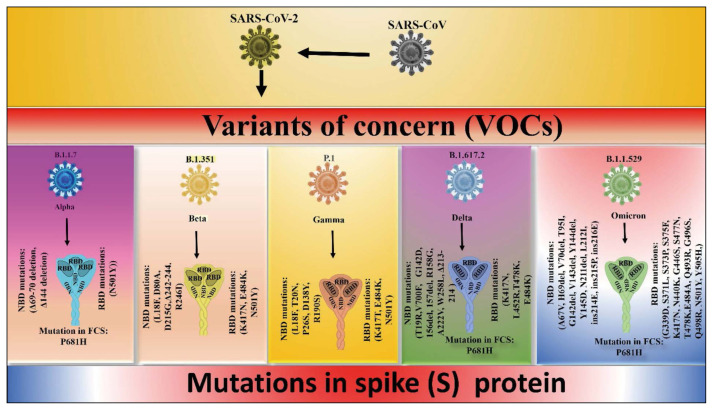
It was originally recognized in the UK on September 20, 2020, and swiftly spread to other parts of the entire planet. The WHO classified it as a SARS-CoV-2 VOC. SARS-CoV-2 contains just one mutation (D614G mutation), however, the alpha version has a total of 23 mutations [20] (Figure 2). According to published data, 14 of these mutations are non-synonymous, whereas the remainder is synonymous [20]. Non-synonymous substitutions are nucleotide mutations that change the amino acid sequence of a protein. A synonymous substitution, on the other hand, is a nucleotide replacement that does not modify the amino acid in the protein. Alterations in the S protein of the virus account for more than 47% of the changes in the alpha form. The findings revealed that the S region of the viral protein has around 9 mutations [20, 21]. The most important mutations in this variation are N501Y, P681H, and D614G, which alter biological processes (Table 1) [20]. The N501Y mutation in the RBD region boosts viral binding to the ACE2 receptor of host cells, whereas the P681H mutation increases virus transmission rate [22]. Another relevant mutation is H69-V70del, which causes immune escape and failure of test kits in this version [23].
Figure 2.
A diagram showing the different genetic mutation in SARS-CoV-2 and its variants of concern (VOC).
According to the literature, this virus has a transmission rate of 40–50% and a faster development rate than the parent virus SARS-CoV-2 [24]. In the case of this variety, there is a substantial danger of death [25]. In this version, the probability of hospitalization and the financial burden on the healthcare system rose. According to the findings, there isn’t much of an influence on vaccine effectiveness against this variation. The vaccine effectiveness of the second dosage of BNT162b2 (Pfizer-BioNTech) and AstraZeneca against this variation was 93.7% and 74.5%, respectively [26]. Another study found that the Moderna vaccine effectiveness in this form was 100% after the second dosage [15]. Furthermore, according to Elisabeth Mahase’s research, Novavax vaccination efficacy against this variation was 86% [27].
Beta variant (B.1.351)
On December 18, 2020, the beta form was identified in South Africa for the first time, and it quickly spread to other regions of the country. The beta version of SARS-CoV-2 has 12 non-synonymous and one deletion gene in contrast to SARS-CoV-2 (Figure 2). The beta variant has nine mutations in the S protein, accounting for around 77% of the total alterations, three of which are in the RBD area (K417N, E484K, and N501Y). E484K and K417N mutations boost viral binding to human ACE2, comparable to the N501Y mutation discovered in the alpha form (Table 1). According to the findings, the mutation increases the likelihood of hospitalization and ICU admission [28]. Arena et al. found that these mutations did not influence the RT-PCR test used to diagnose COVID-19 illness in patients who were infected with the beta version [29]. According to one study, the beta variation was 50% more transmissible than the parent virus [30].
Several vaccines were shown to be effective against this variation. In verified beta variant infected patients, the second dose of BNT162b2 (Pfizer-BioNTech) showed 72% vaccine effectiveness and 100% vaccine effectiveness against symptomatic patients [31]. Chemaitelly et al. found that the Moderna vaccine had vaccine effectiveness of 96.4% against the beta variant [15].
Gamma variant (P.1 or B.1.1.28.1)
It was identified in Brazil in December 2020, and it has 17 non-synonymous mutations and 4 synonymous mutations [32]. The S protein has a profound number of mutations (Figure 2). The mutation N501Y is found in all three variations, while the Beta variants have mutations L18F, K417T, E484K, and D614G (Table 1). Three mutations have been discovered in the RBD area. These mutations improved the binding affinity of human ACE2 as well as the transmission rate [32, 33]. Immune escape, viral load, and virus reinfection are all increased by the mutation [34]. According to the statistics, this variation causes a rapid increase in cases in Brazil, which leads to higher hospitalization, infectiousness, and illness severity [35]. In patients infected with the gamma form, the Moderna COVID-19 vaccine effectiveness was shown to be 77% [36].
Delta variant (B.1.617.2)
In October 2020, the delta version was discovered in the Indian state of Maharashtra, and it quickly spread to other areas of the world. It possesses 17 mutations, including four changes in the RBD of the S protein region (K417N, L452R, T478K, E484K) and two amino acid deletions in the S1 subunit of S protein (E156del, F157del) [37] (Figure 2 and Table 1). The D614G mutation, in which aspartic acid is substituted by glycine at the C-terminus and was also discovered in other variations such as Alpha, Beta, and Gamma, improves the virus’s infectivity [38]. Another key mutation, L452R, improves the virus’s binding to the ACE2 receptor, increasing the rate of transmission [39,40]. The increasing data suggested that these changes are responsible for the virus variants immunological escape [41].
According to Bruxvoort et al., the (mRNA-1273) Moderna COVID-19 vaccine was 86.7% effective in patients infected with the delta version and 98.4% effective in patients infected with the alpha form [42]. The other research found that the Pfizer-BioNTech COVID-19 vaccination (BNT162b2) and AstraZeneca (Chadox1n Cov-19) vaccines were effective in symptomatic patients infected with the delta form at 88% and 67%, respectively [43]. Bernal et al found that after two doses of the AstraZeneca vaccine, the effectiveness was 67.0% and 74.5% in alpha and delta patients, respectively [26]. Furthermore, the Pfizer-BioNTech vaccine demonstrated 88.0% and 93.7% vaccine effectiveness in alpha and beta infected patients after two doses, respectively [26]. In delta variant infected patients, the BBV152 (Covaxin) vaccine achieved 65.2% vaccine effectiveness [44]. Furthermore, Desai et al. found that Covaxin had roughly 50% vaccination efficacy in symptomatic delta variant patients in another research [38].
