Abstract
Introduction:
The diagnosis of CLL/SLL relies on flow cytometric immunophenotyping. Increasing emphasis is being placed on precise detection of the minimal residual disease. Following antigen recommendations of ERIC and ESCCA’s Harmonization Project, we validated a 14-color assay for the characterization CD5+ lymphoproliferative neoplasms and CLL MRD with a sensitivity of at least 10−4.
Methods:
The assay was designed based on ERIC/ESCCA recommended antigens with the addition of CD40 for alternate gating when CD19 expression is reduced. Lower limit of quantitation/lower limit of detection, assay procedural precision, linearity, and limit of blank were established. 52 CD5+ B-cell lymphoproliferative neoplasms (41 CLL/ 11 non-CLL) and 29 normal samples were used for parallel evaluation. Automated cluster identification and quantitation of CLL clones in MRD setting was performed using Barned-Hutt SNE. Separation analysis between CLL and non-CLL phenotypes was performed by PCA and bh-SNE.
Results:
Separation ratios for each antigen exceeded ERIC/ESCCA guidelines. Precision was <20% at LLOQ (0.01%). The limit of blank was <10 /500000 cells. Concordance between the 14-color and legacy assay (Deming regression y=1.01x, r2=0.99) was seen. All 20 samples with MRD levels 0.5–0.006% (median 0.04%) showed an abnormal cell cluster by bh-SNE, with concordant results between manual and automated quantitation (y=x, r2=1). CLL cases clustered together and away from mantle cell lymphoma by bh-SNE and PCA with outlier atypical phenotype CLL cases posing diagnostic challenges by both manual and automated analysis.
Conclusion:
The 14-color CD5+ LPD assay provides a robust standardization platform for MRD and disease characterization using both manual and automated analysis.
Keywords: CLL, MRD, machine learning, mantle cell
Introduction
Chronic lymphocytic leukemia/small lymphocytic lymphoma (CLL/SLL) is the most common leukemia in the western world, both by incidence and prevalence [1, 2]. Additionally, 4–20% of adults over age 40 carry monoclonal CLL-like proliferations (MBL), which have a variable but an overall relatively low risk of progression to CLL/SLL[3, 4]. A proportion of MBL patients are at higher risk for progression and require at least intermittent monitoring[5–7]. The majority of CLL/SLL patients require treatment during their lifetime[8]. Unfortunately, persistent disease and relapses are common and multiple lines of treatment are frequently required. Because the patients tend to be monitored closely before and after the beginning of treatment and the remitting/relapsing disease course, CLL/SLL evaluations tend to form a substantial part of a clinical workload in typical clinical hematopathology and flow cytometry practices. Recently, CLL MRD measurement emerged as a critical tool in managing and sequencing therapies [9–14]. An efficient and precise approach to CLL evaluation is therefore of a paramount importance. In tertiary referral centers, laboratories are frequently called to perform diagnostic workup and follow up with samples from patients previously evaluated and treated elsewhere. In such settings, the ideal assay would therefore distinguish CLL/SLL from other CD5+ lymphoproliferative neoplasms, even when the original phenotype is perturbed by therapy, and at the same time would provide a sensitive MRD evaluation within the same assessment.
From the diagnostic perspective, many cases of CLL/SLL do not pose significant challenges to expert laboratories. Typical cases show a recognizable phenotype (CD5 positive, CD20 dim, CD23 positive, with dim surface immunoglobulin expression) and typical morphologic appearance[15]. These cases can be diagnosed with limited flow cytometry panels (4-color), combined with streamlined morphologic evaluation[8]. However, a significant proportion of CLL/SLL cases do not show these features to the extent of confident separation of CLL/SLL from other CD5 lymphoproliferative neoplasms or subsets of normal CD5+ B cells. This issue becomes particularly acute in minimal residual disease or low-level involvement evaluation scenarios[16–18]. Over time additional immunophenotypic features of CLL/SLL such as down regulation of CD38, CD81, and CD79b expression, increased expression of CD43 and ROR1, as well as expression of CD200 have been demonstrated. This led to the establishment of a more comprehensive phenotype for precise diagnosis of CLL/SLL by enhancing resolution from other CD5+ LPDs (summarized in [17]). Expansion of CLL immunophenotypic panels has also gained a new urgency due to multiple evolving therapies that either target CD19 (the main gating marker), reduce CD19 expression, and/or alter other key immunophenotypic features which are important for disease classification[19, 20]. Validating alternative backup gating antigens and approaches for robust classification is needed.
Multiple MRD assays for CLL/SLL have been reported, ranging from simple 4-color to 10-color assays. In general, a sensitivity of 10−4 to 10−5 can be achieved[17, 21–24]. Good correlation with molecular genetic studies for B cell receptor rearrangement can be seen at least down to 10−4[23]. While more sensitive evaluation can be achieved, it is not clear at this time what threshold below 10−4 provides clinical utility for either changing of therapy or outcome prognostication. Most large trials have used cutoff of 10−4 for this purpose[12]. The target sensitivity of 10−4 also allows for rapid flow cytometry testing without cell concentration and therefore may be a good target point from the standpoint of efficiency and reproducibility.
The increasing number of antigens that have been shown to enhance CLL/SLL assessment has led to reduced opportunities for harmonized assessment of CLL within small flow cytometry panels. The European Research Initiative on CLL (ERIC) & European Society for Clinical Cell Analysis (ESCCA) harmonization initiative investigators surveyed flow cytometry laboratories and developed a list of 14 required and recommended antigens [17]. It is noted that not all the antigens have been evaluated in prospective studies and few of the laboratories appeared to use all of them routinely. Nonetheless, we felt that the consensus approach incorporating well-established CLL-defining antigens is a promising platform for a CD5+ B-LPD assay that can gain wide utilization.
Analytic variability of the manual analysis tends to be the highest contributor to imprecision in population quantitation and is a barrier to assays standardization even when panels of antigens are otherwise harmonized or standardized. We thought to address the problem by using increasingly common clustering tools for multiparametric analysis. We attempted to improve the enumeration precision with the freely available MATLAB package cyt3[25] using the Barnes-Hutt stochastic neighbor embedding (bh-SNE) for the enumeration of CLL MRD clusters even at a very low level.
