Abstract
The flagellar motor/switch complex, consisting of the three proteins FliG, FliM, and FliN, plays a central role in bacterial motility and chemotaxis. We have analyzed FliG, using 10-amino-acid deletions throughout the protein and testing the deletion clones for their motility and dominance properties and for interaction of the deletion proteins with the MS ring protein FliF. Only the N-terminal 46 amino acids of FliG (segments 1 to 4) were important for binding to FliF; consistent with this, an N-terminal fragment consisting of residues 1 to 108 bound FliF strongly, whereas a C-terminal fragment consisting of residues 109 to 331 did not bind FliF at all. Deletions in the region from residues 37 to 96 (segments 4 to 9), 297 to 306 (segment 30), and 317 to 326 (segment 32) permitted swarming, though not at wild-type levels; all other deletions caused paralyzed or, more commonly, nonflagellate phenotype. Except for those near the N terminus, deletions had a dominant negative effect on wild-type cells.
Of the three proteins that make up the flagellar motor/switch of Salmonella (27), FliG can be distinguished in two regards: it is the component that is directly associated with the MS ring (3) and thus to the external rotating components such as the filament, and it is the component that interacts with the Mot proteins to develop torque (8, 12). FliM, in contrast, is responsible for interaction with CheY-P and initiating the switching event (1, 15, 19, 24); the role of FliN remains mysterious.
The physical presence of the motor/switch complex was shown by electron microscopy, which revealed the presence of a ring- or bell-like structure (the C ring) extending into the cytoplasm from the bottom of the basal body MS ring, a structure that is embedded in the cytoplasmic membrane (4, 10).
During the course of an extensive mutational analysis of the switch genes, we discovered two fusion proteins made up of the MS ring proteins FliF and FliG, arranged in the order N-FliF-FliG-C. The mutant carrying one of these fusion proteins (called the full-length fusion protein because the frameshift at the junction resulted in a net loss of only four amino acids) could assemble flagella and swim at almost wild-type levels, strongly indicating that in the wild-type cell these two proteins must be in close physical proximity and function together within the assembled structure. Immunoelectron microscopy on basal bodies from the full-length fusion mutant showed that FliG is located on the cytoplasmic face of the MS ring (3). The second FliF-FliG fusion protein (called the deletion-fusion protein) was missing 56 amino acids from the C terminus of FliF and 94 amino acids from the N terminus of FliG. The strain carrying this deletion-fusion protein was also able to form flagella and swim, although much more poorly than the wild-type strain or the mutant carrying the full-length fusion protein. Recent cryoelectron microscopic studies show that the basal body of the deletion-fusion mutant has a smaller-diameter C ring than the wild-type or full-length fusion structure (22).
We have reported previously analyses of the FliM protein in which we systematically deleted 10-amino-acid segments throughout the protein and looked at the physiological consequences in terms of flagellar assembly, motility, and switching (23) and the binding of FliM to other components (24). Related studies have been published by Tang et al. (21), who used glutathione-S-transferase (GST) fusions and His-tagged proteins, and by Marykwas et al. (14), who used the two-hybrid system in yeast.
Here we have applied the scanning deletion approach to FliG, with special emphasis on its interaction with the MS ring protein FliF.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth media.
Salmonella strains and plasmids used are listed in Table 1. All restriction enzymes and T4 DNA ligase were purchased from New England Biolabs (Beverly, Mass.). Luria medium (LM) used for growth of cells and soft tryptone motility (TM) plates are described in reference 23. SOC medium (18) was used for recovery of cells after electroporation. Ampicillin at 100 μg/ml or chloramphenicol at 25 μg/ml was added as needed. Isopropyl-β-d-thiogalactopyranoside (IPTG) was used at a final concentration of 1 mM for induction and overproduction of proteins.
TABLE 1.
Strains and plasmids used
| Strain or plasmid | Relevant genotype or property | Reference or source |
|---|---|---|
| Salmonella strains | ||
| SJW1103 | Wild type | 28 |
| MKM1 | fliG null strain | This study |
| MKM6 | fliM null strain | This study |
| Plasmids | ||
| pMAK705 | Temperature-sensitive origin of replication, chloramphenicol resistance | 5 |
| pAMH3 | Salmonella fliFGHIJ(K) genes in pBR322 (source of fliG for PCR) | 11 |
| pAMH5 | Salmonella fliLMNO(P) genes in pBR322 (source of fliM for PCR) | 7 |
| pFFF1300 | His-FLAG-fliF in pET19b | 2 |
| pGMK1000 | fliG (wild type) in pUC18 | This study |
| pGΔ1 to pGΔ32 | fliG deletion alleles in pUC18 | This study |
| pGMK3000 | fliG (wild type) in pET19b | This study |
| pGMK3100 | fliG (amino acids 1 to 108, GN) in pET19b | This study |
| pGMK3200 | fliG (amino acids 109 to 331, GC) in pET19b | This study |
| pGETΔ1 to pGETΔ12, pGETΔ21, pGETΔ22, and pGETΔ27 to pGETΔ31 | fliG deletion alleles in pET19b | This study |
Construction of fliG null strain.
