Keywords: degradomics, mass spectrometry, metalloprotease, protease, proteoglycans
Abstract
Proteoglycans are composite molecules comprising a protein backbone, i.e., the core protein, with covalently attached glycosaminoglycan chains of distinct chemical types. Most proteoglycans are secreted or attached to the cell membrane. Their specialized structures, binding properties, and biophysical attributes underlie diverse biological roles, which include modulation of tissue mechanics, cell adhesion, and the sequestration and regulated release of morphogens, growth factors, and cytokines. As an irreversible post-translational modification, proteolysis has a profound impact on proteoglycan function, abundance, and localization. Proteolysis is required for molecular maturation of some proteoglycans, clearance of extracellular matrix proteoglycans during tissue remodeling, generation of bioactive fragments from proteoglycans, and ectodomain shedding of cell-surface proteoglycans. Genetic evidence shows that proteoglycan core protein proteolysis is essential for diverse morphogenetic events during embryonic development. In contrast, dysregulated proteoglycan proteolysis contributes to osteoarthritis, cardiovascular disorders, cancer, and inflammation. Proteolytic fragments of perlecan, versican, aggrecan, brevican, collagen XVIII, and other proteoglycans are associated with independent biological activities as so-called matrikines. Yet, proteoglycan proteolysis has been investigated to only a limited extent to date. Here, we review the actions of proteases on proteoglycans and illustrate their functional impact with several examples. We discuss the applications and limitations of strategies used to define cleavage sites in proteoglycans and explain how proteoglycanome-wide proteolytic mapping, which is desirable to fully understand the impact of proteolysis on proteoglycans, can be facilitated by integrating classical proteoglycan isolation methods with mass spectrometry-based proteomics.
INTRODUCTION
Proteoglycans are composite molecules comprising a protein core with covalently attached glycosaminoglycan (GAG) side chains (1). They function in a variety of molecular networks and act in distinct pathways via the intermolecular interactions that are determined both by the characteristics of core proteins and the GAG chains. The GAG chains are chemically classified according to their disaccharide composition as heparan sulfate (HS), chondroitin sulfate/dermatan sulfate (CS/DS), or keratan sulfate (KS). This distinction is the basis for their classification as heparan sulfate proteoglycans (HSPGs), CSPGs, or KSPGs, although hybrid proteoglycans that carry more than one type of GAG also occur (1). Proteoglycans were also comprehensively classified according to a taxonomy that integrates their cellular and subcellular location, overall gene/protein homology, and the presence of specific protein modules in the core proteins (2). Some proteoglycans such as aggrecan and versican have numerous GAG chains, whereas the majority, including agrin, perlecan, small leucine-rich proteoglycans, syndecans, and glypicans may have only one to three GAG chains. Some are part-time proteoglycans, where the GAG chain, instead of being a constitutive feature, may be a regulated, even infrequent modification. Via distinct spatial distribution and cell-matrix localization, individual proteoglycans occupy distinct niches in the extracellular matrix (ECM), at the cell-matrix interface, and some primarily reside intracellularly [e.g., serglycin, which unusually, can carry heparin or CS/DS chains and SV2, a KSPG in synaptic vesicles (3)]. Most HSPGs are closely approximated to the cell surface since they are inserted into the cell membrane or located in the basement membrane zone, whereas the majority of CSPGs are located in interstitial ECM, where they may be spatially further from the cell. Although both HS and CS chains bind to a variety of morphogens and growth factors, HSPGs may have a more significant role in cellular regulation because of their proximity to the cell surface. CSPGs clearly also have diverse regulatory roles in addition to their structural roles in the ECM, such as via binding to specific morphogens or Toll-like receptors (4–7).
Proteoglycan functions are influenced by the composition, abundance, and charge of the GAG chains, as well as specific sequence features and domain composition of the core proteins. Proteoglycan core proteins are diverse, both in molecular size and cellular context, comprising secreted proteins, single-pass membrane proteins, and glycosylphosphatidylinositol (GPI) anchored proteins, and additional molecular diversity may be contributed by alternative splicing. The known proteoglycans include several molecules with alternative identities as collagens (e.g., collagen IX, collagen XV, and collagen XVIII) (8–10), secreted metalloproteases (ADAMTS7 and ADAMTS12) (11, 12), an endosulfatase (Sulf-2) (13), and Kunitz-type protease inhibitors such as bikunin (14) and appican (15, 16). Mass spectrometry (MS)-based glycopeptide analysis is contributing to discovery of additional proteoglycans, so this unusual molecular category is likely to grow further (17).
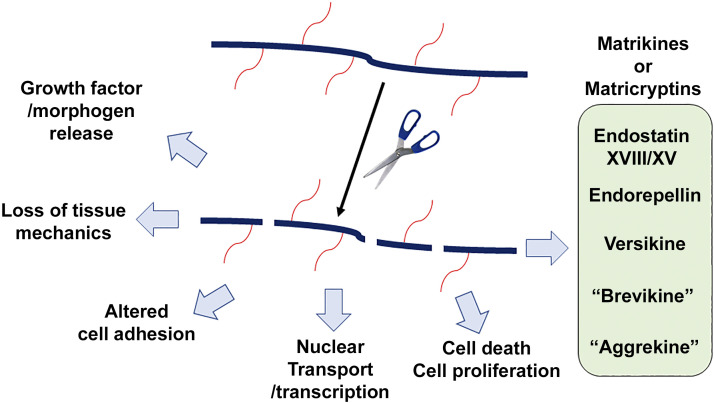
The subject of this review is the irreversible post-translational modification of proteoglycans by proteolysis of their core proteins. Dysregulated proteolysis is a prominent feature of many disease states such as cancer, inflammation, and degenerative disorders, yet its impact on proteoglycan function is relatively thinly explored, because of a paucity of detailed mapping of cleavages in proteoglycans and the lack of assignment of observed proteoglycan cleavages to specific proteases. Many proteolytic events are likely to be inactivating, leading to loss of function of the proteoglycan (Fig. 1). However, proteolysis may also activate the function of proteoglycans and other molecules via precursor maturation steps, as well as by constitutive cleavages that do not excise a precursor propeptide region, but may be necessary to relieve unfavorable structural constraint or expose essential binding sites. Ectodomain shedding of cell-surface proteoglycans can relocate the ectodomain to more distal locations, and the shed form of at least one membrane-anchored proteoglycan (syndecan-1) is endocytosed and redirected to the nucleus (18). Moreover, proteolytic processing can extend the inherent molecular diversity of proteoglycans by generation of fragments with new functional properties. Since most proteoglycans exist in the ECM, their fragments are considered as “matrikines” or “matricryptins” (19) (Fig. 1).
Figure 1.
Examples of the biological impact of proteoglycan proteolysis. The cartoon shows proteolysis of a proteoglycan [the solid black line represents the core protein, red lines the glycosaminoglycan (GAG) chains] by a protease (represented by scissors), and the potential functional impact. The box at right illustrates previously identified proteolytic products with independent bioactivities, which contribute to the effects shown on the left. Since the terms aggrekine and brevikine were coined anew for this review, they are shown in quotation marks.
Proteases from all known classes of secreted and cell-surface proteases act on proteoglycans, although metalloproteases, serine, and cysteine proteases are the most prominently featured. Secreted proteases primarily target ECM proteoglycans, whereas membrane-bound proteases are implicated in ectodomain shedding, although this distinction has exceptions. Here, we use several examples of proteolysis of specific proteoglycans to illustrate the diverse proteolytic mechanisms, impact of proteolysis at the molecular level, and functional consequences of proteoglycan proteolysis. In the closing section of the review, we discuss an approach to comprehensively map proteolytic cleavages in proteoglycans that will be necessary to expand the knowledge on this topic.
PROTEOLYSIS OF AGGRECAN AND OTHER CARTILAGE PROTEOGLYCANS IS A MAJOR PATHOGENIC MECHANISM IN OSTEOARTHRITIS
Cartilage is enriched in several proteoglycans, especially the large aggregating proteoglycan aggrecan, which forms giant complexes with hyaluronan that endow the tissue with compression resistance (20). Aggrecan is also essential in neurogenesis (21), regulates neural crest cell migration along with versican (22), and is a key component of perineuronal nets, believed to be the basis for loss of neuronal plasticity in the mature central nervous system (23). Several small proteoglycans including biglycan, decorin, epiphycan, and fibromodulin that interact with collagens, matrix glycoproteins, and cell membrane components are found in cartilage and functionally similar nucleus pulposus of the intervertebral disk and undergo proteolysis therein (24–27). Although osteoarthritis (OA) is a disease of all joint components, proteolytic cartilage destruction is recognized as a major contributor to its pathogenesis and was extensively investigated in this context. Aggrecan is the classic hyaluronan-binding CS/DS/KS PG, a class that includes versican, brevican, and neurocan, which are collectively referred to as lecticans or hyalectans. They have a similar multimodular structure, with two conserved globular domains (G1 and G3) flanking a GAG-rich central domain. Aggrecan uniquely has an additional globular domain, G2, which resides between the G1 and GAG-bearing domains. The unstructured segment between the G1 and G2 domains is termed the inter-globular domain (IGD) and it is significant with respect to proteolysis, since cleavage here releases the entire GAG-bearing region comprising a KS-rich domain, along with two chondroitin sulfate-rich domains (CS1 and CS2) between G2 and G3. The IGD is attacked by matrix metalloproteinases (MMPs), cathepsins, and aggrecanases, the last being members of the ADAMTS family, which has a major role in cartilage breakdown (28, 29). The MMP/cathepsin cleavage occurs at the Asn341-Phe342 peptide bond [in the (human) sequence context VDIPEN341-F342FGVGGE] (30). Aggrecanases cleave downstream of the MMP site at the Glu373-Ala374 peptide bond (sequence context NITEGE373-A374RGSVI) and products of this cleavage were identified in synovial fluids from patients with OA, joint injury, and inflammatory joint disease (31). N-linked KS in the IGD, most likely at position Asn368, confers susceptibility to aggrecanases (32). The C-terminal fragment resulting from cleavages in the IGD carries most of the GAG content in aggrecan and its release is frequently quantified by chemical and immunological assays for sulfated GAG chains (33, 34). Aggrecan fragmentation may also be associated with the turnover of hyaluronan (HA) since in organ culture of normal bovine articular cartilage slices, the turnover kinetics of newly synthesized, radiolabeled aggrecan and HA were found to be comparable (35). In addition to the IGD site, human aggrecan is cleaved by the principal aggrecanases ADAMTS4 (aggrecanase-1) and ADAMTS5 (aggrecanase-2) at Glu1545-Gly1546, Glu1714-Gly1715, Glu1819-Ala1820, and Glu1919-Leu1920, whereas bovine aggrecan is preferentially cleaved at Glu1667-Gly1668, followed by Glu1480-Gly1481, Glu373-Ala374 in the IGD and at Glu1771-Ala1772 and Glu1871-Leu1872 in the CS2 region (36). Although ADAMTS4 and ADAMTS5 have broadly similar activities against aggrecan, ADAMTS5 was at least 1,000-fold more active than ADAMTS4 under physiological conditions (37). Catalytic domains of ADAMTS4 and ADAMTS5 without the downstream ancillary domains, which are required for substrate binding lack activity against aggrecan, and their specific C-terminal modules have an important role in binding to the GAGs; moreover, glycosylation of aggrecan is important for cleavage by aggrecanases (37, 38). Proteolysis by dual action of ADAMTS proteases and MMPs/cathepsin generates a 32-mer aggrecan peptide which could be termed an “aggrekine,” since it interacts with Toll-like receptors on dorsal root ganglia sensory neurons, inciting joint pain (5, 39).
Biglycan, a member of the class I family of small leucine-rich repeat proteoglycans (SLRPs), has CS/DS chains attached at Ser42 and Ser47. During biglycan synthesis, the astacin-like metalloprotease bone morphogenetic protein-1 (BMP-1), as well as the related proteases mammalian Tolloid and mammalian Tolloid-like 1 (mTLL-1) cleave off an N-terminal propeptide (40). ADAMTS4 and -5 cleave bovine and human biglycan near the beginning of their fifth leucine-rich repeat domain at Asn187-Cys188; in contrast, decorin was not cleaved by these aggrecanases (41), but clearly also undergoes catabolism in OA cartilage (42). Soluble biglycan has potent proinflammatory effects and roles in cancer (43, 44), and it is possible that biglycan fragments could act similarly, such as in the context of inflammation occurring in osteoarthritis. Although lumican peptides and recombinant fragments such as lumcorin and lumikine have antitumor activity and inhibit the key tumor protease MT1-MMP/MMP14 (45, 46), it remains unclear whether the specific fragments occur endogenously via proteolysis.
Analysis of proteolysis in OA has typically investigated activities of candidate proteases against intact cartilage or individual cartilage components. Recently, an analysis of OA cartilage used the mass spectrometry-based terminomics strategy Terminal Amine Isotopic Labeling of Substrates (TAILS) (47) to identify sites of proteolysis in cartilage components in an unbiased fashion (42). The analysis identified 118 cleavage sites in aggrecan of which 41 sites were located in the G1 domain, seven sites in the IGD, 36 sites in G2, 2 sites in CS1, seven sites in CS2, and 23 sites in G3 domain (42). The relative paucity of cleavage sites detected in the GAG-bearing region is explained by factors that are discussed in the final section of the review. The analysis also identified numerous sites in fibromodulin and several cleavage sites in biglycan, decorin, lumican, lubricin, and versican (42). Among several proteases also identified in this analysis, the serine protease HtrA1 was consistently the most abundant (42). Previously, an HtrA1-specific cleavage site at Val356-Thr357, occurring between the IGD aggrecanase and MMP cleavage sites was characterized in detail and a neo-epitope antibody detecting the cleavage showed greater staining in OA cartilage (48). Digestion of nondiseased articular cartilage with HtrA1 identified several cleavage sites in fibromodulin, PRELP, aggrecan, and biglycan, many of which were present in the OA degradome (42). Furthermore, selective digestion of biglycan with HtrA1 identified eight specific cleavage sites (42). Thus, in addition to ADAMTS proteases, MMPs, and cathepsin, HtrA1 has emerged as a major mediator of proteoglycan degradation in OA cartilage, and these activities may also be relevant to proteoglycan degradation in the vasculature, where HtrA1 is expressed by smooth muscle cells and in the eye, where it is implicated in age-related macular degeneration (49–52).