Methods:
Samples:
Samples were obtained from the Memorial Sloan Kettering (MSKCC) flow cytometry laboratory under the institutional review board approved protocol. The diagnosis of each neoplastic entity was performed by an MSKCC hematopathologist and was carried out through integrated flow cytometry, cytomorphology, molecular genetics, and cytogenetic analysis as appropriate according to WHO classification[26]
Antibodies and reagents:
Table 1 summarizes antibodies used for 14-color and legacy 2-tube CLL assay.
Table 1:
Antobodies used for staining
| 14-color CLL MRD Panel | Clone/Vendor | 8-color B cell legacy panel | Clone/Vendor2 | 6-color CLL add-on panel | Clone/Vendor3 |
|---|---|---|---|---|---|
|
| |||||
| CD79b BB515 | BD/3A2-2E7 | sKappa FITC | Dako/polyclonal | FMC7 FITC | BD/FMC7 |
| CD5 PE | BD/L17F12 | sLambda PE | Dako/polyclonal | CD23 PC7 | Coulter/9P25 |
| ROR1 PE/DAZZLE 594 | Biolegend/2A2 | CD10 PC5 | Coulter/ALB1 | CD200 APC | eBioscience Dx/OX104 |
| CD23 BB700 | BD/M-L233 | CD20 PC7 | Coulter/B9E9 | CD45 APC-H7 | BD/2D1 |
| CD19 PC7 | Coulter/J3-119 | CD38 APC | BD/HB7 | CD19 BV421 | BD/HIB19 |
| CD81 APC | Coulter/JS64 | CD22 APC-A700 | Coulter/SJ10.1H11 | CD5 BV510 | Biolegend/L17F12 |
| CD20 APC/FIRE 750 | Biolegend/2H7 | CD45 APC-H7 | BD/2D1 | ||
| CD200 BV421 | Biolegend/OX104 | CD19 BV421 | BD/HIB19 | ||
| CD40 BV480 | BD/5C3 | CD5 BV510 | Biolegend/L17F12 | ||
| CD43 BV605 | BD/1G10 | ||||
| CD38 BV786 | BD/HIT2 | ||||
| CD3 BUV395 | BD/SK7 | ||||
| CD10 BUV737 | BD/HI10a | ||||
| CD45 BUV805 | BD/H130 | ||||
Cell processing/staining and acquisition:
Flow cytometry staining for initial method evaluation was performed by standard protocols[27] using simultaneous ammonium chloride lysis and fixation. Briefly 100 microliters (uL) of blood containing 0.5 to 1.5 million cells were stained with a cocktail of antibodies for 15 min at room temperature, followed by ammonium chloride lysis with 0.025% formaldehyde for 15 min. Cells were washed with phosphate-buffered saline with bovine serum albumin and sodium azide (PBA) and the cell pellets were resuspended 100uL PBA. Fluorescence minus one and single stains were performed using the same methodology. The legacy assay was acquired on standardized BD FACS Canto-10 flow cytometers, while the 14-color assay was acquired on BD Fortessa-X20 flow cytometers equipped with four lasers (see supplemental table 1 for detailed configuration)
Flow cytometry analysis:
Manual analysis was performed using Woodlist software (generous gift of Dr. Brent Wood, University of Washington).
For bh-SNE analysis, CD19 positive B cell populations were first identified in Woodlist and exported as compensated listmode files.
For merged sample MRD analysis MRD positive CLL samples were merged with cyt3 MATLAB (MathWorks, Natick, MA) package, bh-SNE was performed on the merged samples and visualized within the software. For individual sample MRD analysis, bh-SNE was performed directly on each sample with CLL-like clusters visualized and gated within the software using bh-SNE display.
For CLL vs. non-CLL phenotype cluster analysis, abnormal B cells (20 cases of CLL and 20 cases of mantle cell lymphoma) were first subsampled for 500 cells each with cyt3 and merged into a single file. Bh-SNE analysis and visualization were performed with the cyt3 package.
Results
Anti-CD40 for gating of mature B cells.
CD40 is expressed at a moderate level by mature B cells, but not by other lymphoid subsets[28]. Its expression on CLL cells is well documented [29]. A subset of monocytes expresses CD40 at a low level, but the population can generally be excluded through scatter gating. To validate the anti-CD40 gating, we evaluated 16 diagnostic and 20 CLL MRD samples (median MRD 0.03% (range 0.004–0.4%))in patients treated with various therapies, including at least nine patients treated with a combination of Bruton tyrosine kinase (BTK) inhibitor, BCL-2 inhibitor and anti-CD20 targeting antibody, for the ability to enumerate CLL cells without using anti-CD19 in gating. Illustration of CD19 vs. CD40 dependent gating is shown in figure 1, panels a and b. Both CD40 mean fluorescence intensity (MFI) and separation ratios increased in the MRD setting compared to diagnostic samples. The mean MFI was 2536(1235–3729) at diagnosis vs. 4050(1697–8242) at MRD, p<0.01. The mean of the MFI ratios between CLL cells and T cells was 10(5–19) at diagnosis and 32(8–54) in the MRD setting (p<0.01), (figure 1 c and d). The MFI of CD40 on CLL cells at diagnosis was comparable to mature normal B cells (p>0.1). Quantitation of CLL cells was not affected by using anti-CD40 instead of anti-CD19 for gating either at diagnosis or MRD time points (figure 1e and f).
