Abstract
Coronavirus disease 2019 (COVID-19), caused by severe acute respiratory syndrome coronavirus type 2 (SARS-CoV-2), has led to a global pandemic. Deep learning (DL) technology and molecular dynamics (MD) simulation are two mainstream computational approaches to investigate the geometric, chemical and structural features of protein and guide the relevant drug design. Despite a large amount of research papers focusing on drug design for SARS-COV-2 using DL architectures, it remains unclear how the binding energy of the protein-protein/ligand complex dynamically evolves which is also vital for drug development. In addition, traditional deep neural networks usually have obvious deficiencies in predicting the interaction sites as protein conformation changes. In this review, we introduce the latest progresses of the DL and DL-based MD simulation approaches in structure-based drug design (SBDD) for SARS-CoV-2 which could address the problems of protein structure and binding prediction, drug virtual screening, molecular docking and complex evolution. Furthermore, the current challenges and future directions of DL-based MD simulation for SBDD are also discussed.
Keywords: Deep learning, Molecular dynamics simulation, Structure-based drug design, SARS-CoV-2
1. Introduction
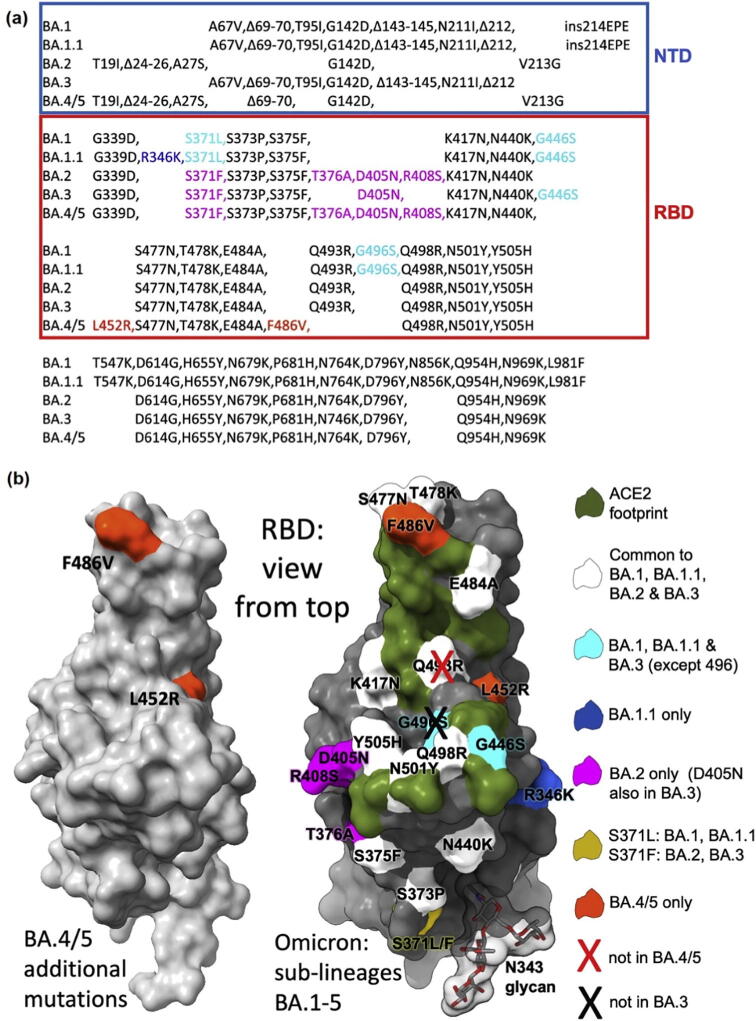
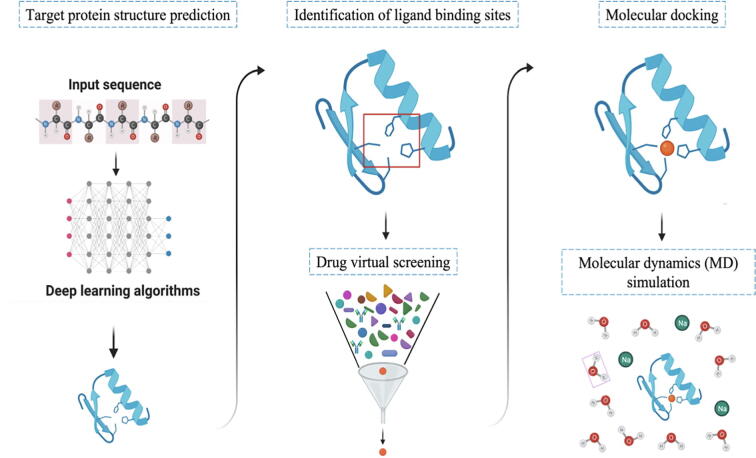
Since the end of 2019, the COVID-19 caused by SARS-CoV-2 has emerged into an unprecedented public health crisis, causing many deaths around the world. By July 2022, more than 566 million confirmed cases and 6.37 million deaths had been reported globally (https://covid19.who.int/). The common symptoms of COVID-19 are pneumonia, shortness of breath, dry cough, tiredness [1], and even some neurological complications [2]. Besides, the coronavirus is continuously mutating, with some new variants being more virulent and transmissible than the original ones. At present, the Omicron is the dominant variant globally and accounts for almost all sequences reported to Global Initiative on Sharing Avian Influenza Data (GISAID) [3]. Up to now, the Omicron variant has emerged several sub-lineages including BA.1, BA.1.1, BA.2, BA.3, BA.4 and BA.5. The BA.2 and BA.3 are highly relative to BA.1 but contain some different mutations in the N-terminal domain (NTD) and receptor binding domain (RBD) of the spike (S) (Fig. 1a). Comparing to BA.1, the BA.1.1 has an additional R346K mutation in the RBD [4]. The S sequences of BA.4 and BA.5 are identical which are speculated evolving from BA.2. The BA.4 and BA.5 show additional mutations in the RBD, including R493Q, L452R and F486V. It is worthy of noting that the R493Q is a reverse mutation of Q493R appeared in other Omicron sub-lineages (Fig. 1) [5]. Rao et al. have analyzed 1.79 million S glycoprotein sequences of SARS-CoV-2 and found that the virus is fine-tuning the S protein with numerous amino acids (AA) insertions and deletions [6]. Facing of the fast mutating, possibly more virulent, transmissible, and cunning virus, new therapeutic molecules are urgently required [7]. However, the traditional experimental drug discovery is usually expensive and time-consuming. Structure-based drug design (SBDD) is a proven highly effective and economical computer-aided design approach that could speed up the drug discovery for SARS-CoV-2 [8], [9], [10]. A typical SBDD starts from the input protein sequence, builds a three-dimensional (3D) structure by structure biology or structure prediction, identifies binding sites, discovers active modulators through virtual screening or de novo design, predicts the protein–protein/ligand docking sites with high accuracy, and lastly simulates the dynamics evolution of the macromolecule (Fig. 2) [11], [12].
Fig. 1.
The mutations of Omicron sub-lineages. (a) The mutations in S of BA.1, BA.1.1, BA.2, BA.3, BA.4 and BA.5 with NTD and RBD indicated; (b) The positions of RBD mutations, with mutations common to all Omicron colored in white, common to BA.1 and BA.1.1 colored in cyan, unique to BA.1.1 colored in blue, and unique to BA.2 colored in magenta. Residue Serine 371 is mutated to Leucine 371 (S371L) in BA.1 and BA.1.1, but mutated to Phenylalanine (S371F) in BA.2 and BA.3. The RBD is plotted by gray surface with the hACE2 footprint colored in dark green [5]. Reproduced with permission. Copyright 2022, Elsevier. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Fig. 2.
The basic steps involved in the SBDD method. The first panel shows target protein structure prediction, at which the sequence is input and, through DL algorithm, the protein structure is obtained. The second panel shows the identification of ligand binding sites, and the next step is drug virtual screening. In the last panel, compounds obtained by molecular docking are synthesized and tested in vitro, and finally MD simulations are performed.
During the past few years, the deep learning (DL) technology has been rapidly implemented into drug discovery. DL is an algorithm based on an artificial neural network (ANN) for data representation learning [13]. Up to now, several DL frameworks, such as deep neural network (DNN) [14], [15], convolutional neural network (CNN) [16], [17], deep confidence network [18], recurrent neural network (RNN) [19], and generative adversarial network (GAN) [20], [21], etc. have been broadly applied in various fields, achieving better results comparing to other computational models [22].
In terms of novel Coronavirus treatment, DL-based computer-aided drug bio-computation can quickly identify drug molecules that effectively prevent infection, showing potentials in finding the cures for COVID-19 [23]. In addition, some new programs of DL such as AlphaFold and its 2nd version (AlphaFold2) have been utilized in 3D structure prediction of protein which show ultra-high accuracy comparable to data collected by cryo-EM [24], [25], [26]. Artificial intelligence (AI) has attracted tremendous interests in the research of SBDD [27], [28], [29], [30], [31]. However, limitations of DL models used in drug design are that they learn through observations, and fail to consider the dynamic interactions of protein–protein/ligand. The molecular dynamics (MD) simulations act a crucial role in studying biological systems, which can be used to reveal different conformations of proteins, evolutions of protein-protein/ligand interactions and spontaneously complex phenomena such as protein folding [32], [33], [34]. In the past decades, more researchers have realized that MD can overcome the major limitations of SBDD, including those limitations routinely appear in ligand docking calculations without sampling the protein conformational rearrangements during ligand binding. The MD simulations also offer opportunities to make sound scientific breakthroughs in COVID-19 research, which contribute to a comprehensive understanding of mechanisms of virus infection and pathogenesis of COVID-19 [35], [36]. In addition, they are effective ways to visualize the essence of protein–protein/ligand interactions and guide the drug discovery and design. For example, MD simulations can be performed to evaluate the stabilities and interactions of the human angiotensin-converting enzyme 2 (hACE2) receptor with screened natural inhibitors to identify novel drug candidates against SARS-CoV-2 [37]. A major drawback of MD is the accuracy of simulations should always rely on proteins with known structures (usually downloaded from the Protein Data Bank (PDB) [38]).
Considering the advantages of both the DL and MD computational methods, DL-based MD simulation for SBDD has been adopted against SARS-CoV-2. This paper firstly reviews the latest applications and research progresses of DL technology in SBDD. Then, the DL-based MD simulation in SBDD for SARS-CoV-2 is highlighted and discussed. At last, we discuss the future direction for computational methods in SBDD. During the COVID-19 global pandemic, we believe this review could offer novel insights into the drug design against SARS-CoV-2.