Omicron (B.1.1.529)
Most scientists and researchers are worried all over the world because of the high number of mutations in the S proteins of the omicron variant. Kim et al. earlier revealed that the omicron variation has roughly 39 mutations in the S protein, 27 of which are novel alterations. Among 39 mutations, 15 mutations were found in the RBD region, which is about 38% of the total mutations of the omicron variant [45] (Figure 2). In the case of the delta variant, the total number of the mutation in the RBD region is only 4, which is much less as compared to the omicron variant. There are about 10 mutations in the Receptor Binding Motif which is a part of the RBD region, and this part interacts with the host ACE2 [3]. The frequency of the mutation in the RBM of S protein is 760 times higher than the whole S gene and mutation in RBM of Omicron is 5 times more as compared to the delta variant [45]. The total amino acids in the S proteins of the SARS-CoV-2 variant is 1273, while in omicron 3 amino acids are less, which is 1270. As shown in Figure-1 RBD region is RBD 319–545, RBM, 438–507, and NTD, is 18–305.
According to certain research, the Omicron variant exhibited a reduced capacity to bind to the ACE2 receptor than the wild-type SARS-CoV-2. Two key mutations in the RBD region of this variation, Q493K and Q498R, diminish the binding affinity with the ACE2 receptor, whereas the N501Y mutation enhances it [3]. H655Y and N679K mutations near the furin cleavage site speed up S cleavage and make the virus more infectious. By enhancing S protein cleavage, one significant mutation, P681H, promotes transmissibility. These alterations resulted in a greater rate of reinfection in previously infected SARS-CoV-2 patients, indicating that the virus is more transmissible [46]. Based on another investigation, the Omicron variant had 13-fold the viral infectivity of the delta version and was 2.8-fold more infectious. According to Chen et al, the Omicron form may evade immunization twice as often as the Delta version [47].
One crucial concern is whether the currently existing vaccines, such as AstraZeneca’s COVID-19, Pfizer’s BioNTech, and Moderna’s COVID-19, are effective against Omicron-infected patients. Some preliminary research suggests that Omicron may evade the immune system and have lower vaccination effectiveness than other variations. Muik et al. found that after two doses of the Pfizer-BioNTech COVID-19 vaccine (BNT162b2), sera exhibited more than 22-fold lower neutralizing titers against Omicron as compared to Wuhan pseudovirus [48]. When compared to two doses, omicron-neutralizing titers were 23-fold higher one month following the third immunization, with titers equivalent to Wuhan-neutralizing titers after two doses [48]. Another study stated that immunization effectiveness was 70% after another dose of Pfizer-BioNTech vaccine during the Proxy Omicron Period and 93% against hospitalization for COVID-19 during the comparator period [49]. Furthermore, another research found that following primary immunization, the vaccine effectiveness of the Pfizer–BioNTech and Moderna vaccines was 55.2% and 36.7%, respectively. We currently lack data or evidence to support the effectiveness of other important vaccines [50].
Many Omicron sub-lineages are currently accessible in numerous countries, including BA.1, BA.1.1, BA.2, BA.4, and BA.5. Evans et al. found that the sub-lineages BA.1 and BA.1.1 successfully evade the immune system, which can be overcome by administering a COVID-19 vaccination booster dosage [51]. BA.3 has around 33 mutations, 31 of which are shared with BA.1 and the remaining two with BA.2 (2). Because it lacks several critical alterations, BA.3 spreads more slowly than BA.1. The BA.4 and BA.5 viruses were originally discovered in South Africa in 2022, and subsequently spread around the world. These two lineages feature important mutations in the RBD area, such as L452R, F486V, and R493Q, which allow them to evade the immune system. On the 8th of May 2022, 37 percent of cases in Portugal belonged to BA.5, and it is projected that this sublineage would become dominant on the 22nd of May 2022. (https://www.ecdc.europa.eu/en/news-events/epidemiological-update-sars-cov-2-omicron-sub-lineages-ba4-and-ba5). The data in this research is only up to May 2022, and it is projected that many additional subvariants will arise in the days following May 2022.
EPIDEMIOLOGY OF SARS-CoV-2 AND ITS VARIANTS OF CONCERN
COVID-19 is a pandemic illness necessitated by the SARS-CoV-2 virus, which the WHO recognized as a pandemic on March 11, 2020 [52]. The first COVID-19 case was discerned in Wuhan, Hubei Province, China, in the latter weeks of December 2019, and it quickly spread to practically all countries [53]. The WHO coined the acronym COVID-19, which stands for Corona Virus Disease 2019, in February 2020. On May 22, 2020, the total number of cases and deaths, according to WHO, were 4,995,996 and 327,821, respectively [54]. According to John Hopkins, the total number of cases globally was 374.75 million as of January 31, 2022, with 5.67 million deaths (https://coronavirus.jhu.edu/).
SARS-CoV-2 disseminated more swiftly than SARS-CoV and the Middle East respiratory coronavirus. The initial instance of SARS-CoV-2 transmission into humans is thought to have been from an animal to a person, followed by local transmission and community transmission. As we know, the initial case was reported from a seafood market in Wuhan, China, therefore its source is likely to be zoonotic [55] (Table 2). SARS-CoV-2 can be conveyed from individual to individual by direct touch, droplets, coughing, sneezing, or conversing with an infected patient [56]. The research also revealed that it may transmit via the air by inhaling respiratory droplets from an infected individual and that the virus can survive for a long period in the air. Furthermore, there is substantial evidence that viruses may enter our bodies via aerosol through breathing [57]. One of the most important modes of SARS-CoV-2 transmission is through the lungs. According to some accounts, it can also be transmitted by contacting contaminated surfaces. This virus is also identified in stool and saliva, indicating that transmission by feces and saliva is feasible, although no cases of transmission via feces and saliva have been documented so far [56]. Because the coronavirus may be found in water and sewage, it is assumed that it can be transferred through these mediums as well. However, there is very little evidence to support this assumption [58]. Although the SARS-CoV-2 strain has been detected in certain animals, there is currently insufficient data to suggest that SARS-CoV-2 transmission through fomites, household pets, and agricultural animals is conceivable [59]. Although viral RNA has been identified from sperm and blood donations, no cases of SARS-CoV-2 transmission have been described in the literature due to sexual or foodborne transmission [56]. According to a meta-analysis conducted by Kotlyar, 2.3% of newborns from COVID-19 infected women showed positive SARS-CoV-2 strain verified by RT-PCR testing, showing that vertical transmission is unlikely [60].
Table 2.