VERSICAN PROTEOLYSIS BY ADAMTS PROTEASES HAS A SIGNIFICANT IMPACT IN THE DEVELOPING EMBRYO AND IN ADULT TISSUE HOMEOSTASIS
The structure, physiological roles of versican, and its proteolysis were discussed comprehensively in prior reviews (53–56). Here, a synopsis of the essential facts and an update on versican proteolysis are presented. Versican is the most widely distributed of the lecticans (57); however, alternative splicing dictates the prevalence of neural and extraneural isoforms (58). In contrast to the N-terminal (G1) and C-terminal (G3) globular domains consistently present in all versican splice isoforms, splicing of two large CS-bearing domains located between the G1 and G3 domains, each encoded by individual exons, can generate four CS-bearing isoforms. V2, which is restricted to the nervous system, contains only the smaller of the two domains, GAGα, whereas V1 (containing the GAGβ domain) and V0 (containing both GAGα and GAGβ domains) have a wider tissue distribution. Less is known about the prevalence of V4, reported in cancer, which contains only the N-terminal-most five CS-chains (59) and V3, which as a non-CS carrying isoform, is not a proteoglycan and is thus irrelevant to the present discussion. Specific ADAMTS cleavage sites were identified near the N-terminus of each GAG domain (Glu405-Gln406 in GAGα, V0/V2 sequence enumeration; Glu441-Ala442 in the GAGβ domain, V1 sequence enumeration) (60, 61), but the GAGβ cleavage site occurring at the Glu441-Ala442 peptide bond is the better studied of the two. As in aggrecan, the versican GAG chains appear to be necessary for optimal proteolysis; indeed, the two most N-terminal CS chains in versican V1 are required for cleavage at the Glu441-Ala442 site (62). Glu441-Ala442 cleavage is undertaken by ADAMTS proteases (ADAMTS1, ADAMTS4, ADAMTS5, ADAMTS9, ADAMTS15, and ADAMTS20) and generates an N-terminal G1 domain-containing fragment named versikine (63) whose C-terminal sequence DPEAAE441 was used to generate a neo-epitope antibody that is widely used to detect cleaved versican (60). Ample evidence exists that this site is cleaved extensively during embryogenesis, and that the ADAMTS proteases have definitive and crucial roles in versican proteolytic turnover, such as in cardiac jelly regression (ADAMTS1), cardiac valve sculpting (ADAMTS5 and ADAMTS9), palate closure (ADAMTS9 and ADAMTS20), skin pigmentation (ADAMTS9 and ADAMTS20), and resorption of interdigital webs during limb development (for review, see Ref. 54). The last-mentioned context was instrumental in discovery of a bioactive role for versikine in apoptosis of interdigit mesenchyme (63). Specifically, investigation of soft tissue syndactyly occurring in mouse gene knockouts lacking ADAMTS5 and ADAMTS20 demonstrated failed web regression accompanied by reduced mesenchymal apoptosis, reduced versican cleavage, and versican accumulation in the webs (63). These observations led to a series of experiments that demonstrated that versikine was required to induce mesenchymal apoptosis during web regression and identified an autoregulatory loop in which mesenchymal cell-derived ADAMTS proteases generated versikine for induction of their own demise (63). The significance of versican cleavage specifically at the Glu441-Ala442 site was recently strengthened by analysis of two independent mouse germline mutations intended to abrogate cleavage at this site, resulting in mouse alleles termed VcanR and VcanAA, each of which consistently demonstrated soft tissue syndactyly (64, 65). Both alleles are viable, although slight early mortality was reported in VcanR/R mice (64). Furthermore, analysis of digit separation in exon-specific mutants with loss of GAGα or GAGβ identified syndactyly only in the latter mutants, providing additional evidence supporting a specific role of versikine, which is specifically generated by cleavage within the latter domain (65).
ADAMTS proteases are not the only proteases that target the versican core protein since MMPs and plasmin are also known to do so (66, 67), but their impact on versican turnover in vivo, if any, is not reflected in specific phenotypes in the respective protease mutants. Using a novel proteomics strategy, ADAMTS1, ADAMTS4, and ADAMTS5 were recently shown to be active at several additional sites in the versican core protein (68). These findings potentially explain why VcanR and VcanAA homozygotes are viable, have relatively mild phenotypes, and do not fully replicate the phenotypes of mouse ADAMTS mutants. Beyond embryogenesis, recent work in VcanR/R mice has identified a role for versican cleavage in skin wound healing and the resolution of intestinal inflammation and damage (69).
ADAMTS proteolysis of versican was implicated in blood cancer, multiple myeloma, and in colorectal cancer (70, 71). Specifically, macrophage-produced versican was cleaved by ADAMTS proteases from tumor stromal cells to generate versikine, which enhanced immune recognition of myeloma cells (71). Versican acts as a danger-associated molecular pattern (DAMP) component that interacts with Toll-like receptors (TLRs), such as TLR2 on alveolar macrophages and promotes the production of inflammatory cytokines. In contrast, versikine elicits distinct transcriptional responses, promotes antitumor function of effector dendritic cells, and enhances intratumoral CD8+ cell infiltration. In the context of mouse models of influenza pneumonia, accumulation of versican as a result of reduced ADAMTS5 activity impedes migration of CD8+ T-cells from the draining lymph nodes, which allows them to fully execute effector functions and clear the infection (72). ADAMTS4 was recently implicated in influenza in humans and mice and defined as a potential drug target acting via cleavage of versican made by lung stromal fibroblasts (73), but it is not yet known if versikine has a role in viral pneumonia. ADAMTS5 was implicated in both aggrecan and versican turnover in the cardiovascular system, since Adamts5 mutant mice show aggrecan accumulation in the aorta (74), and ADAMTS5 turnover of proteoglycans was implicated in lipoprotein retention and vascular wall remodeling in atherosclerosis, vascular grafts, and aortic aneurysms (75–77).
IMPACT OF BREVICAN AND NEUROCAN PROTEOLYSIS IN THE NERVOUS SYSTEM
Brevican exists both as a secreted and GPI-anchored HA-binding CSPG. It is cleaved at the Glu395-Ser396 peptide bond, which occurs in a similar sequence and positional context as the well-studied ADAMTS cleavage sites in aggrecan and versican, to generate an N-terminal 50-kDa fragment and a 90-kDa C-terminal fragment (78). Cleavage was strongly associated with intracranial invasiveness of a glioma cell line, and conversely, gliosarcoma cells where the cleavage did not occur were noninvasive (79). Using a neoepitope antibody (anti-QEAVESE, named B50), which reacted specifically with the 50-kDa fragment, gene expression analysis, digestion with recombinant ADAMTS4 and specific inhibitors, ADAMTS4 was identified as the protease that was responsible for the cleavage (79) and proteolytic cleavage was also shown to be essential for glioma tumor cell invasion (80, 81). The site is also cleaved by ADAMTS5, but not ADAMTS1 (82). Site-specific mutation of the cleavage site was used to generate a variant resistant to ADAMTS-processing (80). The cleavage-resistant variant reduced invasiveness of glioma cells suggesting that one of the two fragments generated may act independently of brevican in promoting invasion (80). Subsequently, it was shown that the N-terminal fragment bound to fibronectin to promote cell migration (83). Thus, ADAMTS cleavage of brevican is functionally of great consequence in glioma invasion. Furthermore, increased brevican cleavage was noted in several regions of the brain after kainite-induced seizures and associated with seizure-induced degenerative lesions (84). Subsequent to a review of the role of brevican processing in neuroplasticity (85), studies have identified reduced brevican peptides in cerebrospinal fluid in traumatic brain injury and hydrocephalus, in association with increased ADAMTS activity (86).
Neurocan is cleaved at the Met368-Leu369 peptide bond to generate an N-terminal fragment, named neurocan 130 and a C-terminal fragment termed neurocan-C (87). Neurocan is cleaved in the adult, but not in juvenile nervous system (87), with neurocan 130 persisting in the perineuronal net, consistent with it containing the N-terminal HA-binding G1 domain (88). Neurocan is cleaved by both ADAMTS4 and ADAMTS12 (89). The precise ADAMTS12 cleavage site is unknown, but on the basis of fragment size, appears to be distinct from cleavage at Glu395-Ser396 (89). The neurocan-C fragment is increased in the cerebrospinal fluid in amyotrophic lateral sclerosis (90). The translational impact of the ability of ADAMTS proteases to cleave proteoglycans such as versican, aggrecan, brevican, and neurocan may lie in their ability to reduce the neurite inhibitory activity of these CSPGs in perineuronal nets via proteolysis and a possible conversion to promigratory effect, demonstrated for example, by ADAMTS4 promotion of functional recovery after spinal cord injury (91).
PROTEOLYSIS OF CELL-SURFACE PROTEOGLYCANS
Syndecans 1–4 and glypicans 1–6 are HSPGs with key roles at the cell surface. Syndecans regulate cell adhesion by acting as coreceptors for integrins and members of both families bind growth factors and cytokines. Whereas syndecans are single-pass transmembrane molecules, glypicans are membrane-bound by a GPI anchor.
A major function of glypicans appears to be regulation of signaling pathways stimulated by Wnts, hedgehogs (Hhs), and fibroblast growth factors (FGFs) by binding to these morphogens/growth factors via the HS chains (92). Glypican-3 has been heavily studied in this context and is essential for tissue morphogenesis and development since mutations in this gene result in Simpson–Golabi–Behmel syndrome, a generalized overgrowth malformation (93). It is a serological marker for early stage hepatocellular cancer because the amino-terminal portion cleaved between Arg358 and Ser359 is specifically detected in the sera of patients with these tumors (94), although the protease responsible is unknown.
Glypicans appear to be subject to endoproteolytic cleavage, specifically described in glypican-3 and -4, by a furin-like convertase during their biosynthesis (95, 96). The cleavage is located at the C-terminal end of the cysteine-rich domain and generates two subunits that remain attached by disulfide bonds. The structural change created by proteolysis has remarkable impact on both the binding properties and GAG-chain sulfation and processing by convertases is required for glypican-3 to suppress Hh signaling (97). Furthermore, glypicans can be shed into the extracellular microenvironment by notum, a cell surface lipase, through cleavage of the GPI anchor (98) and glypican-4 is released from astrocytes by proteolytic shedding by ADAM9 (99).
All syndecans have been found to be proteolytically shed from the cell surface (100), and the mechanisms and impact of shedding on wound healing and cancer were previously reviewed in-depth (101). The shed ectodomains resulting from MMP, thrombin, ADAMTS, or ADAM activity, can inhibit the activity of syndecans by competition, and subsequent to ectodomain shedding intramembrane proteolysis may release the intracellular domain with new signaling properties (101). Syndecan-2 ectodomain shedding inhibits angiogenesis by regulating endothelial cell migration (102). Syndecan-1 shed from multiple myeloma cells is endocytosed by bone marrow stromal cells and trafficked to nuclei in an HS-dependent manner, where it binds to and inhibits histone acetyltransferases, influencing gene transcription. In contrast to classical ectodomain shedding, where entire ectodomains are released, however, ADAMTS1 and ADAMTS4 cleaved off only the N-terminal 6–7 kDa. Although the cleaved peptide bond was not identified, the site of cleavage was judged to be near the N-terminal HS attachment site, but unrelated to ADAMTS1 binding to HS, since cleavage also occurred in cells that do not attach HS to core proteins (103). ADAMTS1 knockdown in vascular endothelial cells demonstrated reduced cell-surface syndecan-4, which was attributed to increased MMP9 (103).
Betaglycan, a single-pass transmembrane CSPG that functions as a high-affinity TGF-β receptor (also designated TGFβRIII), undergoes juxtamembrane cleavage by plasmin, granzyme B, the zinc metalloproteases MT1-MMP (MMP14), and BMP1/Tolloid-like proteases as well as intramembrane γ-secretases with significant impact on TGFβ signaling and tumorigenesis (104–108). The BMP-1 cleavage sites were determined using Edman sequencing and N-terminomics (see the final section for explanation of these approaches) (105).
Agrin is a widely expressed HSPG comprising a 220 kDa core protein with two HS/CS chains. Agrin has two transcriptional start sites and thus two alternative N-termini. The short form is localized in the brain as a transmembrane protein (109), whereas the longer form is secreted and is localized in the ECM (110), where it binds to laminin (111). Agrin knockout mice die at birth from asphyxiation, because of its requirement for proper assembly of the neuromuscular junction (112). It is associated with many diseases, including in the liver, where it accumulates in cirrhosis and hepatocellular carcinoma (113). Agrin is expressed in the brain and accumulates in Alzheimer’s disease (114) and in the renal glomerular basement membrane (115). Agrin has an important role in mechanotransduction in keratinocytes and liver cancer cells, and in the former, one mechanosensory role is to upregulate the protease MMP12 (116, 117). Administration of agrin in animal models of myocardial infarction reactivated cardiomyocyte proliferation and ultimately led to cardiac repair (118). Hence, agrin proteolysis has a high potential functional significance, but only limited knowledge is presently available.
Neurotrypsin, a serine protease expressed in the nervous system, cleaves agrin at two C-terminal sites, an α site (Arg995-Ala996) and β site (Lys1754-Ser1755), to generate 90 and 22 kDa fragments (119). The two cleavage sites have distinct sequences, with the α site having a conserved Arg residue at position P1 (the residue immediately upstream of the cleavage site using the nomenclature of Schechter and Berger for protease cleavage sites (120)] and Glu at the P2 position, whereas the β site has a conserved Lys at the P1 position and Glu at the P2 position (119). Mutagenesis of the cleavage sites showed that neurotrypsin preferentially bound the P2 Glu residue in both neurotrypsin cleavage sites (119). Neither site was cleaved in neurotrypsin-deficient mice, suggesting that in vivo, cleavage of neural agrin at these sites was strictly dependent on neurotrypsin (119). Agrin is additionally susceptible to matrix metalloproteinase (MMP) activity. MMP3 treatment resulted in loss of agrin staining in synaptic basal lamina (121), and subsequent work showed MMP3 localization to the neuromuscular junction and direct cleavage of agrin by MMP3 (122). Moreover, altered neuromuscular junction structure and function, with increased acetylcholine receptors, junctional folds, and agrin immunoreactivity in Mmp3 null mice support the hypothesis that synaptic activity may induce MMP3 activation to remove agrin from the synaptic basal lamina (122).
