Abstract
FliE is a flagellar basal body protein of Salmonella whose detailed location and function have not been established. A mutant allele of fliE, which caused extremely poor flagellation and swarming, generated extragenic suppressors, all of which mapped to flgB, one of four genes encoding the basal body rod; the fliE flgB pseudorevertants were better flagellated and swarmed better than the fliE parent, especially when the temperature was reduced from 37 to 30°C. Motility of the pseudorevertants in liquid culture was markedly better than motility on swarm plates; we interpret this to mean that reduced flagellation is less deleterious at low viscous loads. Overproduction of the mutant FliE protein improved the motility of the parental fliE mutant and its pseudorevertants, though not to wild-type levels. Overproduction of suppressor FlgB (but not wild-type FlgB) in the fliE mutant also resulted in improved motility. The second-site FlgB mutation by itself had no phenotype; cells swarmed as well as wild-type cells. When overproduced, wild-type FliE was dominant over FliE-V99G, but the reverse was not true; that is, overproduced FliE-V99G was not negatively dominant over wild-type FliE. We conclude that the mutant protein has reduced probability of assembly but, if assembled, functions relatively well. Export of the flagellar protein FlgD, which is known to be FliE dependent, was severely impaired by the FliE-V99G mutation but was significantly improved in the suppressor strains. The FliE mutation, V99G, was close to the C terminus of the 104-amino-acid sequence; the suppressing mutations in FlgB were all either G119E or G129D, close to the C terminus of its 138-amino-acid sequence. Affinity blotting experiments between FliE as probe and various basal body proteins as targets and vice versa revealed strong interactions between FliE and FlgB; much weaker interactions between FliE and other rod proteins were observed and probably derive from the known similarities among these proteins. We suggest that FliE subunits constitute a junction zone between the MS ring and the rod and also that the proximal rod structure consists of FlgB subunits.
This study concerns the flagellar protein FliE, a poorly understood component of the Salmonella flagellar basal body, which is a complex structure containing subunits of many proteins. They include, in addition to FliE, the protein of the cytoplasmic membrane MS ring (FliF), the four proteins of the rod (FlgB, FlgC, FlgF, and FlgG), and the proteins of the periplasmic P ring (FlgI) and outer membrane L ring (FlgH). FliL, an integral membrane protein, is also found to associate with the basal body (22). With milder isolation protocols, one of the motor/switch proteins (FliG) remains attached to the MS ring, as does a peripheral structure called the cytoplasmic or C ring, consisting of the other two motor/switch proteins (FliM and FliN) (4, 8). The integral membrane motor proteins MotA and MotB are believed to be associated with the basal body (9), interacting with FliG (26). Finally, there is growing evidence that the integral membrane components of the type III flagellar protein export apparatus (FlhA, FlhB, FliO, FliP, FliQ, and FliR) are basal body components, probably located in a patch of membrane at the center of the MS ring (3, 14). Thus, depending on the definition, there could be as many as 20 basal body components.
The fliE gene itself has some interesting characteristics (Fig. 1): it is the only gene in its transcriptional unit, it is divergent from the two other flagellar operons in its region (region IIIb), and it is located at one end of a large, mostly noncoding region between flagellar regions IIIa and IIIb (17, 21). The former two characteristics are suggestive of a somewhat specialized function for the fliE gene product. FliE was shown by Müller et al. (17) to be located in the basal body; based on data obtained in a prior study (7), its stoichiometry was estimated at about nine subunits per basal body. It is a small protein (104 amino acids, deduced molecular mass of 11,114 Da) and lacks the terminal hydrophobic heptad repeats that are characteristic of the axial family of flagellar proteins (5, 6). Also, in contrast to the axial proteins, whose predicted α-helical regions are for the most part confined to their termini, FliE has a high predicted α-helical content throughout most of its sequence (mean predicted content of 74 mol%).
FIG. 1.
Schematic representation (not to scale) of flagellar regions I and IIIb of the Salmonella chromosome, with emphasis on the two genes, flgB and fliE (bold letters), that are the subject of this study. Transcriptional units (operons) are indicated by arrows. flgB is the first gene in the flgBCDEFGHIJ operon. Operons not referred to in the text are indicated by dashed arrows. n/c, noncoding region flanking region IIIb.
The axial proteins form a long, continuous, hollow cylindrical structure consisting of the rod, hook, hook-filament junction proteins, filament, and filament cap. Not only is this structure important for the function of the flagellum as a motor organelle but its central channel or lumen is the physical pathway by which axial protein subunits reach their assembly destination, the tip of the growing flagellum. The component substructures are all built with the following common theme. The subunits lie on the so-called basic helix of a cylindrical lattice. This underlying local helical symmetry (not to be confused with the macroscopic helicity of the flagellar filament) means that, in principle, subunits could be added indefinitely like the steps of a helical staircase. This is in contrast to the substructures of the basal body such as the MS ring, which have closed annular symmetry and thus a fixed number of subunits (thought to be about 26 in the case of the MS ring protein, FliF [7]).
The rod and MS ring appear to abut each other closely. How do substructures with fundamentally different symmetries join together? A specialized zone might facilitate the junction. FliE seems a possible candidate for construction of such a junction zone. In a study of the morphological assembly of the flagellum, Kubori et al. (11) found that flagellar precursors, all of which contained the MS ring protein, contained FliE only if they also contained the rod proteins, suggesting that the rod and FliE might form a complex.
If FliE lies on the periplasmic side of the MS ring, is it a substrate for the type III flagellar export pathway, as are the axial proteins? The answer is almost certainly yes, although difficulties were experienced in its detection in export assays (14) (difficulties which were also experienced in the case of the rod proteins, whose periplasmic location is well established). FliE was found to interact with some, but not all, of the known components of the export apparatus in affinity blots (15). It also interacts with FlgJ, a periplasmic muramidase (18) (unpublished data). Like all known substrates of the flagellar export pathway, FliE does not undergo signal peptide cleavage. FliE, however, possesses the unique property of being necessary for the efficient export of other substrates (14). This property could be explained (at least in general terms) if it is the first, and physically most proximal, of the export substrates.