A PCR fragment was synthesized, using pAMH3 as the template DNA, with a BamHI site at the 5′ end and a HindIII site at the 3′ end, and with about 1,000 bp on either side of fliG to allow for complementarity during the resolution step. This FliG deletion contains only the first 4 amino acids at the N terminus and last 10 amino acids at the C terminus, and it retains the natural ribosome binding site for fliH immediately downstream of fliG. The deletion fragment, constructed according to a protocol similar to that described by Williams et al. (25) using an MJ MiniCycler (MJ Research, Watertown, Mass.) and Taq polymerase (Sigma-Aldrich, St. Louis, Mo.), was ligated into pMAK705. The ligation mixture was concentrated by ethanol precipitation and used to electroporate SK6600 (5) (E. coli Pulser; Bio-Rad Laboratories, Hercules, Calif.). After incubation in SOC medium at 30°C for 1 h, cells were plated on LM–Cm (chloramphenicol)–IPTG–5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside and incubated at 30°C. White colonies were selected and grown, their plasmids were purified using a QIAprep spin plasmid kit (Qiagen, Valencia, Calif.), and the construction was verified by restriction digestion and sequencing (Sequenase kit; U.S. Biochemical, Cleveland, Ohio).
A clone carrying the fliG deletion was electroporated into JR501 (r− m+) (17), treated as described above, and incubated at 30°C on LM-Cm. Plasmid was purified and electroporated into SJW1103, and cointegrates were selected. The procedure of Hamilton et al. (5) was followed for resolution of the plasmid and generation of the desired deletion on the chromosome. Chromosomal DNA was prepared (26) from clones selected as Cms at 30°C. The region around the fliG gene was amplified and sequenced to verify the presence of the desired fliG deletion. The resultant fliG null strain was called MKM1. (A similar procedure was carried out to construct a fliM null strain using pAMH5 as the template DNA. The final construction encoded a FliM deletion having four amino acids at the N terminus and nine amino acids at the C terminus. This strain was named MKM6.)
Construction of FliG scanning deletion mutants.
fliG deletion mutants were generated using the protocol developed by Toker et al. (23). Outside PCR primers had an XbaI site near the 3′ end of fliF and a HindIII site near the 5′ end of fliH for cloning into pUC18. The wild-type fliG gene was in plasmid pGMK1000. Subcloning into pET19b was carried out using PCR to introduce an NdeI site at the 5′ end of fliG and a BamHI site at the 3′ end of the gene (after the stop codon), using the pUC-based deletion plasmid as the template DNA. Mutant clones in both pUC18 and pET19b were sequenced to verify the constructions and to establish that there were no errors introduced by the PCR amplification.
Construction of truncated FliG mutants.
Wild-type, N-terminal, and C-terminal versions of FliG were constructed and cloned into pET19b, using NdeI and BamHI restriction sites engineered into the PCR primers. The N-terminal version of FliG (GN, encoded by pGMK3100) contained amino acids 1 to 108, and the C-terminal version (GC, encoded by pGMK3200) contained amino acids 109 to 331. Wild-type FliG was in plasmid pGMK3000.
Swarm plates and motility studies.
For complementation studies, freshly transformed cells were spotted on soft TM-Amp (ampicillin) plates and incubated at 30°C. For studies of motility and flagellation, freshly transformed cells were grown with vigorous shaking overnight in LM-Amp at 30°C, diluted 1:100 in fresh LM-Amp, and grown at 30°C with vigorous shaking for about 6 h. Samples from the wild-type culture were monitored for motility. When the cells appeared to be at maximum motility (usually in late exponential phase), the samples were diluted 1:10 into motility medium (10 mM phosphate buffer [pH 7], 0.1 mM EDTA) and observed by high-intensity dark-field light microscopy (13).
Antibodies.
Polyclonal antibodies against FliG, FliM, and FliN were a gift from K. Oosawa and S.-I. Aizawa, Teikyo University, Utsunomiya, Japan. Monoclonal anti-FliF was described in reference 6. Monoclonal anti-FLAG M2 was purchased from Sigma-Aldrich, anti-GST was purchased from Amersham Pharmacia (Piscataway, N.J.), INDIA-His-probe-HRP (horseradish peroxidase) and rabbit anti-goat conjugated to HRP were purchased from Pierce (Rockford, Ill.), and HRP-conjugated goat anti-rabbit and goat anti-mouse antibodies were purchased from Bio-Rad.
Immunoblotting.
Plasmids containing mutant fliG alleles were transformed into MKM1, and immunoblotting was performed to test for protein production and/or stability as described by Toker et al. (23). Anti-FliG was used at a dilution of 1:50,000 and detected with an ECL (enhanced chemiluminescence) immunodetection kit (Amersham International, Little Chalfont, United Kingdom).
Purification of His-FLAG-FliF protein.