MMP1, -7, -12, and -14 (MT1-MMP) cleavage sites in agrin were determined by Edman degradation sequencing (123). Agrin is cleaved by MMP1 after Asn141 and His1822, by MMP7 after Lys154 and Thr1831, by MMP12 after Ser162 and Thr1831, and by MMP14 after Asn141 and Lys154. An additional agrin cleavage site was uncovered for MMP3 at Thr1650. These sites are each distinct from the observed neurotrypsin cleavages. Furthermore, MMP12 cleavage of agrin abolished binding to laminin, adversely affecting the neuromuscular junction, providing further support for a potential neuroregulatory impact of MMPs (123). Therefore, multiple MMPs can cleave agrin at differing sites that potentially disrupt binding to other ECM components and disrupt or mitigate its function. However, the impact of the various cleavages in vivo remains to be determined conclusively.
Among other neural proteoglycans, receptor-type protein tyrosine phosphatase β (RPTPβ, also known as PTPzeta) is a CSPG whose ectodomain is termed phosphacan. Phosphacan is cleaved by ADAMTS4 (91), potentially making additional contribution to its neurotropic effect provided by brevican and neurocan cleavage, and by plasmin, MMPs, and ADAM17 at several sites, some of which were identified using synthetic peptides as substrates (124, 125). NG2, a transmembrane neural CSPG that is also expressed by vascular smooth muscle cells, is similarly cleaved by several proteases resulting in generation of a soluble ectodomain, with MMP9 being implicated in this effect by several studies (126–131). ADAM10 juxtamembrane cleavage of the NG2 ectodomain is followed by intramembrane cleavage by γ-secretase, resulting in release of an intracellular domain to which regulatory effects on gene expression were attributed (132). Intriguingly, NG2 serves as a receptor for a key Clostridium difficile virulence factor, TcdB, which binds to its ectodomain, and the soluble ectodomain had a protective impact on cells (133). Thus, proteolytic shedding of the ectodomain has the potential to modulate pathogenicity of this potentially lethal infection.
ADAMTS7 AND ADAMTS12—DUAL EXISTENCE AS PROTEASES AND PROTEOGLYCANS
Among the 19 ADAMTS proteases, ADAMTS7 and ADAMTS12, are highly homologous, with an identical domain structure and conserved sequence, which includes a mucin-like domain with attached CS-chains, rendering them proteoglycans, and they may have arisen by duplication of a primordial gene that also encoded a mucin-like domain and GAG-attachment sites (11, 12, 134). They are coexpressed in many tissues, including tendons, and can compensate for each other via transcriptional adaptation (11). Adamts7 mutant, Adamts12 mutant, and Adamts7/Adamts12 double-mutant mice are viable, yet the last mentioned have heterotopic tendon ossification and abnormal collagen fibrils, implying a role for these protease-proteoglycans in connective tissue assembly and homeostasis (11). ADAMTS7 was implicated in atherosclerosis and coronary artery disease in humans, with subsequent validation obtained in mutant mice (135). Like most ADAMTS proteases, ADAMTS7 is secreted and activated via furin processing after Arg220, which was shown to occur at the cell surface (12). Notably, activation by furin was perturbed by a nonsynonymous variant in the propeptide, Ser214 to Pro, associated with atherosclerosis susceptibility, which impaired furin excision of the propeptide; this variant was associated with reduced proteolytic processing of thrombospondin-5 and decreased smooth muscle cell migration (136). The Pro variant is thus predicted to be protective in coronary artery disease (136). ADAMTS7 substrates were identified in two studies, which identified cleavage sites in the proteoglycans agrin, sulf-2, perlecan, and serglycin (137, 138). Also identified were 40 autocatalytic cleavage sites, mostly in the ADAMTS7 propeptide and spacer module, which have the potential to regulate its activity by affecting activation and substrate binding, respectively. The most robust cleavages occurred at two sites in the spacer, specifically, Glu729-Val730 and Glu732-Ala733 (137). These cleavages remove a substantial portion of the C-terminal ancillary domain and thus could have a significant effect on cleavage site efficacy and specificity.
ENDOSTATIN AND ENDOREPELLIN, PROTEOGLYCAN FRAGMENTS WITH BROAD BIOLOGICAL IMPACT
Endostatin and endorepellin are C-terminal fragments generated by proteolysis of collagen XVIII and perlecan, respectively. Endostatin is a powerful antiangiogenic molecule (139, 140) with additional roles in autophagy, fibrosis, regulation of Wnt signaling, and synaptogenesis (141–147). In part, the basis for these diverse roles could be the numerous known interactions of endostatin, including with other collagens, matricellular proteins, integrins, VEGF-R2, and GAGs (148). Collagen XVIII is a multiplexin-class collagen and HSPG containing 10 collagenous domains that are separated and flanked by noncollagenous domains, three of which contain HS-chains, although endostatin itself has no GAGs (140). The collagen XVIII C-terminal region contains a trimerization domain that binds perlecan and laminin to embed it in basement membranes. Beyond this trimerization domain is a linker and the noncollagenous 1 domain (NC1), which when released, is known as endostatin. The three collagen XVIII isoforms, which differ in the composition of the N-terminal half, nevertheless contain the HS chains as well as the NC1/endostatin domain (140).
The linker between the trimerization domain and endostatin is susceptible to attack by numerous proteases (149), which liberate the NC1 domain, generating several endostatin isoforms with distinct N-termini. Initial work suggested two-step proteolysis, in which cleavage by a metalloprotease generated an intermediate form before generation of canonical endostatin by neutrophil and pancreatic elastase or cathepsin L (150). Treatment of recombinant collagen XVIII NC1 with a variety of proteases from different classes confirmed that cathepsin L and elastases efficiently cleaved NC1 into endostatin fragments that differed from each other by 1 amino acid (149) and were slightly larger than the canonical 25 kDa endostatin fragment (139, 151). Cathepsin K and D also cleaved off NC1 but the fragments were much larger and generated at slower rates (149). Several MMPs, including MMP-3, -9, -12, -13, and -20 created the intermediate fragment within 5 h of incubation, whereas MMP-1, -2, -8, and -14 showed lower efficiency (149, 152). The aspartic protease cathepsin D also has activity toward NC1 but generates a 31 kDa product. The varying cleavage sites and the resulting distinct N-termini are consistent with observations that different circulating forms of endostatin exist (153) but nevertheless share antiangiogenic activity. Interestingly, cathepsins and elastase degraded endostatin within 2–20 h after its generation (149). The collagen XVIII homolog collagen XV, which is also an HSPG, has an endostatin-like NC1 domain, but the preceding hinge region is less sensitive to proteolysis than collagen XVIII (154). Endostatin-XV has similar binding partners in ECM as endostatin-XVIII, but is not as well studied as the latter.
Perlecan/HSPG2, whose core protein is the largest among all proteoglycans, typically contains 3 HS chains at its N-terminus, but in some cell types, CS or KS chains replace one of the HS chains, and an additional GAG chain may be attached to the C-terminal domain V (155, 156). Its five designated domains, i.e., domains I to V from N- to C-terminus, each have a modular structure and interact with a variety of other ECM components, growth factors, and other molecules to influence diverse biological processes in many contexts (for review, see Refs. 156 and 157). Perlecan is abundant in basement membranes and chondrocyte pericellular matrix, where it is proposed to serve a mechanosensory role (158). Because of its large size and multimodular structure, it is susceptible to the action of many proteases. Perlecan was initially shown to be cleaved by plasmin, MMP3, and rat collagenase following heparanase treatment (although this was not necessarily needed for digestion by each protease) resulting in the release of basic fibroblast growth factor (159). Domain IV is sensitive to proteolytic degradation and its cleavage by MMP7 promoted prostate cancer cell line invasiveness in vitro (160, 161). An 81-kDa fragment arising from proteolytic release of domain V, termed endorepellin, which itself binds to endostatin, was extensively investigated to define its origins and effects (162). The action of endorepellin in inhibiting endothelial migration was comparable with endostatin, and in some experiments was even greater, yet these two angiogenesis inhibitors essentially lost their activities upon binding to each other (162). The initial discovery of the activity of domain V/endorepellin also identified an Asp-N proteolytic activity that cleaved off the LG3 module of human perlecan, which possesses most of the angiostatic effects on vascular endothelium; the cleavage of LG3 at the Asn4196-Asp4197 peptide bond was later attributed to Tolloid-like proteases (162, 163).
The biological impact, clinical relevance, and mechanisms of endorepellin and other angiostatic proteoglycan fragments were previously extensively reviewed, hence only one key translational and mechanistic aspect is provided as an example here, illustrating the diverse effects on the vasculature (2, 164–166). Persistently high domain V fragment levels were observed after stroke, attesting to the presence of the appropriate protease activities that cleave off domain V (167). Perlecan-deficient mice have larger infarcts and less reparative angiogenesis than wild-type littermates after stroke (168, 169). Analysis of cerebral ischemia using an ex vivo bioassay has suggested that perlecan is sensitive to proteolysis by MMPs and cathepsins B and L in the ischemic tissue (170). Systemic administration of recombinant endorepellin in a mouse model of stroke provided neuroprotection and enhanced angiogenesis, enabling neural repair and rescued stroke-affected motor function (168). Perlecan-deficient mice had fewer neuroblastic cells after stroke, but recombinant domain V fragment was able to restore healing (167). Perlecan domain V enhanced PDGF-BB-induced phosphorylation of PDGFRβ, SHP-2, and FAK in part through integrin α5β1 and promoted pericyte migration after cerebral ischemia (169). In contrast, the LG1-LG2 and LG3 fragments of perlecan domain V inhibit angiogenesis by interacting with α2β1 integrin, reducing endothelial attachment to fibronectin and collagen I and the migration and proliferation of vascular endothelium. Endorepellin is proapoptotic and prevents collagen-induced tube formation by endothelial cells (162).
MAPPING PROTEOLYSIS WITHIN THE PROTEOGLYCANOME
These examples discussed illustrate the enormous impact of proteoglycan proteolysis in disease and embryonic development. How were these cleavage sites identified? How may one undertake future discovery of similarly significant proteolytic events in proteoglycans most effectively? Until the adoption of protein mass spectrometry (MS), identification of protein cleavage sites relied on N-terminal sequencing by Edman degradation, in which the protein sequence was determined by step-wise cleavage of N-terminal residues and their identification (171). This technique is by no means redundant because MS-based approaches also have their limitations, especially with regard to proteoglycan analysis (see below). Edman degradation requires isolation of a single protein species (such as by its purification, or gel electrophoresis followed by transfer to a polyvinylidene fluoride membrane) for accurate sequencing and provides a single N-terminal peptide sequence per protein species, whereas MS can handle samples of greater complexity, with higher throughput and greater sensitivity to simultaneously define multiple cleavage sites in a single experiment. Furthermore, Edman degradation is limited by the resolution of gels, and challenged further when the protein fragments are <4 kDa, typically requires greater amounts of protein than MS, will not work if the N-terminal residue is chemically modified, such as by naturally occurring acetylation, and disulfide-bonded Cys residues are generally not sequenced unless chemically modified before analysis (172). At least five to six amino acid residues should be sequenced for reliable identification by Edman degradation, especially in proteins with repetitive sequences.
MS, typically as liquid chromatography-tandem MS (LC-MS/MS), is deployed in a variety of strategies for identifying cleavage sites (173, 174). LC-MS/MS experiments traditionally utilize bottom-up proteomics, colloquially known as shotgun proteomics, in which the protein(s) of interest is digested into peptides by proteases such as trypsin, which have a high cleavage site fidelity (trypsin reliably cleaves downstream of Arg/Lys residues and only rarely elsewhere). Subsequently, the peptides are injected into the mass spectrometer and their sequence is determined by fragmentation during tandem MS. Peptides that clearly have arisen from cleaved ends of proteins in the sample will have nontryptic N-or C-termini (i.e., if cleaved by proteases without trypsin-like specificity). Rarely, both N- and C-termini are nontryptic. Semitryptic and nontryptic peptides indicate preexisting cleavages in the protein occurring before trypsin digest for shotgun proteomics. Martin et al. (68) developed a quantitative label-free z-score-based approach that ranked abundance of semitryptic peptides and identified 27 sites of versican cleavage by ADAMTS1, -4, and -5. The known Glu441-Ala442 site, consistently identified as a site cleaved by each protease in these experiments, provided a benchmark for ranking the other, previously unknown cleavages. Several novel cleavage sites were detected with similar z-scores as peptides reporting cleavage at Glu441-Ala442, demonstrating that these ADAMTS proteases each cleaved versican at additional sites. Interestingly, despite sharing the Glu441-Ala442 site, each protease had different preferred sites. The novel ADAMTS1, ADAMTS4, and ADAMTS5 cleavages occurred throughout the full-length proteoglycan sequence but were predominantly located in the GAGβ-region in two distinct clusters, one located centrally, and the other toward the C-terminus (68). In contrast, control versican digests (i.e., performed with inactive proteases) demonstrated tryptic peptides with higher abundance than in the digested sample that contained within them the inferred cleavage sites, supporting cleavage site identification (68). The findings affirmed a previously noted preference for cleavage by these ADAMTS proteases after Glu residues (60) and also indicated a preference for hydrophobic residues at the P1′ position [the residue immediately downstream of the cleavage site using the nomenclature of Schechter and Berger for protease cleavage sites (68, 120)]. However, this approach, albeit technically straightforward, will not detect the activity of proteases with trypsin-like specificity, a drawback that can be circumvented by employing “working” proteases with different specificity from trypsin, such as chymotrypsin, Glu-C, and Asp-N. The numbers of informative semitryptic peptides detected will be proportionately lower in mixtures having a complex protein composition (high proteome complexity) and this approach is thus best reserved for analysis of sites in purified proteins.