Figure 1:
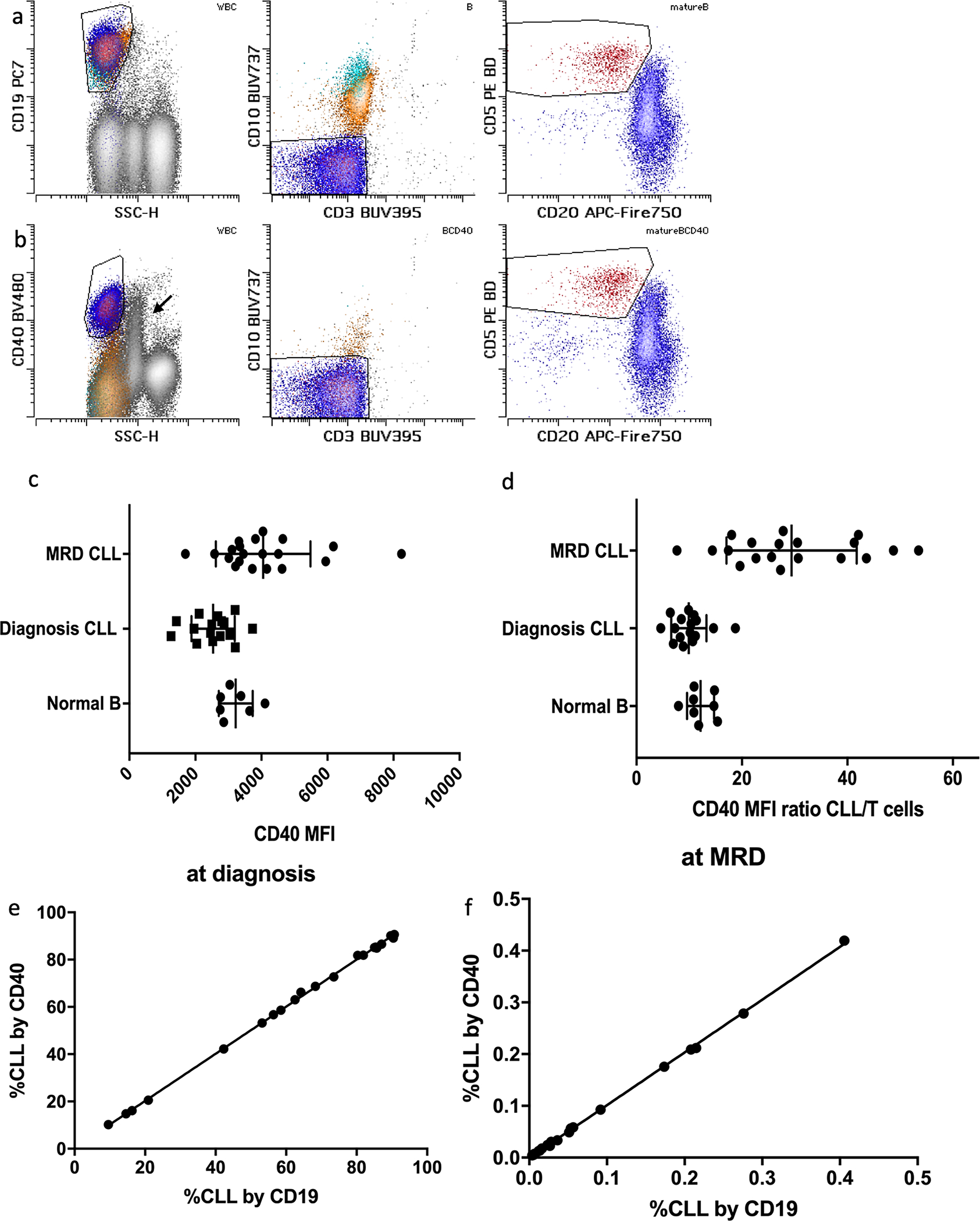
Examples of CD19(a) and CD40(b)-dependent gating are shown. Mature B cells are shown in dark blue, while immature (CD10+) intermediate and early B cells are shown in yellow and cyan respectively. CLL population is colored red. Note expression of CD40 on monocytes (arrow) and lack of expression of CD40 on early and most of the intermediate B cells. CD40 MFI (c) and MFI ratios (d) are shown. CLL MFI at diagnosis were comparable to normal mature B cells, but variably increased in MRD setting. Comparison of CLL population quantitation in diagnostic (e) and MRD settings (f) is shown. No significant proportionate or constant biases were observed.
Separation indices for other antigens
Panel design aimed to optimize separation between negatively and positively staining populations, and when needed (such as for CD20, CD79b, and CD81), dynamic ranges of the positive parameters. We aimed to match or exceed recommended separation ratios from ERIC/ESSCA harmonization project. Calculation of the separation ratios was performed on 7 normal marrow and peripheral blood samples. Table 2 summarizes the separation ratios and controls used for the antigens in the context of the total 14 antigen panel. CD38 and CD45 ratios were not formally measured due to a lack of stable and useful positive and negative controls within the peripheral blood and marrow samples. Visual examination confirmed adequate brightness and appropriate staining patterns for the reagents. An example of CLL MRD analysis showing common alterations (reduced CD20, CD81, CD79a, expression of CD43 and ROR1) within the panel is shown in figure 2a. Three fluorescence minus one and three single stain experiments revealed no antibody interference (data not shown).
Table 2:
Separation ratios in 14-color CLL tube
| 14-color CLL | Positive Population | Negative Population | Separation Ratio | Range(min-max) | ERIC/ESCCA recommedation |
|---|---|---|---|---|---|
|
| |||||
| CD79b BB515 | CD20+ B cells | CD3+ T cells | 21 | 2–63 | ≥15(30) |
| CD5 PE | CD3+ T cells | CD19+ B cells | 131 | 85–213 | ≥30(65) |
| ROR1 PE/DAZZLE 594 | B-progenitors | T cells | 8 | 6–10 | ≥5 |
| CD23 BB700 | CD23+ B cells | T cells | 17 | 7–32 | ≥5 |
| CD19 PC7 | CD20+ B cells | CD3+ T cells | 76 | 41–116 | ≥10 |
| CD81 APC | CD3+ T cells | Granulocytes | 20 | 12–32 | ≥12(20) |
| CD20 APC/FIRE 750 | CD19+ B cells | CD3+ T cells | 29 | 5–63 | ≥10(20) |
| CD200 BV421 | CD19+ B cells | CD3+ T cells | 8 | 6–12 | ≥5 |
| CD40 BV480 | B cells | T cells | 12 | 8–15 | N/A |
| CD43 BV605 | CD3+ T cells | CD20+ B cells | 50 | 14–127 | ≥15(40) |
| CD3 BUV395 | T cells | CD19+ B cells | 154 | 37–238 | N/A |
| CD10 BUV737 | Granulocytes | T cells | 12 | 5–20 | ≥10 |
Figure 2:
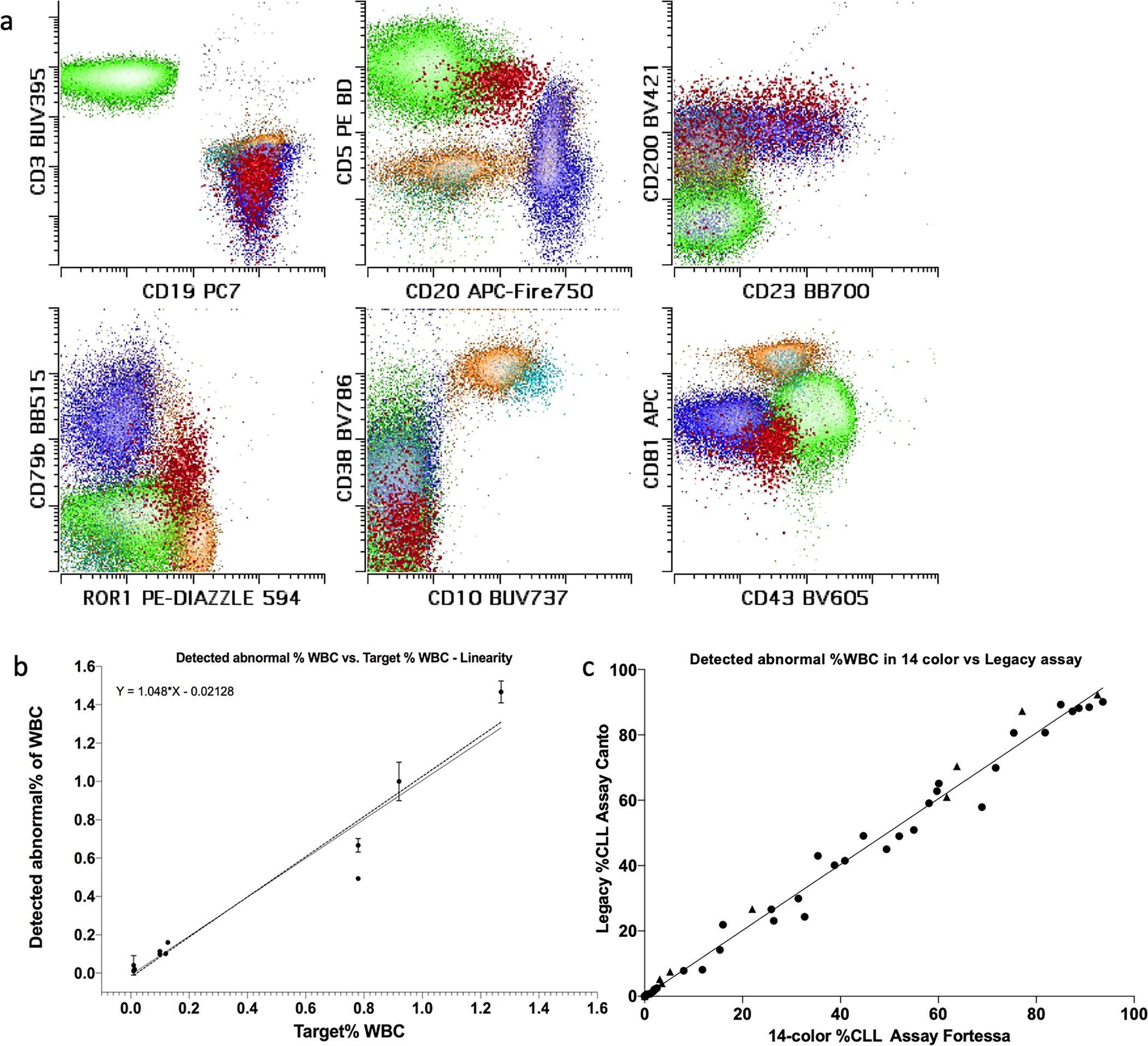
Example of lymphoid population appearance in the 14-color CLL tube is shown in panel (a). T cells are shown in green for comparison. Mature B cells are shown in dark blue, while early and intermediate immature B cells are shown in cyan and yellow respectively. CLL population (emphasized) in MRD setting is shown in red. Note aberrantly decreased expression of CD20, CD79b and CD81, with aberrant increased expression of CD43 and ROR1 on CLL population. Also note that both CD43 and ROR1 are normally expressed by a subset of immature B cells, requiring CD10 and/or CD38, and/or CD81 to separate them from CLL cells. Data for evaluation of linearity and lower limit of quantitation (LLOQ) is shown in panel (b). Triplicate analysis at proposed LLOQ of 0.01% was performed on 5 samples. While dilution from 1% to 0.01% was performed on 3 samples in triplicate. Deming regression equation is shown. Data for parallel testing by legacy 8-color assay vs. 14-color assay is shown in panel (c), no proportionate or constant biases over the analytical measurement range were seen.