Plasmid pFFF1300 (2) containing N-terminally His-FLAG-tagged FliF was transformed into Escherichia coli BL21(DE3)pLysS (20). A single colony of freshly transformed cells was grown overnight in 10 ml of LM-Amp-Cm at 37°C, 2 ml was transferred to 100 ml of the same medium (prewarmed) and grown for 1.5 to 2 h at 37°C (to an optical density at 600 nm [OD600] of 0.5), and IPTG was added to a final concentration of 1 mM. Growth was continued for an additional 3 h at 37°C. Cells were harvested and frozen at −20°C until needed. Then 200-ml aliquots of frozen cells were thawed and resuspended in B-PER reagent (Pierce), and FliF was prepared from the inclusion body pellet as instructed by the manufacturer. The final pellet was solubilized in binding buffer (BB) (20 mM Tris [pH 8.0], 5 mM imidazole, 500 mM NaCl)–1% N-tetradecyl-N,N-dimethyl-3-ammonio-1-propanesulfonate detergent (SB14; Sigma-Aldrich) and applied to a 5-ml Ni-nitrilotriacetic acid column (Qiagen) equilibrated with BB–0.2% SB14. After a 10-ml wash with BB–0.2% SB14 and a 10-ml wash with the same buffer plus 20 mM imidazole, the His-FLAG-FliF was eluted from the column in 15 ml of buffer containing 100 mM EDTA, 20 mM Tris (pH 8.0), 500 mM NaCl, and 0.2% SB14. Fractions containing FliF (as visualized by silver-stained gels) were pooled and dialyzed against three changes of 1 liter each of 20 mM Tris (pH 8.0)–1 mM EDTA. The dialyzed material was concentrated from about 7 ml to 2 ml in a Centriprep 30 concentrator (Amicron, Beverly, Mass.). The purified His-FLAG-FliF was tested by immunoblotting using either anti-FLAG or anti-FliF antibody. Both antibodies detected the purified protein.
Purification of N-terminally His-tagged FliM and FliN.
N-terminally His-tagged FliM (His-FliM) cloned into pTrc99A was constructed by Anne Toker (unpublished data). After transformation of BL21(DE3), His-FliM production was induced using IPTG at 1 mM as described above. Frozen cells from 200 ml of culture were suspended in 5 ml of B-PER and treated for purification of inclusion bodies. The inclusion body pellet was dissolved in 2 ml of 6 M guanidine-HCl and dialyzed against 1 liter (with three changes) of Tris-buffered saline (TBS)–1 M guanidine-HCl.
N-terminally His-tagged FliN, purified as described elsewhere (24), was a gift from Anne Toker.
Affinity blotting.
The affinity blotting procedure of Toker and Macnab (24) was followed, using (i) anti-FliF antibody or anti-FLAG M2 antibody to detect interactions with FliF and (ii) anti-FliG, anti-FliM, or anti-FliN antibody to detect interactions with FliG, FliM, or FliN. pGMK3000, pGMK3100, and pGM3200 or the deletion plasmids pGETΔ1, etc., were transformed into BL21(DE3)pLysS, and fresh colonies were grown in LM-Amp-Cm to an OD600 of about 0.5; the cells were then induced with 1 mM IPTG for 3 h at 37°C, and 0.5 OD unit of each sample was washed and resuspended in 100 μl of protein sample buffer. After sodium dodecyl sulfate-polyacrylamide gel electrophoresis on a 12.5% gel, the samples were transferred to nitrocellulose membranes and standard immunoblotting procedures were followed except that an additional step was added: 100 μl of purified protein per 10 ml of TBS–0.1% Tween 20 was added to the blots and left shaking overnight at room temperature. Then conventional immunoblotting using the ECL protocol was performed.
RESULTS
Construction and complementation properties of a Salmonella fliG null strain.
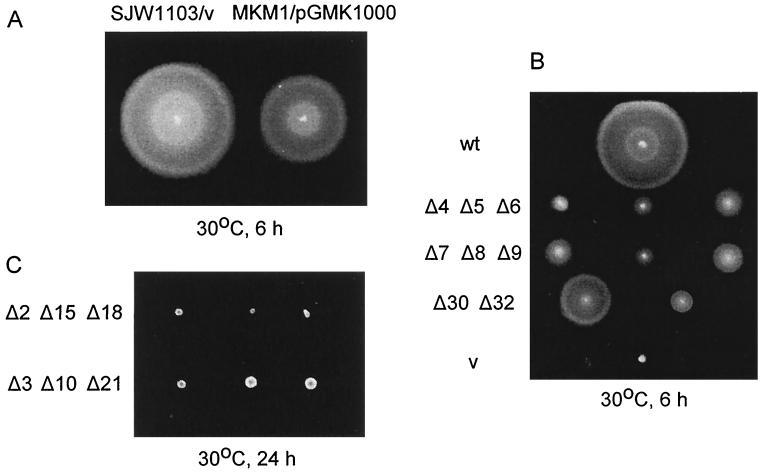
Although several fliG in-frame deletion mutants existed in Salmonella (8), there were none with a deletion covering most of the gene. We constructed a Salmonella fliG null strain, MKM1, which encodes a product consisting of only 4 amino acids at the N terminus and 10 amino acids at the C terminus (see Materials and Methods). MKM1 transformed with the pUC-based plasmid pGMK1000 carrying the wild-type fliG gene swarmed about 70% as well as the wild-type strain SJW1103 transformed with the vector alone (Fig. 1A).
FIG. 1.
Complementation of the Salmonella fliG null strain MKM1 with fliG deletion alleles in pUC18. A freshly transformed colony was spotted on soft TM-Amp plates and incubated at 30°C for the times indicated. (A) Comparison of SJW1103 (wild-type fliG)/pUC18 and MKM1/pGMK1000 (wild-type fliG). (B) MKM1 transformed with plasmids containing those fliG alleles (GΔ4, etc.; abbreviated as Δ4, etc.) that resulted in significant swarming. wt (wild type), pGMK1000; v (vector), pUC18. (C) MKM1 transformed with plasmids containing representative fliG alleles that resulted in negligible swarming after the standard 6-h incubation (not shown); after 24 h, some of these (bottom row) showed some very limited spreading.