This table depicted an epidemiological investigation of SARS-CoV2 and its variations of concern (VOC).
| SARS-CoV-2 (Wuhan strain) | Alpha variant | Beta | Gamma | Delta | Omicron | References | |
|---|---|---|---|---|---|---|---|
| Lineage | – | B.1.1.7 | B.1.351 | P.1 | B.1.617.2 | B.1.1.529 | WHO website |
| Country first detected | China | United Kingdom | South Africa | Brazil, | India | Multiple countries | WHO website |
| Year and month first detected | December, 2019 | September, 2020 | May, 2020 | November, 2020 | October, 2020 | November, 2020 | WHO website |
| Mean R0 | 3.28 | 2.27 | – | – | 5.08 | – | [2, 53, 62, 70–73] |
| % Re or Rt relative to non-VOC | – | 29% | 25% | 38% | 97% | – | [73] |
| Mean Incubation time in days | 5 days | 3 | 5 days | 5 days | 4 days | 4 days | [74–83] |
| % Hospitalization | 67.8% | 11.0% | 19.3%/20%/69.3% | 20.0% | 27.3% | 41.3% | [61, 66, 69] |
| % ICU admission | 42% | 1.4% | 2.3%/3.1%/36% | 2.1% | 4.9% | 18.5% | [61, 66, 69] |
| % Death | 19.7% | 2.0% | 5.2%/0.2%/25.5% | 3.9% | 0.7% | 2.7% | [61, 66, 69] |
| Fully vaccinated patients (After 14 days of second dose) | |||||||
| % Hospitalization | – | – | Decreased16.3% | – | Decreased (12.1%) | – | [64, 66] |
| % ICU admission | – | – | Decreased 0.0% | – | Decreased 1.1% | – | [64, 66] |
| % Death | – | – | Decreased 0.0% | – | Decreased 0.0% | – | [64, 66] |
| Fully vaccinated patients | |||||||
| Transmission | Decreased | Decreased | Decreased | – | Decreased | – | [84–93] |
In September 2020, lineage B.1.1.7 (‘UK variation’), also known as the Alpha variant, was discovered as a novel VOC in the United Kingdom (UK) (Table 2). It later spread throughout Europe and the rest of the world. According to a surveillance investigation, the Alpha variant was present in Belgium at 7.1% from January 4 to January 10, 2021, and at 90.3% from May 3 to May 16, 2021 [61]. When compared to other variations, alpha variants have a transmissibility rate of more than 75%, according to research by Leung et al. [62]. Another study found that alpha variants have a 45% to 71% higher probability of transmission than non-VOCs [63]. The secondary was found to be 1.31 times greater than other non-VOC instances in research conducted in Canada [28]. According to the ECDC, alpha variations had a higher percentage of hospitalization, ICU (Intensive Care Unit) admission, and mortality than non-VOCs at 11.0%, 1.4%, and 2.0%, respectively [61]. The findings revealed that in the alpha version, hospitalization cases at a younger age (mean age: 63 years) are greater than in non-VOCs (mean age: 69 years) [61].
In May 2020, the beta form (B.1.351) was discovered for the first time in South Africa. Later, it spread to about 98 nations. It began in Nelson Mandela Bay, South Africa, and has since spread to practically every European country [64]. According to an article published by Nature Journal in 2021, the effective reproduction number in South Africa went over 1 at the end of October, and the number of cases was thirteen thousand per week and seven thousand fatalities per week in the middle of July [65]. When compared to the alpha variation, the beta variant had a greater percentage of hospitalizations (19.3%) [61]. Also, as compared to the alpha form, the percentage of ICU admission in hospitals and mortality was 2.3% and 5.2%, respectively [61]. According to another study conducted by Abu-Raddad and his research group, the percentage of beta variant hospitalizations and ICU admissions was 20% and 3.1%, respectively [66]. According to research conducted in Qatar, the percentages of hospitalization, ICU admission, and mortality for beta variants were 20%, 3.%, and 0.2%, respectively [66]. Hospitalization (16.3%), ICU admission (0.0%), and mortality (0.0%) were all dramatically decreased after 14 days after getting the second dose of immunization, according to the same study group [64].
According to the World Health Organization, the gamma variety was first discovered in Brazil in November of 2020 [67]. In November 2020, there were 75.5 cases per 100,000 people, which increased to 397 cases per 100,000 people in February 2021 in Brazil [67]. The percentage of hospitalizations in the gamma variation (P.1) was 20.0%, which is greater than the Alpha and beta variants. The gamma variation had a greater rate of ICU admissions than the alpha variant, at 2.1% [61]. However, the fatality rate was 3.9%, which was practically identical to the beta variation [61]. The gamma variation’s transmissibility rate was 2.6 times greater than the preceding version, indicating that it was more transmissible than the alpha and beta variants [68]. According to a study published by Faria et al., the gamma variation has 1.4–2.2 times greater transmission than non-gamma lineages. Hospitalization, ICU admission, and fatality rates were 20%, 2.1%, and 3.9%, respectively, according to the statistics [61].
According to a study published in JAMA Internal Medicine, the percentages of hospitalization, ICU admission, and mortality for the delta variation in Qatar were 27.3%, 4.9%, and 0.7%, respectively [66]. Furthermore, for the delta version, hospitalization, ICU admission, and death were 12.1%, 1.1%, and 0.0% after 14 days after second dose immunization, respectively [66].
According to a recent study, the omicron version resulted in a considerable reduction in hospitalization and ICU admissions. The hospitalization, ICU admission, and death rates in the first wave (SARS-CoV-2) were 67.8%, 42 %, and 19.7%, respectively, according to this study done in South Africa [69]. Hospitalization, ICU admission, and death were found to be 69%, 36%, and 25.5%, respectively, during the second wave (Beta variation). Furthermore, the hospitalization, ICU admission, and mortality rates were 69.3%, 29.9%, and 29.1%, respectively, during the third wave (delta variation). However, there was a significant reduction in hospitalization, ICU admission, and mortality with the fourth wave (Omicron version) (November 15 to December 7, 2021), which were 41.3%, 18.5%, and 2.7%, respectively [69]. SARS-CoV-2 N501Y mutant strains early transmissibility evaluation in the UK, October to November 2020.
Coronavirus transmissibility is determined by the basic reproduction number (R0), which represents the number of new infections in an epidemic induced by the original case in a community [70]. To put it another way, it’s the average number of secondary transmissions from a single infected individual. The effective reproduction number (Rt or Re) is the number of infected people in a partially susceptible population that may be infected by a single ill individual at any given time. In other words, the effective reproduction number (Rt,) predicts the likelihood of an epidemic spreading at a given period. The R0 is critical for reducing coronavirus transmission and assessing the efficiency of public health initiatives [71]. An R0 value of less than one implies that coronavirus transmission or new infections are diminishing, and the outbreak will be over shortly. Furthermore, if R0 is greater than 1, infections are more likely to proliferate, making it difficult to control a pandemic/epidemic in this condition. The R0 value for SARS-CoV-2 pertaining to the WHO is 1.4 to 2.5, with an average of 1.95, although according to another study, the mean R0 value for SARS-CoV-2 is 3.28 [72]. All five VOCs (Alpha, Beta, Gamma, Delta and Omicron) demonstrated a better Rt than SARS-CoV-2. According to the research, Delta variant Rt is extremely high when compared to other VOCs. Campbell et al found that the mean Rt of VOCs was 29%, 25%, 38%, and 97% for Alpha, Beta, gamma, and delta, respectively [73]. However, the Rt value of the omicron version is unknown at this time. According to certain research, the R0 of the alpha variant was 75–78%, and the Rt was 1.1–2.8 [62]. According to research done by Pan et in Wuhan, China, the Rt value was 3.82 during the early stages of the outbreak (January 24, 2020). When severe public health actions were implemented, the cases began to decline, and Rt reached 1.0 (February 6, 2020), before falling further in the following days to less than 0.3 (March 1, 2020) [70, 71].