Thus, a variety of lines of evidence suggest that FliE may be located between the MS ring and the rod. We report here genetic and biochemical evidence that strongly supports this hypothesis and indicates that FlgB constitutes the most proximal rod structure.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
The strains and plasmids used in this study are listed in Table 1. Plasmid vectors used were pET19b (Novagen), pTrc99A (Pharmacia), and pUC18 (New England Biolabs). Luria-Bertani (LB) medium, soft tryptone agar plates, and M9-Casamino Acids medium are described in reference 14.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| E. coli strains | ||
| XL1-Blue | Recipient for cloning experiments | Stratagene |
| BL21(DE3)pLysS | Overproduction of proteins from pET-based plasmid | Novagen |
| HMS174(DE3)pLysS | Overproduction of proteins from pET-based plasmid | Novagen |
| Salmonella strains | ||
| SJW1103 | Wild-type strain for motility and chemotaxis | 25 |
| SJW1110 | fliE-V99G | This study |
| MY7001 | fliE-V99G flgB1 | This study |
| MY7002 | fliE-V99G flgB2 | This study |
| SJW1190 | Wild type | This study |
| SJW1191 | flgB1 | This study |
| SJW1192 | flgB2 | This study |
| SJW501 | flgA::Tn10 | This study |
| SJW503 | flgE::Tn10 | This study |
| SJW720 | flgG::Tn10 | This study |
| SJW91 | flhA::Tn10 | This study |
| SJW1572 | ΔfliE-fliK | 24 |
| SJW1110-flgA | fliE flgA | This study |
| SJW1110-flgG | fliE flgG | This study |
| SJW1110-flhA | fliE flhA | This study |
| SJW1525 | flgB | 19 |
| SJW156 | flgD | 19 |
| SJW1371 | ΔfliE | 19 |
| Plasmids | ||
| pET-FLAG-19b | Cloning vector | 2 |
| pMM1001 | pTrc99A/FliE | This study |
| pMM1006 | pTrc99A/FliE-V99G | This study |
| pMM1003 | pET19b/His-FLAG-FliE | 15 |
| pMM1005 | pET19b/His-FLAG-FliE-V99G | This study |
| pMM1101 | pTrc99A/FlgB | This study |
| pMM1106 | pTrc99A/FlgB1 | This study |
| pMM1108 | pTrc99A/FlgB2 | This study |
| pMM1102 | pET19b/His-FlgB | 15 |
| pMM1105 | pET19b/His-FlgB1 | This study |
| pMM1107 | pET19b/His-FlgB2 | This study |
| pMM1202 | pET19b/His-FlgC | This study |
| pMM1302 | pET19b/His-FlgF | This study |
| pMM204 | pET19b/His-FlgG | This study |
| pMM1601 | pET19b/His-FlgE | 15 |
| pMM711 | pET19b/His-FlgD | 15 |
| pMM1002 | pET19b/His-FliE | 15 |
| pMM601 | pET19b/His-MotAC | 15 |
Strain construction and mapping of suppressor mutations.
Suppressor mutations in pseudorevertants of SJW1110 (fliE) were mapped by phage P22-mediated transduction. The four strains used as recipients were SJW1572 (ΔfliE-fliK), SJW1110-flgA (fliE flgA), SJW1110-flgG (fliE flgG), and SJW1110-flhA (fliE flhA). The latter three double mutants were constructed as follows. flgA::Tn10, flgG::Tn10, and flhA::Tn10 were transferred into SJW1110 from SJW501 (flgA::Tn10), SJW720 (flgG::Tn10), and SJW91 (flhA::Tn10), respectively, by P22(Int)-mediated transduction. From tetracycline-resistant (Tetr) transductants, Tets clones were selected by the method of Bochner et al. (1). To confirm the constructions of these strains, they were subjected to complementation tests with representative fliE, flgA, flgG, and flhA mutants.
The approximate positions of the suppressor mutations in region I were determined by three-point crosses. First, flgA::Tn10 or flgE::Tn10 was transferred into a pseudorevertant (e.g., MY7002; see Results) from SJW501 (flgA::Tn10) or SJW503 (flgE::Tn10). As it was possible that in some of the Tetr transductants the suppressor mutation was replaced by wild-type DNA sequence accompanying the Tn10 factor, 50 Tetr transductants each (described as the MY7002-flgA::Tn10 group and the MY7002-flgE::Tn10 group) were reserved. Their genotypes were fliE sup flgA::Tn10 and fliE sup flgE::Tn10, respectively. The test consisted of three transductional crosses: from MY7002-flgA::Tn10 to SJW1110-flgG, from MY7002-flgE::Tn10 to SJW1110-flgA, and from MY7002-flgE::Tn10 to SJW1110-flgG. The cross from MY7002-flgA::Tn10 to SJW1110-flgA was not carried out because of the overlap of their flgA mutations.
Isolated second-site mutations from MY7001 (fliE flgB1) and MY7002 (fliE flgB2) were prepared by transducing region I of these strains into SJW191 (ΔflgA-flgI); these transductants were named SJW1191 (flgB1) and SJW1192 (flgB2), respectively. In transductions with SJW1110-flgA (fliE flgA) and SJW1110-flgG (fliE flgG) as recipients, they yielded the pseudorevertant phenotype, confirming that they carried the suppressor mutations. A control strain, SJW1190, was constructed by transducing wild-type DNA from region I into SJW191 and exhibited wild-type swarming.
PCR and cloning.
Synthetic primers containing restriction sites were synthesized using a model 392 DNA/RNA synthesizer (Applied Biosystems). PCR was carried out using an MJ MiniCycler (MJ Research) and Taq DNA polymerase (Sigma). PCR products were prepared from agarose gels with a QIAquick gel extraction kit (Qiagen). Restriction enzymes and T4 DNA ligase (New England Biolabs) were used according to the manufacturers' recommendations. Recombinant plasmids were purified with a QIAprep spin plasmid kit (Qiagen).
DNA sequencing.
DNA sequencing was carried out with modified T7 DNA polymerase Sequenase (U.S. Biochemical) and various synthetic primers.
Purification of N-terminally His-FLAG-tagged FliE and FliE-V99G.