ΔFliG mutant phenotypes on semisolid agar and in liquid medium.
We constructed pUC-based plasmids encoding FliG with various deletion segments. The first deletion segment of FliG (GΔ1) started at amino acid 7 and continued through residue 16. The remaining deletion segments followed in order, i.e., GΔ2 (amino acids 17 to 26), GΔ3 (27 to 36), etc., finishing with GΔ32 (317 to 326), virtually at the end of the 331-amino-acid protein. MKM1 was transformed with these plasmids and examined for swarming in semisolid agar plates. GΔ4 to GΔ9, GΔ30, and GΔ32 supported some degree of swarming, ranging from 20% (e.g., GΔ8) to 60% (GΔ30) of the wild-type level (Fig. 1B). FliG with deletion segments GΔ1 to GΔ3, GΔ10 to GΔ29, and GΔ31 failed to complement MKM1 after the usual incubation time of 6 h on swarm plates; however, after prolonged (24-h) incubation, some (GΔ3, GΔ10, and GΔ21) had slightly larger colonies than the rest (Fig. 1C).
By dark-field microscopy, mutants with deletion segments GΔ4 to GΔ9, GΔ30, and GΔ32 showed various degrees of motility from vigorous wild-type (GΔ4, GΔ6 to GΔ8, and GΔ30) to clockwise switch-biased (GΔ5, GΔ9, and GΔ32). Those with GΔ10, GΔ18, GΔ19, GΔ24, GΔ25, GΔ28, GΔ29, and GΔ31 were flagellate but paralyzed. Those with GΔ11, GΔ13, GΔ16, GΔ17, GΔ20, GΔ21 and GΔ27 were nonflagellate, although an occasional cell with flagella could be seen. Those with GΔ1 to GΔ3, GΔ12, GΔ14, GΔ15, GΔ22 to GΔ23 and GΔ26 were nonflagellate.
Thus, the immediate N terminus and several 10-amino-acid segments throughout the rest of the protein are more or less essential for flagellar assembly. Many other segments can be deleted while sustaining flagellar assembly but not rotation. Only relatively short regions close to the N and C termini are dispensable for motility.
Lack of function of a given deletion protein could derive from protein instability. This issue will be addressed below.
Dominance studies.
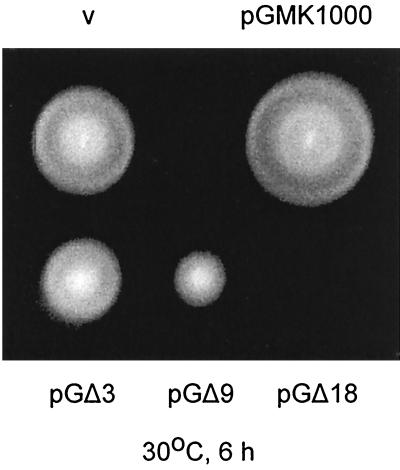
Swarm assays were carried out in the Salmonella wild-type strain, SJW1103, to test for the effect of the mutant protein on the wild-type flagellar motor. Representative results are shown in Fig. 2.
FIG. 2.
Dominance tests of the scanning deletion fliG mutants in Salmonella wild-type strain SJW1103. A freshly transformed colony was spotted on soft TM-Amp plates and incubated at 30°C for 6 h. v (vector), pUC18; pGMK1000, wild-type FliG; pGΔ3, pGΔ9, and pGΔ18, plasmids encoding deletion FliG proteins that exhibit mild, strong, and total negative dominance, respectively.
The wild-type FliG protein exhibited a slight positive multicopy effect; i.e., SJW1103/pGMK1000 swarmed slightly better than SJW1103/pUC18. All deletion mutants exhibited negative dominance to some degree. For GΔ1 to GΔ4 and GΔ30, the effect was mild (swarming at 60 to 90% of that of SJW1103 transformed with vector). GΔ5 to GΔ9, GΔ11, GΔ26, GΔ31, and GΔ32 swarmed at 25 to 50% of the wild-type level. With GΔ10, GΔ12 to GΔ25 and GΔ27 to GΔ29, swarming was almost totally inhibited. We also examined free-swimming cells by dark-field microscopy for dominance effects, with similar results.
Thus, strong negative dominance was seen throughout much of the FliG sequence, from GΔ10 through GΔ29, with only two exceptions. We conclude that proteins with short deletions in essentially all of the sequence except the terminal regions can still bind to other components of the system, such as FliF or the other switch proteins, but do so in a destructive fashion that interferes with productive binding of wild-type FliG.
The examples where dominance effects were relatively mild fell into two categories: those where the deletion proteins themselves retained a high degree of function (GΔ4 to GΔ9, GΔ30, and GΔ32) and those where they did not (GΔ1 to GΔ3, GΔ11, GΔ26, and GΔ31). The former category is fairly easy to explain, but the latter category raises the possibility that the mutant protein was not dominant because it was unstable.