Assay sensitivity and cell recovery using spiked samples
To establish the analytical sensitivity of the new CLL MRD assay, dilution experiments were performed. Five separate samples (three normal bone marrow aspirates and two normal peripheral bloods) were spiked with samples containing abnormal mature B cells with varying immunophenotypes at close to proposed LLOQ of 0.01% (0.01–0.013, median of 0.011, target range of 31–44 cells, median of 37) and stained in triplicate. The 14-color CLL MRD assay detected a median of 0.011% (0.009% – 0.02%) of abnormal mature B cells within white blood cell (WBC) compartment, representing a median of 34 (32–72) abnormal mature B cells, with a median precision of 8.5% (3.4%–12.9%) CV of abnormal mature B cells within WBC compartment and with a median precision of 11% (3.7%–17.8%) CV of the abnormal mature B cell absolute count. The median percentage recovery of the abnormal mature B cells of each triplicate was 91% (83% - 160%) with a median of 340,236 (301,102 – 365,028) WBCs analyzed. This data is summarized in supplemental table 2. No more than 5 cells were detected in the “CLL gates” in the negative samples used for spike-in based both on the phenotype of the positive spike-in samples and a separate blinded analysis for any phenotype compatible with CLL by manual review.
Three of the samples above were also used to evaluate assay recoveries at disease levels of approximately 1% and 0.1%. The data is summarized in figure 2b. No significant deviation from linearity was observed with the three levels of CLL cell dilution and precision of <20% was maintained at each point. Deming regression showed a slope of 1.048 (95% interval: 0.7171 to 1.378) and an intercept of −0.02128 (−0.08814 to 0.04558).
To evaluate the limit of blank/the limit of detection, 20 samples from patients without a history of CLL/SLL and negative for CLL/SLL by in-house assay were pooled together with positive samples and were blindly evaluated for the presence of CLL phenotype. None of the negative samples were judged as positive by evaluators at a level of 10 or more abnormal cells.
Parallel analysis
The assay performance was compared to the prior lab analysis using a split-sample approach (see methods section for description of the older method). Overall, 72 routine laboratory samples were analyzed (42 samples were positive for CLL or CLL-like monoclonal B cell lymphocytosis (range 0.02–90% WBC), 11 had other CD5 positive B cell neoplasms (range 0.02–92% WBC), 29 were negative for a neoplastic B cell population). Perfect qualitative concordance (72/72 samples concordant for the presence or absence of disease) in the blinded analysis was achieved. The evaluation of quantitative concordance is shown in figure 2c. Deming regression showed a slope of 1.01 (95% interval: 0.98– 1.04) and an intercept of 0 (−0.8 1to 0.82).