Ten milliliters of overnight culture of Escherichia coli BL21(DE3)pLysS carrying either pMM1003 or pMM1005 (which encodes the N-terminally His-FLAG-tagged FliE or FliE-V99G, respectively, on pET19b) was inoculated into 200 ml of LB containing ampicillin and incubated at 37°C with shaking until the cell density reached an optical density at 600 nm of 0.6 to 0.8. Then isopropyl-β-d-thiogalactopyranoside (IPTG) was added to a final concentration of 1 mM, and incubation was continued for another 5 h. The cells were collected by centrifugation and stored at −20°C. The cells were thawed, suspended in 20 ml of binding buffer (20 mM Tris-HCl [pH 8.0], 500 mM NaCl) and sonicated (model 450 Sonifier; Branson Ultrasonics Corp.). These lysates were centrifuged at low speed to pellet undisrupted cells and inclusion bodies. Both His-FLAG-FliE and His-FLAG-FliE-V99G were found to be in the pellet fraction as inclusion bodies and were suspended in 5 ml of binding buffer. Then 5 ml of 6 M guanidine HCl was added to 5 ml of the suspension of inclusion bodies. After rotation at 4°C for 1 h and centrifugation (13,000 × g, 10 min, 4°C) to remove undisrupted cells, this solution was dialyzed overnight against 1 liter of binding buffer with two changes. After centrifugation to remove any reaggregated products, the supernatant was loaded onto a Ni-nitrilotriacetic acid agarose column (bed volume, 3 ml) preequilibrated with 9 ml of binding buffer. After washing with 18 ml of binding buffer containing 60 mM imidazole, the tagged FliE proteins were eluted with 18 ml of a linear 100 to 600 mM imidazole gradient and were found to be purified almost to homogeneity. They were then dialyzed overnight against 1 liter of 50 mM Tris-HCl (pH 8.0)–100 mM NaCl with two changes.
Preparation of the periplasmic contents and culture supernatants.
Periplasmic contents and culture supernatants were prepared as described elsewhere (14).
Immunoblotting and affinity blotting.
Immunoblotting with anti-FlgD antibody and anti-β-lactamase antibody (as a control for cell fractionation) was carried out as described elsewhere (14). Affinity blotting was carried out as described previously (15).
RESULTS
Extragenic suppressors of a fliE mutation map between the flgA and flgE genes in flagellar region I.
In a suppression analysis of the fliE gene of Salmonella, most of the examples we encountered were intragenic and will be described elsewhere. However, one fliE mutant, SJW1110, gave rise to extragenic suppressors also.
Cells from 200 colonies of SJW1110 on nutrient agar were streaked on 8% nutrient gelatin agar (24). During 4 days of incubation at 37°C, 177 pseudorevertant clones, which formed small swarms emerging from the streak, were isolated. Of these, nine produced swarms of the pseudorevertant type in crosses with SJW1110-flgG (second mutation in flagellar region I) but not in crosses with SJW1110-flhA (second mutation in region II) or SJW1110-fliK (second mutation in region IIIa); thus, these nine pseudorevertants have their suppressor mutations in flagellar region I. The remaining 168 pseudorevertants produced swarms only in crosses with SJW1572 (ΔfliE-fliK), showing that their suppressor mutations were in flagellar region IIIb and could have been intragenic.
Flagellar region I, at 23 min, is far from the location of fliE, which is in flagellar region IIIb at 40 min (Fig. 1). It consists mostly of genes encoding structural proteins of the flagellar apparatus, including the basal body rod and ring proteins, hook protein, hook-capping protein, and hook-filament junction proteins.
To determine the approximate positions of the suppressor mutations in region I, three-point crosses were carried out (see Materials and Methods). Representative results are given for one of the pseudorevertants, MY7002, in Table 2. For all nine pseudorevertants, the results showed that the second-site mutation lay between flgA (a gene whose product is responsible for P-ring formation) and flgE (the hook protein gene) (Fig. 1).
TABLE 2.
Results of three-point crosses to establish the location of SJW1110 extragenic suppressorsa
| Donor | Recipient | No. of donor clones that produced swarms/50 donor clones |
|---|---|---|
| MY7002-flgA::Tn10 | SJW1110-flgG | 8 |
| MY7002-flgE::Tn10 | SJW1110-flgA | 15 |
| MY7002-flgE::Tn10 | SJW1110-flgG | 0 |
The nine extragenic suppressor mutants turned out to have just two distinct mutations. MY7002 is an example of the second group. Cross results for all nine extragenic suppressors were similar.
The suppressor mutations are at the 3′ end of the proximal rod gene flgB.
We suspected the mutations were most likely to lie within one of the three proximal rod genes: flgB, flgC, or flgF. DNA sequence analysis confirmed that this was the case and established that all nine examples corresponded to just two mutations near the 3′ end of flgB: the nucleotide changes were G356A (two examples) and G386A (seven examples). We sequenced the entire flgB and flgC genes and established that these were the sole mutations present in the region. The corresponding pseudorevertants were named MY7001 (fliE flgB1) and MY7002 (fliE flgB2).
At the amino acid level, the two mutations in FlgB correspond to the substitutions G119E and G129D, respectively. Thus, both mutations replace a glycine residue by an acidic residue, and they lie within 10 amino acids of each other and within 19 amino acids of the C terminus. However, in view of the mutation being suppressed (see below), steric issues may be more important than introduction of a negative charge.
The parental mutation is V99G, near the C terminus of FliE.
DNA sequence analysis of the mutation in the parental fliE strain SJW1110 revealed that it was a missense mutation, T296G at the nucleotide level, which corresponds to the substitution V99G at the amino acid level, almost at the C terminus of the 104-amino-acid protein. We refer to this mutant protein as FliE-V99G.
Motility of SJW1110, its pseudorevertants and isolated second-site suppressor mutants.
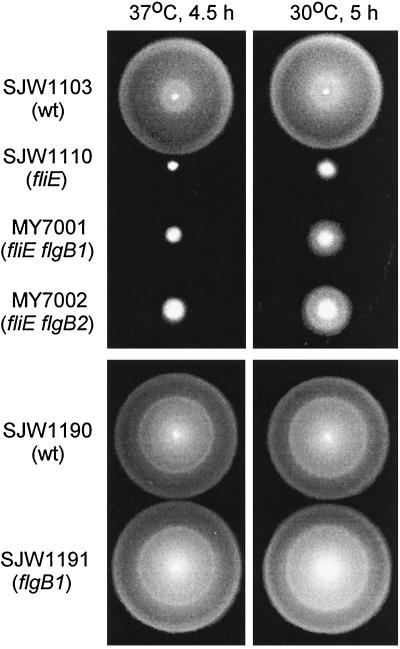
The fliE parent, SJW1110, swarmed very poorly at 37°C but slightly better at 30°C (Fig. 2). The pseudorevertants MY7001 and MY7002 both showed a much more pronounced temperature sensitivity, swarming considerably better than SJW1110 at 30°C (Fig. 2).