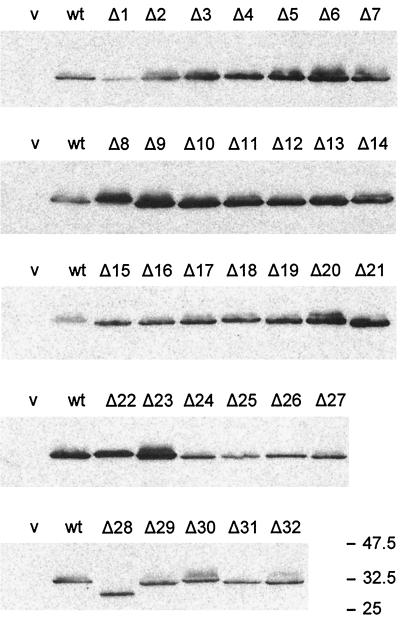
Cellular levels of mutant FliG proteins.
To test whether mutant FliG proteins conferring nonflagellate phenotype or failing to exert dominance effects were stable, we performed immunoblots on whole cell extracts of all of the ΔFliG clones (in pUC18) transformed into the fliG null strain MKM1 (Fig. 3). In the null background, mutants carrying deletion segments GΔ1 and GΔ24 to GΔ32 produced smaller amounts of FliG than MKM1 transformed with pGMK1000, which encodes wild-type FliG; GΔ28 appeared smaller than the other deletion proteins and was probably being degraded, since it sometimes gave a faint band at the same position as the other deletion proteins. All other deletion mutants produced levels of FliG equal to or greater than the level of wild-type FliG carried on a plasmid. Similar results regarding stability of the deletion proteins was obtained with wild-type strain SJW1103 as the host (data not shown), with the exception that here the level of GΔ29, like that of GΔ28, was low; in no case, however, was the level of mutant FliG produced from the pUC-based plasmid as low as the chromosomal level of wild-type FliG in SJW1103 cells transformed with vector alone. We conclude that, with the possible exceptions of GΔ28 and GΔ29, the levels of all deletion proteins were high enough that they cannot be the explanation for either impaired function or lack of strong negative dominance.
FIG. 3.
Cellular levels of FliG, as measured by immunoblotting using polyclonal anti-FliG antibody, in the fliG null strain MKM1 transformed with the plasmids shown. v (vector), pUC18; wt (wild type), pGMK1000; Δ1, etc., pGΔ1, etc. Molecular mass markers are indicated on the right in kilodaltons.
Affinity blotting using purified FliF protein.
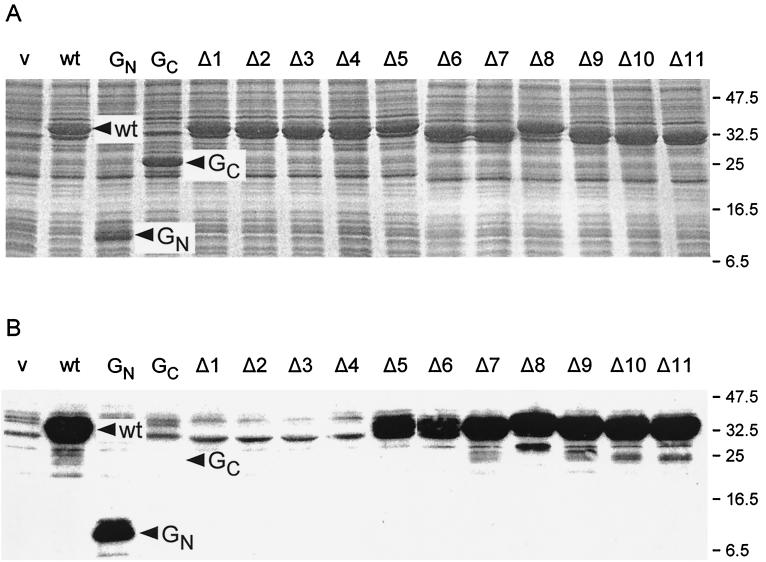
We next wanted to examine the question of binding between the MS ring proteins FliF and FliG. Affinity blotting was carried out with purified N-terminally His-FLAG-tagged FliF (see Materials and Methods) as the probe and various versions of FliG, overexpressed from pET19b-based vectors, as targets. We first did this with full-length FliG (encoded by plasmid pGMK3000) and two N- and C-terminal fragments (encoded by plasmids pGMK3100 and pGMK3200, respectively). The N-terminal fragment (GN) consisted of amino acids 1 to 108, while the C-terminal fragment (GC) consisted of amino acids 109 to 331. Coomassie blue-stained gels of whole cells of E. coli BL21(DE3)pLysS transformed with these plasmids and grown in the presence of IPTG established that all three proteins were present at similar levels (Fig. 4A). In affinity blots, using either monoclonal anti-FliF antibody (Fig. 4B) or commercial monoclonal anti-FLAG antibody (data not shown), His-FLAG-FliF bound strongly to both wild-type FliG and GN but did not bind at all to GC.
FIG. 4.