The incubation period is defined as the interval between exposure and the development of symptoms. SARS-CoV-2 has a median incubation time of 5.7 days, with some studies claiming it to be 5.4 days (range 2–14) days [74, 75]. According to Dhouib et al’s meta-analysis, the average incubation period was 6.2 days [76]. According to Feng and his colleagues, the average incubation time of SARS-CoV-2 was 5.2 days [77]. Severe acute respiratory syndrome (SARS) had a mean incubation time of 5 days (range 2–10 days) in Canada in 2003, according to studies [78]. According to certain research, the mean incubation time of MERS-CoV in South Korea (6.9 days) was longer than in Saudi Arabia (5.0 days), and the overall mean incubation period was 5–7 days (range, 2–14 days) [79, 80]. In comparison to the wild strain (5 days), the average incubation time of the alpha variant was shorter (3 days) [81]. According to Grant et al. in 2021, the incubation period for the delta variation was about 4 days, whereas the incubation period for the beta and gamma variants was around 5 days [82]. In the S gene target failure (SGTF) instances involving the omicron variation (BA.1), the average incubation duration was around 3.2 days [83].
Several investigations were conducted to examine how vaccinations affected SARS-CoV-2 and VOC transmission. Many studies in the literature have found that viral loads in vaccinated patients are lower (high Ct value) than in unprotected ones. According to research by Hsu et al., the rate of transmission in vaccinated patients was three times lower than in unprotected patients [84]. They discovered that Ct values in vaccinated patients were much greater than in unvaccinated people. According to recent data, virus loads in fully vaccinated persons were lower than in unvaccinated people, indicating a lower transmission rate in vaccinated patients [85]. Two doses of the Pfizer vaccine, according to research done in Israel, dramatically limit transmission among household contacts [86]. In a separate trial conducted in England, home transmissions were decreased by 40 to 50% in vaccinated patients [87]. The second research was conducted in Denmark, which found a 42% decrease in transmission of the delta variation [88]. More data advocates that immunization lowers the rate of COVID-19 transmission in patients. The Ct value was high after 12–37 days after the first dose of vaccination, indicating a decreased virus load and transmission rate [89]. From August to September 2021, research in the Netherlands found that two doses of vaccination were effective against transmission by 40% [90]. A study conducted by Eyre et al. showed that vaccination significantly reduced the transmission of alpha and beta variants [91]. The findings revealed that index patients with alpha and beta variants injected with the second dose vaccination had higher Ct values (lower viral load), indicating a lower transmission rate [91]. However, as compared to the alpha form, the delta variant had a lower Ct value, indicating that it had a greater transmission rate [91].
CLINICAL FEATURES OF COVID-19 AND ITS VARIANTS OF CONCERN
COVID-19 symptoms began to manifest after incubation period of the virus. Research conducted by Huang et al, which was published in the Lancet journal in 2020, indicated that the majority of patients who had COVID-19 were men (73%), with the remainder being female [92]. The total number of patients in this research, both male and female, was 41. The most prevalent symptoms of COVID-19 are fever (98%), cough (76%), and myalgia or tiredness (44%). Sputum production (28%) was the least prevalent symptom, followed by headache (8%), hemoptysis (5%), and diarrhea (3%). About 55% of the patients exhibited dyspnea, and 63% of the patients had lymphopenia. The chest CT revealed pneumonia in every single one of the 41 patients (100%). Hypoxemia is some of the additional symptoms. Many respiratory-related clinical symptoms, such as rhinorrhea, nausea, sneezing, sore throat, acute cardiac damage, and acute respiratory distress syndrome, were also observed with COVID-19 infection [93]. A 33-year-old woman’s CT scan revealed several ground-glass opacities in both lungs [92]. COVID-19 individuals experienced leucopenia and lymphopenia, according to blood tests. The D-dimer and prothrombin time of patients admitted to the ICU were greater. Furthermore, aspartate aminotransferase and troponin I (hs-cTnI) levels were elevated in roughly 37% and 12% of patients, respectively [92].
Another research with a larger sample size found that patients with wild-type COVID-19 had cough, fever, and exhaustion in 83%, 81%, and 38% of cases, respectively [94]. Furthermore, expectoration, chest discomfort, diarrhea, dyspnea, stomach pain, and vomiting were reported in 17%, 19%, 14%, 14%, 13%, and 11% of patients, respectively. In this investigation, however, no patients with COVID-19 wild type were identified to have a throat infection. Cough, fever, and weariness were seen in 50%, 36%, and 23% of delta-infected patients respectively. Expectoration, sore throat, chest discomfort, diarrhea, dyspnea, stomach pain, and vomiting were reported in approximately 12%, 25%, 2%, 12%, 25%, 2%, 7%, 1%, 1%, and 1% of patients infected with the delta variant, respectively [94].
In Singapore, 829 patients took part in research to learn more about the clinical aspects of SARS-CoV-2 and their variants of concerns [95]. According to the statistics, patients with SARS-CoV-2 infection had a fever, cough, dyspnea, sore throat, and nasal congestion/or rhinorrhea in 69%, 64%, 11%, 41%, and 32%, respectively. In addition, approximately 38% developed pneumonia, 11% required additional oxygen, 6% required ICU hospitalization, and 1% of SARS-CoV-2 infected patients died. Fever, cough, dyspnea, sore throat, nasal congestion, and/or rhinorrhea were reported by 58%, 49%, 5%, 26%, and 19% of Alpha variant infected individuals, respectively. Furthermore, around 16% got pneumonia, 5% required additional oxygen, 4% required ICU hospitalization, and no Alpha-infected patients died [95]. Fever, cough, dyspnea, sore throat, nasal congestion, and/or rhinorrhea were seen in 42%, 30%, 3%, 18%, and 18% of Beta variant infected patients, respectively [95]. In addition, roughly 9% of patients got pneumonia, 3% required additional oxygen, 0% required ICU care, and none of the Alpha-infected patients died. Fever, cough, dyspnea, sore throat, nasal congestion, and/or rhinorrhea were seen in 72%, 46%, 19%, 34%, and 16% of Delta variant infected patients, respectively [95]. In addition, almost 49% got pneumonia, 28% required additional oxygen, 4% required ICU hospitalization, and 1% of Delta-infected patients died [95].
