Abstract
Purpose
This research concentrated on the biological effects and special mechanism of circ_0003221 in bladder cancer (BLCA).
Materials and Methods
The level quantification by reverse transcription-quantitative polymerase chain reaction was administrated for circ_0003221, microRNA-892b (miR-892b) and 24-dehydrocholesterol reductase (DHCR24). The biological behaviors were assessed by EDU assay and colony formation assay for proliferation, and transwell assay for cell motility. Glycolytic metabolism was tested using the commercial kits. DHCR24 protein level and cell markers were measured through western blot. The analysis of interaction potential was conducted via dual-luciferase reporter assay and pull-down assay. Circ_0003221 was implemented via tumor xenograft assay in vivo.
Results
Abnormal circ_0003221 upregulation was affirmed in BLCA. BLCA cell proliferation, motility and glycolysis were impeded after circ_0003221 level was knocked down. MiR-892b was identified as a target for circ_0003221. Reduction of miR-892b relieved si-circ_0003221-induced anti-tumor response in BLCA cells. In addition, miR-892b targeted DHCR24 and circ_0003221/miR-892b could regulate the level of DHCR24. The effects of si-circ_0003221 were also counteracted by DHCR24 overexpression.
Conclusions
The current evidence elucidated circ_0003221 targeted miR-892b to elevate the DHCR24 level, thus accelerating cell development and glycolytic metabolism of BLCA cells.
Keywords: 24-dehydrocholesterol reductase, Bladder cancer, Circ_0003221, miR-892b
Graphical Abstract
INTRODUCTION
Bladder cancer (BLCA) is a prevalent malignancy with highly aggressive and invasive potentials [1]. There are 550,000 new cases of BLCA and nearly 170,000 deaths around the world annually [2,3]. BLCA patients have reduced survival rates with frequent recurrences and metastasis [4]. The treatment for BLCA has significant advances in recent years, and non-coding RNAs (ncRNAs) have been used as important molecular targets in BLCA [5,6]. The exploration of pathogenesis and the discovery of biomolecules may contribute to developing specific diagnostic tools and therapeutic strategies for BLCA [6].
Circular RNAs (circRNAs) are endogenous ncRNAs with regulatory roles in cancer progression [7]. Dysregulated circRNAs are associated with carcinogenesis and treatment of BLCA [8]. CircRNAs act as cancer regulators and novel biomarkers through sponging microRNAs (miRNAs) to affect gene expression [9]. Circ_0003221 accelerated cell malignant phenotypes of cervical cancer via regulating cytoplasmic polyadenylation element-binding protein 4 by playing the sponge effect on miR-758-3p [10]. Also, circ_0003221 was displayed to lead to the promotion of BLCA cell migration and proliferation [11]. The functional mechanism of circ_0003221 in BLCA has never been researched.
MiRNAs are implicated in many cellular processes of BLCA by inducing the expression effects on special targets [12]. MicroRNA-892b (miR-892b) has been indicated to inhibit cell metastasis via downregulating PRAPDC1A in breast cancer and targeting lysophosphatidic acid receptor 1 in nasopharyngeal cancer [13,14]. Additionally, miR-892b resulted in anti-tumor effects on BLCA cells partly by blocking the expression of matrix metallopeptidase 9 (MMP9) [15]. It is unknown about other targets of miR-892b in BLCA.
The issued study reported that 24-dehydrocholesterol reductase (DHCR24) participated in cell invasiveness in urothelial carcinoma [16]. DHCR24 also enhanced BLCA cell proliferation ability and regulated the oncogenesis-related glycolysis [17]. However, whether the function of miR-892b is correlated to the oncogenic DHCR24 is unclear. Moreover, the association between circ_0003221 and DHCR24 remains to be explored.
This report was to investigate the roles of circ_0003221 in regulating cell behaviors and glycolytic metabolism of BLCA cells. More importantly, we intended to research the circ_0003221/miR-892b/DHCR24 axis underlying the function of circ_0003221.
MATERIALS AND METHODS
1. Patient samples and cells
BLCA samples (n=55) and normal adjacent controls (n=55) were collected from 55 BLCA patients during transurethral resection of bladder tumor surgery at Chongqing University Three Gorges Hospital. Then samples were conserved at -80℃ and used for isolation of total RNA or protein. Those patients with preoperative treatment were excluded from this study. All protocols were approved by the Ethics Committee of Chongqing University Three Gorges Hospital and followed the Declaration of Helsinki. Written informed consent was obtained from all enrolled patients.
Normal SV-HUC-1 and BLCA cells (5637, T24) were bought from BeNa Culture Collection (Beijing, China), then maintained in Dulbecco’s modified eagle medium (DMEM, Hyclone, Logan, UT, USA) with penicillin/streptomycin (1%, Sigma, St. Louis, MO, USA) and fetal bovine serum (10%, Serapro, Naila, Germany). Cells were cultured in a 37℃ and 5% CO2 incubator, and sub-culture was performed every three days.
2. RNA or vector transfection
Circ_0003221 of small interfering (si) RNA (si-circ_0003221), mimic or inhibitor of miR-892b (miR-892b, anti-miR-892b), and control RNAs (si-NC, miR-NC, anti-NC) were synthesized by RIBOBIO (Guangzhou, China). DHCR24 coding sequence was inserted into the pEXP-RB-Mam vector (RIBOBIO) and the pEXP-RB-Mam-DHCR24 vector was named as DHCR24. The 5637 and T24 cells with 70% confluence were performed with transfection of RNAs and vectors following the specification of Lipofectamine™ 3000 (Invitrogen, Carlsbad, CA, USA).