Automated separation of MRD populations.
Manual analysis introduces operator bias in the enumeration of cell subsets. The problem is likely amplified in the enumeration of small populations. This poses problems for assay harmonization between laboratories and even for tracking small populations within the same laboratory. In an attempt to overcome this limitation, we compared manual expert gating to CLL-like cluster identification using visual(vi)-SNE plots post bh-SNE analysis. 20 samples with low level (0.005–0.5%) MRD were analyzed by either manual analysis gated by an expert analyst or via bh-SNE within the cyt3 package by distinct CLL-like cluster on vi-SNE plots. Examples of manual and automated vi-SNE gating are shown in figures 3a and 3b. A separate CLL cluster was invariably present in vi-SNE plots for each MRD sample. Quantitation of the CLL-like cluster on vi-SNE and by manual analysis was highly similar, with Deming regression showing a slope of 1.01 (95% interval: 0.98–1.04) and an intercept of 0.00 (−0.001–0.003). As expected, higher levels of disagreement were seen with decreasing levels of MRD positivity (figure 3c). Interestingly, a separation between the normal residual mature and immature cell clusters, plasma cells, and spurious events, from CLL cells in vi-SNE was enhanced by CD79b, CD81, ROR1, CD38, CD40, and CD10 (data not shown). In particular, CD40 was useful in identifying small numbers of CLL cells vs. spurious ROR1+, CD20-, CD5-positive non-CLL events.
Figure 3:
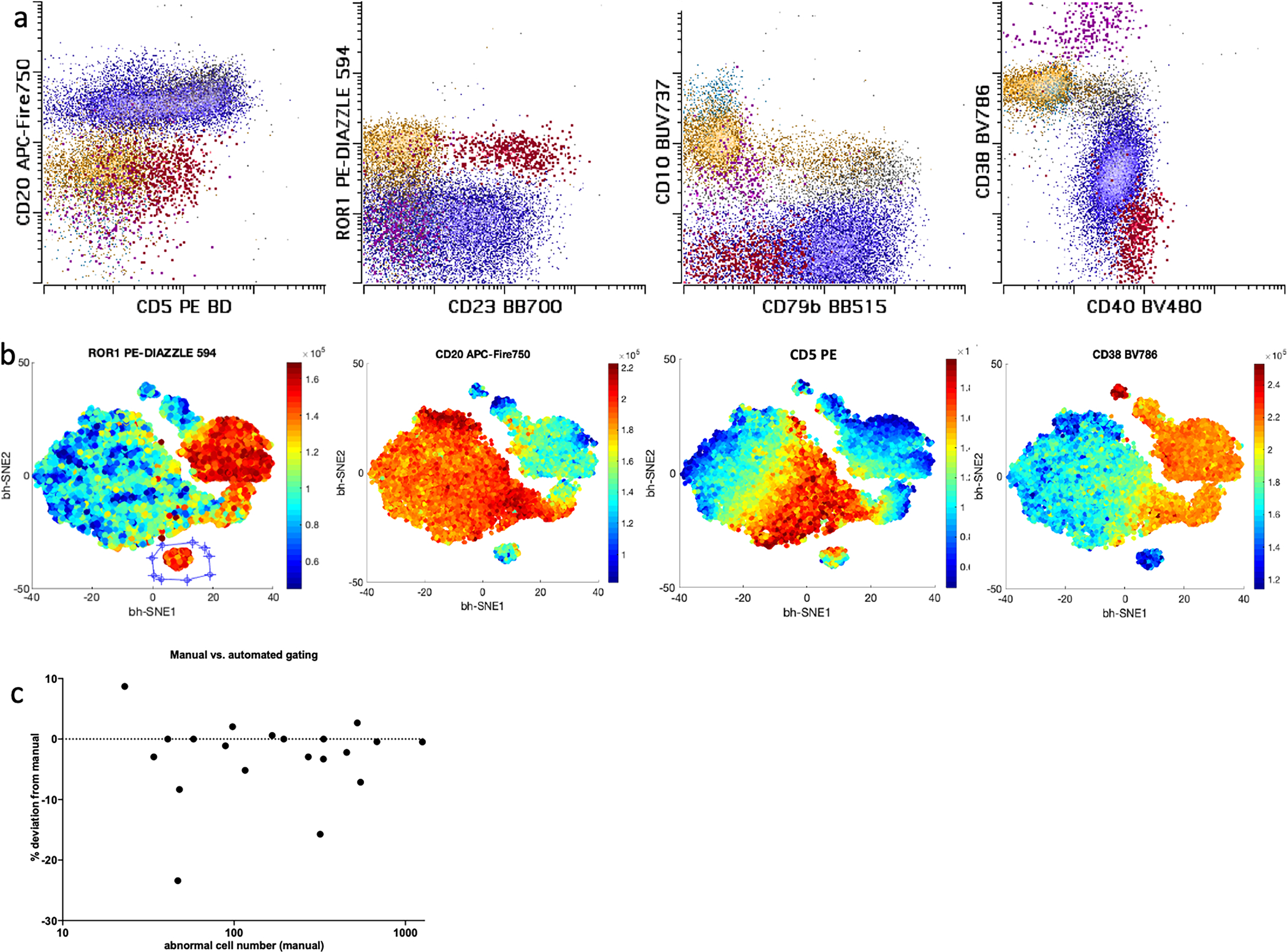
Example of CLL gating on manual (a) vs. vi-SNE assisted analysis is shown (b). Same sample is represented. Only B cells and plasma cells (content of CD19 vs. side scatter gate) are shown. In the manual gate mature B cells are shown in dark blue, while early and intermediate B cells are shown in cyan and yellow respectively. Plasma cells are shown in violet. CLL population (emphasized) in MRD setting is shown in red. Note that gating CLL on manual analysis would require several sequential gates. vi-SNE plots (b) are colored by relative intensity of the marker as labeled at the top of the plot. Conversely CLL population (separate cluster gated on ROR1 plot) could be easily separated with objective gating on vi-SNE plot showing all B cells. Quantitation differences between expert manual and vi-SNE assisted gating of CLL in MRD setting ranging from (0.005–0.5%) is shown in panel c. Percent differences in cell quantitation are plotted. As expected, lower number of cells resulted in slightly higher disagreement between the methods.