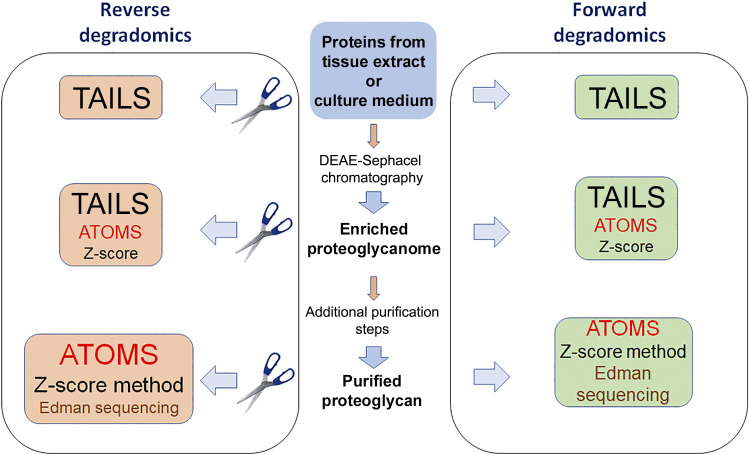
These shortcomings led to development of N- and C-terminomics strategies in which protein termini in complex mixtures or digests of purified proteins are first chemically modified or labeled with adducts or isobars of known mass at the protein level (to identify the cleaved fragments subsequently in a trypsin-digested protein mixture). Because the analysis can be conducted on a proteome-wide (-omics) scale it is referred to as degradomics; forward degradomics refers to de novo identification of cleaved termini in complex protein mixtures without reference to a protease. Reverse degradomics refers to terminomics delineation of cleavages resulting from the activity of a specific protease that is added to a protein or protein mixture or is undertaken in cells or tissues lacking a specific protease gene (Fig. 2). An attractive feature of reverse degradomics methods is the ability to use stable isotope tags of differential mass or reporter content to label termini, which permits multiplexing, greatly reducing sample handling error and MS run-to-run variation while facilitating statistical analysis and relative peptide quantitation (175).
Figure 2.
Strategies for identification of proteoglycan cleavages. The starting material (center) for degradomics investigations could be complex samples such as tissue extract, culture medium, or an enriched proteoglycan mixture without further characterization or individual purified proteoglycans. In forward degradomics, cleavage sites existing in the samples are determined de novo without reference to any protease, using a single label to tag and block protein N-termini. For reverse degradomics, the samples are treated with a protease of choice to specifically determine its proteoglycan targets, using inactive protease as a control; next, the protein N-termini are labeled with tags of different mass, and the samples are combined for further analysis. The colored boxes illustrate the recommended methods appropriate to the samples with order of preference indicated by relative text size, i.e., a proteome-scale method such as Terminal Amine Isotopic Labeling of Substrates (TAILS) is best suited for highly complex samples; Edman degradation or z-score proteomics is suited for single protein bands or molecular species; multiple cleavage sites in a single protein or proteoglycan mixture of limited complexity are best identified by Amine Terminal Oriented Mass Spectrometry (ATOMS). The z-score method would be accessible to most laboratories, but has limited applicability for complex samples, and will not detect trypsin-like cleavages. These strategies and their limitations and the preferred applications of each are explained in the text.
N-terminomics is presently more widely used than C-terminomics since N-termini are more readily reactive (174). One of the more widely used N-terminomics techniques is termed Terminal Amine Isotopic Labeling of Substrates (TAILS) (47). Following labeling of protein N-termini (which blocks their further reactivity), the sample is digested with trypsin, and labeled peptides are enriched by negative selection, using a hyperbranched polyglycerol aldehyde polymer to remove reactive N-termini (47). Thus, peptides containing cleaved protein termini are preferentially brought forward for MS. TAILS is used both for forward degradomics and reverse degradomics. In a variation of TAILS, named Amino-Terminal Oriented Mass Spectrometry of Substrates (ATOMS), the labeled/blocked peptides do not undergo enrichment. ATOMS is preferred when determining cleavage sites in a single protein substrate, for example, one digested with a single protease of interest (172) (Fig. 2). TAILS and ATOMS have each been used for identifying cleavage sites in cartilage proteoglycans (42). Proteoglycan substrates and cleavage sites were also revealed in TAILS experiments that undertook broad, unbiased searches for substrates of specific proteases. For example, a reverse degradomics strategy used TAILS to seek ADAMTS2 and ADAMTS14 substrates both in cultured fibroblast ECM and in mouse skin, identifying decorin, biglycan, and lumican as prospective ADAMTS2 and ADAMTS14 substrates and perlecan and versican as specific ADAMTS14 substrates, with delineation of specific cleavage sites (176, 177). HtrA1 digestion of cartilage in a reverse degradomics approach demonstrated numerous cleavage sites in aggrecan, biglycan, decorin, and fibromodulin, and the cleavage sites in biglycan were validated by direct cleavage of recombinant biglycan using recombinant HtrA1 and application of ATOMS (42).
Degradomics analyses have shown relatively few cleavages in the GAG-bearing regions of versican and aggrecan. In part, it could be that these regions are innately resistant to proteolysis because of their surface carbohydrate, but they are also likely to be relatively resistant to detection by MS. Martin et al. (68) observed that the GAGβ domain of V1 versican had a lower prevalence of Arg residues, suggesting that the peptides resulting from versican (and aggrecan) digestion could be larger than in most proteins and may conceivably exceed the mass/charge windows typically used in MS. The variable carbohydrate modifications in these regions are likely to render the peptides unrecognizable by spectrum-analysis software, which seeks peptides of fixed mass/charge ratio. The net negative charge of some proteoglycan core proteins resulting from a higher-than-normal frequency of negatively charged amino acids and lower frequency of Arg/Lys residues could also be unfavorable for peptide detection by MS. For these reasons, even though Edman degradation is relatively limited in its current usage, it still has an important role to play in defining N-termini of proteoglycan fragments. However, these limitations of MS-based approaches do not apply to the majority of proteoglycans, which may lack the high GAG- and O-linked oligosaccharide content of aggrecan and versican and more frequently contain only 1–3 GAG chains clustered in a short sequence.
Conclusions and Future Prospects
This review has used a few examples of the impact of proteolysis on molecular structure, the roles of specific proteases, and the biological consequences of proteolysis to illustrate that proteolysis of proteoglycans is both widespread and consequential. The key points of this review are that proteolysis can be activating as well as inactivating, that diverse groups of proteases are involved, and that the resulting proteolytic fragments can have distinct roles. Whereas specific proteolytic cleavages occurring in aggrecan, versican, collagen XVIII, and perlecan and the proteases responsible have received much attention, much less is known about proteolysis of other proteoglycans and their derivative matrikines. Although the cellular signaling impact of cell-surface proteoglycans has received much attention, much less is known about the proteases responsible for cleavages and the specific fragments generated.
MS has revolutionized investigation of degradomes, revealing a hitherto unanticipated scale of cleavages where it has been applied and providing tantalizing views of a mostly unexplored world. A strategy that will be very productive in identifying proteoglycan cleavages in tissues is to combine enrichment of proteoglycans with MS applications such as the z-score method, TAILS, or ATOMS (76) (Fig. 2). Specifically, enriching tissue proteoglycans by ion-exchange chromatography based on their net negative charge improves the likelihood of their detection by mass spectrometry in complex samples (by reducing overall proteome complexity and “expanding” the proteoglycanome at the expense of the nonproteoglycan components) (178). Tissue proteoglycans that are enriched in this way (76) can provide a library of substrates for digestion with any protease of interest to identify its cleavage sites (Fig. 2). Indeed, neural proteoglycans were previously isolated using diethylaminoethyl (DEAE)-Sepharose chromatography for digestion by ADAMTS4, but only candidate substrate cleavages (brevican, neurocan, and phosphacan) were subsequently examined by Western blotting (91), whereas application of TAILS would likely have revealed additional cleavages. If a sufficiently large number of cleavage sites are determined for a protease, another benefit of an -omics approach is the ability to ascertain definitive sequence preferences around cleavage sites using bioinformatics (179). Combining forward and reverse degradomics can causally implicate specific proteases in the generation of cleaved peptide bonds identified in tissues (42). This strategy could be informative for uncovering proteoglycan degradomes of tissues such as the vasculature and brain, where proteoglycans are functionally critical, but constitute a minor fraction of their proteomes. In tissues such as cartilage, which has a high proteoglycan content and is cell-poor relative to other tissues, proteoglycan enrichment may not be necessary. Subsequent to uncovering proteolytic events, neo-epitope antibodies that specifically recognize the cleaved fragments but not intact proteoglycans are now well established as useful research and clinical biomarker tools (33, 36, 48, 60, 61, 65, 180). The bioactivity of individual proteoglycan fragments could be systematically ascertained, although this is likely to be a herculean task given the plethora of cleavage sites that occur. Nonetheless, where importance of a proteolytic cleavage is revealed by genetic, biochemical, or biomarker evidence, investigation of the activity of proteolytic fragments could identify targetable disease pathways. The significance of endostatin, endorepellin, and versikine suggests that a potentially large number of potential matrikines may exist and await exploration.
GRANTS
This work was supported by National Institute of Arthritis and Musculoskeletal and Skin Diseases Grant AR078498 and the Arthritis Foundation.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
S.S.A. prepared figures; T.J.M., S.B., D.R.M., and S.S.A. drafted manuscript; T.J.M., S.B., D.R.M., and S.S.A. edited and revised manuscript; T.J.M., S.B., D.R.M., and S.S.A. approved final version of manuscript.
ACKNOWLEDGMENTS
This article is part of the special collection "Deciphering the Role of Proteoglycans and Glycosaminoglycans in Health and Disease." Liliana Schaefer, MD, served as Guest Editor of this collection.
REFERENCES
- 1. Lindahl U, Couchman J, Kimata K, Esko JD. Proteoglycans and sulfated glycosaminoglycans. In: Essentials of Glycobiology, edited by Varki A, Cummings RD, Esko JD, Stanley P, Hart GW, Aebi M, Darvill AG, Kinoshita T, Packer NH, Prestegard JH, Schnaar RL, Seeberger PH.. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press, 2015, p. 207–221. [Google Scholar]
- 2. Iozzo RV, Schaefer L. Proteoglycan form and function: a comprehensive nomenclature of proteoglycans. Matrix Biol 42: 11–55, 2015. doi: 10.1016/j.matbio.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Scranton TW, Iwata M, Carlson SS. The SV2 protein of synaptic vesicles is a keratan sulfate proteoglycan. J Neurochem 61: 29–44, 1993. doi: 10.1111/j.1471-4159.1993.tb03535.x. [DOI] [PubMed] [Google Scholar]
- 4. Le Jan S, Hayashi M, Kasza Z, Eriksson I, Bishop JR, Weibrecht I, Heldin J, Holmborn K, Jakobsson L, Soderberg O, Spillmann D, Esko JD, Claesson-Welsh L, Kjellen L, Kreuger J. Functional overlap between chondroitin and heparan sulfate proteoglycans during VEGF-induced sprouting angiogenesis. Arterioscler Thromb Vasc Biol 32: 1255–1263, 2012. doi: 10.1161/ATVBAHA.111.240622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Miller RE, Ishihara S, Tran PB, Golub SB, Last K, Miller RJ, Fosang AJ, Malfait AM. An aggrecan fragment drives osteoarthritis pain through Toll-like receptor 2. JCI Insight 3: e95704, 2018. doi: 10.1172/jci.insight.95704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nandadasa S, O’Donnell A, Murao A, Yamaguchi Y, Midura RJ, Olson L, Apte SS. The versican-hyaluronan complex provides an essential extracellular matrix niche for Flk1(+) hematoendothelial progenitors. Matrix Biol 97:40–57, 2021. doi: 10.1016/j.matbio.2021.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Schaefer L, Dikic I. Autophagy: instructions from the extracellular matrix. Matrix Biol 100–101: 1–8, 2021. doi: 10.1016/j.matbio.2021.06.002. [DOI] [PubMed] [Google Scholar]