3. Reverse transcription-quantitative polymerase chain reaction assay
Tissues or cells were harvested for RNA isolation using TransZol reagent (TransGen, Beijing, China). Afterwards, EasyScript® All-in-One First-Strand cDNA Synthesis SuperMix for qPCR (TransGen) and TransStart® Green qPCR SuperMix (TransGen) were applied for reverse transcription and expression analysis according to the instruction books. The used specific primers were exhibited in Table 1. β-actin (circ_0003221, DHCR24) and U6 (miR-892b) were selected as the endogenous references, followed by data calculation through the 2-ΔΔCt method [18].
Table 1. Primer sequences used for reverse transcription-quantitative polymerase chain reaction.
| Name | Primer sequence (5′–3′) |
|---|---|
| circ_0003221 | Forward: GGCGATCATACTGGGAGATG |
| Reverse: TGTGATTCAAGTTGGGGTCA | |
| miR-892b | Forward: GCCGAGCACTGGCTCCTTTC |
| Reverse: CTCGTATCCAGTGCAGGGT | |
| 24-Dehydrocholesterol reductase | Forward: ATGGCAGCTTTGTGCGATG |
| Reverse: ACGCAGCTTGACGTACTTCT | |
| β-actin | Forward: AGCGAGCATCCCCCAAAGTT |
| Reverse: GGGCACGAAGGCTCATCATT | |
| U6 | Forward: CTCGCTTCGGCAGCACA |
| Reverse: AACGCTTCACGAATTTGCGT |
4. EDU assay
Forty-eight-well plates were inoculated with 4×104 5637 and T24 cells, followed by cell transfection for 48 hours. The proliferation ability was examined according to the user’s manual of EDU Cell Proliferation Kit (Sigma). Diamidine phenylindole (DAPI, Sigma) was applied to stain cell nuclei. EDU and DAPI merged cells (EDU positive) were analyzed under the fluorescence microscope (Olympus, Tokyo, Japan).
5. Colony formation assay
The 5637 and T24 cells were seeded into the 12-well plates with 5×104/well, then incubated at 37℃ for two weeks. Then colony staining was performed with 0.1% crystal violet (Sigma), and colony number was calculated using the ImageJ software (National Institutes of Health, Bethesda, MD, USA).
6. Transwell assay
Cell migration or invasion detection was carried out in transwell chamber (Corning Inc., Corning, NY, USA). The upper chamber was seeded with 1×105 cells in migration assay, while the equal number of cells were pipetted into matrigel (Corning Inc.) coated chamber in invasion assay. After the addition of cell medium into the lower chamber, the chamber was incubated in the 37℃ incubator for 24 hours. After staining with crystal violet (Sigma), migrated and invaded cells were counted via the inverted microscope (Olympus).
7. Glucose consumption and lactate production assays
Cell culture supernatants of 5637 and T24 were added into the 48 well-plates, respectively. Subsequently, Glucose Assay Kit-WST® for glucose consumption and Lactate Assay Kit-WST® for lactate production were performed as per the instruction books of Dojindo Molecular Technologies Inc. (Kumamoto, Japan).
8. Western blot
After protein acquisition through Radioimmunoprecipitation assay lysis buffer (Sigma) and quantification by BCA Protein Kit (Sigma), western blot was implemented as previously stated [19]. The proteins were examined by incubation of primary antibodies at 4℃ overnight, including Cyclin D1 (#55506, 1:1000), matrix metalloproteinase 2 (MMP2; #40994, 1:1000), pyruvate kinase M2 (PKM2; #4053, 1:1000), DHCR24 (#2033, 1:1000), and β-actin (#4970, 1:1000). Whereafter, anti-rabbit IgG secondary antibody (#7054, 1:3000) was incubated at 25℃ for 1 hour. All antibodies were bought from Cell Signaling Technology (Boston, MA, USA). Then the special blots on the membranes were exhibited through SignalFire™ Elite ECL Reagent (Cell Signaling Technology) and data were analyzed employing the ImageJ software.
9. Dual-luciferase reporter assay
The pmirGLO (Promega, Madison, WI, USA) was used as a basic luciferase vector. The original circ_0003221 or DHCR24 3’UTR sequence was considered as the wild-type (WT) sequence, which contained the miR-892b binding sites. The mutant-type (MUT) sequences were referred to circ_0003221 or DHCR24 3’UTR sequence after miR-892b sites were mutated. Then the recombinant circ_0003221 WT, circ_0003221 MUT, DHCR24 3’UTR WT and DHCR24 3’UTR MUT vectors were constructed. The 5637 and T24 cells were carried out by co-transfection with miR-892b or miR-NC and circ_0003221 vectors or DHCR24 3’UTR vectors. Cells were incubated at 37℃ for 48 hours, followed by detecting luciferase activity via Dual-luciferase Reporter Detection Kit (Promega).
10. RNA pull-down assay
After transfection of biotin-coupled miRNA (Bio-miR-892b, Bio-miR-NC, RIBOBIO), cell lysates were harvested for incubation of streptavidin magnetic beads (Thermo Fisher Scientific, Waltham, MA, USA). Then RNA was extracted for circ_0003221 quantification via reverse transcription-quantitative polymerase chain reaction (RT-qPCR).
11. Tumor xenograft assay
BALB/c male nude mice (Vital River Laboratory Animal Technology, Beijing, China) were injected with 1.5×106 stably transfected 5637 cells, with 6 mice in sh-NC or sh-circ_0003221 group. The volume (length×width2/2) of each tumor was monitored every week, and tumors were dissected from sacrificed mice after 5 weeks. After tumor tissues were weighed, total RNA or protein was isolated for level analysis. Ki67 (Abcam, Cambridge, MA, USA, ab15580) protein was examined via the immunohistochemistry (IHC) analysis. Animal Ethical Committee of Chongqing University Three Gorges Hospital has provided approval for this animal assay.