Automated separation CLL vs. not CLL samples
While most CLL samples can be easily classified with limited immunophenotypic markers, additional phenotyping may be helpful in difficult cases. Moreover, multiple abnormal CD5 positive populations may be seen within the same sample. The panel allowed for a separation of coincident CD5-positive B cell neoplasms as illustrated in figure 4a and 4b. We also formally evaluated the ability of the 14-color panel to discriminate between CLL and MCL cases by cluster analysis. 20 CLL and 20 MCL cases with varying phenotypes were subsampled equally (500 abnormal events each), merged together and analyzed by bh-SNE (see figure 4c) and principal component analysis (4d). As expected, a majority of CLL cases formed a single cluster separated from MCL. However, outlier cases for all categories with a small degree of overlap by principal component analysis were still evident. The outlier CLL cases had atypical phenotypes including relatively bright CD20, lack of CD200, dim to absent CD5 and/or absent CD23 (some of the features are illustrated in figure 4e and 4f). Outlier MCL case lacked CD20, likely post therapy. While cluster analysis is helpful for classification in the majority of cases, the need for integrative diagnosis persists for a small proportion of CD5+ cases, despite extensive phenotyping.
Figure 4:
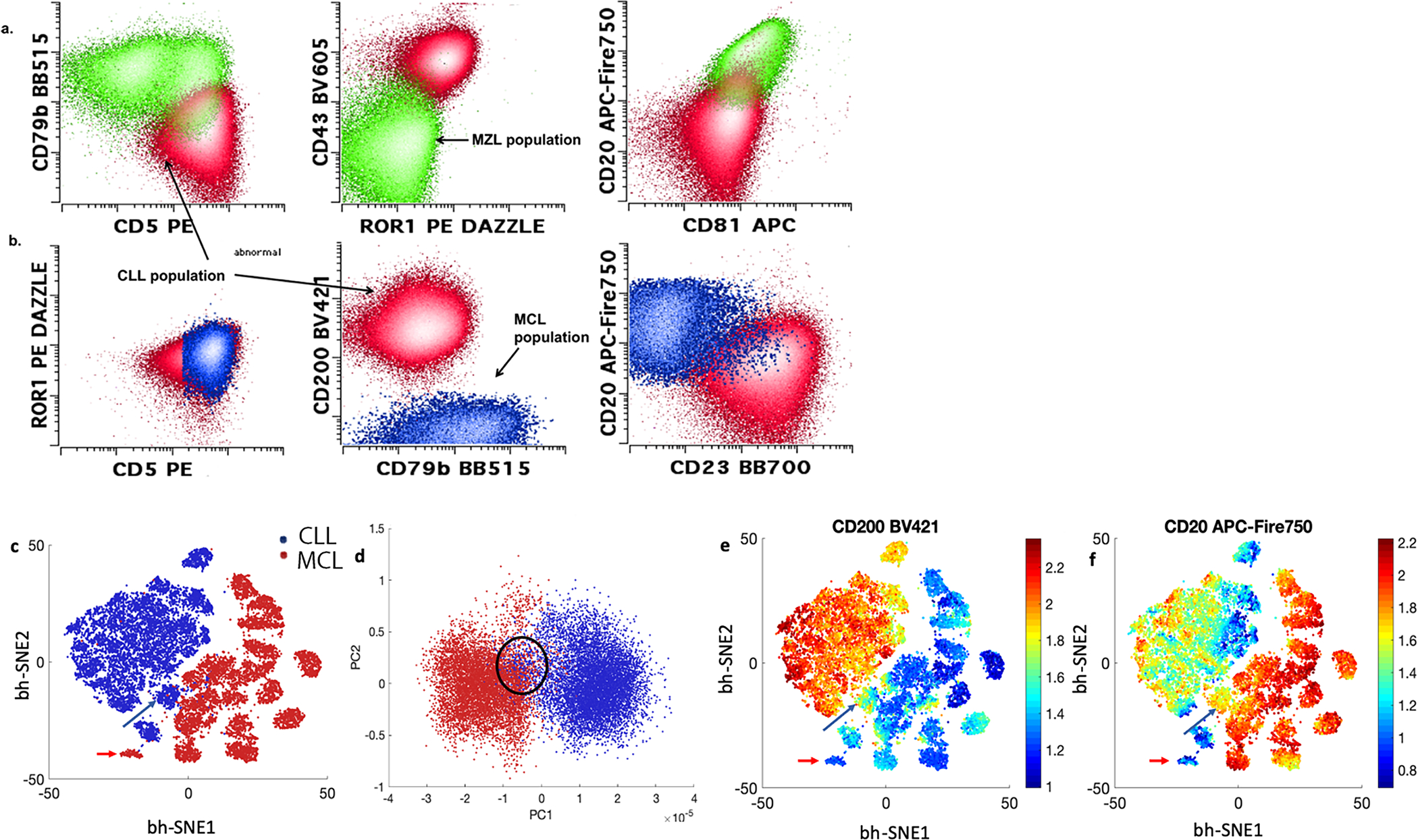
Examples of co-occurring CD5 positive populations within the same samples are shown in panels a and b. The 14-color CLL panel allowed for complete separation between CD5+ marginal zone lymphoma (green) from CLL population (red) (panel a) and mantle cell lymphoma (blue) from CLL (red) (panel b). Bh-SNE-assisted analysis could be helpful in separating mantle cell lymphoma (red) from CLL (blue), 20 cases of CLL and 20 case of mantle cell lymphoma were subsampled for 500 cells each, merged and analyzed by bh-SNE (c) and PCA (d). While majority of cases clustered together with the correct disease group. Outlier or borderline cases could be seen; see mantle cell lymphoma (red arrows) and CLL (blue arrows) in bh-SNE plot c, and circled in PCA plot d. These cases showed atypical phenotypes (loss of CD20 for mantle cell lymphoma), loss of CD200 and relatively bright expression of CD20 for CLL (see e and f)
Discussion
We evaluated a 14-color flow cytometry assay for CLL and other CD5-positive B cell neoplasms. While 13+ color instrumentation is not common in clinical laboratories, the equipment landscape has been rapidly changing. From the practical perspective, the assay addresses a persistent key barrier to assay standardization arising due to preferences for an inclusion of specific antigens that are proven to have utilities in a diagnostic setting. The consensus approach to panel selection may decrease these barriers.