The researchers conducted a retrospective comparative study on 180 Egyptian COVID-19 patients to see if there was a link between laboratory findings and illness severity and mortality risk. Patients with COVID-19 who have dyspnea have a greater risk of illness or death [96]. COVID-19 individuals with COPD and diabetes had an increased chance of COVID-19 severity. Further research conducted by Fouad et al revealed that the COVID-19 patients admitted to ICU were primarily elderly, smokers, hypertensive, and diabetic [97].
According to a study conducted by Costa and his research group, gamma-infected patients had 75% coryza, 74% headache, 73% cough, 54% sore throat, 51% myalgia, 44% asthenia, 30% fever, 25% hyposmia/anosmia, 21% dysgeusia, 20% gastrointestinal tract symptoms, and 10% dyspnea. Furthermore, among gamma-infected patients, 2% were admitted to the hospital, 1% are hospitalized in the ICU, and 1% died [98].
Kim et al. studied the clinical aspects of omicron variations in 2022, finding that 47.5% of the 40 patients were asymptomatic and 52.5% were symptomatic [99]. Sore throat (25%), fever (20%), headache (15%), cough (12.5%), sputum (12.5%), runny nose/coryza (10%), myalgia (5%), fatigue/weakness (2.5%), and loss of smell were among the omicron patients symptoms (2.5%). The results of the chest CT imaging revealed that roughly 85% of the patients had no involvement, indicating that the infection was largely lower respiratory infections, while the remaining 15% had lung infiltrations. The majority of omicron patients were asymptomatic or had minor symptoms and did not require oxygen. Another research found no difference in illness severity or mortality in omicron patients. Furthermore, anosmia/ageusia symptoms were seen in 1.3% of the patients [100].
CONCLUSIONS
Because of various mutations in the S protein and RBD area, the omicron has recently expanded to all regions of the world, with decreased severity but a high transmission rate. Omicron BA.1 and BA.2 are two sublineages of Omicron that vary from one another owing to a genetic mutation. The sublineage BA.2 is the most frequent Omicron sublineage today, and it is more transmissible than BA.1. We think that the current comparative investigation of the epidemiology, genetic alterations, and clinical manifestations of SARS-CoV-2, VOC, and their sublineage will undoubtedly aid in the control of coronavirus transmission, diagnostic testing, prevention, and treatment of infected patients.
Acknowledgements
The author would like to thank the Deanship of Scientific Research at Shaqra University for supporting this work.
Footnotes
Declaration of competing interest
The authors declare that they have no known financial conflicts of interest.
Funding
None
REFERENCES
- 1.Aleem A, Samad AB, Slenker AK. InStatPearls [Internet] StatPearls Publishing; 2022. Emerging variants of SARS-CoV-2 and novel therapeutics against coronavirus (COVID-19) [PubMed] [Google Scholar]
- 2.Zhang L, Jackson CB, Mou H, et al. SARS-CoV-2 spike-protein D614G mutation increases virion spike density and infectivity. Nat Commun. 2020;11( 1):1–9. doi: 10.1038/s41467-020-19808-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu L, Zhou L, Mo M, et al. SARS-CoV-2 Omicron RBD shows weaker binding affinity than the currently dominant Delta variant to human ACE2. Signal Transduct Target Ther. 2022;7( 1):1–3. doi: 10.1038/s41392-021-00863-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lai C-C, Shih T-P, Ko W-C, Tang H-J, Hsueh PR. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus disease-2019 (COVID-19): The epidemic and the challenges. Int J Antimicrob Agents. 2020;55( 3):105924. doi: 10.1016/j.ijantimicag.2020.105924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang MY, Zhao R, Gao LJ, Gao X-F, Wang DP, Cao JM. SARS-CoV-2: structure, biology, and structure-based therapeutics development. Front Cell Infect Microbiol. 2020:724. doi: 10.3389/fcimb.2020.587269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khailany RA, Safdar M, Ozaslan M. Genomic characterization of a novel SARS-CoV-2. Gene reports. 2020;19:100682. doi: 10.1016/j.genrep.2020.100682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xie Y, Karki CB, Du D, et al. Spike proteins of SARS-CoV and SARS-CoV-2 utilize different mechanisms to bind with human ACE2. Front Mol Biosci. 2020:392. doi: 10.3389/fmolb.2020.591873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xia S, Liu M, Wang C, et al. Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Res. 2020;30( 4):343–355. doi: 10.1038/s41422-020-0305-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lan J, Ge J, Yu J, et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581( 7807):215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 10.Walls AC, Park Y-J, Tortorici MA, Wall A, McGuire AT, Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181( 2):281–292e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson BA, Xie X, Kalveram B, et al. Furin cleavage site is key to SARS-CoV-2 pathogenesis. BioRxiv. 2020 [Google Scholar]
- 12.Laha S, Chakraborty J, Das S, Manna SK, Biswas S, Chatterjee R. Characterizations of SARS-CoV-2 mutational profile, spike protein stability and viral transmission. Infect Genet Evol. 2020;85:104445. doi: 10.1016/j.meegid.2020.104445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gram MA, Nielsen J, Schelde AB, et al. Vaccine effectiveness against SARS-CoV-2 infection, hospitalization, and death when combining a first dose ChAdOx1 vaccine with a subsequent mRNA vaccine in Denmark: A nationwide population-based cohort study. PLoS Med. 2021;18( 12):e1003874. doi: 10.1371/journal.pmed.1003874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chadeau-Hyam M, Wang H, Eales O, et al. SARS-CoV-2 infection and vaccine effectiveness in England (REACT-1): a series of cross-sectional random community surveys. Lancet Respir Med. 2022;10( 4):355–366. doi: 10.1016/S2213-2600(21)00542-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chemaitelly H, Yassine HM, Benslimane FM, et al. mRNA-1273 COVID-19 vaccine effectiveness against the B. 1.1. 7 and B. 1.351 variants and severe COVID-19 disease in Qatar. Nat Med. 2021;27( 9):1614–1621. doi: 10.1038/s41591-021-01446-y. [DOI] [PubMed] [Google Scholar]