FIG. 2.
Swarming of the fliE mutant SJW1110, its pseudorevertants MY7001 (fliE flgB1) and MY7002 (fliE flgB2), and strains constructed with the second-site mutations only (SJW1191 [flgB1] and SJW1192 [flgB2] [not shown]). Cells were spotted on semisolid tryptone agar plates and incubated at the restrictive temperature (37°C) or the permissive temperature (30°C) for the times indicated. wt, wild type.
We observed by high-intensity dark-field microscopy the free-swimming motility of SJW1110, MY7001, and MY7002 cells grown at 30°C. Only about 20 to 30% of SJW1110 cells could swim in liquid medium, and their swimming was much slower and more ragged than that of the wild-type strain SJW1103; swimming cells had only one or two flagella (wild-type cells typically have five or more flagella). About 80 to 90% of the pseudorevertants MY7001 and MY7002 were motile and swam at about 70% of wild-type speed; they typically had three to four flagella. Thus, free-swimming motility of the pseudorevertants was less impaired than swarming, probably because reduced flagellar number is less deleterious under low-load conditions.
Using P22-mediated transduction, we constructed strains containing only the second-site alleles, flgB1 and flgB2 (i.e., the fliE allele was wild type). These strains, SJW1191 (Fig. 2) and SJW1192 (not shown), swarmed at essentially wild-type levels and showed no temperature sensitivity; thus, there is no phenotype associated with the suppressor mutations alone.
Complementation, dominance, and multicopy properties of the fliE-V99G mutation.
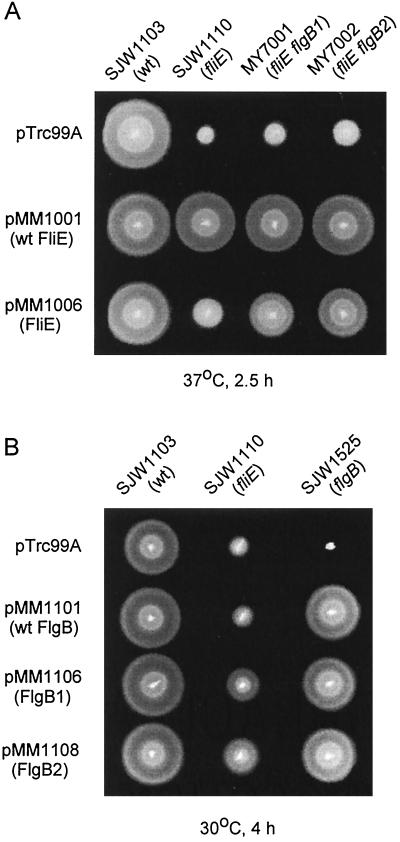
We cloned the wild-type and mutant fliE alleles into pTrc99A to give plasmids pMM1001 and pMM1006, respectively. These were introduced into SJW1103 (wild type), SJW1110 (fliE), MY7001 (fliE flgB1), and MY7002 (fliE flgB2), and the resulting transformants were inoculated onto soft tryptone agar plates and incubated at both 30 and 37°C; the data for 37°C are shown in Fig. 3A.
FIG. 3.
(A) Swarming of SJW1103 (wild type [wt]), SJW1110 (fliE) and its pseudorevertants MY7001 and MY7002 transformed with pTrc99A (vector control), pMM1001 (wild-type FliE), and pMM1006 (FliE-V99G). Cells were spotted on semisolid tryptone agar plates and incubated at 37°C for 2.5 h; similar results were obtained at 30°C except that the suppression of the SJW1110 defect by pMM1006 was more pronounced. (B) Swarming of SJW1103 (wild type), SJW1110 (fliE), and null flgB mutant SJW1525, transformed with pTrc99A (vector control), pMM1101 (wild-type FlgB), pMM1106 (FlgB1), and pMM1108 (FlgB2). Cells were spotted on semisolid tryptone agar plates and incubated at 30°C for 4 h.
pMM1001 fully complemented all of the hosts. The fact that MY7001 and MY7002 were complemented shows that wild-type FliE is dominant and confirms the observation that the suppressor flgB mutations by themselves have no phenotype.
pMM1006 failed to exert a negative dominance effect on wild-type SJW1103 cells, indicating that although (judging by the pseudorevertant phenotype) the mutant FliE can be exported and can assemble, it does not compete effectively with wild-type FliE in these regards. pMM1006 exerted a positive multicopy effect on the swarming motility of SJW1110, seen clearly at 37°C (Fig. 3A) and even more pronounced at 30°C (not shown). This suggests that, though FliE-V99G assembles into the growing flagellar structure inefficiently, overproduction increases the probability of assembly. At both temperatures, but especially at 37°C, MY7001 and MY7002 carrying pMM1006 swarmed better than SJW1110 carrying the same plasmid, indicating that FliE-V99G coassembles more efficiently with mutant FlgB than with wild-type FlgB.
We examined the motility in liquid medium of transformed cells grown at the more stringent temperature of 37°C. Only about 30% of untransformed MY7001 and MY7002 cells were motile, and their swimming was significantly slower than when they were grown at 30°C. Transformation with pMM1006, however, resulted in extremely vigorous motility, indistinguishable from wild-type motility. Thus, motility is improved by FliE-V99G overproduction as well as by suppression by FlgB1 or FlgB2.
Complementation, dominance and multicopy properties of the suppressor flgB mutations.