2. Basic steps of SBDD
2.1. Target protein structure prediction by DL
With the rapid development of structural analysis techniques such as X-ray and nuclear magnetic resonance, more and more protein structures have been solved and stored in the PDB [39]. However, the structures of many target proteins have not been solved yet due to the limitations of experimental techniques. Obtaining the accurate structure of a protein is essential to understand its biological function [40].
In the initial study, some researchers use Swiss model to do homology modeling of protein structure with the target sequence as input (http://swissmodel.expasy.org/interactive) [41]. But this online model has some deficiencies, which is impossible to model proteins with insertion of short AA chains or without highly similar templates [42], [43]. Another online modeling server, the iterative threading assembly refinement (I-TASSER) (http://zhanglab.ccmb.med.umich.edu/I-TASSER/) predicts protein structure based on the sequence-to-structure-to-function paradigm [44]. It uses AA sequence information to generate 3D atomic models from multiple threading alignments and iterative structural assembly simulations. The function of the protein is then inferred by structurally matching the 3D models with other known proteins. Although I-TASSER can provide disulfide bonding modes, secondary and tertiary structures, and functional annotations on ligand binding sites, but it takes a quite long time to build complex structures [45]. In 2020, DeepMind unveiled its AlphaFold2, a DL-based structure prediction method that ranked the first in protein structure prediction in 14th Critical Assessment of Techniques for Protein Structure Prediction (CASP14) competition [46]. AlphaFold2 is now freely accessible with novel neural network architectures that have improved the accuracy from its earlier version AlphaFold. The full realization of AlphaFold2 mainly consists of the neural network EvoFormer and the structure module [24], [47]. The EvoFormer uses two transformers and one clear communication channel between them. Each head is specialized for one particular type of data, such as a multiple sequence alignment (MSA), and a representation of pairwise interactions between AA. The information of the contiguous representation which allows for regular exchange of information and iterative refinement is also incorporated. The structure module uses the first part of MSA, as well as the pair features obtained by calculation, and initializes all the residual frames from the coordinate origin and calculates the updated backbone frames. Finally, the specific 3D atomic coordinates are predicted [24]. Later, researchers have independently reproduced many ideas of AlphaFold2 and implemented in the so-called RoseTTAFold [48]. Evans et al. have released AlphaFold-multimer, a refined version of AlphaFold2 for the prediction of protein complexes [49], generalizing the use of AlphaFold2 and RoseTTAFold which are usually for single chain prediction. Most recently, Mirdita et al. have developed ColabFold, which offers accelerated prediction of protein structures and complexes based on fast homology search of many-against-many sequence searching N(MMseqs2) by AlphaFold2 or RoseTTAFold [50]. The fast development of DL technology has provided more opportunities for accurate protein structure prediction. SARS-CoV-2 is a single-stranded RNA virus with a genome of about 30 kb [51], [52]. In addition to the 4 structural proteins (S, nucleocapsid (N), membrane (M), and envelope (E)), the SARS-CoV2 genome encodes 16 non-structural proteins (NSPs 1–16) which are essential for virus replication, eliciting the immune response and representing targets to develop future prophylactic and therapeutic approaches against COVID-19 [53]. Yang et al. have predicted the structures of S, M, and N proteins of the Omicron variant using AlphaFold2, and investigated the effects of mutations on the S1, NTD and RBD domains of S protein [54]. The high-precision structures of M and N proteins obtained by AlphaFold2 could provide a basis for understanding the replication and propagation characteristics of Omicron. Robertson et al. have evaluated the consistency of the models generated by AlphaFold2 by atomic pairs measured with residual dipole coupling (RDC) in solution [55]. They have found that the AlphaFold2 models are entirely consistent with the experimental RDC data for most proteins. Yang et al. have adopted AlphaFold2 to predict the structures of S proteins according to the sequences of the mutants and successfully predicted the S proteins of 10 major variants of SARS-CoV-2, including the Original, Alpha, Beta, Gamma, Delta, Epsilon, IOTA, Kappa, Lambda and 21H strains [56]. Gupta et al. have combined the cryo-EM with AlphaFold2 to obtain the atomic model of full-length SARS-COV-2 non-structural protein 2 (NSP2) [57], which reveals a highly-conserved zinc ion-binding site, suggesting a role for NSP2 in RNA binding. Through the mapping of emerging mutations from variants of SARS-CoV-2 on the resulting structures, the potential host-NSP2 interaction regions can be observed. These studies demonstrate that DL is more likely to be locally accurate for domain structure prediction, sufficient for global structure prediction and to offer comprehensive structure modeling strategies combined with experimental constraints.
The current literature on target protein structure prediction by DL mainly uses AlphaFold2 and analogical methods mentioned above. Although the structure prediction of a protein by DL could provide some essential resources for speculating its function, experimental works are still needed to further confirm the result. Besides, AlphaFold2 and other DL computational approaches have intrinsic defects. For example, AlphaFold2 predicts protein structures according to protein structures (training data) in the PDB, but many of these structures are not actually in their folded states. That is, the proteins could only correctly fold into specific structures upon binding to other proteins, substrates or metal ions, or assemble into large complexes. We are still facing the problems of the difficulties of analyzing protein functions, and the high costs of protein structure determinations through experiments. To address those problems, further studies in accurate determination of protein structures are necessary [58].
2.2. Identification of ligand binding sites
A typical SBDD procedure involves the development of potential drug molecules or ligands which can form stable complexes with a given receptor at its binding sites. A prerequisite is to find out the druggable and functionally relevant binding sites on the 3D structure of the protein [59]. Information about binding sites is also required for specific docking. The binding sites could be traditionally identified by site-directed mutation studies or X-ray crystal structures of target protein [60]. Compared with traditional methods, DL-based models can be trained in a fully data-driven manner with little expert knowledge to predict the binding sites more quickly and accurately [61].
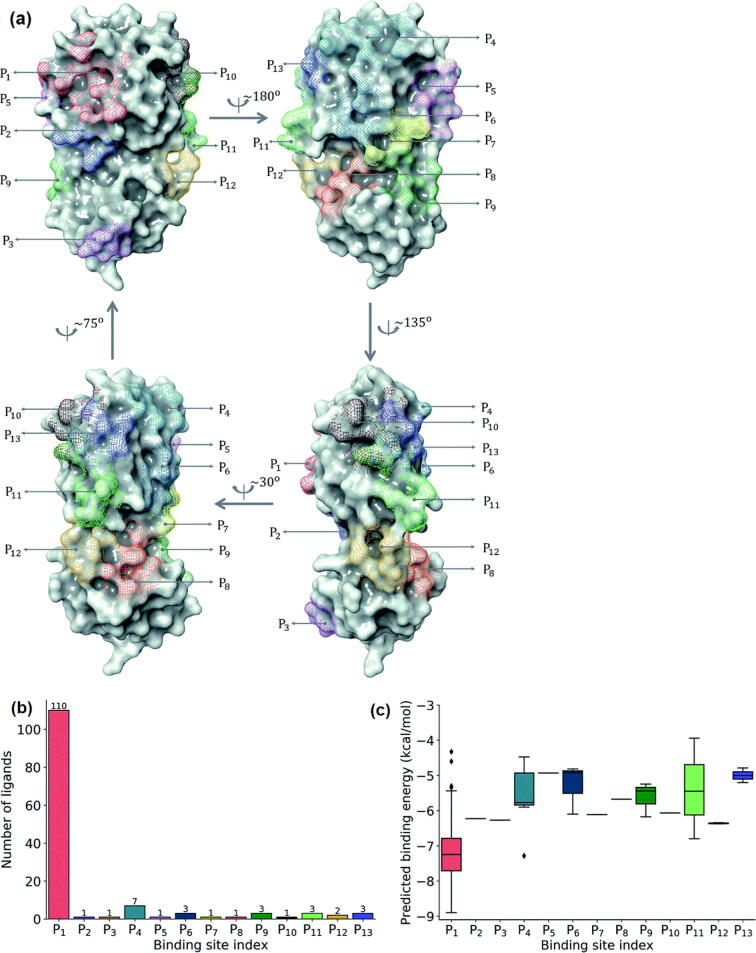
Nazem et al. have used an improved U-Net model based on the dice loss function to accurately predict binding sites of SARS-CoV-2 proteins [59]. The performance of the model on independent test datasets and SARS-CoV-2 shows that the segmentation model could predict binding sites more accurate than the recently released DL model DeepSite [62]. Nguyen et al. have integrated algebraic topology and DL (MathDL) to provide a reliable ranking of the binding affinities for 137 SARS-CoV-2 3-chymosin-like cysteine protease (3CLpro) inhibitors. The 3CLpro is an essential molecular target of SARS-CoV-2 [63]. They have reported 13 distinct binding pockets of the SARS-CoV-2 3CLpro which are denoted by Pi, i = 1, 2, …, 13, as illustrated in Fig. 3a. Among all the 13 binding pockets, P1 is the most common binding region of the SARS-CoV-2 3CLpro, which attracts around 80 % of ligands in the data set of 137 complexes while the binding pockets P2, P3, P5, P7, P8, and P10 are the least common binding sites consisting of only one ligand (Fig. 3b). In addition, the binding pocket P1 with the lowest media binding energy value has been found as the best region on the SARS-CoV-2 3CLpro for inhibitor design (Fig. 3c)[64]. Li et al. have developed the ligand neural network (l-Net), a novel graphic-generating model for the end-to-end design of chemically and conformationally efficient 3D molecules with high drug similarity [65]. Later, l-Net has been combined with Monte Carlo tree search for SBDD. The drawbacks of l-Net include limited versatility, requirement of expert knowledge, and heavily dependence on feature engineering. Then, Li et al. have introduced the DeepLigBuilder, a new drug design method based on DL which could generate 3D molecular structures of the binding sites of the target protein [65]. In a case study of SARS-CoV-2 3CLpro inhibitor design, DeepLigBuilder has recommended a list of drug-like compounds with novel chemical structures, high predictive affinity, and similar binding characteristics to known inhibitors by combining deep generation models with atomic-level interaction assessments.
Fig. 3.
(a) All binding site pockets observed from 137 inhibitors in SARS-CoV-2; (b) Distribution of 137 ligands across 13 distinct binding sites; (c) Box plot of predicted binding energies (kcal/mol) of all inhibitors in each binding site [64]. Reproduced with permission. Copyright 2020, Royal Society of Chemistry.