- 16.Walter EB, Talaat KR, Sabharwal C, et al. Evaluation of the BNT162b2 COVID-19 vaccine in children 5 to 11 years of age. N Engl J Med. 2022;386( 1):35–46. doi: 10.1056/NEJMoa2116298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pilishvili T, Gierke R, Fleming-Dutra KE, et al. Effectiveness of mRNA Covid-19 vaccine among US health care personnel. N Engl J Med. 2021;385( 25):e90. doi: 10.1056/NEJMoa2106599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barda N, Dagan N, Cohen C, et al. Effectiveness of a third dose of the BNT162b2 mRNA COVID-19 vaccine for preventing severe outcomes in Israel: an observational study. The Lancet. 2021;398( 10316):2093–2100. doi: 10.1016/S0140-6736(21)02249-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saciuk Y, Kertes J, Shamir Stein N, Ekka Zohar A. Effectiveness of a third dose of BNT162b2 mRNA vaccine. J Infect Dis. 2022;225( 1):30–33. doi: 10.1093/infdis/jiab556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mohammadi M, Shayestehpour M, Mirzaei H. The impact of spike mutated variants of SARS-CoV2 [Alpha, Beta, Gamma, Delta, and Lambda] on the efficacy of subunit recombinant vaccines. Braz J Infect Dis. 2021:25. doi: 10.1016/j.bjid.2021.101606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang T-J, Yu P-Y, Chang Y-C, et al. Effect of SARS-CoV-2 B. 1.1. 7 mutations on spike protein structure and function. Nat Struct Mol Biol. 2021;28( 9):731–739. doi: 10.1038/s41594-021-00652-z. [DOI] [PubMed] [Google Scholar]
- 22.Liu Y, Liu J, Plante KS, et al. The N501Y spike substitution enhances SARS-CoV-2 infection and transmission. Nature. 2022;602( 7896):294–299. doi: 10.1038/s41586-021-04245-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fiorentini S, Messali S, Zani A, et al. First detection of SARS-CoV-2 spike protein N501 mutation in Italy in August, 2020. Lancet Infect Dis. 2021;21( 6):e147. doi: 10.1016/S1473-3099(21)00007-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Washington NL, Gangavarapu K, Zeller M, et al. Emergence and rapid transmission of SARS-CoV-2 B. 1.1. 7 in the United States. Cell. 2021;184( 10):2587–2594e7. doi: 10.1016/j.cell.2021.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Challen R, Brooks-Pollock E, Read JM, Dyson L, Tsaneva-Atanasova K, Danon L. Risk of mortality in patients infected with SARS-CoV-2 variant of concern 202012/1: matched cohort study. Brit Med J. 2021;372:n579. doi: 10.1136/bmj.n579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bernal JL, Andrews N, Gower C, et al. Effectiveness of Covid-19 vaccines against the B.1.617.2 (Delta) variant. N Engl J Med. 2021;385:585–594. doi: 10.1056/NEJMoa2108891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mahase E. Covid-19: Novavax vaccine efficacy is 86% against UK variant and 60% against South African variant. Brit Med J. 2021;372:n296. doi: 10.1136/bmj.n296. [DOI] [PubMed] [Google Scholar]
- 28.Funk T, Pharris A, Spiteri G, et al. Characteristics of SARS-CoV-2 variants of concern B. 1.1. 7, B. 1.351 or P. 1: data from seven EU/EEA countries, weeks 38/2020 to 10/2021. Eurosurveillance. 2021;26( 16):2100348. doi: 10.2807/1560-7917.ES.2021.26.16.2100348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arena F, Pollini S, Rossolini GM, Margaglione M. Summary of the available molecular methods for detection of SARS-CoV-2 during the ongoing pandemic. In J Mol Sci. 2021;22( 3):1298. doi: 10.3390/ijms22031298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Choi JY, Smith DM. SARS-CoV-2 variants of concern. Yonsei Med J. 2021;62( 11):961–968. doi: 10.3349/ymj.2021.62.11.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Singer SR, Angulo FJ, Swerdlow DL, et al. Effectiveness of BNT162b2 mRNA COVID-19 vaccine against SARS-CoV-2 variant Beta (B. 1.351) among persons identified through contact tracing in Israel: A prospective cohort study. EClinicalMedicine. 2021;42:101190. doi: 10.1016/j.eclinm.2021.101190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Davies NG, Abbott S, Barnard RC, et al. Estimated transmissibility and impact of SARS-CoV-2 lineage B. 1.1. 7 in England. Science. 2021;372(6538):eabg3055. doi: 10.1126/science.abg3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Naveca F, Nascimento V, Souza V, et al. Phylogenetic relationship of SARS-CoV-2 sequences from Amazonas with emerging Brazilian variants harboring mutations E484K and N501Y in the Spike protein. Virological Org. 2021;1:1–8. [Google Scholar]
- 34.Naveca F, Costa CD, Nascimento V, Souza V, et al. Three SARS-CoV-2 reinfection cases by the new Variant of Concern (VOC) 1/501Y.V3. Res Sq. 2021 [Google Scholar]
- 35.Sabino EC, Buss LF, Carvalho MP, et al. Resurgence of COVID-19 in Manaus, Brazil, despite high seroprevalence. The Lancet. 2021;397( 10273):452–455. doi: 10.1016/S0140-6736(21)00183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Charmet T, Schaeffer L, Grant R, et al. Impact of original, B. 1.1. 7, and B. 1.351/P. 1 SARS-CoV-2 lineages on vaccine effectiveness of two doses of COVID-19 mRNA vaccines: Results from a nationwide case-control study in France. The Lancet Regional Health-Europe. 2021;8:100171. doi: 10.1016/j.lanepe.2021.100171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tatsi E-B, Filippatos F, Michos A. SARS-CoV-2 variants and effectiveness of vaccines: a review of current evidence. Epidemiol Infect. 2021:1–24. doi: 10.1017/S0950268821002430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang L, Jackson CB, Mou H, et al. The D614G mutation in the SARS-CoV-2 spike protein reduces S1 shedding and increases infectivity. BioRxiv. 2020 [Google Scholar]
- 39.Christie A, Brooks JT, Hicks LA, et al. Guidance for implementing COVID-19 prevention strategies in the context of varying community transmission levels and vaccination coverage. Morb Mort Wkly Rep. 2021;70( 30):1044. doi: 10.15585/mmwr.mm7030e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen J, Wang R, Wang M, Wei GW. Mutations strengthened SARS-CoV-2 infectivity. J Mol Biol. 2020;432( 19):5212–5226. doi: 10.1016/j.jmb.2020.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McCallum M, Walls AC, Sprouse KR, et al. Molecular basis of immune evasion by the Delta and Kappa SARS-CoV-2 variants. Science. 2021;374( 6575):1621–1626. doi: 10.1126/science.abl8506. [DOI] [PubMed] [Google Scholar]
- 42.Bruxvoort KJ, Sy LS, Qian L, et al. Effectiveness of mRNA-1273 against delta, mu, and other emerging variants of SARS-CoV-2: test negative case-control study. Brit Med J. 2021:375. doi: 10.1136/bmj-2021-068848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Deb P, Molla M, Ahmed M, Saif-Ur-Rahman K, Das MC, Das D. A review of epidemiology, clinical features and disease course, transmission dynamics, and neutralization efficacy of SARS-CoV-2 variants. Egypt J Bronchol. 2021;15( 1):1–14. [Google Scholar]