(A) Coomassie blue-stained gels of whole cell protein from E. coli BL21(DE3)pLysS transformed with pET-based plasmids expressing various alleles of fliG and grown in the presence of IPTG. (B) Affinity blot of the same samples using purified N-His-FLAG-FliF as a probe. v (vector), pET19b; wt, pGMK3000 (wild-type FliG); GN, pGMK3100 (N-terminal fragment of FliG, residues 1 to 108); GC, pGMK3200 (C-terminal fragment of FliG, residues 109 to 331); Δ1, etc., pGETΔ1, etc. Molecular mass markers are indicated on the right in kilodaltons.
The analysis was then extended to the FliG deletion proteins. The results for GΔ1 to GΔ11 are shown in Fig. 4. The Coomassie blue-stained gel in Fig. 4A established that all deletion proteins were being overproduced to essentially the same extent. The affinity blot in Fig. 4B showed that GΔ1 to GΔ4 were not recognized at all by FliF, whereas the remainder of the deletion proteins, GΔ5 to GΔ11 were recognized as strongly as wild-type FliG or GN. Cells transformed with all of the other pET-based plasmids that we constructed (GΔ12, GΔ21, GΔ22, and GΔ27 to GΔ31) also resulted in full recognition by FliF (data not shown); it seems likely that this would be true also of those segments we did not test, all of which lay in the C-terminal two-thirds of the FliG sequence.
Affinity blotting of FliG proteins using FliM and FliN as probes.
We attempted affinity blotting using purified His-FliM protein but found no evidence for binding to wild-type FliG or to any of the FliG truncation or deletion proteins; this could be due to the pronounced tendency of FliM to form inclusion bodies when present at high concentrations, as has been noted by ourselves and others (16, 29). Purified His-FliN showed no binding to wild-type FliG or to any of the FliG truncation or deletion proteins; likewise, purified His-FliG showed no binding to overproduced FliN. Toker and Macnab (24), using FliG as the probe and FliM variants as targets, found that FliG bound to FliM, and specifically to its N-terminal two-thirds. Tang et al. (21) found that the N-terminal two-thirds of FliG bound to FliM and to FliN; sequence toward the end of the fragment used was essential for the binding.
DISCUSSION
In this study, we constructed systematic 10-amino-acid deletions of the motor/switch protein FliG and examined the consequences in terms of flagellation, motility, dominance over wild-type FliG, cellular protein levels, and binding to the MS ring protein FliF. The results are summarized in schematic form in Fig. 5.
FIG. 5.
Schematic illustration of various properties of FliG as manifested by the deletion mutants used in this study. FliG is 331 amino acids long. A total of 32 deletion segments were used, each 10 amino acids in length. Segment 1 consisted of amino acids 7 to 16, and the remainder were contiguous, finishing with segment 32 (amino acids 317 to 326). The qualitative shading scale goes from white (zero function) through light gray and dark gray to black (full function). The segments indicated by hatching were not tested for FliF binding.
A number of lines of evidence indicate that of the three motor/switch proteins (FliG, FliM, and FliN), FliG is the one most closely and directly associated with the basal body MS ring, an annular structure made up of about 26 subunits of the FliF protein (9).
We had found previously that a full-length in-frame fusion protein of FliF-FliG was able to function almost normally in flagellar assembly and motility (3). This was the first indication that in the wild-type cell, the C terminus of FliF and the N terminus of FliG were normally positioned close to each other and that N-terminal sequence of FliG was likely to be important in the binding interaction. This inference is supported by our finding in the present study that GN binds as well as the wild type to FliF, whereas GC does not bind at all (Fig. 4B).
We had also encountered previously a deletion-fusion version of FliF-FliG that showed some limited function in terms of motor rotation and switching even though it was missing the first 94 amino acids at the N terminus of FliG as well as 56 residues at the C terminus of FliF (3). This result could be interpreted in two ways. Either sequence essential for binding of FliG to FliF does not commence until fairly far in from the FliG N terminus, or covalent linkage in the deletion-fusion protein substantially replaces the normal noncovalent association between the two proteins. The present study clearly indicates that the latter interpretation is the correct one. Deletion of any of the first four 10-amino-acid segments of FliG completely abolished its ability to bind FliF in affinity blot assays, whereas deletion of any subsequent segment left FliG binding to FliF intact. Segments lacking the immediate N-terminal sequence of FliG showed only mild negative dominance, further supporting the importance of that sequence for binding to FliF.
This leads to the interesting conclusion that a sequence of only around 40 amino acids suffices to provide an interaction strong enough to survive the shear forces generated during flagellar rotation. This description may be misleading, however, since there are likely to be about 26 such interactions per flagellum, and if lateral FliF-FliF interactions and FliG-FliG interactions are strong, the overall binding energy between FliF and FliG may be much greater. FliF-FliF interactions are certainly strong, since the MS ring is a stable particle; there is evidence from coisolation experiments that FliG does interact with itself (21), although it is difficult to estimate how strong these interactions are.
The less than wild-type level of function in the deletion-fusion protein could then derive from several causes: a linkage that, though covalent, lacks a total of 150 amino acids is unlikely to position FliF and FliG with perfect wild-type geometry. Also 10-amino-acid deletions 4 through 9 resulted in partially impaired function in swarm tests (Fig. 1B), and so in the deletion-fusion protein, sequence that is important for function if not for binding sequence is lost. FliGΔ4 seems to be a marginal case in this regard: in spite of no apparent binding to FliF in affinity assays (Fig. 4B), it did sustain some degree of swarming (Fig. 1B), although cells were very poorly flagellated; this may be an instance where cooperativity of binding gives a slight degree of function. Finally, loss of C-terminal sequence of FliF could have consequences of its own.