Next we cloned the wild-type flgB gene and the suppressor alleles, flgB1 and flgB2, into pTrc99A to give pMM1101, pMM1106, and pMM1108, respectively. These plasmids were introduced into wild-type SJW1103, SJW1110 (fliE-V99G), and a flgB null strain, SJW1525, and the transformants were inoculated onto the soft tryptone agar plates and incubated at 30°C for 4 h (Fig. 3B).
pMM1101 (wild-type FlgB) did not affect the wild-type strain and complemented the flgB null mutant but failed to complement SJW1110; thus, even in multicopy, wild-type FlgB could not compensate for the FliE-V99G defect. pMM1106 (FlgB1) and pMM1108 (FlgB2), however, suppressed to a considerable degree the FliE-V99G defect of SJW1110, indicating that these mutant FlgB proteins were dominant over wild-type FlgB in their interaction with FliE-V99G. FlgB1 and FlgB2 also complemented the flgB null mutant, confirming the lack of a phenotype associated with these suppressor alleles in isolation.
When the host was the null fliE mutant SJW1371, no swarming occurred with any of the transformants (data not shown), ruling out the possibility that suppression by FlgB was occurring by a bypass mechanism.
FliE-V99G inhibits the export of FlgD.
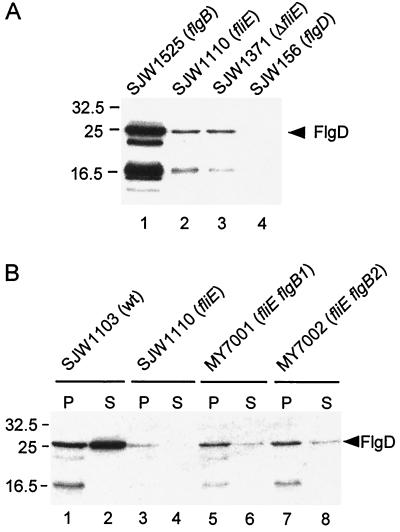
In a previous study, a fliE deletion mutant (SJW1371) failed to export detectable amounts of the hook-capping protein FlgD into the periplasm (14). To examine whether the mutation FliE-V99G affects FlgD export, we prepared the periplasmic contents from SJW1110 grown at 30°C and performed immunoblotting with anti-FlgD antibody. Only a very small amount of FlgD was detected in the periplasm from SJW1110 (Fig. 4A, lane 2), about the same as with the fliE deletion mutant SJW1371 (lane 3). FlgD was strongly detected in the periplasmic fraction from a flgB mutant (lane 1), as shown previously (14).
FIG. 4.
Effects of FliE mutations on export of the hook-capping protein, FlgD, assayed by immunoblotting using polyclonal antibody against FlgD. (A) Periplasmic contents of the strains shown. Lane 1, SJW1525 (flgB), positive control; lane 2, SJW1110 (fliE), parent of the pseudorevertants isolated in this study; lane 3, SJW1371 (ΔfliE null mutant); lane 4, SJW156 (flgD), negative control. Lower bands represent degradation products of FlgD, which is susceptible to proteolysis in the periplasm. (B) Periplasmic contents (P) and culture supernatants (S) of the strains shown. Lanes 1 and 2, wild type (wt); lanes 3 and 4, SJW1110 (fliE); lanes 5 and 6 (MY7001) and 7 and 8 (MY7002), extragenic suppressors of SJW1110 with second-site mutations in flgB. Note the absence of FlgD degradation in the culture supernatants. Molecular mass markers are shown on the left in kilodaltons.
We next examined the effects of the flgB second-site suppressor mutations on FlgD export (Fig. 4B). The amount of FlgD in the periplasm from MY7001 (lane 5) and MY7002 (lane 7), though much lower than the wild-type level (lane 1), was higher than that from SJW1110 (lane 3). The difference between wild-type and mutant strains was even more pronounced when it came to export into the culture supernatant. With the wild-type strain, large amounts were exported (lane 2); with the pseudorevertants, small amounts were detected (lanes 6 and 8), and with the fliE parent, none was detected (lane 4). Thus, the second-site mutations suppress to some extent the failure of FliE-V99G to export FlgD. These export data, and especially those for the culture supernatants, are consistent with the severely and moderately reduced extent of flagellation and motility of the parent and pseudorevertants, respectively.
Affinity blotting analysis of FliE and the basal body rod proteins.
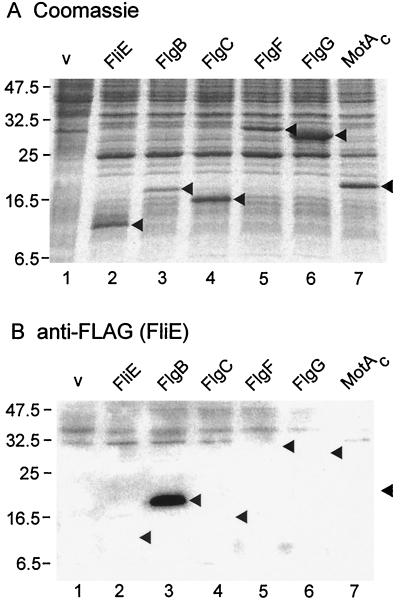
Using purified N-terminally His-FLAG-tagged FliE as a probe, we performed affinity blotting against the following proteins: (i) the rod proteins FlgB, FlgC, FlgF, and FlgG; (ii) FliE itself; (iii) the hook protein FlgE and the hook-capping protein FlgD; and (iv) as a negative control, MotAC, the cytoplasmic loop region of MotA (15). Whole cell proteins were prepared from E. coli HMS174(DE3)pLysS carrying pET19b-based plasmids encoding N-terminally His-tagged versions of these proteins. These tagged proteins, and also the His-FLAG-tagged FliE being used as the probe, were functional in complementation tests (data not shown). The whole cell samples were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (Fig. 5A), transferred onto nitrocellulose membranes, probed with N-His-FLAG-FliE, and then immunoblotted with anti-FLAG-antibody (Fig. 5B).
FIG. 5.
(A) Coomassie blue-stained gels after SDS-PAGE of whole cell proteins from E. coli HMS174(DE3)pLysS carrying pET19b-based plasmids overproducing the following export substrates under induction with 1 mM IPTG: lane 1, pET19b (vector [v]); lane 2, pMM1002 (FliE); lane 3, pMM1102 (FlgB); lane 4, pMM1202 (FlgC); lane 5, pMM1302 (FlgF); lane 6, pMM204 (FlgG); lane 7, pMM601 (MotAC, negative control). The positions of these overproduced proteins are indicated by arrowheads. (B) Affinity blotting of the same samples as in panel A, using purified His-FLAG-tagged FliE as the probe, and detection of the probe with monoclonal anti-FLAG antibody. The positions of the target proteins are indicated by arrowheads. Molecular mass markers are shown on the left in kilodaltons.