- 44.Ella R, Reddy S, Blackwelder W, et al. Efficacy, safety, and lot-to-lot immunogenicity of an inactivated SARS-CoV-2 vaccine (BBV152): interim results of a randomised, double-blind, controlled, phase 3 trial. The Lancet. 2021;398( 10317):2173–2184. doi: 10.1016/S0140-6736(21)02000-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim S, Nguyen TT, Taitt AS, et al. SARS-CoV-2 Omicron mutation is faster than the chase: multiple mutations on spike/ACE2 Interaction Residues. Immune Netw. 2021;21(6) doi: 10.4110/in.2021.21.e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pulliam JR, van Schalkwyk C, Govender N, et al. Increased risk of SARS-CoV-2 reinfection associated with emergence of the Omicron variant in South Africa. MedRxiv. 2021 doi: 10.1126/science.abn4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen J, Wang R, Gilby NB, Wei GW. Omicron variant (B.1.1.529): infectivity, vaccine breakthrough, and antibody resistance. J Chem Inf Model. 2022;62( 2):412–422. doi: 10.1021/acs.jcim.1c01451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Muik A, Lui BG, Wallisch A-K, et al. Neutralization of SARS-CoV-2 Omicron by BNT162b2 mRNA vaccine–elicited human sera. Science. 2022:eabn7591. doi: 10.1126/science.abn7591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Collie S, Champion J, Moultrie H, Bekker L-G, Gray G. Effectiveness of BNT162b2 vaccine against omicron variant in South Africa. N Engl J Med. 2022;386( 5):494–496. doi: 10.1056/NEJMc2119270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hansen CH, Schelde AB, Moustsen-Helms IR, et al. Vaccine effectiveness against SARS-CoV-2 infection with the Omicron or Delta variants following a two-dose or booster BNT162b2 or mRNA-1273 vaccination series: A Danish cohort study. medRxiv. 2021 [Google Scholar]
- 51.Evans JP, Zeng C, Qu P, et al. Neutralization of SARS-CoV-2 Omicron sub-lineages BA.1, BA.1.1, and BA.2. Cell Host Microbe. 2022 doi: 10.1016/j.chom.2022.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cucinotta D, Vanelli M. WHO declares COVID-19 a pandemic. Acta Bio Medica: Atenei Parmensis. 2020;91( 1):157. doi: 10.23750/abm.v91i1.9397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhu N, Zhang D, Wang W, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382( 8):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sharma A, Tiwari S, Deb MK, Marty JL. Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2): a global pandemic and treatment strategies. Int J Antimicrob Agents. 2020;56( 2):106054. doi: 10.1016/j.ijantimicag.2020.106054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Meyerowitz EA, Richterman A, Gandhi RT, Sax PE. Transmission of SARS-CoV-2: a review of viral, host, and environmental factors. Ann Intern Med. 2021;174( 1):69–79. doi: 10.7326/M20-5008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang W, Xu Y, Gao R, et al. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA. 2020;323( 18):1843–1844. doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Santarpia JL, Herrera VL, Rivera DN, et al. The infectious nature of patient-generated SARS-CoV-2 aerosol. MedRxiv. 2020 [Google Scholar]
- 58.Peiris JSM, Chu C-M, Cheng VC-C, et al. Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia: a prospective study. The Lancet. 2003;361( 9371):1767–1772. doi: 10.1016/S0140-6736(03)13412-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shi J, Wen Z, Zhong G, et al. Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS–coronavirus 2. Science. 2020;368( 6494):1016–1020. doi: 10.1126/science.abb7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kotlyar AM, Grechukhina O, Chen A, et al. Vertical transmission of coronavirus disease 2019: a systematic review and meta-analysis. Am J Obstet Gynecol. 2021;224( 1):35–53e3. doi: 10.1016/j.ajog.2020.07.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Butt AA, Dargham SR, Chemaitelly H, et al. The global transmission of new coronavirus variants. Environmental research. JAMA Intern Med. 2022;182( 2):197–205. doi: 10.1001/jamainternmed.2021.7949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Curran J, Dol J, Boulos L, et al. Transmission characteristics of SARS-CoV-2 variants of concern Rapid Scoping Review. medRxiv. 2021 doi: 10.1136/bmjopen-2021-055781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Buchan SA, Tibebu S, Daneman N, et al. Increased household secondary attacks rates with variant of concern severe acute respiratory syndrome coronavirus 2 index cases. Clin Infect Dis. 2022;74( 4):703–706. doi: 10.1093/cid/ciab496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tegally H, Wilkinson E, Giovanetti M, et al. Detection of a SARS-CoV-2 variant of concern in South Africa. Nature. 2021;592( 7854):438–443. doi: 10.1038/s41586-021-03402-9. [DOI] [PubMed] [Google Scholar]
- 65.Maslo C, Friedland R, Toubkin M, Laubscher A, Akaloo T, Kama B. Characteristics and outcomes of hospitalized patients in South Africa during the COVID-19 Omicron wave compared with previous waves. JAMA. 2022;327( 6):583–584. doi: 10.1001/jama.2021.24868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhao Y, Huang J, Zhang L, Chen S, Gao J, Jiao H. The global transmission of new coronavirus variants. Environ Res. 2022;206:112240. doi: 10.1016/j.envres.2021.112240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Coutinho RM, Marquitti FMD, Ferreira LS, et al. Model-based estimation of transmissibility and reinfection of SARS-CoV-2 P. 1 variant. Commun Med. 2021;1( 1):1–8. doi: 10.1038/s43856-021-00048-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Faria NR, Mellan TA, Whittaker C, et al. Genomics and epidemiology of the P. 1 SARS-CoV-2 lineage in Manaus, Brazil. Science. 2021;372( 6544):815–821. doi: 10.1126/science.abh2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.da Silva JF, Esteves RJ, Siza C, et al. Cluster of SARS-CoV-2 Gamma Variant Infections, Parintins, Brazil, March 2021. Emerg Infect Dis. 2022;28( 1):262. doi: 10.3201/eid2801.211817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pan A, Liu L, Wang C, et al. Association of public health interventions with the epidemiology of the COVID-19 outbreak in Wuhan, China. JAMA. 2020;323( 19):1915–1923. doi: 10.1001/jama.2020.6130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Inglesby TV. Public health measures and the reproduction number of SARS-CoV-2. JAMA. 2020;323( 21):2186–2187. doi: 10.1001/jama.2020.7878. [DOI] [PubMed] [Google Scholar]