Deletion mutant proteins GΔ5 to GΔ9, which displayed significant but much less than wild-type motility in the FliG null background (Fig. 1B), all interfered considerably with the function of the wild-type version of FliG, resulting in a loss of motility of about 50% in dominance experiments. Therefore, we conclude that these deletion proteins compete with wild-type FliG for incorporation into the motor structure (via their essentially wild-type FliF binding ability) and thereby cause reduced motility because these functions are impaired.
Except for a short region near the C terminus of FliG, the remainder of the deletion alleles were characterized by having either no or extremely reduced ability to assemble flagella and no ability to rotate the few flagella they might have had. Nonetheless, they showed wild-type FliF binding and strong negative dominance. Thus, they had the ability to interfere with wild-type FliG function, specifically with regard to its ability to sustain flagellar assembly.
This is likely to be at least in part a failure to bind FliM and FliN. Although we did not detect FliG-FliM or FliG-FliN interactions in this study, it is clear from other work that they exist. Tang et al. (21) have shown by coisolation that GST-FliM and GST-FliN fusions can both bind FliG; a FliG fragment consisting of residues 1 to 245 can bind to both GST-FliM and GST-FliN, whereas a fragment consisting of residues 1 to 226 cannot bind either. Toker and Macnab (24), investigating the properties of FliM, found that a fragment consisting of about the N-terminal two-thirds of the sequence bound to FliG, but that a slightly shorter fragments failed to do so.
In an analysis of spontaneous fliG mutations (8), we found that extensive in-frame deletions permitted flagellar assembly, the two largest being of residues 77 to 126 (corresponding to segments 8 to 12) and 99 to 145 (approximately segments 10 to 14). Thus, they extend substantially beyond the last fully flagellate, paralyzed FliG deletion allele (GΔ10). We interpret these data to mean that FliG sequence essential for FliM and FliN binding does not commence until approximately position 145. At the C-terminal end we had found several appreciable deletions starting at about position 280; this is in agreement with the data of Tang et al. (21) that C-terminal sequence is not essential for FliM or FliN binding.
FliG has been identified as the switch protein most directly involved in torque generation (12). Even when overproduced, mutant FliG proteins associated with Mot− phenotype never result in motility; this contrasts with the situation in fliM or fliN, where overproduction of the mutant proteins does restore motility. Lloyd et al. (12) found that the N-terminal two-thirds of FliG are sufficient for flagellar assembly. Our results agree with and further refine this observation, demonstrating that the N-terminal 46 amino acids are necessary and sufficient for binding to FliF, thus mounting the motor onto the MS ring of the basal body.
ACKNOWLEDGMENTS
We thank Brian Lane, Priyadarshan Gupta, and Stacey Denenberg for help in constructing the deletion mutations, Anne Toker for the gift of purified His-FliG and His-FliN proteins, and Kenji Oosawa and Shin-Ichi Aizawa for the gift of antibodies against FliG, FliM, and FliN.
This research was supported by USPHS grant GM40335.
REFERENCES
- 1.Bren A, Eisenbach M. The N terminus of the flagellar switch protein, FliM, is the binding domain for the chemotactic response regulator, CheY. J Mol Biol. 1998;278:507–514. doi: 10.1006/jmbi.1998.1730. [DOI] [PubMed] [Google Scholar]
- 2.Fan F, Ohnishi K, Francis N R, Macnab R M. The FliP and FliR proteins of Salmonella typhimurium, putative components of the type III flagellar export apparatus, are located in the flagellar basal body. Mol Microbiol. 1997;26:1035–1046. doi: 10.1046/j.1365-2958.1997.6412010.x. [DOI] [PubMed] [Google Scholar]
- 3.Francis N R, Irikura V M, Yamaguchi S, DeRosier D J, Macnab R M. Localization of the Salmonella typhimurium flagellar switch protein FliG to the cytoplasmic M-ring face of the basal body. Proc Natl Acad Sci USA. 1992;89:6304–6308. doi: 10.1073/pnas.89.14.6304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Francis N R, Sosinsky G E, Thomas D, DeRosier D J. Isolation, characterization and structure of bacterial flagellar motors containing the switch complex. J Mol Biol. 1994;235:1261–1270. doi: 10.1006/jmbi.1994.1079. [DOI] [PubMed] [Google Scholar]