Consistent with the genetic suppression result, FliE bound strongly to FlgB (lane 3). There was no evidence for binding to FliE itself (lane 2), FlgC (lane 4), FlgF (lane 5), FlgG (lane 6), the negative control MotAC (lane 7), or FlgE or FlgD (data not shown). After longer exposures, weak interaction of FliE with itself and with FlgC could be seen (data not shown).
The FliE-V99G mutation results in decreased swarming and substrate export, whereas in combination with the FlgB1 or FlgB2 mutations, swarming and substrate export are recovered to a considerable degree (Fig. 2 and 4). It was possible that these physiological effects might be reflected in binding assays. However, wild-type and mutant FliE bound equally well to wild-type and mutant FlgB (data not shown).
DISCUSSION
Genetic and biochemical data both demonstrate that FliE interacts with the rod protein FlgB.
Information about the basal body protein FliE has been much more fragmentary than that about most of the other basal body components such as the ring and rod proteins. From two independent experimental approaches, we conclude that FliE interacts strongly with one of the proximal rod proteins, FlgB. Thus, its location in the basal body is very likely to be proximal to the rod, as we discuss further below.
The first approach was genetic. A particular fliE mutant (SJW1110) gave rise to extragenic suppressors as well as a number of intragenic ones. The extragenic suppressor mutations all lay in one of the proximal rod proteins, FlgB, and were either G119E or G129D of the 138-amino-acid sequence. Possible implications of the mutations and their location in the sequence will be discussed below. The second approach, which used the technique of affinity blotting (15), demonstrated a strong interaction between FliE and FlgB and a much weaker interaction between FliE and either FlgC or FliE itself.
FlgB is probably the most proximal of the four rod proteins.
FlgG constitutes the most distal rod structure. The evidence derives from a mutation in the MS ring protein FliF that results in the so-called rod-fragile phenotype (20) whereby hook-filaments are readily shed in viscous media. Attached to the proximal end of the hooks is a partial rod structure, consisting of the rod protein FlgG.
Until now, the remaining three rod proteins—FlgB, FlgC, and FlgF—have been grouped together as proximal rod proteins without any differentiation as to order. Our data suggest that FlgB constitutes the most proximal rod structure. This leaves FlgC and FlgF as intermediate rod proteins, but again without any differentiation as to order. A weak argument—the order of the genes within the operon—suggests that the order may be FlgB-FlgC-FlgF-FlgG.
FliE may consist of separate regions for transport and for structural interactions with the rod.
We have shown previously that FliE is a basal body protein which is necessary for the export of rod/hook substrates. For technical reasons, we were unable to detect its own export but strongly suspected that it was an export substrate. Our finding in the present study—that FliE and the proximal rod protein FlgB interact—further supports this supposition since it is hard to imagine why there would be such an interaction in the cytoplasm, and it is also hard to imagine FliE, which is tenaciously associated with the basal body, having a cytoplasmic location. If FliE is exported, it should have regions of its sequence that constitute the recognition signal for its export by the flagellar apparatus. Other exported flagellar proteins (e.g., flagellin and hook protein) are recognized by N-terminal sequence (10, 12). Consistent with this, FliE-V99G, with its mutation almost at the C terminus, was able to be exported, as judged by its ability to support the export of other substrates such as FlgD and to support motility, especially if overproduced and especially if in combination with the suppressor forms of FlgB.
FliE-V99G was not, however, able to support FlgD export at wild-type levels. This could have several explanations. It itself might be exported less efficiently than wild-type FliE; it might be less stable in the periplasm; or it might interact less productively with FliF to promote export (see below). Export of FlgD was improved by the presence of the suppressor FlgB protein (Fig. 4B). This suggests that productive interaction between FliE and FlgB either stabilizes FliE or ensures maximum efficiency of export. However, it should be noted that in a wild-type FliE background, FlgB is not required for export (14).
We tentatively suggest that the N-terminal region of FliE is important for its own export and that its C-terminal region is important for structural interactions with the proximal rod.
Why is FliE needed for the export of other flagellar proteins, and is this its primary role?
In our study of the flagellar export pathway (14), we found that export of one substrate was independent of export of another (provided they belong to the same specificity type, i.e., rod/hook type or filament type). FliE was an exception to this rule, since its export was a prerequisite for export of other substrates in significant amounts. Why should this be so? It suggests that FliE is, at least in a formal sense, a component of the export apparatus as well as a substrate. Its putative position immediately following the MS ring, whose pore is believed to contain the membrane components of the export apparatus, is uniquely different from the locations of all of the axial components (rod proteins, hook protein, flagellin, etc.) which provide the lumen for diffusion of export substrates to their final destination but are not in the immediate vicinity of the export apparatus proper. We suggest that a FliF-FliE interaction may alter the conformation of the MS ring in a manner that permits the export apparatus to deliver its substrates to the periplasm. Interestingly, the central region of the MS ring may undergo a substantial conformation change, depending on physical conditions (23).
In spite of the fact that FliE is necessary for export of other substrates to proceed normally, we suspect that this may not be its sole or even primary function; it could be an incidental function deriving from its physical location. An equivalent situation exists on the cytoplasmic face of the MS ring, where FliG and the C ring are necessary for flagellar protein export even though their primary function is in motor rotation and switching.
It is also worth noting that a very low basal level of export can occur even in the absence of FliE (Fig. 4A, lane 3; the mutation in strain SJW1371 is a total deletion of the gene). This argues against a vital role of FliE in export.
We propose that the primary role of FliE may be as a structural adapter between the annular symmetry of the MS ring and the helical symmetry of the rod and all subsequent axial structures.
If this is the case, we propose that FliE not be called a rod protein, since it differs in so many ways from the rod and other axial proteins. Instead, we propose tentatively calling it an (MS ring)-rod junction protein, acknowledging that its association with the MS ring has not yet been experimentally demonstrated.
Given the proposed location of FliE between the MS ring protein FliF and the rod protein FlgB, it seemed plausible that examples might be found of FliE suppressors lying in FliF. Thus far, attempts to identify such suppressors have been unsuccessful. FliF did interact with FliE in an affinity blot assay; however, since it also interacted with a wide range of other export substrates, we are uncertain of the significance of this result.