12. Statistical analysis
Data were obtained after three independent experiments with three parallels, then data (the mean±standard deviation) was analyzed by SPSS 22.0 (IBM Corp., Armonk, NY, USA). The difference was compared utilizing Student’s t-test and analysis of variance by Tukey’s test. There was a significant difference if p<0.05.
RESULTS
1. Circ_0003221 expression was upregulated in BLCA
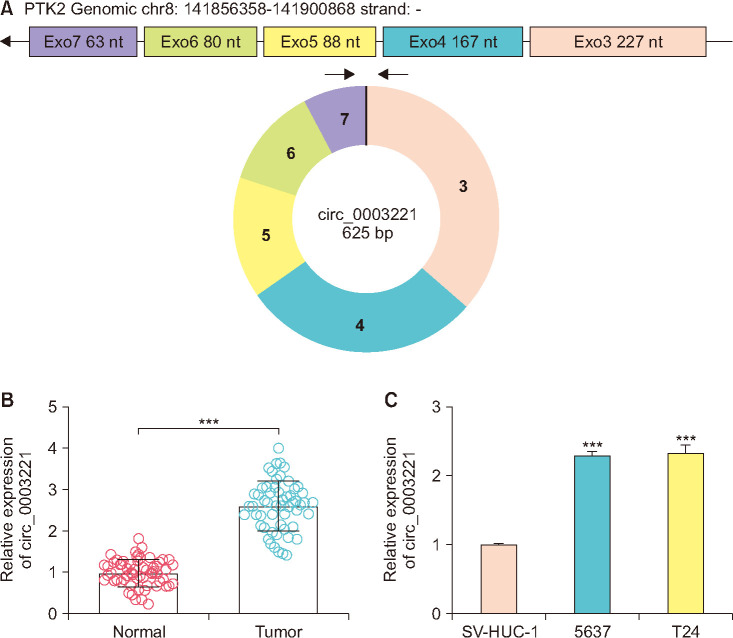
Circ_0003221 was an exonic circRNA (exon 3-7) originating from protein tyrosine kinase 2 gene, and it locates on the chr8:141856358-141900868 (Fig. 1A). The RT-qPCR results revealed that circ_0003221 level was increased in 55 BLCA tissues (Fig. 1B) and 5637/T24 cells (Fig. 1C), contrasted with normal samples and SV-HUC-1 cells. The upregulation of circ_0003221 was validated in BLCA.
Fig. 1. Circ_0003221 expression was upregulated in bladder cancer (BLCA). (A) Genomic information of circ_0003221. Circ_0003221 was quantified using reverse transcription-quantitative polymerase chain reaction in BLCA tissues (B) and 5637/T24 cells (C), as well as the negative controls. nt, nucleotide. ***p<0.001.
2. BLCA cell proliferation, motility and glycolysis were impeded after circ_0003221 knockdown
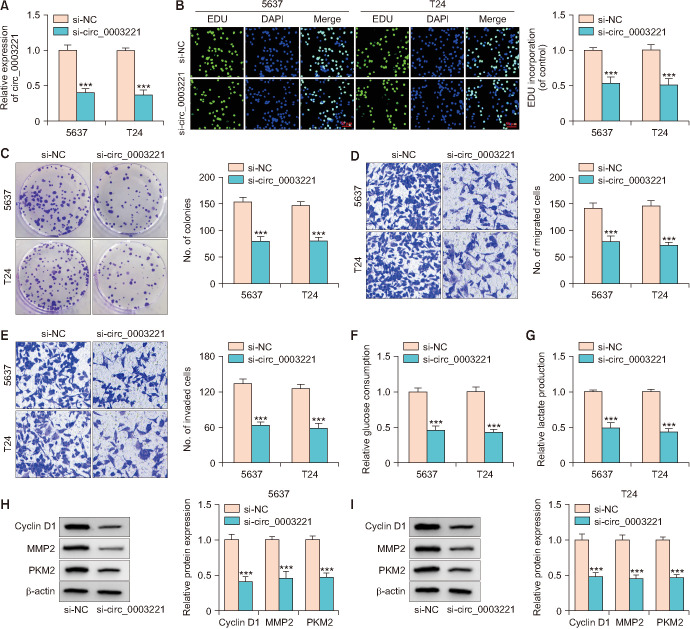
The function exploration of circ_0003221 was implemented in si-circ_0003221-transfected 5637 and T24 cells. Circ_0003221 level was significantly inhibited by si-circ_ 0003221 transfection, contraposed to si-NC transfection (Fig. 2A). The proliferation ability by EDU assay (Fig. 2B) and colony formation assay (Fig. 2C) was suppressed after reduction of circ_0003221. Cell detection by transwell assay displayed that migration (Fig. 2D) and invasion (Fig. 2E) capacities were reduced, with the silence of circ_0003221. Glucose consumption (Fig. 2F) and lactate production (Fig. 2G) were inhibited by siRNA circ_0003221, indicating that circ_0003221 downregulation blocked the glycolytic metabolism. Silencing circ_0003221 resulted in protein inhibition of Cyclin D1 (cell proliferation marker), MMP2 (cell invasion marker) and PKM2 (glycolysis marker) in 5637 and T24 cells (Fig. 2H, I). All in all, circ_0003221 functioned as an oncogenic regulator in BLCA.
Fig. 2. Bladder cancer cell proliferation, motility and glycolysis were impeded after circ_0003221 knockdown. (A) Reverse transcription-quantitative polymerase chain reaction was performed for quantifying circ_0003221 in 5637 and T24 cells with si-NC or si-circ_0003221 transfection. EDU assay or colony formation assay (B, C) and transwell assay (D, E) were performed for examination of proliferation and migration/invasion (crystal violet, ×100 magnification). Glucose consumption (F) and lactate production (G) through kits were performed for glycolysis evaluation. (H, I) Western blot was performed for examining Cyclin D1, MMP2 and PKM2. DAPI, diamidine phenylindole. ***p<0.001.