- 5.Hamilton C M, Aldea M, Washburn B K, Babitzke P, Kushner S R. New method for generating deletions and gene replacements in Escherichia coli. J Bacteriol. 1989;171:4617–4622. doi: 10.1128/jb.171.9.4617-4622.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Homma M, Aizawa S-I, Dean G E, Macnab R M. Identification of the M-ring protein of the flagellar motor of Salmonella typhimurium. Proc Natl Acad Sci USA. 1987;84:7483–7487. doi: 10.1073/pnas.84.21.7483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Homma M, Iino T, Macnab R M. Identification and characterization of the products of six region III flagellar genes (flaAII.3 through flaQII) of Salmonella typhimurium. J Bacteriol. 1988;170:2221–2228. doi: 10.1128/jb.170.5.2221-2228.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Irikura V M, Kihara M, Yamaguchi S, Sockett H, Macnab R M. Salmonella typhimurium fliG and fliN mutations causing defects in assembly, rotation, and switching of the flagellar motor. J Bacteriol. 1993;175:802–810. doi: 10.1128/jb.175.3.802-810.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jones C J, Macnab R M, Okino H, Aizawa S-I. Stoichiometric analysis of the flagellar hook-(basal-body) complex of Salmonella typhimurium. J Mol Biol. 1990;212:377–387. doi: 10.1016/0022-2836(90)90132-6. [DOI] [PubMed] [Google Scholar]
- 10.Khan I H, Reese T S, Khan S. The cytoplasmic component of the bacterial flagellar motor. Proc Natl Acad Sci USA. 1992;89:5956–5960. doi: 10.1073/pnas.89.13.5956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kihara M, Homma M, Kutsukake K, Macnab R M. Flagellar switch of Salmonella typhimurium: gene sequences and deduced protein sequences. J Bacteriol. 1989;171:3247–3257. doi: 10.1128/jb.171.6.3247-3257.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lloyd S A, Tang H, Wang X, Billings S, Blair D F. Torque generation in the flagellar motor of Escherichia coli: evidence of a direct role for FliG but not FliM or FliN. J Bacteriol. 1996;178:223–231. doi: 10.1128/jb.178.1.223-231.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Macnab R M. Examination of bacterial flagellation by dark-field microscopy. J Clin Microbiol. 1976;4:258–265. doi: 10.1128/jcm.4.3.258-265.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marykwas D L, Schmidt S A, Berg H C. Interacting components of the flagellar motor of Escherichia coli revealed by the two-hybrid system in yeast. J Mol Biol. 1996;256:564–576. doi: 10.1006/jmbi.1996.0109. [DOI] [PubMed] [Google Scholar]
- 15.Mathews M A A, Tang H L, Blair D F. Domain analysis of the FliM protein of Escherichia coli. J Bacteriol. 1998;180:5580–5590. doi: 10.1128/jb.180.21.5580-5590.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oosawa K, Ueno T, Aizawa S-I. Overproduction of the bacterial flagellar switch proteins and their interactions with the MS ring complex in vitro. J Bacteriol. 1994;176:3683–3691. doi: 10.1128/jb.176.12.3683-3691.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ryu J, Hartin R J. Quick transformation in Salmonella typhimurium LT2. BioTechniques. 1990;8:43–44. [PubMed] [Google Scholar]
- 18.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 19.Sockett H, Yamaguchi S, Kihara M, Irikura V M, Macnab R M. Molecular analysis of the flagellar switch protein FliM of Salmonella typhimurium. J Bacteriol. 1992;174:793–806. doi: 10.1128/jb.174.3.793-806.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Studier F W, Moffatt B A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 21.Tang H, Braun T F, Blair D F. Motility protein complexes in the bacterial flagellar motor. J Mol Biol. 1996;261:209–221. doi: 10.1006/jmbi.1996.0453. [DOI] [PubMed] [Google Scholar]
- 22.Thomas D R, Morgan D G, DeRosier D J. Rotational symmetry of the C ring and a mechanism for the flagellar rotary motor. Proc Natl Acad Sci USA. 1999;96:10134–10139. doi: 10.1073/pnas.96.18.10134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Toker A S, Kihara M, Macnab R M. Deletion analysis of the FliM flagellar switch protein of Salmonella typhimurium. J Bacteriol. 1996;178:7069–7079. doi: 10.1128/jb.178.24.7069-7079.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Toker A S, Macnab R M. Distinct regions of bacterial flagellar switch protein FliM interact with FliG, FliN, and CheY. J Mol Biol. 1997;273:623–634. doi: 10.1006/jmbi.1997.1335. [DOI] [PubMed] [Google Scholar]
- 25.Williams A W, Yamaguchi S, Togashi F, Aizawa S-I, Kawagishi I, Macnab R M. Mutations in fliK and flhB affecting flagellar hook and filament assembly in Salmonella typhimurium. J Bacteriol. 1996;178:2960–2970. doi: 10.1128/jb.178.10.2960-2970.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Woo T H S, Cheng A F, Ling J M. An application of a simple method for the preparation of bacterial DNA. BioTechniques. 1992;13:696–698. [PubMed] [Google Scholar]
- 27.Yamaguchi S, Aizawa S-I, Kihara M, Isomura M, Jones C J, Macnab R M. Genetic evidence for a switching and energy-transducing complex in the flagellar motor of Salmonella typhimurium. J Bacteriol. 1986;168:1172–1179. doi: 10.1128/jb.168.3.1172-1179.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yamaguchi S, Fujita H, Sugata K, Taira T, Iino T. Genetic analysis of H2, the structural gene for phase-2 flagellin in Salmonella. J Gen Microbiol. 1984;130:255–265. doi: 10.1099/00221287-130-2-255. [DOI] [PubMed] [Google Scholar]
- 29.Zhao R, Schuster S C, Khan S. Structural effects of mutations in Salmonella typhimurium flagellar switch complex. J Mol Biol. 1995;251:400–412. doi: 10.1006/jmbi.1995.0443. [DOI] [PubMed] [Google Scholar]