- 8. Halfter W, Dong S, Schurer B, Cole GJ. Collagen XVIII is a basement membrane heparan sulfate proteoglycan. J Biol Chem 273: 25404–25412, 1998. doi: 10.1074/jbc.273.39.25404. [DOI] [PubMed] [Google Scholar]
- 9. Li D, Clark CC, Myers JC. Basement membrane zone type XV collagen is a disulfide-bonded chondroitin sulfate proteoglycan in human tissues and cultured cells. J Biol Chem 275: 22339–22347, 2000. doi: 10.1074/jbc.M000519200. [DOI] [PubMed] [Google Scholar]
- 10. McCormick D, van der Rest M, Goodship J, Lozano G, Ninomiya Y, Olsen BR. Structure of the glycosaminoglycan domain in the type IX collagen-proteoglycan. Proc Natl Acad Sci USA 84: 4044–4048, 1987. doi: 10.1073/pnas.84.12.4044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mead TJ, McCulloch DR, Ho JC, Du Y, Adams SM, Birk DE, Apte SS. The metalloproteinase-proteoglycans ADAMTS7 and ADAMTS12 provide an innate, tendon-specific protective mechanism against heterotopic ossification. JCI Insight 3: e92941, 2018. doi: 10.1172/jci.insight.92941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Somerville RP, Longpre JM, Apel ED, Lewis RM, Wang LW, Sanes JR, Leduc R, Apte SS. ADAMTS7B, the full-length product of the ADAMTS7 gene, is a chondroitin sulfate proteoglycan containing a mucin domain. J Biol Chem 279: 35159–35175, 2004. doi: 10.1074/jbc.M402380200. [DOI] [PubMed] [Google Scholar]
- 13. El Masri R, Seffouh A, Roelants C, Seffouh I, Gout E, Pérard J, Dalonneau F, Nishitsuji K, Noborn F, Nikpour M, Larson G, Crétinon Y, Friedel-Arboleas M, Uchimura K, Daniel R, Lortat-Jacob H, Filhol O, Vivès RR. Extracellular endosulfatase Sulf-2 harbors a chondroitin/dermatan sulfate chain that modulates its enzyme activity. Cell Rep 38: 110516, 2022. doi: 10.1016/j.celrep.2022.110516. [DOI] [PubMed] [Google Scholar]
- 14. Lord MS, Melrose J, Day AJ, Whitelock JM. The inter-α-trypsin inhibitor family: versatile molecules in biology and pathology. J Histochem Cytochem 68: 907–927, 2020. doi: 10.1369/0022155420940067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Beckmann AM, Glebov K, Walter J, Merkel O, Mangold M, Schmidt F, Becker-Pauly C, Gütschow M, Stirnberg M. The intact Kunitz domain protects the amyloid precursor protein from being processed by matriptase-2. Biol Chem 397: 777–790, 2016. doi: 10.1515/hsz-2015-0263. [DOI] [PubMed] [Google Scholar]
- 16. Pangalos MN, Shioi J, Efthimiopoulos S, Wu A, Robakis NK. Characterization of appican, the chondroitin sulfate proteoglycan form of the Alzheimer amyloid precursor protein. Neurodegeneration 5: 445–451, 1996. doi: 10.1006/neur.1996.0061. [DOI] [PubMed] [Google Scholar]
- 17. Noborn F, Nikpour M, Persson A, Sihlbom C, Nilsson J, Larson G. A glycoproteomic approach to identify novel proteoglycans. Methods Mol Biol 2303: 71–85, 2022. doi: 10.1007/978-1-0716-1398-6_7. [DOI] [PubMed] [Google Scholar]
- 18. Kovalszky I, Hjerpe A, Dobra K. Nuclear translocation of heparan sulfate proteoglycans and their functional significance. Biochim Biophys Acta 1840: 2491–2497, 2014. doi: 10.1016/j.bbagen.2014.04.015. [DOI] [PubMed] [Google Scholar]
- 19. Ricard-Blum S, Vallet SD. Fragments generated upon extracellular matrix remodeling: biological regulators and potential drugs. Matrix Biol 75-76: 170–189, 2019. doi: 10.1016/j.matbio.2017.11.005. [DOI] [PubMed] [Google Scholar]
- 20. Hascall VC, Heinegard D. Aggregation of cartilage proteoglycans. I. The role of hyaluronic acid. J Biol Chem 249: 4232–4241, 1974. [PubMed] [Google Scholar]
- 21. Hayes AJ, Melrose J. Aggrecan, the primary weight-bearing cartilage proteoglycan, has context-dependent, cell-directive properties in embryonic development and neurogenesis: aggrecan glycan side chain modifications convey interactive biodiversity. Biomolecules 10: 1244, 2020. doi: 10.3390/biom10091244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Perissinotto D, Iacopetti P, Bellina I, Doliana R, Colombatti A, Pettway Z, Bronner-Fraser M, Shinomura T, Kimata K, Morgelin M, Lofberg J, Perris R. Avian neural crest cell migration is diversely regulated by the two major hyaluronan-binding proteoglycans PG-M/versican and aggrecan. Development 127: 2823–2842, 2000. doi: 10.1242/dev.127.13.2823. [DOI] [PubMed] [Google Scholar]
- 23. Rowlands D, Lensjø KK, Dinh T, Yang S, Andrews MR, Hafting T, Fyhn M, Fawcett JW, Dick G. Aggrecan directs extracellular matrix-mediated neuronal plasticity. J Neurosci 38: 10102–10113, 2018. doi: 10.1523/JNEUROSCI.1122-18.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Heinegard D, Saxne T. The role of the cartilage matrix in osteoarthritis. Nat Rev Rheumatol 7: 50–56, 2011. doi: 10.1038/nrrheum.2010.198. [DOI] [PubMed] [Google Scholar]
- 25. Melrose J, Fuller ES, Roughley PJ, Smith MM, Kerr B, Hughes CE, Caterson B, Little CB. Fragmentation of decorin, biglycan, lumican and keratocan is elevated in degenerate human meniscus, knee and hip articular cartilages compared with age-matched macroscopically normal and control tissues. Arthritis Res Ther 10: R79, 2008. doi: 10.1186/ar2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Melrose J, Smith SM, Fuller ES, Young AA, Roughley PJ, Dart A, Little CB. Biglycan and fibromodulin fragmentation correlates with temporal and spatial annular remodelling in experimentally injured ovine intervertebral discs. Eur Spine J 16: 2193–2205, 2007. doi: 10.1007/s00586-007-0497-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shu CC, Flannery CR, Little CB, Melrose J. Catabolism of fibromodulin in developmental rudiment and pathologic articular cartilage demonstrates novel roles for MMP-13 and ADAMTS-4 in C-terminal processing of SLRPs. Int J Mol Sci 20: 579, 2019. doi: 10.3390/ijms20030579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lark MW, Bayne EK, Flanagan J, Harper CF, Hoerrner LA, Hutchinson NI, Singer II, Donatelli SA, Weidner JR, Williams HR, Mumford RA, Lohmander LS. Aggrecan degradation in human cartilage. Evidence for both matrix metalloproteinase and aggrecanase activity in normal, osteoarthritic, and rheumatoid joints. J Clin Invest 100: 93–106, 1997. doi: 10.1172/JCI119526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Stanton H, Melrose J, Little CB, Fosang AJ. Proteoglycan degradation by the ADAMTS family of proteinases. Biochim Biophys Acta 1812: 1616–1629, 2011. doi: 10.1016/j.bbadis.2011.08.009. [DOI] [PubMed] [Google Scholar]
- 30. Fosang AJ, Neame PJ, Hardingham TE, Murphy G, Hamilton JA. Cleavage of cartilage proteoglycan between G1 and G2 domains by stromelysins. J Biol Chem 266: 15579–15582, 1991. [PubMed] [Google Scholar]
- 31. Sandy JD, Flannery CR, Neame PJ, Lohmander LS. The structure of aggrecan fragments in human synovial fluid. Evidence for the involvement in osteoarthritis of a novel proteinase which cleaves the Glu 373-Ala 374 bond of the interglobular domain. J Clin Invest 89: 1512–1516, 1992. doi: 10.1172/JCI115742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Poon CJ, Plaas AH, Keene DR, McQuillan DJ, Last K, Fosang AJ. N-linked keratan sulfate in the aggrecan interglobular domain potentiates aggrecanase activity. J Biol Chem 280: 23615–23621, 2005. doi: 10.1074/jbc.M412145200. [DOI] [PubMed] [Google Scholar]
- 33. Hughes CE, Caterson B, Fosang AJ, Roughley PJ, Mort JS. Monoclonal antibodies that specifically recognize neoepitope sequences generated by ‘aggrecanase’ and matrix metalloproteinase cleavage of aggrecan: application to catabolism in situ and in vitro. Biochem J 305: 799–804, 1995. doi: 10.1042/bj3050799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Little CB, Flannery CR, Hughes CE, Mort JS, Roughley PJ, Dent C, Caterson B. Aggrecanase versus matrix metalloproteinases in the catabolism of the interglobular domain of aggrecan in vitro. Biochem J 344: 61–68, 1999. [PMC free article] [PubMed] [Google Scholar]
- 35. Ng CK, Handley CJ, Preston BN, Robinson HC. The extracellular processing and catabolism of hyaluronan in cultured adult articular cartilage explants. Arch Biochem Biophys 298: 70–79, 1992. doi: 10.1016/0003-9861(92)90095-e. [DOI] [PubMed] [Google Scholar]
- 36. Tortorella MD, Pratta M, Liu RQ, Austin J, Ross OH, Abbaszade I, Burn T, Arner E. Sites of aggrecan cleavage by recombinant human aggrecanase-1 (ADAMTS-4). J Biol Chem 275: 18566–18573, 2000. doi: 10.1074/jbc.M909383199. [DOI] [PubMed] [Google Scholar]
- 37. Gendron C, Kashiwagi M, Lim NH, Enghild JJ, Thogersen IB, Hughes C, Caterson B, Nagase H. Proteolytic activities of human ADAMTS-5: comparative studies with ADAMTS-4. J Biol Chem 282: 18294–18306, 2007. [Erratum in J Biol Chem 282: 28296, 2007]. doi: 10.1074/jbc.M701523200. [DOI] [PubMed] [Google Scholar]
- 38. Tortorella M, Pratta M, Liu RQ, Abbaszade I, Ross H, Burn T, Arner E. The thrombospondin motif of aggrecanase-1 (ADAMTS-4) is critical for aggrecan substrate recognition and cleavage. J Biol Chem 275: 25791–25797, 2000. doi: 10.1074/jbc.M001065200. [DOI] [PubMed] [Google Scholar]
- 39. Lees S, Golub SB, Last K, Zeng W, Jackson DC, Sutton P, Fosang AJ. Bioactivity in an Aggrecan 32-mer fragment is mediated via Toll-like receptor 2. Arthritis Rheumatol 67: 1240–1249, 2015. doi: 10.1002/art.39063. [DOI] [PubMed] [Google Scholar]
- 40. Scott IC, Imamura Y, Pappano WN, Troedel JM, Recklies AD, Roughley PJ, Greenspan DS. Bone morphogenetic protein-1 processes probiglycan. J Biol Chem 275: 30504–30511, 2000. doi: 10.1074/jbc.M004846200. [DOI] [PubMed] [Google Scholar]
- 41. Melching LI, Fisher WD, Lee ER, Mort JS, Roughley PJ. The cleavage of biglycan by aggrecanases. Osteoarthritis Cartilage 14: 1147–1154, 2006. doi: 10.1016/j.joca.2006.05.014. [DOI] [PubMed] [Google Scholar]
- 42. Bhutada S, Li L, Willard B, Muschler G, Piuzzi N, Apte SS. Forward and reverse degradomics defines the proteolytic landscape of human knee osteoarthritic cartilage and the role of the serine protease HtrA1. Osteoarthritis Cartilage 30: 1091–1102, 2022. doi: 10.1016/j.joca.2022.02.622. [DOI] [PubMed] [Google Scholar]
- 43. Zeng-Brouwers J, Beckmann J, Nastase MV, Iozzo RV, Schaefer L. De novo expression of circulating biglycan evokes an innate inflammatory tissue response via MyD88/TRIF pathways. Matrix Biol 35: 132–142, 2014. doi: 10.1016/j.matbio.2013.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Diehl V, Huber LS, Trebicka J, Wygrecka M, Iozzo RV, Schaefer L. The role of decorin and biglycan signaling in tumorigenesis. Front Oncol 11: 801801, 2021. doi: 10.3389/fonc.2021.801801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pietraszek K, Brézillon S, Perreau C, Malicka-Błaszkiewicz M, Maquart FX, Wegrowski Y. Lumican-derived peptides inhibit melanoma cell growth and migration. PLoS One 8: e76232, 2013. doi: 10.1371/journal.pone.0076232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pietraszek-Gremplewicz K, Karamanou K, Niang A, Dauchez M, Belloy N, Maquart FX, Baud S, Brézillon S. Small leucine-rich proteoglycans and matrix metalloproteinase-14: Key partners? Matrix Biol 75–76: 271–285, 2019. doi: 10.1016/j.matbio.2017.12.006. [DOI] [PubMed] [Google Scholar]
- 47. Kleifeld O, Doucet A, Auf Dem Keller U, Prudova A, Schilling O, Kainthan RK, Starr AE, Foster LJ, Kizhakkedathu JN, Overall CM. Isotopic labeling of terminal amines in complex samples identifies protein N-termini and protease cleavage products. Nat Biotechnol 28: 281–288, 2010. doi: 10.1038/nbt.1611. [DOI] [PubMed] [Google Scholar]
- 48. Chamberland A, Wang E, Jones AR, Collins-Racie LA, LaVallie ER, Huang Y, Liu L, Morris EA, Flannery CR, Yang Z. Identification of a novel HtrA1-susceptible cleavage site in human aggrecan: evidence for the involvement of HtrA1 in aggrecan proteolysis in vivo. J Biol Chem 284: 27352–27359, 2009. doi: 10.1074/jbc.M109.037051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Klose R, Prinz A, Tetzlaff F, Weis EM, Moll I, Rodriguez-Vita J, Oka C, Korff T, Fischer A. Loss of the serine protease HTRA1 impairs smooth muscle cells maturation. Sci Rep 9: 18224, 2019. doi: 10.1038/s41598-019-54807-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Tom I, Pham VC, Katschke KJ Jr, Li W, Liang WC, Gutierrez J, Ah Young A, Figueroa I, Eshghi ST, Lee CV, Kanodia J, Snipas SJ, Salvesen GS, Lai P, Honigberg L, van Lookeren Campagne M, Kirchhofer D, Baruch A, Lill JR. Development of a therapeutic anti-HtrA1 antibody and the identification of DKK3 as a pharmacodynamic biomarker in geographic atrophy. Proc Natl Acad Sci USA 117: 9952–9963, 2020. doi: 10.1073/pnas.1917608117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tsuchiya A, Yano M, Tocharus J, Kojima H, Fukumoto M, Kawaichi M, Oka C. Expression of mouse HtrA1 serine protease in normal bone and cartilage and its upregulation in joint cartilage damaged by experimental arthritis. Bone 37: 323–336, 2005. doi: 10.1016/j.bone.2005.03.015. [DOI] [PubMed] [Google Scholar]