3. Circ_0003221 targeted miR-892b in BLCA cells
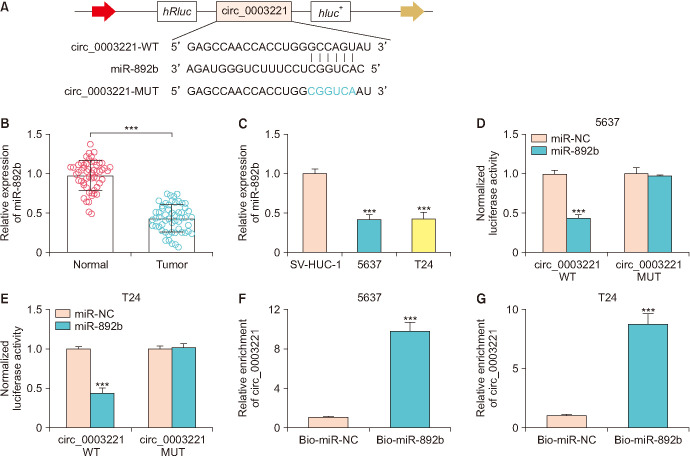
Circinteractome predicted that circ_0003221 sequence could bind to the miR-892b sequence, and the binding sites were shown in Fig. 3A. RT-qPCR analysis manifested that miR-892b was evidently reduced in BLCA tissues and cells contrasted with normal controls (Fig. 3B, C). Transfection of miR-892b has repressed the luciferase activity of circ_0003221 WT, while no effect was observed in luciferase activity of circ_0003221 MUT plasmid (Fig. 3D, E). Also, biotin-coupled miR-892b assay exhibited that circ_0003221 was pulled down by miR-892b in 5637 and T24 cells (Fig. 3F, G). Thus, miR-892b acted as a miRNA target of circ_0003221.
Fig. 3. Circ_0003221 targeted miR-892b in bladder cancer (BLCA) cells. (A) Circinteractome was used to analyze circ_0003221 and miR-892b binding sites. (B, C) MiR-892b level was quantified using reverse transcription-quantitative polymerase chain reaction in BLCA. Circ_0003221 and miR-892b interaction analysis was administrated using dual-luciferase reporter assay (D, E) and RNA pull-down assay (F, G). WT, wild-type; MUT, mutant-type. ***p<0.001.
4. The si-circ_0003221-induced tumor inhibition was attenuated by miR-892b downregulation in BLCA cells
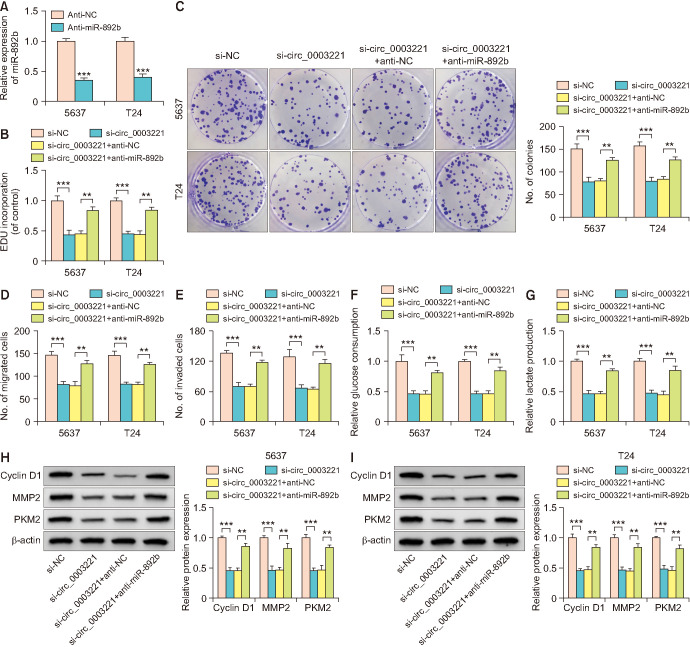
The miR-892b level was downregulated in 5637 and T24 cells with anti-miR-892b transfection, by comparison with anti-NC group (Fig. 4A). The suppressive effects on proliferation (Fig. 4B, C), migration or invasion (Fig. 4D, E) by si-circ_0003221 were abated by anti-miR-892b in 5637 and T24 cells. In addition, miR-892b inhibitor abolished the si-circ_0003221-mediated reduction of glucose consumption (Fig. 4F) and lactate production (Fig. 4G). Western blot demonstrated that si-circ_0003221-evoked Cyclin D1, MMP2 and PKM2 protein downregulation was mitigated by anti-miR-892b (Fig. 4H, I). Circ_0003221 served as a tumor promoter via sponging miR-892b.
Fig. 4. The si-circ_0003221-induced tumor inhibition was attenuated by miR-892b downregulation in bladder cancer cells. (A) Transfection efficiency of anti-miR-892b was assessed via reverse transcription-quantitative polymerase chain reaction in 5637 and T24 cells. Cell proliferation and motility were evaluated using EDU assay (B) or colony formation assay (C) and transwell migration or invasion assay (D, E) in si-NC, si-circ_0003221, si-circ_0003221+anti-NC and si-circ_0003221+anti-miR-892b groups. Glycolysis was analyzed using glucose uptake (F), lactate production (G) and protein detection (H, I). **p<0.01, ***p<0.001.