It is worthy to note that SARS-CoV-2 has undergone several mutations since its emergence and is still evolving rapidly [66]. In particular, the S protein has been profoundly mutated with RBD as a primary domain showing many mutant clusters [67], [68]. The virus may accumulate further mutations at the RBD site to improve its interaction with hACE2 and escalate its infectivity. Predicting potential sites of non-synonymous mutations and the evolution of protein structural modifications that lead to drug tolerance are critical problems regarding the treatments of new mutants. Padhi and Tripathi have used a computational high-throughput interface-based protein design strategy to identify mutation hot spots and potential adaptation characteristics in the binding sites of 3CLPro. They have found that several mutants exhibit reduced binding affinity to drugs Boceprevir and Telaprevir, out of which hotspot residues having a strong tendency to undergo positive selection have been identified. The results indicate that these drugs have larger footprints in the mutational landscape of 3CLpro and hence encompass the highest potential for positive selection and adaptation [69].
These state-of-the-art models for SBDD demonstrate the capability of computational approaches especially the incorporation of DL methods in predicting protein binding sites and its potential in accelerating drug design and discovery for COVID-19. However, a majority of them have been limited by the expressivity of the handcrafted features and the availability of similar proteins. In addition, some of the DL methods are surprisingly failed in the identification and ranking of binding sites accurately. Recently, Tubiana et al. have introduced ScanNet, an end-to-end, interpretable geometric DL model that learns features directly from 3D structures of proteins. It builds representations of atoms and amino acids based on the spatial-chemical arrangements of their neighbors. The ScanNet is trained for detecting protein–protein and protein-antibody binding sites, demonstrating high accuracy in presenting folding of unseen protein and interpreting the filters learned. They have successfully predicted epitopes of the SARS-CoV-2 S protein and validated known antigenic regions [70]. ScanNet is also demonstrated easily to be generalized to other classes of binding sites with sufficient available training data. Extension to partner-specific binding prediction and guiding molecular docking is a promising future direction in SBDD.
2.3. Compound library preparation-drug virtual screening
DL-aided virtual screening by using the 3D structures of target protein and small molecule drugs together with their binding could help to achieve very large-scale drug screening from chemical libraries [71]. The DL methods are typically data type dependent and have been extensively used in drug prediction and design. For example, text mining techniques and graphics-based approaches are used for drug reuse, while autoencoder approaches are largely helpful in predicting drug possibilities, drug target relationships, and generation of new drug molecules. Graph Convolutional Network like DL and molecule transformer-drug target interaction (MT-DTI) methods have been proved to be successful in predicting available antiviral drugs that are effective against SARS-CoV-2. AI/DL subsets, homology modeling, virtual screening and molecular docking are the most commonly used SARS-COV-2 drug reuse approaches to identify potentially effective drugs for the treatment of COVID-19 infection [72]. Up to now, hundreds of potential drugs have been discovered by the AI/DL technology [73], [74], [75], [76], [77], saving more time and effort for drug development than the traditional experimental methods.
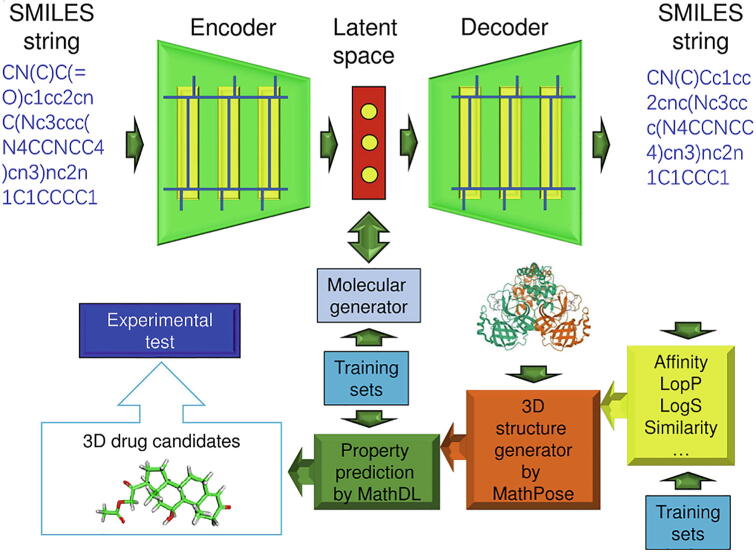
Joshi et al. have used DL methods to conduct virtual screening of natural compounds to find effective drugs against COVID-19 [78]. In their work, DL is used to predict 3CLpro inhibitor in CHEMBL3927 dataset, and the predicted models have been developed and evaluated based on coefficient of determination (R2), mean absolute error (MAE), mean square Error (MSE), root mean squared error (RMSE) and loss. Karki et al. have introduced the application of an end-to-end DNN framework called SSnet, which is used to repurpose approved drugs from a large drug library [79]. They have firstly screened a small library of approved drugs from DrugBank and ZINC by SSnet to identify compounds that have high-binding affinities. The results are cross-validated against traditional drug docking algorithm using the Autodock Vina scoring function. Then the SSnet approach has been extended to a library of 750,000 compounds in BindingDB, with the compounds that have poor predicted binding capacity discarded. In this way, potential binding agents can be identified to target hACE2 for possible COVID-19 treatments. Nand et al. have used the DL prediction model, drug similarity screening and molecular docking to screen 1528 anti-HIV-1 compounds, and finally screened out 41 compounds against 3CLpro. Considering the IC50 values of known inhibitors, a DL model has been constructed to re-screen the 41 compounds and resulted in 22 hit compounds. Finally, 2 out of the 22 hit compounds have been screened out as potential targets of 3CLpro, which could be used as drugs to treat SARS-CoV-2. Gao et al. have developed a generative network complex (GNC) platform to design new drug candidates for treatment of SARS-CoV-2, as shown in Fig. 4. The first component of the platform is a generative network including an encoder, a latent space, a molecule generator, and a decoder. The simplified molecular input line entry specification (SMILES) strings are the input to generative network to generate novel molecules, which are fed into the second component of GNC, a 2D fingerprint-based DNN, to re-evaluate their druggable properties. The next component is the MathPose model which is used to predict the 3D structure information of the compounds selected by the 2D fingerprint-based DNN. The bioactivities of those compounds are further estimated by the structure-based DL model named MathDL. The druggable properties predicted by the last component of GNC are used as an indicator to select the promising drug candidates [80]. Kumari and Subbarao have proposed a DL model based on a CNN architecture which could be used to predict the inhibitory activity of 3CLpro against unknown compounds during virtual screening for SARS-CoV-2 [81]. The descriptors which represent the chemical molecules are input into the CNN framework training model to predict active compounds, and could be further used to develop new targeted anti-SARS-CoV-2 compounds. Ahmed et al. have integrated a CNN to find the spatial relationship between input and output data to help predict the affinity of protein binding to multiple ligands in the general family without docking postures or complex user inputs [82]. They have proposed to predict ligands by using the structures of target proteins (PDB format) and ligands (SDF format) as inputs for target binding affinities. The CNN has been used to learn representations from hidden layers and affinity prediction tasks can be extracted from inputs. In comparison to some widely used methods, their work shows better performance for predicting high-resolution protein crystals and non-peptide ligands. In detail, they have used two levels (atomic level and composite level) for feature extraction and compared their performance using the same network. The algorithm is relied on sensitive binding cavity detection which uses mathematical morphology to find deep and shallow pockets (if any) in a given protein. The coordinates of the predicted binding cavity of the protein are placed around the center of mass of the ligand and rotated to various combinations, while the resultant 4D tensor is further processed with CNN. The dataset has approximately more than 5000 complexes including complexes that are not part of PDBbind. The ligand set they have used also represents a diverse set and is one of the highlights of their approach. Joshi et al. have developed a domain aware generation framework called 3D-scaffold which takes the 3D coordinates of the required scaffold as input, and the 3D coordinates of the new treatment candidate scaffold as output, while always preserving the required scaffold in the generated structure [83]. It shows good performance on SARS-CoV-2 3CLpro and NSP15 targets. Most importantly, their DL model performs well with relatively small 3D structural training data and could quickly learn to generalize new scaffolds, highlighting their potential applications in generating target-specific candidates. Wang et al. have adopted a directed message passing neural network which could learn the structure–activity behaviors from a collection of anti-beta-CoV active and inactive compounds to successfully construct a broad-spectrum anti-viral compound prediction model for new active drugs against SARS-CoV-2. After that, they have applied transfer learning to fine-tune the model with the newly reported SARS-COV-2 drugs, generating a specific SARS-COV-2 predictive model called COVIDVS-3. The COVIDVS-3 has been proved capable of screening a large compound library with 4.9 million drug-like molecules from ZINC15 database and recommending a list of potential anti-SARS-CoV-2 compounds for further experimental tests [84]. The experimental validation has demonstrated that the COVIDVS-3 is highly efficient and can be used to screen large compound databases containing millions or more compounds, successfully accelerating the drug discovery process for COVID-19. Gentile et al. have developed an AI-powered virtual screening pipe, which uses deep docking with the Autodock GPU, Glide SP, FRED, ICM and QuickVina2 programs to screen 40 billion molecules that target SARS-CoV-2 3CLpro [85]. Their findings have provided a new starting point for the lead-to-lead optimization campaign for 3CLpro and encouraged the developments of fully automated AI-based drug discovery protocols. Budak et al. have successfully used structural information of molecules and proteins to prepare a list of repurposed drug candidates from FDA-approved drugs by a graph neural network-based graph early fusion affinity (GEFA) model. The Tanimoto/jaccard similarity analysis is conducted on data sets from public databases DrugBank and PubChem, and a list of similar drugs have been prepared by comparing the drugs used in the treatment of COVID-19 with the drugs used in the treatment of other diseases [86]. Kang et al. have analyzed RNA-seq datasets using various bioinformatics methods (e.g., gene ontology, protein–protein interaction-based networks) to profile the upregulation of molecular pathways and analyze the gene enrichment of normal human bronchial epithelial (NHBE) cells infected with SARS-CoV-2. The results suggest that COVID-19 is similar to acute mode of chronic obstructive pulmonary disease (COPD) caused by SARS-CoV-2 infection and the drug Tiotropium may be effective for patients with COVID-19 [87]. Ting et al. have identified basic infections of pathogenesis by comparing core signaling pathways between COVID-19-ARDS (acute respiratory distress syndrome) and non-viral-ARDS. The DNN of the Drug-target interaction model (DNN-DTI) has been trained in advance through the drug-target interaction database to predict drug candidates with identified biomarkers. These predicted drug candidates have been further narrowed down as potential multimolecular drugs through a drug design specification filter [88]. Hu et al. have proposed a new framework called AI model and enzymological experiments (AIMEE) to identify inhibitors of 3CLpro against SARS-COV-2. Based on two rounds of experiments, interpretability of the central model in AIMEE, and mapping of the DL extracted features to the domain knowledge of chemical properties, a commercially available compound has been selected and proven to be an activity-based probe of 3CLpro [89]. Zeng et al. have reported an integrative, network-based DL methodology to identify repurposable drugs for COVID-19. Specifically, they have built a comprehensive knowledge graph which includes 15 million edges across 39 types of relationships connecting drugs, diseases, proteins/genes, pathways, and expression collecting from 24 million PubMed publications. Using such network-based DL methodology, they have successfully identified 41 repurposable drugs whose therapeutic associations with COVID-19 can be validated by transcriptomic and proteomics analysis in SARS-CoV-2 infected human cells [90]. Yuvaraj et al. have designed a DNN model that can accurately sense the protein–ligand interactions of specific drugs and make decision on which drug produces effective interactions against SARS-COV-2 [91].