- 72.Liu Y, Gayle AA, Wilder-Smith A, Rocklöv J. The reproductive number of COVID-19 is higher compared to SARS coronavirus. J Travel Med. 2020;27(2):taaa021. doi: 10.1093/jtm/taaa021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Campbell F, Archer B, Laurenson-Schafer H, et al. Increased transmissibility and global spread of SARS-CoV-2 variants of concern as at June 2021. Eurosurveillance. 2021;26( 24):2100509. doi: 10.2807/1560-7917.ES.2021.26.24.2100509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Salzberger B, Buder F, Lampl B, et al. Epidemiology of SARS-CoV-2. Infection. 2021;49( 2):233–239. doi: 10.1007/s15010-020-01531-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yang L, Dai J, Zhao J, Wang Y, Deng P, Wang J. Estimation of incubation period and serial interval of COVID-19: analysis of 178 cases and 131 transmission chains in Hubei province. China Epidemiol Infect. 2020:148. doi: 10.1017/S0950268820001338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dhouib W, Maatoug J, Ayouni I, et al. The incubation period during the pandemic of COVID-19: a systematic review and meta-analysis. Syst Rev. 2021;10( 1):1–14. doi: 10.1186/s13643-021-01648-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li Q, Guan X, Wu P, et al. Early transmission dynamics in Wuhan, China, of novel coronavirus–infected pneumonia. N Engl J Med. 2020;382( 13):1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Varia M, Wilson S, Sarwal S, et al. Investigation of a nosocomial outbreak of severe acute respiratory syndrome (SARS) in Toronto, Canada. CMAJ. 2003;169( 4):285–292. [PMC free article] [PubMed] [Google Scholar]
- 79.Virlogeux V, Fang VJ, Park M, Wu JT, Cowling BJ. Comparison of incubation period distribution of human infections with MERS-CoV in South Korea and Saudi Arabia. Sci Rep. 2016;6( 1):1–7. doi: 10.1038/srep35839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lauer SA, Grantz KH, Bi Q, et al. The incubation period of coronavirus disease 2019 (COVID-19) from publicly reported confirmed cases: estimation and application. Ann Intern Med. 2020;172( 9):577–582. doi: 10.7326/M20-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Homma Y, Katsuta T, Oka H, et al. The incubation period of the SARS-CoV-2 B1. 1.7 variant is shorter than that of other strains. J Infect. 2021;83( 2):e15–e17. doi: 10.1016/j.jinf.2021.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Grant R, Charmet T, Schaeffer L, et al. Impact of SARS-CoV-2 Delta variant on incubation, transmission settings and vaccine effectiveness: Results from a nationwide case-control study in France. The Lancet Regional Health-Europe. 2022;13:100278. doi: 10.1016/j.lanepe.2021.100278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kremer C, Braeye T, Proesmans K, André E, Torneri A, Hens N. Observed serial intervals of SARS-CoV-2 for the Omicron and Delta variants in Belgium based on contact tracing data, 19 November to 31 December 2021. medRxiv. 2022 doi: 10.3201/eid2808.220220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hsu L, Grüne B, Buess M, et al. COVID-19 Breakthrough infections and transmission risk: real-world data analyses from Germany’s largest public health department (Cologne) Vaccines. 2021;9( 11):1267. doi: 10.3390/vaccines9111267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Vitiello A, Ferrara F, Troiano V, La Porta R. COVID-19 vaccines and decreased transmission of SARS-CoV-2. Inflammopharmacology. 2021;29( 5):1357–1360. doi: 10.1007/s10787-021-00847-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Prunas O, Warren JL, Crawford FW, et al. Vaccination with BNT162b2 reduces transmission of SARS-CoV-2 to household contacts in Israel. Science. 2022:eabl4292. doi: 10.1126/science.abl4292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Harris RJ, Hall JA, Zaidi A, Andrews NJ, Dunbar JK, Dabrera G. Impact of vaccination on household transmission of SARS-COV-2 in England. medRxiv. 2021 doi: 10.1056/NEJMc2107717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lyngse FP, Mølbak K, Denwood M, et al. Effect of Vaccination on household transmission of SARS-CoV-2 Delta VOC. medRxiv. 2022 doi: 10.1038/s41467-022-31494-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Levine-Tiefenbrun M, Yelin I, Katz R, et al. Initial report of decreased SARS-CoV-2 viral load after inoculation with the BNT162b2 vaccine. Nat Med. 2021;27( 5):790–792. doi: 10.1038/s41591-021-01316-7. [DOI] [PubMed] [Google Scholar]
- 90.de Gier B, Andeweg S, Backer JA, et al. Vaccine effectiveness against SARS-CoV-2 transmission to household contacts during dominance of Delta variant (B. 1.617. 2), the Netherlands, August to September 2021. Euro Surveill. 2021;26( 44):2100977. doi: 10.2807/1560-7917.ES.2021.26.44.2100977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Eyre DW, Taylor D, Purver M, et al. Effect of Covid-19 vaccination on transmission of alpha and delta variants. N Engl J Med. 2022;386( 8):744–756. doi: 10.1056/NEJMoa2116597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. The Lancet. 2020;395( 10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lei J, Li J, Li X, Qi X. CT imaging of the 2019 novel coronavirus (2019-nCoV) pneumonia. Radiology. 2020;295( 1):18. doi: 10.1148/radiol.2020200236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hu Z, Huang X, Zhang J, Fu S, Ding D, Tao Z. Differences in clinical characteristics between Delta Variant and Wild-Type SARS-CoV-2 Infected Patients. Front Med. 2021:8. doi: 10.3389/fmed.2021.792135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ong SW, Chiew CJ, Ang LW, et al. Clinical and virological features of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants of concern: a retrospective cohort study comparing B.1.1.7 (Alpha), B.1.351 (Beta), and B.1.617.2 (Delta) Clin Infect Dis. 2021:ciab721. doi: 10.1093/cid/ciab721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Taha SI, Samaan SF, Shata AK, Baioumy SA, Abdalgeleel SA, Youssef MK. Baseline characteristics and outcomes of 180 Egyptian COVID-19 patients admitted to quarantine hospitals of Ain Shams University: a retrospective comparative study. Afro-Egypt J Infect Endem Dis. 2021;11( 3):295–305. [Google Scholar]
- 97.Fouad SH, Allam MF, Ibrahim S, et al. ICU admission of COVID-19 patients: identification of risk factors. Egypt J Anaesth. 2021;37( 1):202–207. [Google Scholar]
- 98.Luna-Muschi A, Borges IC, de Faria E, et al. Clinical features of COVID-19 by SARS-CoV-2 Gamma variant: A prospective cohort study of vaccinated and unvaccinated healthcare workers. J Infect. 2022;84( 2):248–288. doi: 10.1016/j.jinf.2021.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kim M-K, Lee B, Choi YY, et al. Clinical Characteristics of 40 Patients Infected With the SARS-CoV-2 Omicron Variant in Korea. J Korean Med Sci. 2022;37(3) doi: 10.3346/jkms.2022.37.e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lee JJ, Choe YJ, Jeong H, et al. Importation and transmission of SARS-CoV-2 B. 1.1. 529 (omicron) variant of concern in Korea, November 2021. J Korean Med Sci. 2021;36(50) doi: 10.3346/jkms.2021.36.e346. [DOI] [PMC free article] [PubMed] [Google Scholar]