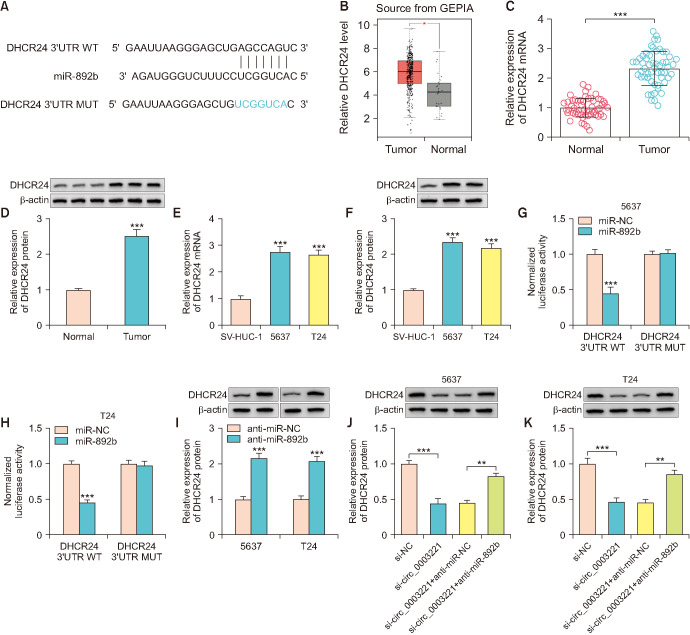
5. Circ_0003221 mediated the DHCR24 expression by targeting miR-892b
Targetscan software showed that SHCR24 3’UTR sequence contained the complementary miR-892b sites (Fig. 5A). GEPIA data (http://gepia.cancer-pku.cn/detail.php) has reported that DHCR24 was highly expressed in tumor tissues of BLCA patients (Fig. 5B). Also, the evidence from RT-qPCR and western blot displayed that DHCR24 was upregulated in our BLCA samples (Fig. 5C, D) and 5637/T24 cells (Fig. 5E, F). Overexpression of miR-892b inhibited luciferase intensity of DHCR24 3’UTR WT group but not DHCR24 3’UTR MUT group (Fig. 5G, H). DHCR24 protein expression was increased by anti-miR-892b transfection contrasted with anti-NC group (Fig. 5I). Additionally, miR-892b inhibitor offset DHCR24 downregulation triggered by si-circ_0003221 (Fig. 5J, K). Taken together, circ_0003221 could regulate DHCR24 via absorbing miR-892b.
Fig. 5. Circ_0003221 mediated the DHCR24 expression by targeting miR-892b. (A) The site analysis of DHCR24 and miR-892b was conducted by Targetscan. (B) The expression level of DHCR24 in GEPIA dataset. DHCR24 level analysis was carried out via reverse transcription-quantitative polymerase chain reaction and western blot in bladder cancer tissues (C, D) and cells (E, F). (G, H) Dual-luciferase reporter assay was administrated for analyzing miR-892b and DHCR24 3’UTR combination in 5637 and T24 cells. (I) Western blot was applied for miR-892b quantification in anti-NC or anti-miR-892b transfected 5637 and T24 cells. (J, K) DHCR24 protein determination was conducted using western blot after si-circ_0003221 and anti-miR-892b co-transfection. DHCR24, 24-dehydrocholesterol reductase; UTR, untranslated region; WT, wild-type; MUT, mutant-type. **p<0.01, ***p<0.001.
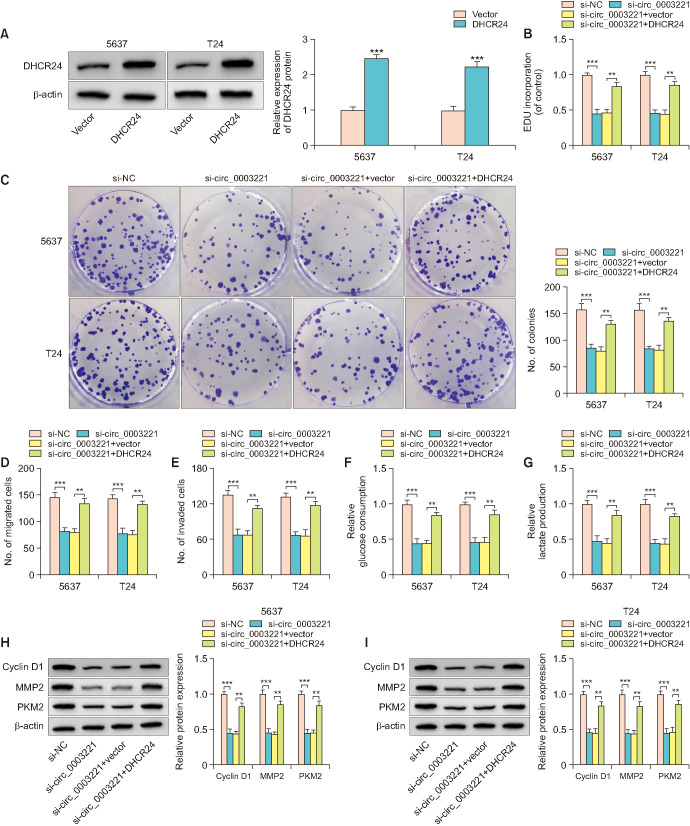
6. DHCR24 overexpression relieved the function of si-circ_0003221 in BLCA cells
DHCR24 overexpression was achieved by transfection of DHCR24 vector in 5637 and T24 cells. As exhibited in Fig. 6A, DHCR24 protein level was enhanced in DHCR24 transfection group compared with vector group. DHCR24 upregulation countervailed the influences of si-circ_0003221 on proliferation (Fig. 6B, C) and motility (Fig. 6D, E). Meanwhile, the si-circ_0003221-induced glycolysis inhibition (Fig. 6F, G) and protein marker suppression (Fig. 6H, I) were lightened following the overexpression of DHCR24. Therefore, the oncogenic DHCR24 was responsible for circ_0003221 function.