Fig. 4.
A schematic illustration of the generative network complex. SMILES strings are encoded into the latent space vectors via a gated RNN-based encoder. A molecule generator is applied on the latent vectors to achieve desirable druggable properties, such as binding affinity, partition coefficient (LogP), similarity, etc. from pre-trained DNNs. The resulting drug-like molecules are then decoded into SMILES strings via an RNN-based decoder. The physical properties of the decoded SMILES strings are examined by multitask DNNs. Potential drug candidates are then input into a MathPose unit to generate 3D structures, which are further validated by a mathematical DL (MathDL) unit to recommend new drug candidates [80]. Reproduced with permission. Copyright 2020, PubMed Central.
As shown above, different DL architectures and databases have been adopted and successfully applied in drug virtual screening against SARS-COV-2. However, one of the major challenges for drug screening from large databases is the growing demand for computational resources, which are usually unaffordable for most labs due to the high computational costs. Therefore, various DL-based docking simulation technologies have been proposed to perform such tasks without a large amount of computing resources. Another major challenge for drug virtual screening is the possibility of generating false positives and incorrect ranking of ligands docked. Different results can be obtained through different virtual screening methods even with the same input. We hope that more and better virtual screening technologies will flourish in future and become the mainstream in SBDD.
2.4. Molecular docking
Molecular docking is a computational technique to study the interaction between a target protein and a ligand at the atomic level, which can be used to predict the ligand conformation as well as its position and orientation within binding sites, and offers assessment of the binding affinity [92]. Various scoring functions [93] have been used to evaluate the binding affinity between a ligand and a receptor. In most cases, the success of molecular docking is dependent on the target and computational methods[94]. In literature, most studies on molecular docking prediction tend to predict molecules that bind protein targets with detectable affinities and solved crystal structures [95]. The real challenge is to calculate the relative binding energy with sufficient accuracy to allow as many true positives as possible in the final selection of compounds. With the assistance of DL technologies, sliding docking (or other docking events) and docking scores can be predicted and achieved quickly and more accurately. Notably, the Drug Design Data Resource (D3R), a community-oriented initiative that collects and uses scientific data to test and advance the computer aided Drug Design technology through community-wide blind prediction challenges, has been established to provide opportunities for prediction of protein–ligand poses, affinity rankings, and relative binding free energies. D3R enables rigorous evaluation of normally used computational techniques and succeeds in reducing costs and accelerating the discovery of new drugs in a range of therapeutic areas. However, it does not have the power to clearly distinguish among most approaches with respect to incorporation of machine learning or choosing from structure-based, ligand-based methods and alchemical free energy methods [96].
Ton et al. have developed a new deep docking platform, deep docking (DD), which could quickly predict docking scores for sliding docking, enabling structure-based virtual screening of billions of purchasable molecules in a short period [97]. Mylonas et al. have reported a method which combines a variety of conformation ligands with ResNET-based CNN scoring feature, including the docking output scores in the evaluation. When tested against the DUD-E dataset (a dataset to help benchmark molecular docking programs by providing challenging decoys), it shows significant performance, especially in early enrichment which exceeds the current benchmarks [98]. Their proposed method is eventually applied to target the emerging COVID-19, and successfully discovers the inhibitors for the SARS-COV-2 S protein-hACE2 interaction. By introducing MolAICal software and combining the advantages of DL model and classical algorithm, Bai et al. have proposed a method to generate 3D drugs in 3D pockets of the target proteins [99]. The MolAICal software is mainly composed of two 3D drug design modules. In the first module of MolAICal, genetic algorithm and DL model are used for drug design and training of FDA-approved drug fragments, and Vinardo score fitting is performed based on PDBbind database (a comprehensive collection of experimentally measured binding affinity data for the protein–ligand complexes deposited in the PDB). In the second module, DL generative model is trained by drug-like molecules from ZINC database while molecular docking is invoked by Autodock Vina automatically. For the drug design of SARS-COV-2, MolAICal is demonstrated capable of generating various and novel ligands with good binding scores to SARS-COV-2 3CLpro. Nguyen et al. have combined mathematics and DL methods to provide a reliable ranking of the binding affinities of 137 SARS-CoV-2 3CLpro inhibitor structures [64]. Anwar et al. have proposed a robust experimental design that combines DL methods with molecular docking experiments to identify the most promising drug candidates from the FDA-approved list of drugs which could be used for the treatment of COVID-19. FDA-approved drugs with the highest KIBA scores (representing drug-protein binding affinities) are selected for molecular docking simulations. The results show that 16 drugs demonstrate the highest predicted inhibitory potential against key SARS-CoV-2 viral proteins. In addition, the highest inhibition of papain-like cysteine protease (PLpro) activity can be seen with rifapentine and Flavin adenine dinucleotide (FAD) disodium which have high predicted KIBA scores and binding affinities [100].
In view of the above-mentioned works, we understand that computational drug virtual screening works as a prefilter to select molecules according to a particular predefined criterion of potentially active drugs against a target protein. The DL-based molecular docking is used to further select and investigate the protein-more potent drugs interactions. The docking-based virtual screening is the best example which performs the selections of drugs from large libraries and anlyzes the binding affinities of drugs at particular regions of the target protein receptor with elucidated 3D structures. The combination of DL technology and molecular docking enables fast selection and ideal binding results on drug design, however, there exists a typical drawback at the present stage. A large number of docking software including X-Score, Autodock Vina, and ChemScore use the empirical scoring functions which are limited to features extracted from structural information and often assume a linear relationship existed between the features and the binding affinity. Therefore, researchers are still committed to search for alternative scoring functions that are able to describe the nonlinear relationships between the features and the binding affinity [101].
2.5. Molecular dynamics (MD) simulation
MD simulations of proteins were first carried out in the late 1970s [102]. This powerful tool is used to predict the position of each atom in a molecular system in time domain based on Newton's laws of motion [103]. MD simulations have been widely used in SBDD processes because the technique can help to investigate dynamic atomic details such as binding, unbinding, and conformational changes of receptors which are not easily available from experimental studies [32], [104]. Furthermore, MD simulations can reveal receptor-ligand interaction dynamics such as association and dissociation, and quantify the thermodynamics, dynamics, and free energy landscape [105].
Chloroquine (CQ) has been a potential effective treatment for COVID-19 [106]. Hydroxychloroquine, a CQ derivative, has been reported as a better inhibitory effect than CQ against SARS-COV-2 [107]. Beura and Chetti have adopted a series of computational methods, including pharmacophore model, molecular docking, MM_GBSA (molecular mechanics with generalized Born and surface area) study, ADME (absorption, distribution, metabolism and excretion) property analysis and MD simulations to study the interactions of CQ and its derivatives with SARS-CoV-2 [67]. The structural properties of the compounds and the interactions between the ligand and receptor have been investigated by the pharmacophore model and molecular docking study. The MM_GBSA study and ADME property analysis have revealed the binding free energy of the protein–ligand complex and the pharmacological properties of the compounds. The optimal synthesized CQ derivative molecule CQD15 has been selected and further simulated by MD to obtain the root-mean-square deviation (RMSD), ligand properties, and protein–ligand contact. Their work offers a complete process of SBDD for SARS-CoV-2. The MD simulation plays a critical role in drug design which allows a more accurate estimate of the thermodynamics and kinetics associated with drug-target recognition and binding [32]. Compared with CQ and hydroxychloroquine, CQD15 shows a better inhibitory effect on SARS-COV-2. The anti-influenza drug, Arbidol, has been reported able to neutralize SARS-CoV-2 [36]. However, the detailed mechanism behind the inhibition remains unclear. Herein, Padhi et al. have presented the atomic insights into the SARS-CoV-2 membrane fusion inhibition mechanism. Analyses based on MD simulations show that Arbidol binds with high affinity and is stable at the RBD/hACE2 interface [36]. In addition, identifying key residues of RBD and hACE2 that interact with Arbidol by MD could open the door to therapeutic strategies and developments of higher efficacy Arbidol derivatives or primary drug candidates.
In drug development, the discovery of lead compounds and subsequent optimization of identified compounds into drug candidates should be followed by target identification or target validation. The goal is to discover and design compounds with good binding affinity and selectivity for the target. Docking and scoring are standard tools for rapidly estimating favorable ligand binding positions and energies. However, as it is mentioned above, the scoring functions of the current docking tools still have many limitations [108]. Using MD to address these limitations allows a more accurate and facilitative assessment of the binding affinity of selected compounds. In addition, MD could help to unravel the atomic details of protein-drug interaction and explain the molecular mechanism behind it.
3. DL-based MD simulation in SBDD for SARS-CoV-2
Predictions of protein–ligand binding has broad biological significance [102], [105]. However, performing such analyses to cover the entire chemical space of small molecules together with complex biological proteins requires tremendous computational power [109]. DL is now playing an increasingly pivotal role in SBDD as described above. In particular, the advances of the use of DL-based MD computational methods have enabled researchers to understand the binding mode, affinity and evolution of atomic system by using appropriate models and algorithms in which the selected model could “learn” the patterns inherent in the input data. Here, we have summarized some representative works in Table 1.
Table 1.
