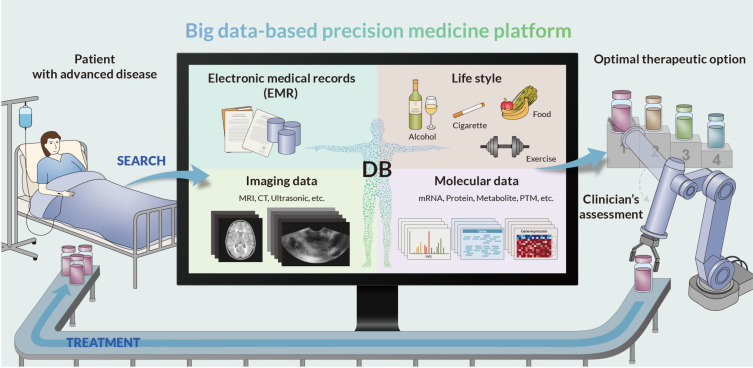
Fig. 1. Schematic overview of big data-based precision medicine platform.
As vast amounts of diverse data have been accumulated in public repositories (Athar et al., 2019; Barrett et al., 2013; Tryka et al., 2014), it has become possible to decode relationships and rules that the data contained. For instance, you will see a list of “people you may know” whenever you open the Facebook application on your mobile. People with significant numbers of common acquaintances with you are chosen to be added to the list based on the social network in the Facebook database. Therefore, the social network data relationships appear pretty accurate. Moreover, people check whether a specific English description is grammatically correct by googling the description. Grammatical correctness of the description can be then determined based on how often the particular description was used in the documents, assuming that the frequently used descriptions are grammatically correct.
Numerous scholars are attempting this big data-based approach to foretell optimal therapeutic options for patients with refractory diseases. Consider that you have gathered the following data for 1 million patients with nonsmall cell lung cancer: 1) electronic medical records; 2) lifestyle information (foods, alcohol, smoking, exercise, etc.); 3) DNA mutations; 4) levels of cancer-related mRNAs, proteins, metabolites, and posttranslational modifications in lung tissue, ascites, blood, and urine samples; and 5) image data from magnetic resonance imaging, computed tomography, and ultrasonic measurement. You then deposited all these data to a database with exploration and search tools. Patients with advanced nonsmall cell lung cancer for whom an optimal therapeutic option could not be determined are then searched against this database. According to the medical, clinical, molecular, and image data of the database, the search will result in the top 100 patients with the most similar characteristics. Finally, an optimal therapeutic option can be determined as the one that demonstrated the best prognosis among therapeutic options employed for the top 100 patients, assuming that the chosen therapeutic option would work best because it worked well for similar patients. Numerous industrial and medical sectors begin to employ this big data-based precision medicine platform.
Although this platform sounds promising, it also has several issues that should be addressed prior to its application in clinical settings. Similar to Google that does not explain why an English description is correct or incorrect when you check the grammar of the description using Google, this approach does not explain why the therapeutic option should work best clinically or mechanistically. Whether the predicted therapeutic option is good or not should be evaluated by clinicians’ experience with and knowledge of the disease. Furthermore, it is unclear how similar top 100 patients should be chosen because the relative importance of the medical, clinical, molecular, and image data is unknown when examining the similarity between patients. Data scientists are currently developing methods to solve these problems to enable the practice of the big data-based precision medicine approach in real clinical settings.
Footnotes
CONFLICT OF INTEREST
The author has no potential conflicts of interest to disclose.
REFERENCES
- Athar A., Füllgrabe A., George N., Iqbal H., Huerta L., Ali A., Snow C., Fonseca N.A., Petryszak R., Papatheodorou I., et al. ArrayExpress update - from bulk to single-cell expression data. Nucleic Acids Res. 2019;47(D1):D711–D715. doi: 10.1093/nar/gky964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett T., Wilhite S.E., Ledoux P., Evangelista C., Kim I.F., Tomashevsky M., Marshall K.A., Phillippy K.H., Sherman P.M., Holko M., et al. NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Res. 2013;41(Database issue):D991–D995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tryka K.A., Hao L., Sturcke A., Jin Y., Wang Z.Y., Ziyabari L., Lee M., Popova N., Sharopova N., Kimura M., et al. NCBI's Database of Genotypes and Phenotypes: dbGaP. Nucleic Acids Res. 2014;42(Database issue):D975–D979. doi: 10.1093/nar/gkt1211. [DOI] [PMC free article] [PubMed] [Google Scholar]