- 52. Urano T, Narusawa K, Kobayashi S, Shiraki M, Horie-Inoue K, Sasaki N, Hosoi T, Ouchi Y, Nakamura T, Inoue S. Association of HTRA1 promoter polymorphism with spinal disc degeneration in Japanese women. J Bone Miner Metab 28: 220–226, 2010. doi: 10.1007/s00774-009-0124-0. [DOI] [PubMed] [Google Scholar]
- 53. Kenagy RD, Plaas AH, Wight TN. Versican degradation and vascular disease. Trends Cardiovasc Med 16: 209–215, 2006. doi: 10.1016/j.tcm.2006.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Nandadasa S, Foulcer S, Apte SS. The multiple, complex roles of versican and its proteolytic turnover by ADAMTS proteases during embryogenesis. Matrix Biol 35: 34–41, 2014. doi: 10.1016/j.matbio.2014.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ricciardelli C, Sakko AJ, Ween MP, Russell DL, Horsfall DJ. The biological role and regulation of versican levels in cancer. Cancer Metastasis Rev 28: 233–245, 2009. doi: 10.1007/s10555-009-9182-y. [DOI] [PubMed] [Google Scholar]
- 56. Islam S, Watanabe H. Versican: a dynamic regulator of the extracellular matrix. J Histochem Cytochem 68: 763–775, 2020. doi: 10.1369/0022155420953922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Bode-Lesniewska B, Dours-Zimmermann MT, Odermatt BF, Briner J, Heitz PU, Zimmermann DR. Distribution of the large aggregating proteoglycan versican in adult human tissues. J Histochem Cytochem 44: 303–312, 1996. doi: 10.1177/44.4.8601689. [DOI] [PubMed] [Google Scholar]
- 58. Dours-Zimmermann MT, Zimmermann DR. A novel glycosaminoglycan attachment domain identified in two alternative splice variants of human versican. J Biol Chem 269: 32992–32998, 1994. [PubMed] [Google Scholar]
- 59. Kischel P, Waltregny D, Dumont B, Turtoi A, Greffe Y, Kirsch S, De Pauw E, Castronovo V. Versican overexpression in human breast cancer lesions: known and new isoforms for stromal tumor targeting. Int J Cancer 126: 640–650, 2010. doi: 10.1002/ijc.24812. [DOI] [PubMed] [Google Scholar]
- 60. Sandy JD, Westling J, Kenagy RD, Iruela-Arispe ML, Verscharen C, Rodriguez-Mazaneque JC, Zimmermann DR, Lemire JM, Fischer JW, Wight TN, Clowes AW. Versican V1 proteolysis in human aorta in vivo occurs at the Glu441- Ala442 bond, a site that is cleaved by recombinant ADAMTS-1 and ADAMTS-4. J Biol Chem 276: 13372–13378, 2001. doi: 10.1074/jbc.M009737200. [DOI] [PubMed] [Google Scholar]
- 61. Westling J, Gottschall PE, Thompson VP, Cockburn A, Perides G, Zimmermann DR, Sandy JD. ADAMTS4 (aggrecanase-1) cleaves human brain versican V2 at Glu405-Gln406 to generate glial hyaluronate binding protein. Biochem J 377: 787–795, 2004. doi: 10.1042/BJ20030896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Foulcer SJ, Nelson CM, Quintero MV, Kuberan B, Larkin J, Dours-Zimmermann MT, Zimmermann DR, Apte SS. Determinants of versican-V1 proteoglycan processing by the metalloproteinase ADAMTS5. J Biol Chem 289: 27859–27873, 2014. doi: 10.1074/jbc.M114.573287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. McCulloch DR, Nelson CM, Dixon LJ, Silver DL, Wylie JD, Lindner V, Sasaki T, Cooley MA, Argraves WS, Apte SS. ADAMTS metalloproteases generate active versican fragments that regulate interdigital web regression. Dev Cell 17: 687–698, 2009. doi: 10.1016/j.devcel.2009.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Islam S, Chuensirikulchai K, Khummuang S, Keratibumrungpong T, Kongtawelert P, Kasinrerk W, Hatano S, Nagamachi A, Honda H, Watanabe H. Accumulation of versican facilitates wound healing: implication of its initial ADAMTS-cleavage site. Matrix Biol 87: 77–93, 2020. doi: 10.1016/j.matbio.2019.10.006. [DOI] [PubMed] [Google Scholar]
- 65. Nandadasa S, Burin Des Roziers C, Koch C, Tran-Lundmark K, Dours-Zimmermann MT, Zimmermann DR, Valleix S, Apte SS. A new mouse mutant with cleavage-resistant versican and isoform-specific versican mutants demonstrate that proteolysis at the Glu(441)-Ala(442) peptide bond in the V1 isoform is essential for interdigital web regression. Matrix Biol Plus 10: 100064, 2021. doi: 10.1016/j.mbplus.2021.100064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Halpert I, Sires UI, Roby JD, Potter-Perigo S, Wight TN, Shapiro SD, Welgus HG, Wickline SA, Parks WC. Matrilysin is expressed by lipid-laden macrophages at sites of potential rupture in atherosclerotic lesions and localizes to areas of versican deposition, a proteoglycan substrate for the enzyme. Proc Natl Acad Sci USA 93: 9748–9753, 1996. doi: 10.1073/pnas.93.18.9748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Passi A, Negrini D, Albertini R, Miserocchi G, De Luca G. The sensitivity of versican from rabbit lung to gelatinase A (MMP-2) and B (MMP-9) and its involvement in the development of hydraulic lung edema. FEBS Lett 456: 93–96, 1999. doi: 10.1016/S0014-5793(99)00929-1. [DOI] [PubMed] [Google Scholar]
- 68. Martin DR, Santamaria S, Koch CD, Ahnström J, Apte SS. Identification of novel ADAMTS1, ADAMTS4 and ADAMTS5 cleavage sites in versican using a label-free quantitative proteomics approach. J Proteomics 249: 104358, 2021. doi: 10.1016/j.jprot.2021.104358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Islam S, Jahan N, Shahida A, Karnan S, Watanabe H. Accumulation of versican and lack of versikine ameliorate acute colitis. Matrix Biol 107: 59–76, 2022. doi: 10.1016/j.matbio.2022.02.004. [DOI] [PubMed] [Google Scholar]
- 70. Hope C, Emmerich PB, Papadas A, Pagenkopf A, Matkowskyj KA, Van De Hey DR, Payne SN, Clipson L, Callander NS, Hematti P, Miyamoto S, Johnson MG, Deming DA, Asimakopoulos F. Versican-derived matrikines regulate Batf3-dendritic cell differentiation and promote T cell infiltration in colorectal cancer. J Immunol 199: 1933–1941, 2017. doi: 10.4049/jimmunol.1700529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Hope C, Foulcer S, Jagodinsky J, Chen SX, Jensen JL, Patel S, Leith C, Maroulakou I, Callander N, Miyamoto S, Hematti P, Apte SS, Asimakopoulos F. Immunoregulatory roles of versican proteolysis in the myeloma microenvironment. Blood 128: 680–685, 2016. doi: 10.1182/blood-2016-03-705780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. McMahon M, Ye S, Izzard L, Dlugolenski D, Tripp RA, Bean AG, McCulloch DR, Stambas J. ADAMTS5 is a critical regulator of virus-specific T cell immunity. PLoS Biol 14: e1002580, 2016. [Erratum in PLoS Biol 17: e3000558, 2019]. doi: 10.1371/journal.pbio.1002580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Boyd DF, Allen EK, Randolph AG, Guo XJ, Weng Y, Sanders CJ, Bajracharya R, Lee NK, Guy CS, Vogel P, Guan W, Li Y, Liu X, Novak T, Newhams MM, Fabrizio TP, Wohlgemuth N, Mourani PM, Wight TN, Schultz-Cherry S, Cormier SA, Shaw-Saliba K, Pekosz A, Rothman RE, Chen KF, Yang Z, Webby RJ, Zhong N, Crawford JC, Thomas PG, PALISI Pediatric Intensive Care Influenza (PICFLU) Investigators. Exuberant fibroblast activity compromises lung function via ADAMTS4. Nature 587: 466–471, 2020. doi: 10.1038/s41586-020-2877-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Dupuis LE, Nelson EL, Hozik B, Porto SC, Rogers-DeCotes A, Fosang A, Kern CB. Adamts5(−/−) mice exhibit altered aggrecan proteolytic profiles that correlate with ascending aortic anomalies. Arterioscler Thromb Vasc Biol 39: 2067–2081, 2019. doi: 10.1161/ATVBAHA.119.313077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Barallobre-Barreiro J, Radovits T, Fava M, Mayr U, Lin WY, Ermolaeva E, Martínez-López D, Lindberg EL, Duregotti E, Daróczi L, Hasman M, Schmidt LE, Singh B, Lu R, Baig F, Siedlar AM, Cuello F, Catibog N, Theofilatos K, Shah AM, Crespo-Leiro MG, Doménech N, Hübner N, Merkely B, Mayr M. Extracellular matrix in heart failure: role of ADAMTS5 in proteoglycan remodeling. Circulation 144: 2021–2034, 2021. doi: 10.1161/CIRCULATIONAHA.121.055732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Cikach FS, Koch CD, Mead TJ, Galatioto J, Willard BB, Emerton KB, Eagleton MJ, Blackstone EH, Ramirez F, Roselli EE, Apte SS. Massive aggrecan and versican accumulation in thoracic aortic aneurysm and dissection. JCI Insight 3: e97167, 2018. doi: 10.1172/jci.insight.97167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Didangelos A, Mayr U, Monaco C, Mayr M. Novel role of ADAMTS-5 protein in proteoglycan turnover and lipoprotein retention in atherosclerosis. J Biol Chem 287: 19341–19345, 2012. doi: 10.1074/jbc.C112.350785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Yamada H, Watanabe K, Shimonaka M, Yamasaki M, Yamaguchi Y. cDNA cloning and the identification of an aggrecanase-like cleavage site in rat brevican. Biochem Biophys Res Commun 216: 957–963, 1995. doi: 10.1006/bbrc.1995.2713. [DOI] [PubMed] [Google Scholar]
- 79. Matthews RT, Gary SC, Zerillo C, Pratta M, Solomon K, Arner EC, Hockfield S. Brain-enriched hyaluronan binding (BEHAB)/brevican cleavage in a glioma cell line is mediated by a disintegrin and metalloproteinase with thrombospondin motifs (ADAMTS) family member. J Biol Chem 275: 22695–22703, 2000. doi: 10.1074/jbc.M909764199. [DOI] [PubMed] [Google Scholar]
- 80. Viapiano MS, Hockfield S, Matthews RT. BEHAB/brevican requires ADAMTS-mediated proteolytic cleavage to promote glioma invasion. J Neurooncol 88: 261–272, 2008. doi: 10.1007/s11060-008-9575-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Zhang H, Kelly G, Zerillo C, Jaworski DM, Hockfield S. Expression of a cleaved brain-specific extracellular matrix protein mediates glioma cell invasion In vivo. J Neurosci 18: 2370–2376, 1998. doi: 10.1523/JNEUROSCI.18-07-02370.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Nakada M, Miyamori H, Kita D, Takahashi T, Yamashita J, Sato H, Miura R, Yamaguchi Y, Okada Y. Human glioblastomas overexpress ADAMTS-5 that degrades brevican. Acta Neuropathol 110: 239–246, 2005. doi: 10.1007/s00401-005-1032-6. [DOI] [PubMed] [Google Scholar]
- 83. Hu B, Kong LL, Matthews RT, Viapiano MS. The proteoglycan brevican binds to fibronectin after proteolytic cleavage and promotes glioma cell motility. J Biol Chem 283: 24848–24859, 2008. doi: 10.1074/jbc.M801433200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Yuan W, Matthews RT, Sandy JD, Gottschall PE. Association between protease-specific proteolytic cleavage of brevican and synaptic loss in the dentate gyrus of kainate-treated rats. Neuroscience 114: 1091–1101, 2002. doi: 10.1016/s0306-4522(02)00347-0. [DOI] [PubMed] [Google Scholar]
- 85. Gottschall PE, Howell MD. ADAMTS expression and function in central nervous system injury and disorders. Matrix Biol 44–46: 70–76, 2015. doi: 10.1016/j.matbio.2015.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Minta K, Brinkmalm G, Thelin EP, Al Nimer F, Piehl F, Tullberg M, Jeppsson A, Portelius E, Zetterberg H, Blennow K, Andreasson U. Cerebrospinal fluid brevican and neurocan fragment patterns in human traumatic brain injury. Clin Chim Acta 512: 74–83, 2021. doi: 10.1016/j.cca.2020.11.017. [DOI] [PubMed] [Google Scholar]
- 87. Matsui F, Watanabe E, Oohira A. Immunological identification of two proteoglycan fragments derived from neurocan, a brain-specific chondroitin sulfate proteoglycan. Neurochem Int 25: 425–431, 1994. doi: 10.1016/0197-0186(94)90018-3. [DOI] [PubMed] [Google Scholar]
- 88. Matsui F, Nishizuka M, Yasuda Y, Aono S, Watanabe E, Oohira A. Occurrence of a N-terminal proteolytic fragment of neurocan, not a C-terminal half, in a perineuronal net in the adult rat cerebrum. Brain Res 790: 45–51, 1998. doi: 10.1016/s0006-8993(98)00009-2. [DOI] [PubMed] [Google Scholar]
- 89. Fontanil T, Mohamedi Y, Moncada-Pazos A, Cobo T, Vega JA, Cobo JL, García-Suárez O, Cobo J, Obaya ÁJ, Cal S. Neurocan is a new substrate for the ADAMTS12 metalloprotease: potential implications in neuropathies. Cell Physiol Biochem 52: 1003–1016, 2019. doi: 10.33594/000000069. [DOI] [PubMed] [Google Scholar]
- 90. Hußler W, Höhn L, Stolz C, Vielhaber S, Garz C, Schmitt FC, Gundelfinger ED, Schreiber S, Seidenbecher CI. Brevican and neurocan cleavage products in the cerebrospinal fluid - differential occurrence in ALS, epilepsy and small vessel disease. Front Cell Neurosci 16: 838432, 2022. doi: 10.3389/fncel.2022.838432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Tauchi R, Imagama S, Natori T, Ohgomori T, Muramoto A, Shinjo R, Matsuyama Y, Ishiguro N, Kadomatsu K. The endogenous proteoglycan-degrading enzyme ADAMTS-4 promotes functional recovery after spinal cord injury. J Neuroinflammation 9: 53, 2012. doi: 10.1186/1742-2094-9-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Filmus J, Capurro M, Rast J. Glypicans. Genome Biol 9: 224, 2008. doi: 10.1186/gb-2008-9-5-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Pilia G, Hughes-Benzie RM, MacKenzie A, Baybayan P, Chen EY, Huber R, Neri G, Cao A, Forabosco A, Schlessinger D. Mutations in GPC3, a glypican gene, cause the Simpson-Golabi-Behmel overgrowth syndrome. Nat Genet 12: 241–247, 1996. doi: 10.1038/ng0396-241. [DOI] [PubMed] [Google Scholar]