Current literature on DL-based MD Simulation in SBDD for SARS-CoV-2.
| Reference | Methods | Objective | Input | Results |
|---|---|---|---|---|
| (Joshi et al., 2021) | DL regression model, molecular docking and MD simulation | To identify potential drugs against SARS-CoV-2 3CLpro | CHEMBL3927 dataset | Two compounds have been identified to be potential hits against 3CLpro.. |
| (Arshia et al., 2021) | DL LSTM generative network via transfer learning, fine-tuning over ten generations, molecular docking and MD simulation | To identify potential inhibitors against SARS-CoV-2 3CLpro | ChEMBL and Moses datasets, SMILES | Four top-ranked ligands have been found to be potential inhibitors against SARS-CoV-2 3CLpro. |
| (Zhang et al., 2020) | DFCNN and DeepBindBC DL models, molecular docking, pocket localized MD simulation, metadynamics and inhouse developed tools | To identify potential drugs targeting SARS-CoV-2 RdRp from drugs available on the market | TargetMol-Approved_Drug_Library | An FDA-approved drug called Pralatrexate that strongly inhibits the replication of SARS-CoV-2 in vitro has been identified. |
| (Casalina et al., 2021) | DL variational autoencoder model and DeepDriveMD simulation | To investigate the mechanism of infectivity of SARS-CoV-2 S protein | Conformational sampling across different systems | Have provided the elucidation of the full glycan shield of S, the role of S glycans in modulating the infectivity of the virus, and the characterization of the flexible interactions between the S and the hACE2. |
| (Lee et al., 2019) | DeepDriveMD simulation | To understand how coupling DL approaches to MD simulation | Villin headpiece protein | Have achieved a performance gain in sampling the folded states by 2.3x and provided quantitative basis to understand how DL drives MD simulation. |
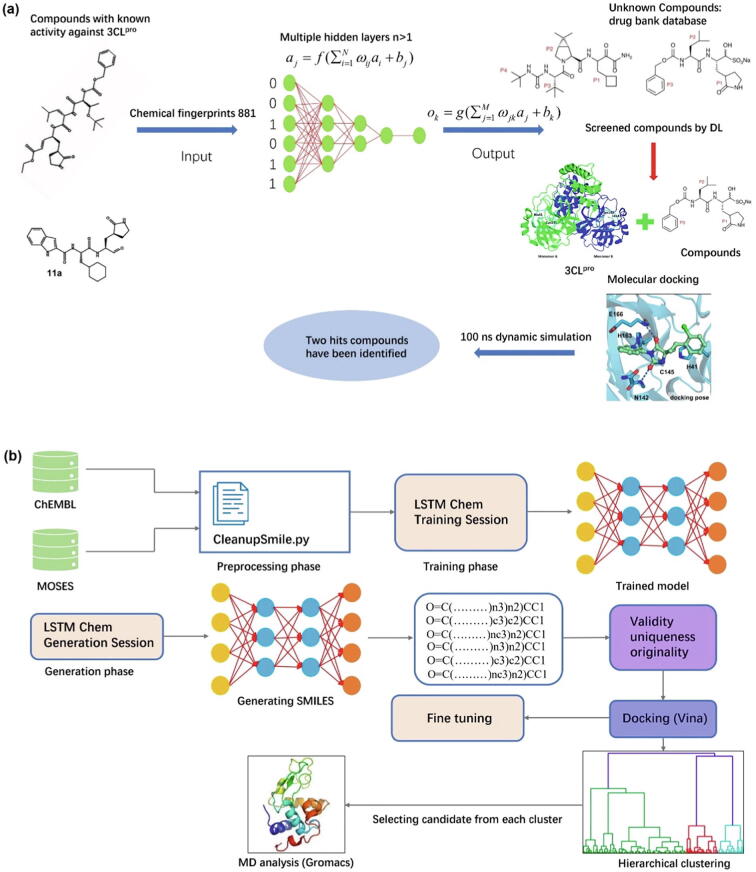
Virus replication is controlled by the coronavirus 3CLpro which is therefore considered a major target and promising opportunity for antiviral discovery with direct acting agents [110]. Joshi et al. have screened 9101 drugs from the drug bank database for SARS-COV-2 3CLPro using DL, molecular docking, and MD simulation techniques [111]. Prior to MD simulations, 500 drugs have been screened by DL regression model and subjected to molecular docking, resulting in ten screened compounds with strong binding affinity. Afterwards, MD simulations have been conducted for five compounds in order to obtain their binding potentials. Two compounds, {4-[(2 s,4e)-2-(1,3-Benzothiazol-2-Yl)-2-(1 h-1,2,3-Benzotriazol-1-Yl)-5-Phenylpent-4-Enyl]Phenyl}(Difluoro)Methylphosphonic Acid and 1-(3-(2,4-dimethylthiazol-5-yl)-4-oxo-2,4-dihydroindeno[1,2-c]pyrazol-5-yl)-3-(4-methylpiperazin-1-yl)urea have been screened as potential hits. Their study suggests two potential drugs that can be tested under experimental conditions to assess efficacy against SARS-COV-2 (Fig. 5a). Arshia et al. have used 2.5 million compounds to train a long short-term memory (LSTM) generative network through transfer learning to identify-four optimal candidates that inhibit 3CLpro in SARS-COV-2 [112]. Fig. 5b shows the block diagram of the network’s training and generation phases, fine-tuning and evaluation sessions for design of potential inhibitors against SARS-COV-2 3CLpro. The datasets used to train the DNN models have been obtained from ChEMBL and ZINC. The SMILE format has collected 2.1 million compound structures from the ChEMBL dataset and 1.9 million molecular structures from Moses, a subset of the ZINC dataset. After removing the molecules containing undesirable atoms or groups, a total of 2.5 million SMILES have been used to train the neural network. The LSTM_Chem network has been used to generate drugs, with its weights trained on ChEMBL. Validity determines whether the generated SMILES are truly valid candidates for a molecule. Uniqueness ensures the uniqueness of the compound within the dataset. Originality guarantees that the generated SMILES are not in any datasets. Then, SMILES that satisfy all three criteria are chosen as eligible molecules. Followingly, molecular docking and MD simulation are conducted. The extensive calculations and statistical analyses of the study indicate that the chosen candidates can be used as potential inhibitors against SARS-COV-2 in computational environments. However, additional in vitro, in vivo, and clinical trials are needed to further prove their true efficacy. Zhang et al. have proposed a hybrid drug screening program based on DL and MD simulations which consists of a dense fully connected neural network (DFCNN) [113], DeepBindBC (https://cbblab.siat.ac.cn/DeepBindBC/index.php) [114], Autodock Vina [115], pocket local MD simulations and metadynamics simulations. DFCNN uses molecular vector data of protein pocket and ligand to estimate the protein–ligand pair as binding or non-binding with a probability value between 0 and 1. DeepBindBC estimates the binding possibility from atom contact information at interaction surface of a modelled 3D protein–ligand complex. DFCNN and DeepBindBC are both DL-based methods. This program could help explore the binding potentials of drugs in TargetMol-Approved_Drug_Library, a drug library containing 1906 of currently available drugs in market. After the predictions by DFCNN and DeepBindBC, 14 candidates have been selected. MD simulations on RNA-dependent RNA polymerase (RdRp)-drug complexes have been adopted to further screen the 14 selected drugs and understand the interactions and stabilities of the complexes. RdRp is believed to be one of the most promising therapeutic targets [116], [117]. Molecules that can bind to the catalytic site of RdRp could potentially interfere the synthesis of viral RNA [118]. Finally, 4 approved drug candidates targeting RdRp have been screened out, and 2 out of 4 (Pralatrexate and Azithromycin) can effectively inhibit SARS-CoV-2 replication in vitro with EC50 values (effective concentration or dose that produces 50 % of the maximum response) of 0.008 μM and 9.453 μM [119].
Fig. 5.
(a) Graphic illustration of drug virtual screening for SARS-COV-2 3CLPro by DL, molecular docking and MD simulation techniques. In the initial stage, 500 drugs have been screened by a DL regression model and subjected to molecular docking, resulting in 10 screened compounds with strong binding affinity. Further, five compounds have been checked for their binding potentials by analyzing MD simulations for 100 ns. In the final stage, two compounds have been screened as potential hits [111]. Reproduced with permission. Copyright 2021, Springer Nature; (b) A block diagram of training and generation phases of the network, fine-tuning and evaluation sessions for design of potential inhibitors against SARS-COV-2 3CLpro. The datasets used to train the DNN models have been obtained from ChEMBL and Moses. The LSTM_Chem network has been used to generate drugs, with its weights trained on ChEMBL. The SMILES that satisfy-three criteria (validity, uniqueness and originality) are retained as eligible molecules. Followingly, molecular docking and MD simulation are conducted [112]. Reproduced with permission. Copyright 2021, Elsevier.
Although DL methods have demonstrated their potential to be efficient by learning from sufficient training data, there are still problems such as overfitting and discrepancies between training data and actual data. Casalino et al. have developed an AI-driven multiscale simulation framework to investigate SARS-CoV-2 S dynamics, revealing the full glycan shield of S protein and discovering that glycans play an active role in infection [120]. In their work, all-atomic MD simulations have been adopted to combine, augment, and extend the available experimental data set to study the structure, dynamics, and function of SARS-CoV-2 S protein. Traditional MD and weighted integrated enhanced sampling methods have been used. Then the simulations have been combined with a DL-based method as an integrated workflow that “drives” sampling from knowledge gained at one scale to another. Specifically, the DL-based method uses a variational autoencoder (VAE), which is developed based on convolution filters on contact graphs (from MD simulations) to analyze simulated data sets over long-time scales and organize them into a small number of conformational states along with biophysically related response coordinates. For SARS-CoV-2 S protein, the intrinsic size of the such simulation presents a significant challenge in scaling DL-based method to elucidate conformational states associated with functions. Therefore, Casalino et al. have further developed the DeepDriveMD [121], an approach that extends the AI-driven multiscale simulation framework to adaptively run MD simulations ensembles to fold small proteins. The DeepDriveMD has also been performed on a S-hACE2 system with 8.5 million atoms. Three conformations have been extracted from the first set of MD simulations and then been used as a starting point for a new round of MD simulations. Such a dl-driven adaptive MD approach has expanded the conformational space explored and described the flexibility of S in the context of hACE2 binding, revealing the effects of internal structure of S on RBD-hACE2 interaction and the infectious mechanism of SARS-CoV-2 S protein. Their work has successfully uncovered many aspects of peak dynamics and function that are currently unavailable from experiments. In addition, it has provided information on the basic mechanism of viral infection and advanced the technical and methodological limits of molecular simulations.
Simulations of biological macromolecules play an important role in understanding the physical basis of many complex processes [122]. However, the simulations of protein folding at the atomic scale remain challenging, even though computing power has improved and specialized architectures have evolved [123]. This is due to the high dimensional nature of protein conformational landscapes and the inability of atomic and MD simulations to adequately sample these landscapes for observation of the dual aspects of folding events [124]. DL-based MD simulation in SBDD can effectively fold small proteins on supercomputer [32]. Compared with traditional MD approaches, DL-based MD simulation in SBDD provides a quantitative basis with improved performance and reduced computing time, showing strong potentials in structural discovery and drug design in COVID-19 research.