Fig. 6. DHCR24 overexpression relieved the function of si-circ_0003221 in bladder cancer cells. (A) DHCR24 protein measurement was carried out through western blot following transfection with Vector or DHCR24. EDU assay (B) or colony formation assay (C) and transwell assay (D, E) were applied to analyze the proliferation ability and cell motility after si-NC, si-circ_0003221, si-circ_0003221+Vector or si-circ_0003221+DHCR24 transfection in 5637 and T24 cells. Glycolysis was tested by glucose uptake (F), lactate production (G) and marker detection (H, I). DHCR24, 24-dehydrocholesterol reductase. **p<0.01, ***p<0.001.
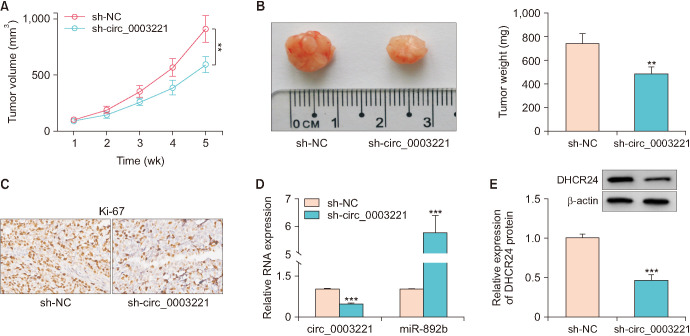
7. Circ_0003221 contributed to tumor growth in vivo by regulating the miR-892b/DHCR24 axis
In xenograft tumor assay, tumor volume and weight were reduced in sh-circ_0003221 group relative to sh-NC group (Fig. 7A, B). IHC assay also demonstrated that Ki-67 protein level was inhibited by circ_0003221 knockdown (Fig. 7C). The expression detection by RT-Qpcr indicated that sh-circ_0003221 induced circ_0003221 downregulation and miR-892b upregulation in tumor tissues (Fig. 7D). Also, DHCR24 protein level was decreased after the knockdown of circ_0003221 (Fig. 7E). Tumor growth could be promoted by circ_0003221 through the miR-892b/DHCR24 axis.
Fig. 7. Circ_0003221 contributed to tumor growth in vivo by regulating the miR-892b/DHCR24 axis. The volume (A) and weight (B) of tumors were measured in sh-NC and sh-circ_0003221 groups. (C) Ki67 protein detection was conducted through IHC assay. Reverse transcription-quantitative polymerase chain reaction and western blot were exploited to measure circ_0003221/miR-892b levels (D) and DHCR24 expression (E). DHCR24, 24-dehydrocholesterol reductase. **p<0.01, ***p<0.001.
DISCUSSION
Targeted molecular therapy has provided a novel perspective for BLCA treatment [20]. The present study showed that circ_0003221 facilitated tumor progression in BLCA by depending on the miR-892b/DHCR24 axis, showing that circ_0003221 might act as a therapeutic target in the clinical practice of BLCA.
CircRNAs are involved in various kinds of cell processes in BLCA. For instance, silence of has-circRNA-403658 reduced cell growth and accelerated apoptosis in BLCA [21]. Overexpression of circRNA-MYLK contributed to angiogenesis and epithelial-mesenchymal transition of BLCA cells [22]. Herein, data analysis revealed that BLCA cell proliferation, and motility were suppressed after circ_0003221 downregulation. Aerobic glycolysis is a well-known metabolic process to support cancer progression, accompanied by glucose uptake and lactate formation [23]. In BLCA cell reprogramme, glucose metabolism is occurred by consuming glucose and producing large amounts of lactate [24]. The research of circRNA is poor in regulating glycolysis of BLCA cells. Recently, circRNA_403658 and circSEMA5A were shown to activate the glycolysis in BLCA cells [21,25]. Our results demonstrated the inhibitory effects of si-circ_0003221 on glucose metabolism. These collective evidences confirmed that circ_0003221 promoted cell progression and glycolysis to function as a carcinogenic factor in BLCA.
Furthermore, circ_0003221 was exhibited to interact with miR-892b in BLCA cells and the regulatory action of si-circ_0003221 was abrogated by miR-892b inhibition. The previous studies have unraveled the miRNA sponge effects of circRNAs in BLCA. circMTO1 repressed BLCA cell metastasis via sponging miR-221 and circ_102336 enhanced tumorigenesis of BLCA by sequestering miR-515-5p [26,27]. Hence, circ_0003221 regulated the biological processes in BLCA cells via acting as a miR-892b sponge. In addition, miR-892b level downregulation and its anti-tumor function in BLCA suggested that miR-892b overexpression might be also a good strategy against BLCA progression.
Through the miRNA sponge mechanism, circRNAs can modulate the levels of different functional genes. For example, circ_0071196 was related to cell development in BLCA by replying on the miR-19b-3p/CIT axis [28]. Circ0001429 promoted cell propagation but reduced cell motility by interacting with miR-205-5p to increase the VEGFA level in BLCA cells [29]. Currently, we validated the target relation between miR-892b and DHCR24. More interestingly, circ_0003221 incurred the upregulation of DHCR24 in BLCA cells via binding to miR-892b and DHCR24 weakened the si-circ_0003221-mediated progression inhibition. It was significant that circ_0003221 affected the biological processes of BLCA cells via regulating DHCR24 expression by targeting miR-892b. The results of animal assay further manifested tumor growth promotion in vivo by circ_0003221. This study still has some limitations. For example, circ_0003221/miR-892b/DHCR24 axis in vivo remains further identification.
Dysregulated circRNAs have been reported to act as biological biomarkers for cancers. Lian et al. [30] discovered that hsa_circ_101555 and hsa_circ_008068 served as diagnostic targets for early-stage lung adenocarcinoma. Herein, circ_0003221 upregulation demonstrated the potential of circ_0003221 as a diagnostic biomolecule in BLCA. Additionally, oncogenic role of circ_0003221 suggested that circ_0003221 downregulation might be used for BLCA treatment. However, it needs further affirmation in clinical practice.