The polarity of the axial structures is probably based on a (C-terminus proximal)-(N-terminus distal) orientation of the individual protein subunits.
From three-dimensional reconstructions and other biophysical evidence concerning the axial structures (most notably the filament), compelling arguments have been made that the principal intersubunit interactions involve α-helical coiled coils between the N terminus of one subunit and the C terminus of the adjacent subunit in the axial direction (13, 16). Thus, the axial structure has a general polarity of the sort N—C••N—C, etc. What is the polarity of these interactions with respect to the overall polarity (cell proximal versus cell distal) of the filament? In such reconstructions, the resolution is still not good enough to give a clear answer to this question (D. DeRosier, D. Morgan, and K. Namba, personal communications). One argument in favor of a (C-terminus proximal)-(N-terminus distal) subunit orientation involves the primary structure of the axial proteins themselves. Substructures like the hook and filament which are bounded on either end by other structures have the hydrophobic heptad repeats that give rise to the coiled coils at both termini, whereas the filament cap protein FliD, which is bounded only on its proximal end (by filament), has only a heptad repeat at its C-terminal end (5).
The two FlgB mutations that suppressed a FliE mutation lay at the C terminus of FlgB, within an extended heptad repeat region (5). Both of these results provide further support for a (C-terminus proximal)-(N-terminus distal) subunit orientation of FlgB and, by extension, other axial proteins.
A model for rod assembly.
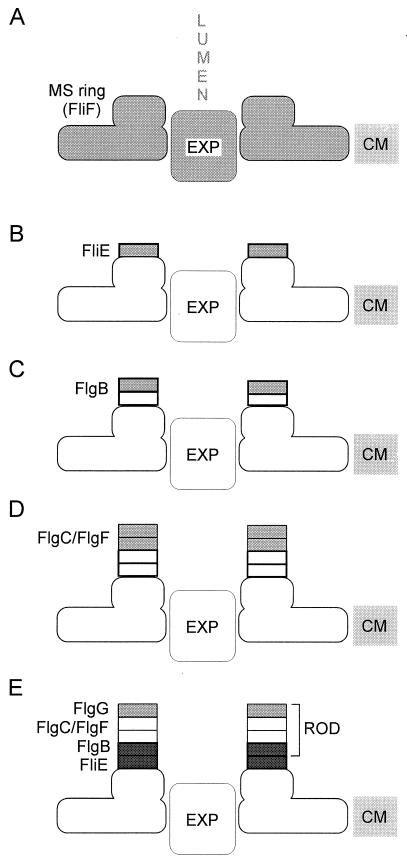
Based on the available evidence, we propose the following model for basal body rod assembly (Fig. 6). FliE is the first flagellar protein to be exported. It associates with the distal end of the MS ring and alters the conformation of the ring and/or the export apparatus such that translocation of other substrates such as the rod proteins is facilitated. Assembly of the rod then proceeds in stages corresponding to the four zones from which it is constructed. First, the FlgB zone assembles onto the FliE zone. Next, the FlgC and FlgF zones assemble in an as yet unknown order. Finally, the distal FlgG zone assembles, the P and L rings nucleate around the rod, and flagellar assembly proceeds to its later stages of hook and filament formation.
FIG. 6.
Proposed pathway for assembly of FliE and the basal body rod. Each incremental structure is indicated in light grey. (A) The process starts with the basal body MS ring, made out of FliF subunits, embedded in the cytoplasmic membrane (CM) and with the flagellar export apparatus (EXP) lying within its central pore. LUMEN represents the central channel that exists within the axial structure as it grows. (B) FliE subunits are exported to the periplasm, where they assemble onto the collar feature of the MS ring. Direct evidence for an interaction between the MS ring and FliE is lacking, but such interaction is postulated because FliE export is a prerequisite for export of other substrates. (C) Subunits of the proximal rod protein FlgB are exported and assemble onto the distal end of the FliE zone. (D) Intermediate rod proteins FlgC and FlgF add (in unknown order) to the FlgB zone. (E) The distal rod protein FlgG adds to the intermediate rod zone; FliE and FlgB, the subjects of this study, are shown in dark grey. A periplasmic muramidase, FlgJ (18), probably also participates in rod assembly but is omitted from the model for simplicity.
ACKNOWLEDGMENTS
We thank Gabriele Miller and May Kihara for technical assistance.
This work was supported by USPHS grants AI12202 and GM40335.