4. Perspective and future direction
4.1. Strengths and challenges
Recognizing the steadily increasing number of positive cases of infection and the lack of approved treatments for SARS-CoV-2, SBDD has emerged as a rapid and reliable technique in pharmaceutical and medical research because it not only saves time, but also helps reduce the cost of designing therapeutic agents [9], [125]. In particular, the DL applications in SBDD can facilitate the discovery of new drugs and reuse of FDA-approved drugs whose safety and side effects have already been known [126]. Due to inherent mutations in the SARS-CoV-2 genome that may hinder disease treatment, applications of DL in SBDD also play a key role in accelerating the process of drug discovery and development against new SARS-CoV-2 variants. MD simulations have been widely used in SBDD because the technique helps to unravel many SARS-CoV-2 atomic details, such as binding, unbinding and conformational changes, at a high resolution that is not normally available from experimental studies [127]. In addition, MD simulations can be used to explore the dynamics of SARS-CoV-2 receptor-ligand interactions and quantify the thermodynamic, kinetic and free energy landscape [128]. In particular, the DL-based MD simulations could undoubtedly inherit the advantages of both DL and MD which could reveal the dynamic evolution of large, complex biological system with selected drugs.
However, the SBDD method used at the current stage still has some limitations. For example, the current drug design method is mainly based on the structures of drug and target biological macromolecules [129]. As mentioned above, many protein structures in the PDB are not actually the folded states of proteins. Some protein structures may only fold upon binding to other proteins, substrates or metal ions, some may only fold when they are chemically modified, and some may fold directly into large complexes such as the ribosome. Moreover, SBDD only considers the interaction between the compound and the target biomolecule but does not consider other interaction modes between the two [129]. Although SBDD is a powerful tool that can provide an intuitive model for scientists to design drugs [11], the results still need to be verified by experiments at the present stage [130].
4.2. Future outlook and concluding thoughts
Research on the pathogenesis of COVID-19 is still ongoing, and the existence of bias, imbalance, and limited data may have a significant impact on the prediction accuracy of DL methods in SBDD [131]. In addition, the increase in COVID-19 positive cases and the lack of approved drugs remain global health issues that require urgent discovery of drugs to prevent and treat the disease [132]. DL prediction has accelerated the virtual identification of structure-based drug target inhibitors for SARS-CoV-2 [133]. The application of DL-based MD simulations in SBDD for SARS-CoV-2 has accelerated the development of new drugs. But the current DL and MD computational methods remain to be further developed. There are two approaches regarding the future developments of these computational methods in SBDD. The first is to continue optimizing the parameters under the existing framework of molecular mechanics, incorporating DL to expand the scope of application, and further introducing the polarization effect of molecular interactions. The second is to introduce a new paradigm, which requires the development of both computational methods and the theory. In this review, we hope to provide the scientific community with a comprehensive review of major new applications of DL methods and DL-based MD simulations in SBDD procedures, as well as their applications in the research of SARS-CoV-2. However, we only focus on protein–ligand problems in SBDD but do not include the protein–protein interactions, which might be the future direction for better designing drugs against SARS-CoV-2.
CRediT authorship contribution statement
Yao Sun: Conceptualization, Writing – original draft, Writing – review & editing. Yanqi Jiao: Writing – original draft. Chengcheng Shi: Writing – review & editing. Yang Zhang: Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The work is supported by the National Natural Science Foundation of China (Ref. 12102113) to Y. S; Guangdong Basic and Applied Basic Research Foundation (2021A1515220115), Project of Educational Commission of Guangdong Province of China (2022KTSCX210) and Opening Funding of the Key Laboratory of Zhejiang Province for Aptamers and Theranostic (2022ASC001) to Y. Z.
Contributor Information
Yao Sun, Email: sunyao0819@hit.edu.cn.
Yang Zhang, Email: zhangyang07@hit.edu.cn.
References
- 1.Wu Y.-C., Chen C.-S., Chan Y.-J. The outbreak of COVID-19: an overview. J Chin Med Assoc. 2020;83(3):217–220. doi: 10.1097/JCMA.0000000000000270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.P. Guadarrama-Ortiz, J. A. Choreño-Parra, C. M. Sánchez-Martínez, F. J. Pacheco-Sánchez, A. I. Rodríguez-Nava, G. García-Quintero, Neurological aspects of SARS-CoV-2 infection: mechanisms and manifestations, Front Neurol, 11 (2020), pp. 1039–1039. [DOI] [PMC free article] [PubMed]
- 3.Wang Q., Anang S., Iketani S., Guo Y., Liu L., Katsamba P.S., et al. Functional properties of the spike glycoprotein of the emerging SARS-CoV-2 variant B.1.1.529. Cell Rep. 2022;39(11) doi: 10.1016/j.celrep.2022.110924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nutalai R., Zhou D., Tuekprakhon A., Ginn H.M., Supasa P., Liu C., et al. Potent cross-reactive antibodies following Omicron breakthrough in vaccinees. Cell. 2022;185(12):2116–2131.e18. doi: 10.1016/j.cell.2022.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tuekprakhon A., Nutalai R., Dijokaite-Guraliuc A., Zhou D., Ginn H.M., Selvaraj M., et al. Antibody escape of SARS-CoV-2 Omicron BA.4 and BA.5 from vaccine and BA.1 serum. Cell. 2022;185(14):2422–2433.e13. doi: 10.1016/j.cell.2022.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.R. S. P. Rao, N. Ahsan, C. Xu, L. Su, J. Verburgt, L. Fornelli, D. Kihara, D. Xu, Evolutionary dynamics of Indels in SARS-CoV-2 spike glycoprotein, Evol Bioinform Online 17 (2021), p. 11769343211064616. [DOI] [PMC free article] [PubMed]
- 7.Lu R.-M., Hwang Y.-C., Liu I.J., Lee C.-C., Tsai H.-Z., Li H.-J., et al. Development of therapeutic antibodies for the treatment of diseases. J Biomed Sci. 2020;27(1) doi: 10.1186/s12929-019-0592-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bajad N.G., Rayala S., Gutti G., Sharma A., Singh M., Kumar A., et al. Systematic review on role of structure based drug design (SBDD) in the identification of anti-viral leads against SARS-Cov-2. Curr Res Pharmacol Drug Discov. 2021;2 doi: 10.1016/j.crphar.2021.100026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gurung A.B., Ali M.A., Lee J., Farah M.A., Al-Anazi K.M. An updated review of computer-aided drug design and its application to COVID-19. BioMed Res Int. 2021;2021 doi: 10.1155/2021/8853056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vlachakis D., Vlamos P. Mathematical multidimensional modelling and structural artificial intelligence pipelines provide insights for the designing of highly specific antiSARS-CoV2 agents. Math Comput Sci. 2021;15(4):877–888. [Google Scholar]
- 11.Batool M., Ahmad B., Choi S. A structure-based drug discovery paradigm. Int J Mol Sci. 2019;20(11) doi: 10.3390/ijms20112783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zheng M., Zhao J., Cui C., Fu Z., Li X., Liu X., et al. Computational chemical biology and drug design: facilitating protein structure, function, and modulation studies. Med Res Rev. 2018;38(3):914–950. doi: 10.1002/med.21483. [DOI] [PubMed] [Google Scholar]
- 13.Zhong G., Wang L.-N., Ling X., Dong J. An overview on data representation learning: from traditional feature learning to recent deep learning. J Financ Data Sci. 2016;2(4):265–278. [Google Scholar]
- 14.Koutsoukas A., Monaghan K.J., Li X., Huan J. Deep-learning: investigating deep neural networks hyper-parameters and comparison of performance to shallow methods for modeling bioactivity data. J Cheminform. 2017;9(1) doi: 10.1186/s13321-017-0226-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lozano-Diez A., Zazo R., Toledano D.T., Gonzalez-Rodriguez J. An analysis of the influence of deep neural network (DNN) topology in bottleneck feature based language recognition. PLoS ONE. 2017;12(8):e0182580. doi: 10.1371/journal.pone.0182580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bazgir O., Zhang R., Dhruba S.R., Rahman R., Ghosh S., Pal R. Representation of features as images with neighborhood dependencies for compatibility with convolutional neural networks. Nat Commun. 2020;11(1) doi: 10.1038/s41467-020-18197-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alzubaidi L., Zhang J., Humaidi A.J., Al-Dujaili A., Duan Y., Al-Shamma O., et al. Review of deep learning: concepts, CNN architectures, challenges, applications, future directions. J Big Data. 2021;8(1) doi: 10.1186/s40537-021-00444-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cortés-Ciriano I., Bender A. Deep confidence: a computationally efficient framework for calculating reliable prediction errors for deep neural networks. J Chem Inf Model. 2019;59(3):1269–1281. doi: 10.1021/acs.jcim.8b00542. [DOI] [PubMed] [Google Scholar]
- 19.Zhou Y. Natural language processing with improved deep learning neural networks. Sci Program. 2022;2022:1–8. [Google Scholar]
- 20.Lan L., You L., Zhang Z., Fan Z., Zhao W., Zeng N., et al. Generative adversarial networks and its applications in biomedical informatics. Front Public Health. 2020;8 doi: 10.3389/fpubh.2020.00164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wei Y., Zhang Z., Wang Y., Xu M., Yang Y., Yan S., et al. Deraincyclegan: rain attentive cyclegan for single image deraining and rainmaking. IEEE Trans Image Process. 2021;30:4788–4801. doi: 10.1109/TIP.2021.3074804. [DOI] [PubMed] [Google Scholar]
- 22.Sarker I.H. Deep learning: a comprehensive overview on techniques, taxonomy, applications and research directions. SN Comput Sci. 2021;2(6) doi: 10.1007/s42979-021-00815-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Asraf A., Islam M.Z., Haque M.R., Islam M.M. Deep learning applications to combat novel coronavirus (COVID-19) pandemic. SN Comput Sci. 2020;1(6) doi: 10.1007/s42979-020-00383-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596(7873):583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Senior A.W., Evans R., Jumper J., Kirkpatrick J., Sifre L., Green T., et al. Improved protein structure prediction using potentials from deep learning. Nature. 2020;577(7792):706–710. doi: 10.1038/s41586-019-1923-7. [DOI] [PubMed] [Google Scholar]
- 26.Bryant P., Pozzati G., Elofsson A. Improved prediction of protein-protein interactions using AlphaFold2. Nat Commun. 2022;13(1) doi: 10.1038/s41467-022-28865-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anighoro A. Deep learning in structure-based drug design. Methods Mol Biol. 2022;2390:261–271. doi: 10.1007/978-1-0716-1787-8_11. [DOI] [PubMed] [Google Scholar]
- 28.Wang D., Cui C., Ding X., Xiong Z., Zheng M., Luo X., et al. Improving the virtual screening ability of target-specific scoring functions using deep learning methods. Front Pharmacol. 2019;10 doi: 10.3389/fphar.2019.00924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sanjeevi M., Hebbar P., Natarajan A., Rashmi S., Rahul C., Mohan A., et al. In: Advances in Protein Molecular and Structural Biology Methods. Tripathi T., Dubey V.K., editors. Academic Press; 2022. Chapter 25 – Methods and applications of machine learning in structure-based drug discovery; pp. 405–437. [Google Scholar]