CONCLUSIONS
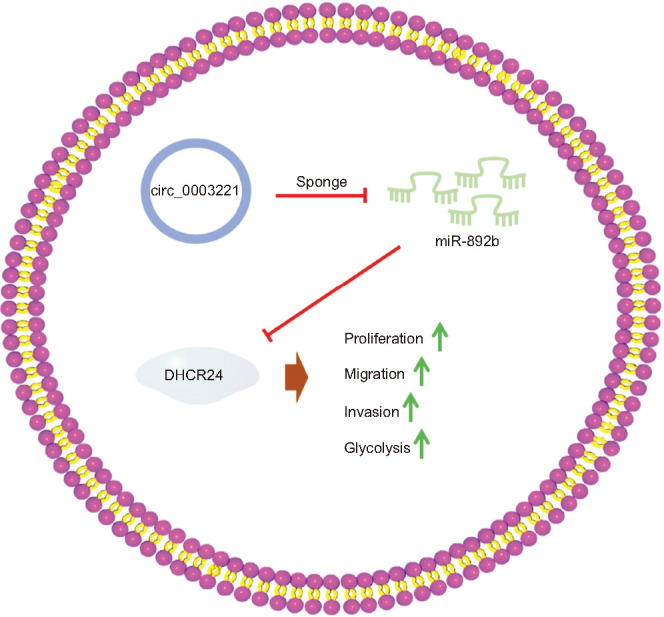
To conclude, circ_0003221 upregulated DHCR24 via sponging miR-892b to promote cell behaviors and glycolytic metabolism of BLCA (Fig. 8). This study identified the oncogenic function of circ_0003221 and the molecular pathway of action in BLCA.
Fig. 8. The graphical abstract of this study. DHCR24, 24-dehydrocholesterol reductase.
Footnotes
CONFLICTS OF INTEREST: The authors have nothing to disclose.
FUNDING: None.
- Research conception and design: Yingchun Jiang and Zongyu Xia.
- Data acquisition: Peng Lu and Yingchun Jiang.
- Statistical analysis: Peng Lu and Yingchun Jiang.
- Data analysis and interpretation: Peng Lu and Yingchun Jiang.
- Drafting of the manuscript: Peng Lu, Yingchun Jiang and Zongyu Xia.
- Critical revision of the manuscript: Peng Lu, Yingchun Jiang and Zongyu Xia.
- Approval of the final manuscript: all authors.
References
- 1.Lenis AT, Lec PM, Chamie K, Mshs MD. Bladder cancer: a review. JAMA. 2020;324:1980–1991. doi: 10.1001/jama.2020.17598. [DOI] [PubMed] [Google Scholar]
- 2.Richters A, Aben KKH, Kiemeney LALM. The global burden of urinary bladder cancer: an update. World J Urol. 2020;38:1895–1904. doi: 10.1007/s00345-019-02984-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Patel VG, Oh WK, Galsky MD. Treatment of muscle-invasive and advanced bladder cancer in 2020. CA Cancer J Clin. 2020;70:404–423. doi: 10.3322/caac.21631. [DOI] [PubMed] [Google Scholar]
- 4.López-Cortés R, Gómez BB, Vázquez-Estévez S, Pérez-Fentes D, Núñez C. Blood-based protein biomarkers in bladder urothelial tumors. J Proteomics. 2021;247:104329. doi: 10.1016/j.jprot.2021.104329. [DOI] [PubMed] [Google Scholar]
- 5.Tran L, Xiao JF, Agarwal N, Duex JE, Theodorescu D. Advances in bladder cancer biology and therapy. Nat Rev Cancer. 2021;21:104–121. doi: 10.1038/s41568-020-00313-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li Y, Li G, Guo X, Yao H, Wang G, Li C. Non-coding RNA in bladder cancer. Cancer Lett. 2020;485:38–44. doi: 10.1016/j.canlet.2020.04.023. [DOI] [PubMed] [Google Scholar]
- 7.Papatsirou M, Artemaki PI, Karousi P, Scorilas A, Kontos CK. Circular RNAs: emerging regulators of the major signaling pathways involved in cancer progression. Cancers (Basel) 2021;13:2744. doi: 10.3390/cancers13112744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng F, Zheng B, Si S, Wang J, Zhao G, Yao Z, et al. The roles of circRNAs in bladder cancer: biomarkers, tumorigenesis drivers, and therapeutic targets. Front Cell Dev Biol. 2021;9:666863. doi: 10.3389/fcell.2021.666863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jiang MP, Xu WX, Hou JC, Xu Q, Wang DD, Tang JH. The emerging role of the interactions between circular RNAs and RNA-binding proteins in common human cancers. J Cancer. 2021;12:5206–5219. doi: 10.7150/jca.58182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xie H, Wang J, Wang B. Circular RNA circ_0003221 promotes cervical cancer progression by regulating miR-758-3p/CPEB4 axis. Cancer Manag Res. 2021;13:5337–5350. doi: 10.2147/CMAR.S311242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu ZQ, Yang MG, Liu HJ, Su CQ. Circular RNA hsa_circ_0003221 (circPTK2) promotes the proliferation and migration of bladder cancer cells. J Cell Biochem. 2018;119:3317–3325. doi: 10.1002/jcb.26492. [DOI] [PubMed] [Google Scholar]
- 12.Parizi PK, Yarahmadi F, Tabar HM, Hosseini Z, Sarli A, Kia N, et al. MicroRNAs and target molecules in bladder cancer. Med Oncol. 2020;37:118. doi: 10.1007/s12032-020-01435-0. [DOI] [PubMed] [Google Scholar]
- 13.Xia W, Liu Y, Cheng T, Xu T, Dong M, Hu X. Extracellular vesicles carry lncRNA SNHG16 to promote metastasis of breast cancer cells via the miR-892b/PPAPDC1A axis. Front Cell Dev Biol. 2021;9:628573. doi: 10.3389/fcell.2021.628573. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 14.Peng J, Liu F, Zheng H, Wu Q, Liu S. Long noncoding RNA ZFAS1 promotes tumorigenesis and metastasis in nasopharyngeal carcinoma by sponging miR-892b to up-regulate LPAR1 expression. J Cell Mol Med. 2020;24:1437–1450. doi: 10.1111/jcmm.14823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shin SS, Park SS, Hwang B, Moon B, Kim WT, Kim WJ, et al. MicroRNA-892b influences proliferation, migration and invasion of bladder cancer cells by mediating the p19ARF/cyclin D1/CDK6 and Sp-1/MMP-9 pathways. Oncol Rep. 2016;36:2313–2320. doi: 10.3892/or.2016.5052. [DOI] [PubMed] [Google Scholar]