While 14-color approach allows for inclusion of all the recommended and required antigens, as well as CD40, we believe the panel may be reduced for use in 8–12 color instruments that are currently common. In particular if diagnostic and follow up panels are separated. While we have not undertaken a rigorous evaluation of the relative utility of each individual antigen in MRD and diagnostic settings our experience with over 300 cases suggests that omission of some antigen may not result in marked performance degradation for manual gating. In the diagnostic setting exclusion of CD10, CD43, ROR1 and CD3 is reasonable, as they contribute little in distinction between CD5 positive B cells neoplasms and are mainly used to exclude other populations in MRD context (CD3, CD10) or highly sensitive MRD detection (CD43, ROR1). In the MRD context there is limited utility to CD200 or CD23 as expression of both can be altered by therapy. In addition, CD81 and CD43, while certainly aberrantly expressed in majority of cases, were rarely used for gating. CD10 may be redundant with bright CD38 and CD81 (if kept) in identification of hematogones. All of these observations need to be confirmed in a rigorous analysis with well-designed shorter panels.
We omitted light chain evaluation in the panel without loss of sensitivity, which is in line with prior observations[23]. CLL-like proliferations are frequently oligoclonal[30, 31] and we found light chains to be of limited utility in this setting in our clinical experience. We allowed attending physicians interpreting the analysis to add light chain analysis as they saw fit. In over 300 cases analyzed since the introduction of the tube in our clinical practice, no add-ons were requested for this reason. Light chain analysis may still be useful when the diagnosis of B cell lymphoproliferative disorder is only suspected to establish a baseline.
Of interest, the markers that were identified as recommended, rather than required by ERIC/ESSCA survey, contributed to CLL cluster formation in automated analysis and therefore should be considered at least for assays with the end goal of utilizing unbiased automated tools for disease enumeration. The added costs of individual antibodies in a large panel may be subsumed by the ease of analysis and lack of need for labor and instrument time associated with add-on tubes for challenging cases. The cost analysis is beyond the scope of the article and is dependent on the setting.
We further validated the use of CD40 for gating of CD5 positive B cell neoplasms. The alternative gating strategy is critical in the age of surface antigen targeting therapy and BTK inhibitors that reduce expression and/or accessibility of common gating targets such as CD19 and CD20. CD22 is dimly expressed in CLL and may be an unreliable gating marker. CD40 expression was not affected by therapies in our study and made CD19-independent gating possible without altering residual disease quantitation. When CD19 is dim rather than negative, or when an increased non-specific binding is present, we found gating on a plot with both CD19 and CD40 more satisfactory (data not shown).
We show that MRD analysis can be streamlined with increased reproducibility by widely available SNE-based tools. Qualitatively, the data is in line with prior observations in B-ALL and AML where MRD-like populations could be detected by vi-SNE[25, 32]. Prior investigations have also shown a high degree of quantitative concordance between manual analysis and vi-SNE in gating immune subsets[33]. However, to our knowledge, this is the first time a full quantitative concordance between SNE and manual gating was demonstrated in an MRD setting. This approach is not confined to a specific software package as such tools are freely available and are increasingly included in commercial flow cytometry analysis software releases. We specifically utilized the cyt3 package for this evaluation because it is freely available and is analysis software independent. Validation and commercialization of fit for purpose software analysis packages with streamlined workflow may speed up their wider adoption and help increase interlaboratory reproducibility.
Our assay was validated with target LLOQ of 10−4 for largely practical efficiency reasons. The very low background of the assay raises a possibility that lower sensitivity limits could be achieved with acquisition of more cells without altering the panel. While the lower LLOQ would require a formal validation, we could confidently identify CLL MRD populations at significantly decreased proportions in a number of samples.
One limitation of the study is lack of clinical outcome data or cross-validation against a test with clinical outcome data such as the one used by ERIC group[23]. We performed extensive analytical validation showing analytical performance in line with ERIC panel. Nonetheless cross-correlating against a test with known clinical predictive parameters or showing its utility in a prospective clinical trial would further strengthen the case for its wide adoption.
In line with prior observations, clustering-based tools had high, but not perfect separation between CD5-positive B cell neoplasms despite extensive phenotypic evaluation[34]. This observation may reflect the overlapping biological properties of atypical cases. For instance, two CLL cases that clustered closer to mantle cell lymphoma due to brighter expression of CD20 and dimmer CD200, showed trisomy 12. The role of a comprehensive evaluation that includes clinical, morphologic, cytogenetic and molecular genetic assessment in the diagnosis of B cell lymphoproliferative neoplasms remains important.
Supplementary Material
Acknowledgments:
Support was provided by grant P30 CA008748 from the Memorial Sloan Kettering Cancer Center.
Footnotes
COI:
M Roshal has received personal fees from Agios, Cellgene and Physicians’ Education Resource, research grants Agios, Cellularity, and Roche. He serves as an advisor and owns equity in Auron Pharmaceuticals. A Dogan has received personal fees from Roche, Corvus Pharmaceuticals, Physicians’ Education Resource, Seattle Genetics, Peerview Institute, Oncology Specialty Group, Takeda, EUSA Pharma and research grants from National Cancer Institute and Roche.