- 30.Kozlovskii I., Popov P. Structure-based deep learning for binding site detection in nucleic acid macromolecules. NAR Genom Bioinform. 2021;3(4) doi: 10.1093/nargab/lqab111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Selvaraj C., Chandra I., Singh S.K. Artificial intelligence and machine learning approaches for drug design: challenges and opportunities for the pharmaceutical industries. Mol Divers. 2021;26(3):1893–1913. doi: 10.1007/s11030-021-10326-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.De Vivo M., Masetti M., Bottegoni G., Cavalli A. Role of molecular dynamics and related methods in drug discovery. J Med Chem. 2016;59(9):4035–4061. doi: 10.1021/acs.jmedchem.5b01684. [DOI] [PubMed] [Google Scholar]
- 33.McCammon J.A., Gelin B.R., Karplus M. Dynamics of folded proteins. Nature. 1977;267(5612):585–590. doi: 10.1038/267585a0. [DOI] [PubMed] [Google Scholar]
- 34.Levitt M., Warshel A. Computer simulation of protein folding. Nature. 1975;253(5494):694–698. doi: 10.1038/253694a0. [DOI] [PubMed] [Google Scholar]
- 35.Padhi A.K., Rath S.L., Tripathi T. Accelerating COVID-19 research using molecular dynamics simulation. J Phys Chem B. 2021;125(32):9078–9091. doi: 10.1021/acs.jpcb.1c04556. [DOI] [PubMed] [Google Scholar]
- 36.Padhi A.K., Seal A., Khan J.M., Ahamed M., Tripathi T. Unraveling the mechanism of arbidol binding and inhibition of SARS-CoV-2: insights from atomistic simulations. Eur J Pharmacol. 2021;894 doi: 10.1016/j.ejphar.2020.173836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Srivastava N., Garg P., Srivastava P., Seth P.K. A molecular dynamics simulation study of the ACE2 receptor with screened natural inhibitors to identify novel drug candidate against COVID-19. PeerJ. 2021;9:e11171. doi: 10.7717/peerj.11171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., et al. The protein data bank. Nucleic Acids Res. 2000;28(1):235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mielke SP, Krishnan VV. Characterization of protein secondary structure from NMR chemical shifts, Prog Nucl Magn Reson Spectrosc, 54 (3-4) (2009), pp. 141–165. [DOI] [PMC free article] [PubMed]
- 40.van Breugel M., Rosa e Silva I., Andreeva A. Structural validation and assessment of AlphaFold2 predictions for centrosomal and centriolar proteins and their complexes. Commun Biol. 2022;5(1) doi: 10.1038/s42003-022-03269-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Waterhouse A., Bertoni M., Bienert S., Studer G., Tauriello G., Gumienny R., et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46(W1):W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Centeno N.B., Planas-Iglesias J., Oliva B. Comparative modelling of protein structure and its impact on microbial cell factories. Microb Cell Factories. 2005;4(1) doi: 10.1186/1475-2859-4-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fiser A. Template-based protein structure modeling. Methods Mol Biol. 2010;673:73–94. doi: 10.1007/978-1-60761-842-3_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roy A., Kucukural A., Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc. 2010;5(4):725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang J, Zhang Y. Protein structure and function prediction using I-TASSER, Curr Protoc Bioinform, 52 (2015), pp. 5.8.1–5.8.15. [DOI] [PMC free article] [PubMed]
- 46.Tunyasuvunakool K., Adler J., Wu Z., Green T., Zielinski M., Žídek A., et al. Highly accurate protein structure prediction for the human proteome. Nature. 2021;596(7873):590–596. doi: 10.1038/s41586-021-03828-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thornton J.M., Laskowski R.A., Borkakoti N. AlphaFold heralds a data-driven revolution in biology and medicine. Nat Med. 2021;27(10):1666–1669. doi: 10.1038/s41591-021-01533-0. [DOI] [PubMed] [Google Scholar]
- 48.Baek M., DiMaio F., Anishchenko I., Dauparas J., Ovchinnikov S., Lee G.R., et al. Accurate prediction of protein structures and interactions using a three-track neural network. Science. 2021;373(6557):871–876. doi: 10.1126/science.abj8754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Evans R, O’Neill M, Pritzel A, Antropova N, Senior A, Green T, et al., Protein complex prediction with AlphaFold-Multimer, bioRxiv, (2022), p. 2021.10.04.463034.
- 50.Mirdita M., Schütze K., Moriwaki Y., Heo L., Ovchinnikov S., Steinegger M. ColabFold: making protein folding accessible to all. Nat Methods. 2022;19(6):679–682. doi: 10.1038/s41592-022-01488-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.S. Kumar, R. Nyodu, V. K. Maurya, S. K. Saxena, Morphology, genome organization, replication, and pathogenesis of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), Coronavirus Disease 2019 (COVID-19), (2020), pp. 23-31.
- 52.Ji D., Juhas M., Tsang C.M., Kwok C.K., Li Y., Zhang Y. Discovery of G-quadruplex-forming sequences in SARS-CoV-2. Brief Bioinform. 2020;22(2):1150–1160. doi: 10.1093/bib/bbaa114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.V’kovski P., Kratzel A., Steiner S., Stalder H., Thiel V. Coronavirus biology and replication: implications for SARS-CoV-2. Nat Rev Microbiol. 2021;19(3):155–170. doi: 10.1038/s41579-020-00468-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang Q., Syed A.A.S., Fahira A., Shi Y. Structural analysis of the SARS-CoV-2 Omicron variant proteins. Research. 2021;2021:9769586. doi: 10.34133/2021/9769586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Robertson A.J., Courtney J.M., Shen Y., Ying J., Bax A. Concordance of x-ray and AlphaFold2 models of SARS-CoV-2 main protease with residual dipolar couplings measured in solution. J Am Chem Soc. 2021;143(46):19306–19310. doi: 10.1021/jacs.1c10588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang Q., Jian X., Syed A.A.S., Fahira A., Zheng C., Zhu Z., et al. Structural comparison and drug screening of spike proteins of ten SARS-CoV-2 variants. Research. 2022;2022:9781758. doi: 10.34133/2022/9781758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gupta M, Azumaya CM, Moritz M, Pourmal S, Diallo A, Merz GE, et al., CryoEM and AI reveal a structure of SARS-CoV-2 Nsp2, a multifunctional protein involved in key host processes, Res Sq, (2021), 10.21203/rs.3.rs-515215/v1.
- 58.Paul D., Sanap G., Shenoy S., Kalyane D., Kalia K., Tekade R.K. Artificial intelligence in drug discovery and development. Drug Discov Today. 2021;26(1):80–93. doi: 10.1016/j.drudis.2020.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nazem F., Ghasemi F., Fassihi A., Dehnavi A.M. 3D U-Net: A voxel-based method in binding site prediction of protein structure. J Bioinform Comput Biol. 2021;19(2):2150006. doi: 10.1142/S0219720021500062. [DOI] [PubMed] [Google Scholar]
- 60.Lionta E., Spyrou G., Vassilatis D.K., Cournia Z. Structure-based virtual screening for drug discovery: principles, applications and recent advances. Curr Top Med Chem. 2014;14(16):1923–1938. doi: 10.2174/1568026614666140929124445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Raschka S., Kaufman B. Machine learning and AI-based approaches for bioactive ligand discovery and GPCR-ligand recognition. Methods. 2020;180:89–110. doi: 10.1016/j.ymeth.2020.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jiménez J., Doerr S., Martínez-Rosell G., Rose A.S., De Fabritiis G. DeepSite: protein-binding site predictor using 3D-convolutional neural networks. Bioinformatics. 2017;33(19):3036–3042. doi: 10.1093/bioinformatics/btx350. [DOI] [PubMed] [Google Scholar]
- 63.Nand M., Maiti P., Joshi T., Chandra S., Pande V., Kuniyal J.C., et al. Virtual screening of anti-HIV1 compounds against SARS-CoV-2: machine learning modeling, chemoinformatics and molecular dynamics simulation based analysis. Sci Rep. 2020;10(1) doi: 10.1038/s41598-020-77524-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nguyen D.D., Gao K., Chen J., Wang R., Wei G.-W. Unveiling the molecular mechanism of SARS-CoV-2 main protease inhibition from 137 crystal structures using algebraic topology and deep learning. Chem Sci. 2020;11(44):12036–12046. doi: 10.1039/d0sc04641h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li Y., Pei J., Lai L. Structure-based de novo drug design using 3D deep generative models. Chem Sci. 2021;12(41):13664–13675. doi: 10.1039/d1sc04444c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rochman ND, Wolf YI, Faure G, Mutz P, Zhang F, Koonin EV. Ongoing global and regional adaptive evolution of SARS-CoV-2, Proc Natl Acad Sci U S A, 118 (29) (2021), p. e2104241118. [DOI] [PMC free article] [PubMed]
- 67.Beura S., Chetti P. In-silico strategies for probing chloroquine based inhibitors against SARS-CoV-2. J Biomol Struct Dyn. 2021;39(10):3747–3759. doi: 10.1080/07391102.2020.1772111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Padhi A.K., Tripathi T. Can SARS-CoV-2 accumulate mutations in the S-protein to increase pathogenicity? ACS Pharmacol Transl Sci. 2020;3(5):1023–1026. doi: 10.1021/acsptsci.0c00113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Padhi A.K., Tripathi T. Targeted design of drug binding sites in the main protease of SARS-CoV-2 reveals potential signatures of adaptation. Biochem Biophys Res Commun. 2021;555:147–153. doi: 10.1016/j.bbrc.2021.03.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tubiana J., Schneidman-Duhovny D., Wolfson H.J. ScanNet: an interpretable geometric deep learning model for structure-based protein binding site prediction. Nat Methods. 2022;19(6):730–739. doi: 10.1038/s41592-022-01490-7. [DOI] [PubMed] [Google Scholar]
- 71.Singh N., Chaput L., Villoutreix B.O. Virtual screening web servers: designing chemical probes and drug candidates in the cyberspace. Brief Bioinform. 2021;22(2):1790–1818. doi: 10.1093/bib/bbaa034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Carpenter K.A., Cohen D.S., Jarrell J.T., Huang X. Deep learning and virtual drug screening. Future Med Chem. 2018;10(21):2557–2567. doi: 10.4155/fmc-2018-0314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang Y., Ye T., Xi H., Juhas M., Li J. Deep learning driven drug discovery: tackling severe acute respiratory syndrome coronavirus 2. Front Microbiol. 2021;12 doi: 10.3389/fmicb.2021.739684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Benarous L., Benarous K., Muhammad G., Ali Z. Deep learning application detecting SARS-CoV-2 key enzymes inhibitors. Clust Comput. 2022 doi: 10.1007/s10586-022-03656-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jin W, Stokes JM, Eastman RT, Itkin Z, Zakharov AV, Collins JJ, Jaakkola TS, Barzilay R. Deep learning identifies synergistic drug combinations for treating COVID-19, Proc Natl Acad Sci U S A, 118 (39) (2021), p. e2105070118. [DOI] [PMC free article] [PubMed]
- 76.Su X., Hu L., You Z., Hu P., Wang L., Zhao B. A deep learning method for repurposing antiviral drugs against new viruses via multi-view nonnegative matrix factorization and its application to SARS-CoV-2. Brief Bioinform. 2021;23(1) doi: 10.1093/bib/bbab526. [DOI] [PubMed] [Google Scholar]
- 77.Azmoodeh SK, Tsigelny IF, Kouznetsova VL. Potential SARS-CoV-2 nonstructural proteins inhibitors: drugs repurposing with drug-target networks and deep learning, Front Biosci (Landmark Ed), 27 (4) (2022), 10.31083/j.fbl2704113. [DOI] [PubMed]
- 78.Joshi T., Joshi T., Pundir H., Sharma P., Mathpal S., Chandra S. Predictive modeling by deep learning, virtual screening and molecular dynamics study of natural compounds against SARS-CoV-2 main protease. J Biomol Struct Dyn. 2021;39(17):6728–6746. doi: 10.1080/07391102.2020.1802341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Karki N., Verma N., Trozzi F., Tao P., Kraka E., Zoltowski B. Predicting potential SARS-COV-2 drugs-in depth drug database screening using deep neural network framework SSnet, classical virtual screening and docking. Int J Mol Sci. 2021;22(4) doi: 10.3390/ijms22041573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gao K, Nguyen DD, Wang R, Wei GW. Machine intelligence design of 2019-nCoV drugs, bioRxiv: the preprint server for biology, (2020), 10.1101/2020.01.30.927889.