REFERENCES
- 1.Bochner B R, Huang H-C, Schieven G L, Ames B N. Positive selection for loss of tetracycline resistance. J Bacteriol. 1980;143:926–933. doi: 10.1128/jb.143.2.926-933.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fan F, Macnab R M. Enzymatic characterization of FliI: an ATPase involved in flagellar assembly in Salmonella typhimurium. J Biol Chem. 1996;271:31981–31988. doi: 10.1074/jbc.271.50.31981. [DOI] [PubMed] [Google Scholar]
- 3.Fan F, Ohnishi K, Francis N R, Macnab R M. The FliP and FliR proteins of Salmonella typhimurium, putative components of the type III flagellar export apparatus, are located in the flagellar basal body. Mol Microbiol. 1997;26:1035–1046. doi: 10.1046/j.1365-2958.1997.6412010.x. [DOI] [PubMed] [Google Scholar]
- 4.Francis N R, Sosinsky G E, Thomas D, DeRosier D J. Isolation, characterization and structure of bacterial flagellar motors containing the switch complex. J Mol Biol. 1994;235:1261–1270. doi: 10.1006/jmbi.1994.1079. [DOI] [PubMed] [Google Scholar]
- 5.Homma M, DeRosier D J, Macnab R M. Flagellar hook and hook-associated proteins of Salmonella typhimurium and their relationship to other axial components of the flagellum. J Mol Biol. 1990;213:819–832. doi: 10.1016/S0022-2836(05)80266-9. [DOI] [PubMed] [Google Scholar]
- 6.Homma M, Kutsukake K, Hasebe M, Iino T, Macnab R M. FlgB, FlgC, FlgF and FlgG. A family of structurally related proteins in the flagellar basal body of Salmonella typhimurium. J Mol Biol. 1990;211:465–477. doi: 10.1016/0022-2836(90)90365-S. [DOI] [PubMed] [Google Scholar]
- 7.Jones C J, Macnab R M, Okino H, Aizawa S-I. Stoichiometric analysis of the flagellar hook-(basal-body) complex of Salmonella typhimurium. J Mol Biol. 1990;212:377–387. doi: 10.1016/0022-2836(90)90132-6. [DOI] [PubMed] [Google Scholar]
- 8.Khan I H, Reese T S, Khan S. The cytoplasmic component of the bacterial flagellar motor. Proc Natl Acad Sci USA. 1992;89:5956–5960. doi: 10.1073/pnas.89.13.5956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Khan S, Dapice M, Reese T S. Effects of mot gene expression on the structure of the flagellar motor. J Mol Biol. 1988;202:575–584. doi: 10.1016/0022-2836(88)90287-2. [DOI] [PubMed] [Google Scholar]
- 10.Kornacker M G, Newton A. Information essential for cell-cycle-dependent secretion of the 591-residue Caulobacter hook protein is confined to a 21-amino-acid sequence near the N-terminus. Mol Microbiol. 1994;14:73–85. doi: 10.1111/j.1365-2958.1994.tb01268.x. [DOI] [PubMed] [Google Scholar]
- 11.Kubori T, Shimamoto N, Yamaguchi S, Namba K, Aizawa S-I. Morphological pathway of flagellar assembly in Salmonella typhimurium. J Mol Biol. 1992;226:433–446. doi: 10.1016/0022-2836(92)90958-m. [DOI] [PubMed] [Google Scholar]
- 12.Kuwajima G, Kawagishi I, Homma M, Asaka J-I, Kondo E, Macnab R M. Export of an N-terminal fragment of Escherichia coli flagellin by a flagellum-specific pathway. Proc Natl Acad Sci USA. 1989;86:4953–4957. doi: 10.1073/pnas.86.13.4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mimori Y, Yamashita I, Murata K, Fujiyoshi Y, Yonekura K, Toyoshima C, Namba K. The structure of the R-type straight flagellar filament of Salmonella at 9 Å resolution by electron cryomicroscopy. J Mol Biol. 1995;249:69–87. doi: 10.1006/jmbi.1995.0281. [DOI] [PubMed] [Google Scholar]
- 14.Minamino T, Macnab R M. Components of the Salmonella flagellar export apparatus and classification of export substrates. J Bacteriol. 1999;181:1388–1394. doi: 10.1128/jb.181.5.1388-1394.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Minamino T, Macnab R M. Interactions among components of the Salmonella flagellar export apparatus and its substrates. Mol Microbiol. 2000;35:1052–1064. doi: 10.1046/j.1365-2958.2000.01771.x. [DOI] [PubMed] [Google Scholar]
- 16.Morgan D G, Owen C, Melanson L A, DeRosier D J. Structure of bacterial flagellar filaments at 11 Å resolution: packing of the α-helices. J Mol Biol. 1995;249:88–110. doi: 10.1006/jmbi.1995.0282. [DOI] [PubMed] [Google Scholar]
- 17.Müller V, Jones C J, Kawagishi I, Aizawa S-I, Macnab R M. Characterization of the fliE genes of Escherichia coli and Salmonella typhimurium and identification of the FliE protein as a component of the flagellar hook-basal body complex. J Bacteriol. 1992;174:2298–2304. doi: 10.1128/jb.174.7.2298-2304.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nambu T, Minamino T, Macnab R M, Kutsukake K. Peptidoglycan-hydrolyzing activity of the FlgJ protein, essential for flagellar rod formation in Salmonella typhimurium. J Bacteriol. 1999;181:1555–1561. doi: 10.1128/jb.181.5.1555-1561.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ohnishi K, Ohto Y, Aizawa S-I, Macnab R M, Iino T. FlgD is a scaffolding protein needed for flagellar hook assembly in Salmonella typhimurium. J Bacteriol. 1994;176:2272–2281. doi: 10.1128/jb.176.8.2272-2281.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Okino H, Isomura M, Yamaguchi S, Magariyama Y, Kudo S, Aizawa S-I. Release of flagellar filament-hook-rod complex by a Salmonella typhimurium mutant defective in the M ring of the basal body. J Bacteriol. 1989;171:2075–2082. doi: 10.1128/jb.171.4.2075-2082.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Raha M, Kihara M, Kawagishi I, Macnab R M. Organization of the Escherichia coli and Salmonella typhimurium chromosomes between flagellar regions IIIa and IIIb, including a large non-coding region. J Gen Microbiol. 1993;139:1401–1407. doi: 10.1099/00221287-139-7-1401. [DOI] [PubMed] [Google Scholar]
- 22.Schoenhals G J, Macnab R M. FliL is a membrane-associated component of the flagellar basal body of Salmonella. Microbiology. 1999;145:1769–1775. doi: 10.1099/13500872-145-7-1769. [DOI] [PubMed] [Google Scholar]
- 23.Suzuki H, Yonekura K, Murata K, Hirai T, Oosawa K, Namba K. A structural feature in the central channel of the bacterial flagellar FliF ring complex is implicated in type III protein export. J Struct Biol. 1998;124:104–114. doi: 10.1006/jsbi.1998.4048. [DOI] [PubMed] [Google Scholar]
- 24.Yamaguchi S, Fujita H, Ishihara A, Aizawa S-I, Macnab R M. Subdivision of flagellar genes of Salmonella typhimurium into regions responsible for assembly, rotation, and switching. J Bacteriol. 1986;166:187–193. doi: 10.1128/jb.166.1.187-193.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yamaguchi S, Fujita H, Taira T, Kutsukake K, Homma M, Iino T. Genetic analysis of three additional fla genes in Salmonella typhimurium. J Gen Microbiol. 1984;130:3339–3342. doi: 10.1099/00221287-130-12-3339. [DOI] [PubMed] [Google Scholar]
- 26.Zhou J, Lloyd S A, Blair D F. Electrostatic interactions between rotor and stator in the bacterial flagellar motor. Proc Natl Acad Sci USA. 1998;95:6436–6441. doi: 10.1073/pnas.95.11.6436. [DOI] [PMC free article] [PubMed] [Google Scholar]