References
- 1.Siegel RL, Miller KD, and Jemal A, Cancer statistics, 2020. CA Cancer J Clin, 2020. 70(1): p. 7–30. [DOI] [PubMed] [Google Scholar]
- 2.Fitzmaurice C, et al. , Global, Regional, and National Cancer Incidence, Mortality, Years of Life Lost, Years Lived With Disability, and Disability-Adjusted Life-years for 32 Cancer Groups, 1990 to 2015: A Systematic Analysis for the Global Burden of Disease Study. JAMA Oncol, 2017. 3(4): p. 524–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shanafelt TD, et al. , Monoclonal B-cell lymphocytosis (MBL): biology, natural history and clinical management. Leukemia, 2010. 24(3): p. 512–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shim YK, et al. , Prevalence of monoclonal B-cell lymphocytosis: a systematic review. Cytometry. Part B, Clinical cytometry, 2010. 78 Suppl 1(Suppl 1): p. S10–S18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kern W, et al. , Monoclonal B-cell lymphocytosis is closely related to chronic lymphocytic leukaemia and may be better classified as early-stage CLL. Br J Haematol, 2012. 157(1): p. 86–96. [DOI] [PubMed] [Google Scholar]
- 6.Parikh SA, Kay NE, and Shanafelt TD, Monoclonal B-cell lymphocytosis: update on diagnosis, clinical outcome, and counseling. Clin Adv Hematol Oncol, 2013. 11(11): p. 720–9. [PubMed] [Google Scholar]
- 7.Criado I, et al. , Low-count monoclonal B-cell lymphocytosis persists after seven years of follow up and is associated with a poorer outcome. Haematologica, 2018. 103(7): p. 1198–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hallek M, et al. , iwCLL guidelines for diagnosis, indications for treatment, response assessment, and supportive management of CLL. Blood, 2018. 131(25): p. 2745–2760. [DOI] [PubMed] [Google Scholar]
- 9.Strati P, et al. , Eradication of bone marrow minimal residual disease may prompt early treatment discontinuation in CLL. Blood, 2014. 123(24): p. 3727–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jain N, et al. , Ibrutinib and Venetoclax for First-Line Treatment of CLL. N Engl J Med, 2019. 380(22): p. 2095–2103. [DOI] [PubMed] [Google Scholar]
- 11.Kater AP, et al. , Fixed Duration of Venetoclax-Rituximab in Relapsed/Refractory Chronic Lymphocytic Leukemia Eradicates Minimal Residual Disease and Prolongs Survival: Post-Treatment Follow-Up of the MURANO Phase III Study. J Clin Oncol, 2019. 37(4): p. 269–277. [DOI] [PubMed] [Google Scholar]
- 12.Thompson M, et al. , Minimal Residual Disease in Chronic Lymphocytic Leukemia in the Era of Novel Agents: A Review. JAMA Oncol, 2018. 4(3): p. 394–400. [DOI] [PubMed] [Google Scholar]
- 13.Thompson PA, et al. , Serial minimal residual disease (MRD) monitoring during first-line FCR treatment for CLL may direct individualized therapeutic strategies. Leukemia, 2018. 32(11): p. 2388–2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thompson PA, et al. , Minimal residual disease undetectable by next-generation sequencing predicts improved outcome in CLL after chemoimmunotherapy. Blood, 2019. 134(22): p. 1951–1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matutes E, et al. , The immunological profile of B-cell disorders and proposal of a scoring system for the diagnosis of CLL. Leukemia, 1994. 8(10): p. 1640–5. [PubMed] [Google Scholar]
- 16.Köhnke T, et al. , Diagnosis of CLL revisited: increased specificity by a modified five-marker scoring system including CD200. Br J Haematol, 2017. 179(3): p. 480–487. [DOI] [PubMed] [Google Scholar]
- 17.Rawstron AC, et al. , Reproducible diagnosis of chronic lymphocytic leukemia by flow cytometry: An European Research Initiative on CLL (ERIC) & European Society for Clinical Cell Analysis (ESCCA) Harmonisation project. Cytometry B Clin Cytom, 2018. 94(1): p. 121–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sorigue M, et al. , Consistency of the Moreau CLL score. J Clin Lab Anal, 2018. 32(5): p. e22376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fraietta JA, et al. , Determinants of response and resistance to CD19 chimeric antigen receptor (CAR) T cell therapy of chronic lymphocytic leukemia. Nat Med, 2018. 24(5): p. 563–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fraietta JA, et al. , Ibrutinib enhances chimeric antigen receptor T-cell engraftment and efficacy in leukemia. Blood, 2016. 127(9): p. 1117–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dowling AK, et al. , Optimization and Validation of an 8-Color Single-Tube Assay for the Sensitive Detection of Minimal Residual Disease in B-Cell Chronic Lymphocytic Leukemia Detected via Flow Cytometry. Lab Med, 2016. 47(2): p. 103–11. [DOI] [PubMed] [Google Scholar]
- 22.Rawstron AC, et al. , Improving efficiency and sensitivity: European Research Initiative in CLL (ERIC) update on the international harmonised approach for flow cytometric residual disease monitoring in CLL. Leukemia, 2013. 27(1): p. 142–9. [DOI] [PubMed] [Google Scholar]
- 23.Rawstron AC, et al. , A complementary role of multiparameter flow cytometry and high-throughput sequencing for minimal residual disease detection in chronic lymphocytic leukemia: an European Research Initiative on CLL study. Leukemia, 2016. 30(4): p. 929–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sartor MM and Gottlieb DJ, A single tube 10-color flow cytometry assay optimizes detection of minimal residual disease in chronic lymphocytic leukemia. Cytometry B Clin Cytom, 2013. 84(2): p. 96–103. [DOI] [PubMed] [Google Scholar]
- 25.Amir el AD, et al. , viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nat Biotechnol, 2013. 31(6): p. 545–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Swerdlow SH, et al. , WHO classification of tumours of haematopoietic and lymphoid tissues. 2017, Lyon: International Agency for Research on Cancer. [Google Scholar]
- 27.Wood BL, Principles of minimal residual disease detection for hematopoietic neoplasms by flow cytometry. Cytometry B Clin Cytom, 2016. 90(1): p. 47–53. [DOI] [PubMed] [Google Scholar]
- 28.Kalina T, et al. , CD Maps—Dynamic Profiling of CD1–CD100 Surface Expression on Human Leukocyte and Lymphocyte Subsets. Frontiers in Immunology, 2019. 10(2434). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee BO, et al. , CD40, but Not CD154, Expression on B Cells Is Necessary for Optimal Primary B Cell Responses. The Journal of Immunology, 2003. 171(11): p. 5707. [DOI] [PubMed] [Google Scholar]
- 30.Brazdilova K, et al. , Multiple productive IGH rearrangements denote oligoclonality even in immunophenotypically monoclonal CLL. Leukemia, 2018. 32(1): p. 234–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sanchez ML, et al. , Incidence and clinicobiologic characteristics of leukemic B-cell chronic lymphoproliferative disorders with more than one B-cell clone. Blood, 2003. 102(8): p. 2994–3002. [DOI] [PubMed] [Google Scholar]
- 32.DiGiuseppe JA, Tadmor MD, and Pe’er D, Detection of minimal residual disease in B lymphoblastic leukemia using viSNE. Cytometry B Clin Cytom, 2015. 88(5): p. 294–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Toghi Eshghi S, et al. , Quantitative Comparison of Conventional and t-SNE-guided Gating Analyses. Front Immunol, 2019. 10: p. 1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Costa ES, et al. , Automated pattern-guided principal component analysis vs expert-based immunophenotypic classification of B-cell chronic lymphoproliferative disorders: a step forward in the standardization of clinical immunophenotyping. Leukemia, 2010. 24(11): p. 1927–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