- 94. Hippo Y, Watanabe K, Watanabe A, Midorikawa Y, Yamamoto S, Ihara S, Tokita S, Iwanari H, Ito Y, Nakano K, Nezu J, Tsunoda H, Yoshino T, Ohizumi I, Tsuchiya M, Ohnishi S, Makuuchi M, Hamakubo T, Kodama T, Aburatani H. Identification of soluble NH2-terminal fragment of glypican-3 as a serological marker for early-stage hepatocellular carcinoma. Cancer Res 64: 2418–2423, 2004. doi: 10.1158/0008-5472.CAN-03-2191. [DOI] [PubMed] [Google Scholar]
- 95. Hagihara K, Watanabe K, Chun J, Yamaguchi Y. Glypican-4 is an FGF2-binding heparan sulfate proteoglycan expressed in neural precursor cells. Dev Dyn 219: 353–367, 2000. doi:. [DOI] [PubMed] [Google Scholar]
- 96. De Cat B, Muyldermans SY, Coomans C, Degeest G, Vanderschueren B, Creemers J, Biemar F, Peers B, David G. Processing by proprotein convertases is required for glypican-3 modulation of cell survival, Wnt signaling, and gastrulation movements. J Cell Biol 163: 625–635, 2003. doi: 10.1083/jcb.200302152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Capurro M, Shi W, Izumikawa T, Kitagawa H, Filmus J. Processing by convertases is required for glypican-3-induced inhibition of Hedgehog signaling. J Biol Chem 290: 7576–7585, 2015. doi: 10.1074/jbc.M114.612705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Traister A, Shi W, Filmus J. Mammalian Notum induces the release of glypicans and other GPI-anchored proteins from the cell surface. Biochem J 410: 503–511, 2008. doi: 10.1042/BJ20070511. [DOI] [PubMed] [Google Scholar]
- 99. Huang K, Park S. Heparan sulfated glypican-4 is released from astrocytes by proteolytic shedding and GPI-anchor cleavage mechanisms. eNeuro 8: ENEURO.0069-21.2021, 2021. doi: 10.1523/ENEURO.0069-21.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Kim CW, Goldberger OA, Gallo RL, Bernfield M. Members of the syndecan family of heparan sulfate proteoglycans are expressed in distinct cell-, tissue-, and development-specific patterns. Mol Biol Cell 5: 797–805, 1994. doi: 10.1091/mbc.5.7.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Manon-Jensen T, Itoh Y, Couchman JR. Proteoglycans in health and disease: the multiple roles of syndecan shedding. FEBS J 277: 3876–3889, 2010. doi: 10.1111/j.1742-4658.2010.07798.x. [DOI] [PubMed] [Google Scholar]
- 102. De Rossi G, Evans AR, Kay E, Woodfin A, McKay TR, Nourshargh S, Whiteford JR. Shed syndecan-2 inhibits angiogenesis. J Cell Sci 127: 4788–4799, 2014. doi: 10.1242/jcs.153015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Rodríguez-Manzaneque JC, Carpizo D, Plaza-Calonge M. D C, Torres-Collado AX, Thai SN-M, Simons M, Horowitz A, Iruela-Arispe ML. Cleavage of syndecan-4 by ADAMTS1 provokes defects in adhesion. Int J Biochem Cell Biol 41: 800–810, 2009. doi: 10.1016/j.biocel.2008.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Boivin WA, Shackleford M, Vanden Hoek A, Zhao H, Hackett TL, Knight DA, Granville DJ. Granzyme B cleaves decorin, biglycan and soluble betaglycan, releasing active transforming growth factor-β1. PLoS One 7: e33163, 2012. [Erratum in PLoS One 7: 2012. doi: 10.1371/journal.pone.0033163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Delolme F, Anastasi C, Alcaraz LB, Mendoza V, Vadon-Le Goff S, Talantikite M, Capomaccio R, Mevaere J, Fortin L, Mazzocut D, Damour O, Zanella-Cléon I, Hulmes DJ, Overall CM, Valcourt U, Lopez-Casillas F, Moali C. Proteolytic control of TGF-β co-receptor activity by BMP-1/tolloid-like proteases revealed by quantitative iTRAQ proteomics. Cell Mol Life Sci 72: 1009–1027, 2015. doi: 10.1007/s00018-014-1733-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Lamarre J, Vasudevan J, Gonias SL. Plasmin cleaves betaglycan and releases a 60 kDa transforming growth factor-β complex from the cell surface. Biochem J 302: 199–205, 1994. doi: 10.1042/bj3020199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Philip A, Hannah R, O’Connor-McCourt M. Ectodomain cleavage and shedding of the type III transforming growth factor-β receptor in lung membranes effect of temperature, ligand binding and membrane solubilization. Eur J Biochem 261: 618–628, 1999. doi: 10.1046/j.1432-1327.1999.00298.x. [DOI] [PubMed] [Google Scholar]
- 108. Velasco-Loyden G, Arribas J, López-Casillas F. The shedding of betaglycan is regulated by pervanadate and mediated by membrane type matrix metalloprotease-1. J Biol Chem 279: 7721–7733, 2004. doi: 10.1074/jbc.M306499200. [DOI] [PubMed] [Google Scholar]
- 109. Burgess RW, Dickman DK, Nunez L, Glass DJ, Sanes JR. Mapping sites responsible for interactions of agrin with neurons. J Neurochem 83: 271–284, 2002. doi: 10.1046/j.1471-4159.2002.01102.x. [DOI] [PubMed] [Google Scholar]
- 110. Burgess RW, Skarnes WC, Sanes JR. Agrin isoforms with distinct amino termini: differential expression, localization, and function. J Cell Biol 151: 41–52, 2000. doi: 10.1083/jcb.151.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Denzer AJ, Brandenberger R, Gesemann M, Chiquet M, Ruegg MA. Agrin binds to the nerve-muscle basal lamina via laminin. J Cell Biol 137: 671–683, 1997. doi: 10.1083/jcb.137.3.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Gautam M, Noakes PG, Moscoso L, Rupp F, Scheller RH, Merlie JP, Sanes JR. Defective neuromuscular synaptogenesis in agrin-deficient mutant mice. Cell 85: 525–535, 1996. doi: 10.1016/s0092-8674(00)81253-2. [DOI] [PubMed] [Google Scholar]
- 113. Tátrai P, Dudás J, Batmunkh E, Máthé M, Zalatnai A, Schaff Z, Ramadori G, Kovalszky I. Agrin, a novel basement membrane component in human and rat liver, accumulates in cirrhosis and hepatocellular carcinoma. Lab Invest 86: 1149–1160, 2006. doi: 10.1038/labinvest.3700475. [DOI] [PubMed] [Google Scholar]
- 114. Verbeek MM, Otte-Höller I, van den Born J, van den Heuvel LP, David G, Wesseling P, de Waal RM. Agrin is a major heparan sulfate proteoglycan accumulating in Alzheimer's disease brain. Am J Pathol 155: 2115–2125, 1999. doi: 10.1016/S0002-9440(10)65529-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Groffen AJ, Ruegg MA, Dijkman H, van de Velden TJ, Buskens CA, van den Born J, Assmann KJ, Monnens LA, Veerkamp JH, van den Heuvel LP. Agrin is a major heparan sulfate proteoglycan in the human glomerular basement membrane. J Histochem Cytochem 46: 19–27, 1998. doi: 10.1177/002215549804600104. [DOI] [PubMed] [Google Scholar]
- 116. Chakraborty S, Njah K, Pobbati AV, Lim YB, Raju A, Lakshmanan M, Tergaonkar V, Lim CT, Hong W. Agrin as a mechanotransduction signal regulating YAP through the hippo pathway. Cell Rep 18: 2464–2479, 2017. doi: 10.1016/j.celrep.2017.02.041. [DOI] [PubMed] [Google Scholar]
- 117. Chakraborty S, Sampath D, Yu Lin MO, Bilton M, Huang CK, Nai MH, Njah K, Goy PA, Wang CC, Guccione E, Lim CT, Hong W. Agrin-Matrix Metalloproteinase-12 axis confers a mechanically competent microenvironment in skin wound healing. Nat Commun 12: 6349, 2021. doi: 10.1038/s41467-021-26717-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Bassat E, Mutlak YE, Genzelinakh A, Shadrin IY, Baruch Umansky K, Yifa O, Kain D, Rajchman D, Leach J, Riabov Bassat D, Udi Y, Sarig R, Sagi I, Martin JF, Bursac N, Cohen S, Tzahor E. The extracellular matrix protein agrin promotes heart regeneration in mice. Nature 547: 179–184, 2017. doi: 10.1038/nature22978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Reif R, Sales S, Hettwer S, Dreier B, Gisler C, Wölfel J, Lüscher D, Zurlinden A, Stephan A, Ahmed S, Baici A, Ledermann B, Kunz B, Sonderegger P. Specific cleavage of agrin by neurotrypsin, a synaptic protease linked to mental retardation. FASEB J 21: 3468–3478, 2007. doi: 10.1096/fj.07-8800com. [DOI] [PubMed] [Google Scholar]
- 120. Schechter I, Berger A. On the size of the active site in proteases. I. Papain. Biochem Biophys Res Commun 27: 157–162, 1967. doi: 10.1016/s0006-291x(67)80055-x. [DOI] [PubMed] [Google Scholar]
- 121. VanSaun M, Werle MJ. Matrix metalloproteinase-3 removes agrin from synaptic basal lamina. J Neurobiol 43: 140–149, 2000. doi:. [DOI] [PubMed] [Google Scholar]
- 122. Werle MJ, VanSaun M. Activity dependent removal of agrin from synaptic basal lamina by matrix metalloproteinase 3. J Neurocytol 32: 905–913, 2003. doi: 10.1023/B:NEUR.0000020631.69804.f5. [DOI] [PubMed] [Google Scholar]
- 123. Patel TR, Butler G, McFarlane A, Xie I, Overall CM, Stetefeld J. Site specific cleavage mediated by MMPs regulates function of agrin. PLoS One 7: e43669, 2012. doi: 10.1371/journal.pone.0043669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Chow JP, Fujikawa A, Shimizu H, Suzuki R, Noda M. Metalloproteinase- and gamma-secretase-mediated cleavage of protein-tyrosine phosphatase receptor type Z. J Biol Chem 283: 30879–30889, 2008. doi: 10.1074/jbc.M802976200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Chow JP, Fujikawa A, Shimizu H, Noda M. Plasmin-mediated processing of protein tyrosine phosphatase receptor type Z in the mouse brain. Neurosci Lett 442: 208–212, 2008. doi: 10.1016/j.neulet.2008.07.028. [DOI] [PubMed] [Google Scholar]
- 126. Asher RA, Morgenstern DA, Properzi F, Nishiyama A, Levine JM, Fawcett JW. Two separate metalloproteinase activities are responsible for the shedding and processing of the NG2 proteoglycan in vitro. Mol Cell Neurosci 29: 82–96, 2005. doi: 10.1016/j.mcn.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 127. Joo NE, Miao D, Bermúdez M, Stallcup WB, Kapila YL. Shedding of NG2 by MMP-13 attenuates anoikis. DNA Cell Biol 33: 854–862, 2014. doi: 10.1089/dna.2014.2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Liu H, Shubayev VI. Matrix metalloproteinase-9 controls proliferation of NG2+ progenitor cells immediately after spinal cord injury. Exp Neurol 231: 236–246, 2011. doi: 10.1016/j.expneurol.2011.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Nishihara T, Remacle AG, Angert M, Shubayev I, Shiryaev SA, Liu H, Dolkas J, Chernov AV, Strongin AY, Shubayev VI. Matrix metalloproteinase-14 both sheds cell surface neuronal glial antigen 2 (NG2) proteoglycan on macrophages and governs the response to peripheral nerve injury. J Biol Chem 290: 3693–3707, 2015. doi: 10.1074/jbc.M114.603431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Nishiyama A, Lin XH, Stallcup WB. Generation of truncated forms of the NG2 proteoglycan by cell surface proteolysis. Mol Biol Cell 6: 1819–1832, 1995. doi: 10.1091/mbc.6.12.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Schultz N, Nielsen HM, Minthon L, Wennström M. Involvement of matrix metalloproteinase-9 in amyloid-β 1-42-induced shedding of the pericyte proteoglycan NG2. J Neuropathol Exp Neurol 73: 684–692, 2014. doi: 10.1097/NEN.0000000000000084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Sakry D, Neitz A, Singh J, Frischknecht R, Marongiu D, Binamé F, Perera SS, Endres K, Lutz B, Radyushkin K, Trotter J, Mittmann T. Oligodendrocyte precursor cells modulate the neuronal network by activity-dependent ectodomain cleavage of glial NG2. PLoS Biol 12: e1001993, 2014. doi: 10.1371/journal.pbio.1001993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Yuan P, Zhang H, Cai C, Zhu S, Zhou Y, Yang X, He R, Li C, Guo S, Li S, Huang T, Perez-Cordon G, Feng H, Wei W. Chondroitin sulfate proteoglycan 4 functions as the cellular receptor for Clostridium difficile toxin B. Cell Res 25: 157–168, 2015. doi: 10.1038/cr.2014.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Huxley-Jones J, Apte SS, Robertson DL, Boot-Handford RP. The characterisation of six ADAMTS proteases in the basal chordate Ciona intestinalis provides new insights into the vertebrate ADAMTS family. Int J Biochem Cell Biol 37: 1838–1845, 2005. doi: 10.1016/j.biocel.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 135. Bauer RC, Tohyama J, Cui J, Cheng L, Yang J, Zhang X, Ou K, Paschos GK, Zheng XL, Parmacek MS, Rader DJ, Reilly MP. Knockout of Adamts7, a novel coronary artery disease locus in humans, reduces atherosclerosis in mice. Circulation 131: 1202–1213, 2015. doi: 10.1161/CIRCULATIONAHA.114.012669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Pu X, Xiao Q, Kiechl S, Chan K, Ng FL, Gor S, Poston RN, Fang C, Patel A, Senver EC, Shaw-Hawkins S, Willeit J, Liu C, Zhu J, Tucker AT, Xu Q, Caulfield MJ, Ye S. ADAMTS7 cleavage and vascular smooth muscle cell migration is affected by a coronary-artery-disease-associated variant. Am J Hum Genet 92: 366–374, 2013. doi: 10.1016/j.ajhg.2013.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Colige A, Monseur C, Crawley JTB, Santamaria S, de Groot R. Proteomic discovery of substrates of the cardiovascular protease ADAMTS7. J Biol Chem 294: 8037–8045, 2019. doi: 10.1074/jbc.RA119.007492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. MacDonald BT, Keshishian H, Mundorff CC, Arduini A, Lai D, Bendinelli K, Popp NR, Bhandary B, Clauser KR, Specht H, Elowe NH, Laprise D, Xing Y, Kaushik VK, Carr SA, Ellinor PT. TAILS identifies candidate substrates and biomarkers of ADAMTS7, a therapeutic protease target in coronary artery disease. Mol Cell Proteomics 21: 100223, 2022. doi: 10.1016/j.mcpro.2022.100223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. O'Reilly MS, Boehm T, Shing Y, Fukai N, Vasios G, Lane WS, Flynn E, Birkhead JR, Olsen BR, Folkman J. Endostatin: an endogenous inhibitor of angiogenesis and tumor growth. Cell 88: 277–285, 1997. doi: 10.1016/s0092-8674(00)81848-6. [DOI] [PubMed] [Google Scholar]