- 16.Lee GT, Ha YS, Jung YS, Moon SK, Kang HW, Lee OJ, et al. DHCR24 is an independent predictor of progression in patients with non-muscle-invasive urothelial carcinoma, and its functional role is involved in the aggressive properties of urothelial carcinoma cells. Ann Surg Oncol. 2014;21 Suppl 4:S538–S545. doi: 10.1245/s10434-014-3560-6. [DOI] [PubMed] [Google Scholar]
- 17.Liu XP, Yin XH, Meng XY, Yan XH, Cao Y, Zeng XT, et al. DHCR24 predicts poor clinicopathological features of patients with bladder cancer: a STROBE-compliant study. Medicine (Baltimore) 2018;97:e11830. doi: 10.1097/MD.0000000000011830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 19.Zhou D, Lin X, Wang P, Yang Y, Zheng J, Zhou D. Circular RNA circ_0001162 promotes cell proliferation and invasion of glioma via the miR-936/ERBB4 axis. Bioengineered. 2021;12:2106–2118. doi: 10.1080/21655979.2021.1932221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bednova O, Leyton JV. Targeted molecular therapeutics for bladder cancer-a new option beyond the mixed fortunes of immune checkpoint inhibitors? Int J Mol Sci. 2020;21:7268. doi: 10.3390/ijms21197268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wei Y, Zhang Y, Meng Q, Cui L, Xu C. Hypoxia-induced circular RNA has_circRNA_403658 promotes bladder cancer cell growth through activation of LDHA. Am J Transl Res. 2019;11:6838–6849. [PMC free article] [PubMed] [Google Scholar]
- 22.Zhong Z, Huang M, Lv M, He Y, Duan C, Zhang L, et al. Circular RNA MYLK as a competing endogenous RNA promotes bladder cancer progression through modulating VEGFA/VEGFR2 signaling pathway. Cancer Lett. 2017;403:305–317. doi: 10.1016/j.canlet.2017.06.027. Erratum in: Cancer Lett 2022;534:215631. [DOI] [PubMed] [Google Scholar]
- 23.Ganapathy-Kanniappan S. Molecular intricacies of aerobic glycolysis in cancer: current insights into the classic metabolic phenotype. Crit Rev Biochem Mol Biol. 2018;53:667–682. doi: 10.1080/10409238.2018.1556578. [DOI] [PubMed] [Google Scholar]
- 24.Afonso J, Santos LL, Longatto-Filho A, Baltazar F. Competitive glucose metabolism as a target to boost bladder cancer immunotherapy. Nat Rev Urol. 2020;17:77–106. doi: 10.1038/s41585-019-0263-6. [DOI] [PubMed] [Google Scholar]
- 25.Wang L, Li H, Qiao Q, Ge Y, Ma L, Wang Q. Circular RNA circSEMA5A promotes bladder cancer progression by upregulating ENO1 and SEMA5A expression. Aging (Albany NY) 2020;12:21674–21686. doi: 10.18632/aging.103971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Y, Wan B, Liu L, Zhou L, Zeng Q. Circular RNA circMTO1 suppresses bladder cancer metastasis by sponging miR-221 and inhibiting epithelial-to-mesenchymal transition. Biochem Biophys Res Commun. 2019;508:991–996. doi: 10.1016/j.bbrc.2018.12.046. [DOI] [PubMed] [Google Scholar]
- 27.Gong P, Xu R, Zhuang Q, He X. A novel circular RNA (hsa_circRNA_102336), a plausible biomarker, promotes the tumorigenesis by sponging miR-515-5p in human bladder cancer. Biomed Pharmacother. 2020;126:110059. doi: 10.1016/j.biopha.2020.110059. [DOI] [PubMed] [Google Scholar]
- 28.Liu Z, Yang Y, Yang Z, Xia S, Lin D, Xiao B, et al. Novel circRNA_0071196/miRNA-19b-3p/CIT axis is associated with proliferation and migration of bladder cancer. Int J Oncol. 2020;57:767–779. doi: 10.3892/ijo.2020.5093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cao W, Zhao Y, Wang L, Huang X. Circ0001429 regulates progression of bladder cancer through binding miR-205-5p and promoting VEGFA expression. Cancer Biomark. 2019;25:101–113. doi: 10.3233/CBM-182380. [DOI] [PubMed] [Google Scholar]
- 30.Lian X, Cao D, Hu X, Mo W, Yao X, Mo J, et al. Circular RNAs hsa_circ_101555 and hsa_circ_008068 as diagnostic biomarkers for early-stage lung adenocarcinoma. Int J Gen Med. 2022;15:5579–5589. doi: 10.2147/IJGM.S367999. [DOI] [PMC free article] [PubMed] [Google Scholar]