- 140. Heljasvaara R, Aikio M, Ruotsalainen H, Pihlajaniemi T. Collagen XVIII in tissue homeostasis and dysregulation—lessons learned from model organisms and human patients. Matrix Biol 57–58: 55–75, 2017. doi: 10.1016/j.matbio.2016.10.002. [DOI] [PubMed] [Google Scholar]
- 141. Hanai J, Gloy J, Karumanchi SA, Kale S, Tang J, Hu G, Chan B, Ramchandran R, Jha V, Sukhatme VP, Sokol S. Endostatin is a potential inhibitor of Wnt signaling. J Cell Biol 158: 529–539, 2002. doi: 10.1083/jcb.200203064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Lin CH, Chen J, Zhang Z, Johnson GV, Cooper AJ, Feola J, Bank A, Shein J, Ruotsalainen HJ, Pihlajaniemi TA, Goligorsky MS. Endostatin and transglutaminase 2 are involved in fibrosis of the aging kidney. Kidney Int 89: 1281–1292, 2016. doi: 10.1016/j.kint.2016.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Yamaguchi Y, Takihara T, Chambers RA, Veraldi KL, Larregina AT, Feghali-Bostwick CA. A peptide derived from endostatin ameliorates organ fibrosis. Sci Transl Med 4: 136ra171, 2012. doi: 10.1126/scitranslmed.3003421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Chau YP, Lin SY, Chen JH, Tai MH. Endostatin induces autophagic cell death in EAhy926 human endothelial cells. Histol Histopathol 18: 715–726, 2003. doi: 10.14670/HH-18.715. [DOI] [PubMed] [Google Scholar]
- 145. Nguyen TM, Subramanian IV, Xiao X, Ghosh G, Nguyen P, Kelekar A, Ramakrishnan S. Endostatin induces autophagy in endothelial cells by modulating Beclin 1 and β-catenin levels. J Cell Mol Med 13: 3687–3698, 2009. doi: 10.1111/j.1582-4934.2009.00722.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Su J, Stenbjorn RS, Gorse K, Su K, Hauser KF, Ricard-Blum S, Pihlajaniemi T, Fox MA. Target-derived matricryptins organize cerebellar synapse formation through α3β1 integrins. Cell Rep 2: 223–230, 2012. doi: 10.1016/j.celrep.2012.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Wang T, Hauswirth AG, Tong A, Dickman DK, Davis GW. Endostatin is a trans-synaptic signal for homeostatic synaptic plasticity. Neuron 83: 616–629, 2014. doi: 10.1016/j.neuron.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Faye C, Chautard E, Olsen BR, Ricard-Blum S. The first draft of the endostatin interaction network. J Biol Chem 284: 22041–22047, 2009. doi: 10.1074/jbc.M109.002964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Ferreras M, Felbor U, Lenhard T, Olsen BR, Delaissé J. Generation and degradation of human endostatin proteins by various proteinases. FEBS Lett 486: 247–251, 2000. doi: 10.1016/s0014-5793(00)02249-3. [DOI] [PubMed] [Google Scholar]
- 150. Wen W, Moses MA, Wiederschain D, Arbiser JL, Folkman J. The generation of endostatin is mediated by elastase. Cancer Res 59: 6052–6056, 1999. [PubMed] [Google Scholar]
- 151. Felbor U, Dreier L, Bryant RA, Ploegh HL, Olsen BR, Mothes W. Secreted cathepsin L generates endostatin from collagen XVIII. EMBO J 19: 1187–1194, 2000. doi: 10.1093/emboj/19.6.1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Heljasvaara R, Nyberg P, Luostarinen J, Parikka M, Heikkilä P, Rehn M, Sorsa T, Salo T, Pihlajaniemi T. Generation of biologically active endostatin fragments from human collagen XVIII by distinct matrix metalloproteases. Exp Cell Res 307: 292–304, 2005. doi: 10.1016/j.yexcr.2005.03.021. [DOI] [PubMed] [Google Scholar]
- 153. Sasaki T, Fukai N, Mann K, Göhring W, Olsen BR, Timpl R. Structure, function and tissue forms of the C-terminal globular domain of collagen XVIII containing the angiogenesis inhibitor endostatin. EMBO J 17: 4249–4256, 1998. doi: 10.1093/emboj/17.15.4249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Sasaki T, Larsson H, Tisi D, Claesson-Welsh L, Hohenester E, Timpl R. Endostatins derived from collagens XV and XVIII differ in structural and binding properties, tissue distribution and anti-angiogenic activity. J Mol Biol 301: 1179–1190, 2000. doi: 10.1006/jmbi.2000.3996. [DOI] [PubMed] [Google Scholar]
- 155. Farach-Carson MC, Carson DD. Perlecan—a multifunctional extracellular proteoglycan scaffold. Glycobiology 17: 897–905, 2007. doi: 10.1093/glycob/cwm043. [DOI] [PubMed] [Google Scholar]
- 156. Melrose J. Perlecan, a modular instructive proteoglycan with diverse functional properties. Int J Biochem Cell Biol 128: 105849, 2020. doi: 10.1016/j.biocel.2020.105849. [DOI] [PubMed] [Google Scholar]
- 157. Lord MS, Tang F, Rnjak-Kovacina J, Smith JGW, Melrose J, Whitelock JM. The multifaceted roles of perlecan in fibrosis. Matrix Biol 68-69: 150–166, 2018. doi: 10.1016/j.matbio.2018.02.013. [DOI] [PubMed] [Google Scholar]
- 158. Guilak F, Hayes AJ, Melrose J. Perlecan in pericellular mechanosensory cell-matrix communication, extracellular matrix stabilisation and mechanoregulation of load-bearing connective tissues. Int J Mol Sci 22: 2716, 2021. doi: 10.3390/ijms22052716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Whitelock JM, Murdoch AD, Iozzo RV, Underwood PA. The degradation of human endothelial cell-derived perlecan and release of bound basic fibroblast growth factor by stromelysin, collagenase, plasmin, and heparanases. J Biol Chem 271: 10079–10086, 1996. doi: 10.1074/jbc.271.17.10079. [DOI] [PubMed] [Google Scholar]
- 160. Grindel BJ, Martinez JR, Pennington CL, Muldoon M, Stave J, Chung LW, Farach-Carson MC. Matrilysin/matrix metalloproteinase-7(MMP7) cleavage of perlecan/HSPG2 creates a molecular switch to alter prostate cancer cell behavior. Matrix Biol 36: 64–76, 2014. doi: 10.1016/j.matbio.2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Tellman TV, Cruz LA, Grindel BJ, Farach-Carson MC. Cleavage of the perlecan-semaphorin 3A-plexin A1-neuropilin-1 (PSPN) complex by matrix metalloproteinase 7/matrilysin triggers prostate cancer cell dyscohesion and migration. Int J Mol Sci 22: 3218, 2021. doi: 10.3390/ijms22063218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Mongiat M, Sweeney SM, San Antonio JD, Fu J, Iozzo RV. Endorepellin, a novel inhibitor of angiogenesis derived from the C terminus of perlecan. J Biol Chem 278: 4238–4249, 2003. doi: 10.1074/jbc.M210445200. [DOI] [PubMed] [Google Scholar]
- 163. Gonzalez EM, Reed CC, Bix G, Fu J, Zhang Y, Gopalakrishnan B, Greenspan DS, Iozzo RV. BMP-1/Tolloid-like metalloproteases process endorepellin, the angiostatic C-terminal fragment of perlecan. J Biol Chem 280: 7080–7087, 2005. doi: 10.1074/jbc.M409841200. [DOI] [PubMed] [Google Scholar]
- 164. Bix G, Fu J, Gonzalez EM, Macro L, Barker A, Campbell S, Zutter MM, Santoro SA, Kim JK, Höök M, Reed CC, Iozzo RV. Endorepellin causes endothelial cell disassembly of actin cytoskeleton and focal adhesions through α2β1 integrin. J Cell Biol 166: 97–109, 2004. [Erratum in J Cell Biol 201: 641, 2013]. doi: 10.1083/jcb.200401150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Chen CG, Iozzo RV. Angiostatic cues from the matrix: endothelial cell autophagy meets hyaluronan biology. J Biol Chem 295: 16797–16812, 2020. doi: 10.1074/jbc.REV120.014391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Poluzzi C, Iozzo RV, Schaefer L. Endostatin and endorepellin: a common route of action for similar angiostatic cancer avengers. Adv Drug Deliv Rev 97: 156–173, 2016. doi: 10.1016/j.addr.2015.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Trout AL, Kahle MP, Roberts JM, Marcelo A, de Hoog L, Boychuk JA, Grupke SL, Berretta A, Gowing EK, Boychuk CR, Gorman AA, Edwards DN, Rutkai I, Biose IJ, Ishibashi-Ueda H, Ihara M, Smith BN, Clarkson AN, Bix GJ. Perlecan domain-V enhances neurogenic brain repair after stroke in mice. Transl Stroke Res 12: 72–86, 2021. doi: 10.1007/s12975-020-00800-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Lee B, Clarke D, Al Ahmad A, Kahle M, Parham C, Auckland L, Shaw C, Fidanboylu M, Orr AW, Ogunshola O, Fertala A, Thomas SA, Bix GJ. Perlecan domain V is neuroprotective and proangiogenic following ischemic stroke in rodents. J Clin Invest 121: 3005–3023, 2011. [Erratum in J Clin Invest 122: 777, 2012]. doi: 10.1172/JCI46358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Nakamura K, Ikeuchi T, Nara K, Rhodes CS, Zhang P, Chiba Y, Kazuno S, Miura Y, Ago T, Arikawa-Hirasawa E, Mukouyama YS, Yamada Y. Perlecan regulates pericyte dynamics in the maintenance and repair of the blood-brain barrier. J Cell Biol 218: 3506–3525, 2019. doi: 10.1083/jcb.201807178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Fukuda S, Fini CA, Mabuchi T, Koziol JA, Eggleston LL Jr, del Zoppo GJ. Focal cerebral ischemia induces active proteases that degrade microvascular matrix. Stroke 35: 998–1004, 2004. doi: 10.1161/01.STR.0000119383.76447.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Edman P. A method for the determination of amino acid sequence in peptides. Arch Biochem 22: 475, 1949. [PubMed] [Google Scholar]
- 172. Doucet A, Overall CM. Amino-terminal oriented mass spectrometry of substrates (ATOMS) N-terminal sequencing of proteins and proteolytic cleavage sites by quantitative mass spectrometry. Methods Enzymol 501: 275–293, 2011. doi: 10.1016/B978-0-12-385950-1.00013-4. [DOI] [PubMed] [Google Scholar]
- 173. Rogers LD, Overall CM. Proteolytic post-translational modification of proteins: proteomic tools and methodology. Mol Cell Proteomics 12: 3532–3542, 2013. doi: 10.1074/mcp.M113.031310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Schilling O, Barre O, Huesgen PF, Overall CM. Proteome-wide analysis of protein carboxy termini: C terminomics. Nat Methods 7: 508–511, 2010. doi: 10.1038/nmeth.1467. [DOI] [PubMed] [Google Scholar]
- 175. Kockmann T, Carte N, Melkko S, Keller UAD, Identification of protease substrates in complex proteomes by iTRAQ-TAILs on a thermo Q exactive instrument. In: Neuromethods, edited by Grant JE, Li H. New York: Springer Science+Business Media, 2016, p. 187–207. [Google Scholar]
- 176. Bekhouche M, Leduc C, Dupont L, Janssen L, Delolme F, Vadon-Le Goff S, Smargiasso N, Baiwir D, Mazzucchelli G, Zanella-Cleon I, Dubail J, De Pauw E, Nusgens B, Hulmes DJ, Moali C, Colige A. Determination of the substrate repertoire of ADAMTS2, 3, and 14 significantly broadens their functions and identifies extracellular matrix organization and TGF-β signaling as primary targets. FASEB J 30: 1741–1756, 2016. doi: 10.1096/fj.15-279869. [DOI] [PubMed] [Google Scholar]
- 177. Leduc C, Dupont L, Joannes L, Monseur C, Baiwir D, Mazzucchelli G, Deroanne C, Colige A, Bekhouche M. In vivo N-terminomics highlights novel functions of ADAMTS2 and ADAMTS14 in skin collagen matrix building. Front Mol Biosci 8: 643178, 2021. doi: 10.3389/fmolb.2021.643178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Koch CD, Apte SS (Editors). Characterization of Proteoglycanomes by Mass Spectrometry. Cham, Switzerland: Springer Nature, 2020, p. 69–82. [Google Scholar]
- 179. Colaert N, Helsens K, Martens L, Vandekerckhove J, Gevaert K. Improved visualization of protein consensus sequences by iceLogo. Nat Methods 6: 786–787, 2009. doi: 10.1038/nmeth1109-786. [DOI] [PubMed] [Google Scholar]
- 180. Mort JS, Flannery CR, Makkerh J, Krupa JC, Lee ER. Use of anti-neoepitope antibodies for the analysis of degradative events in cartilage and the molecular basis for neoepitope specificity. Biochem Soc Symp 70: 107–114, 2003. doi: 10.1042/bss0700107. [DOI] [PubMed] [Google Scholar]