- 81.Kumari M., Subbarao N. Deep learning model for virtual screening of novel 3C-like protease enzyme inhibitors against SARS coronavirus diseases. Comput Biol Med. 2021;132 doi: 10.1016/j.compbiomed.2021.104317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ahmed A, Mam B, Sowdhamini R. DEELIG: A deep learning approach to predict protein-ligand binding affinity, Bioinform Biol Insights, 15 (2021), p. 11779322211030364. [DOI] [PMC free article] [PubMed]
- 83.Joshi R.P., Gebauer N.W.A., Bontha M., Khazaieli M., James R.M., Brown J.B., et al. 3D-Scaffold: a deep learning framework to generate 3D coordinates of drug-like molecules with desired scaffolds. J Phys Chem B. 2021;125(44):12166–12176. doi: 10.1021/acs.jpcb.1c06437. [DOI] [PubMed] [Google Scholar]
- 84.Wang S., Sun Q., Xu Y., Pei J., Lai L. A transferable deep learning approach to fast screen potential antiviral drugs against SARS-CoV-2. Brief Bioinform. 2021;22(6) doi: 10.1093/bib/bbab211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gentile F., Fernandez M., Ban F., Ton A.-T., Mslati H., Perez C.F., et al. Automated discovery of noncovalent inhibitors of SARS-CoV-2 main protease by consensus deep docking of 40 billion small molecules. Chem Sci. 2021;12(48):15960–15974. doi: 10.1039/d1sc05579h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Budak C., Mençik V., Gider V. Determining similarities of COVID-19 – lung cancer drugs and affinity binding mode analysis by graph neural network-based GEFA method. J Biomol Struct Dyn. 2021:1–13. doi: 10.1080/07391102.2021.2010601. [DOI] [PubMed] [Google Scholar]
- 87.Kang K., Kim H.H., Choi Y. Tiotropium is predicted to be a promising drug for COVID-19 through transcriptome-based comprehensive molecular pathway analysis. Viruses. 2020;12(7) doi: 10.3390/v12070776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ting C.T., Yeh S.J., Chen B.S. COVID-19-related versus non-viral acute respiratory distress syndrome: comparison of upper airway molecular pathway and drug discovery design based on systems biology and deep learning methods. International Automatic Control Conference (CACS) 2021;2021:1–7. [Google Scholar]
- 89.Hu F., Wang L., Hu Y., Wang D., Wang W., Jiang J., et al. A novel framework integrating AI model and enzymological experiments promotes identification of SARS-CoV-2 3CL protease inhibitors and activity-based probe. Brief Bioinform. 2021;22(6) doi: 10.1093/bib/bbab301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zeng X., Song X., Ma T., Pan X., Zhou Y., Hou Y., et al. Repurpose open data to discover therapeutics for COVID-19 using deep learning. J Proteome Res. 2020;19(11):4624–4636. doi: 10.1021/acs.jproteome.0c00316. [DOI] [PubMed] [Google Scholar]
- 91.Yuvaraj N., Srihari K., Chandragandhi S., Raja R.A., Dhiman G., Kaur A. Analysis of protein-ligand interactions of SARS-CoV-2 against selective drug using deep neural networks. Big Data Min Anal. 2021;4(2):76–83. [Google Scholar]
- 92.Meng X.Y., Zhang H.X., Mezei M., Cui M. Molecular docking: a powerful approach for structure-based drug discovery. Curr Comput-aided Drug Des. 2011;7(2):146–157. doi: 10.2174/157340911795677602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Guedes I.A., Pereira F.S.S., Dardenne L.E. Empirical scoring functions for structure-based virtual screening: applications, critical aspects, and challenges. Front Pharmacol. 2018;9 doi: 10.3389/fphar.2018.01089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Thafar M., Raies A.B., Albaradei S., Essack M., Bajic V.B. Comparison study of computational prediction tools for drug-target binding affinities. Front Chem. 2019;7 doi: 10.3389/fchem.2019.00782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rudling A., Orro A., Carlsson J. Prediction of ordered water molecules in protein binding sites from molecular dynamics simulations: the impact of ligand binding on hydration networks. J Chem Inf Model. 2018;58(2):350–361. doi: 10.1021/acs.jcim.7b00520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Parks C.D., Gaieb Z., Chiu M., Yang H., Shao C., Walters W.P., et al. D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies. J Comput-Aided Mol Des. 2020;34(2):99–119. doi: 10.1007/s10822-020-00289-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ton A.T., Gentile F., Hsing M., Ban F., Cherkasov A. Rapid identification of potential inhibitors of SARS-CoV-2 main protease by deep docking of 1.3 billion compounds. Mol Inform. 2020;39(8):e2000028. doi: 10.1002/minf.202000028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.S. Mylonas, A. Axenopoulos, S. Katsamakas, I. Gkekas, K. Stamatopoulos, S. Petrakis, P. Daras, Deep learning-assisted pipeline for virtual screening of ligand compound databases: application on inhibiting the entry of SARS-CoV-2 into human cells, 2020 IEEE 20th International Conference on Bioinformatics and Bioengineering (BIBE), Cincinnati, OH, USA., 2020, pp. 132–139.
- 99.Bai Q., Tan S., Xu T., Liu H., Huang J., Yao X. MolAICal: a soft tool for 3D drug design of protein targets by artificial intelligence and classical algorithm. Brief Bioinform. 2020;22(3) doi: 10.1093/bib/bbaa161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Anwaar M.U., Adnan F., Abro A., Khan R.A., Rehman A.U., Osama M., et al. Combined deep learning and molecular docking simulations approach identifies potentially effective FDA approved drugs for repurposing against SARS-CoV-2. Comput Biol Med. 2022;141 doi: 10.1016/j.compbiomed.2021.105049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.McNutt A.T., Francoeur P., Aggarwal R., Masuda T., Meli R., Ragoza M., et al. GNINA 1.0: molecular docking with deep learning. J Cheminform. 2021;13(1) doi: 10.1186/s13321-021-00522-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hollingsworth S.A., Dror R.O. Molecular dynamics simulation for all. Neuron. 2018;99(6):1129–1143. doi: 10.1016/j.neuron.2018.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.J. Meller, Molecular dynamics, In eLS, (Ed.), (2001), 10.1038/npg.els.0003048.
- 104.Rath S.L., Padhi A.K., Mandal N. Scanning the RBD-ACE2 molecular interactions in Omicron variant. Biochem Biophys Res Commun. 2022;592:18–23. doi: 10.1016/j.bbrc.2022.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Pantsar T., Poso A. Binding affinity via docking: fact and fiction. Molecules. 2018;23(8) doi: 10.3390/molecules23081899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Rebeaud M.E., Zores F. SARS-CoV-2 and the use of chloroquine as an antiviral treatment. Front Med. 2020;7 doi: 10.3389/fmed.2020.00184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Liu J., Cao R., Xu M., Wang X., Zhang H., Hu H., et al. Hydroxychloroquine, a less toxic derivative of chloroquine, is effective in inhibiting SARS-CoV-2 infection in vitro. Cell Discov. 2020;6(1) doi: 10.1038/s41421-020-0156-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.A. Sethi, K. Joshi, K. Sasikala, M. Alvala, Molecular docking in modern drug discovery: principles and recent applications, in: V. Gaitonde, P. Karmakar, A. Trivedi (Eds.), Drug Discovery and Development-New Advances, IntechOpen, 2020, 10.5772/intechopen.85991.
- 109.Reymond J.-L. The chemical space project. Acc Chem Res. 2015;48(3):722–730. doi: 10.1021/ar500432k. [DOI] [PubMed] [Google Scholar]
- 110.Vandyck K., Deval J. Considerations for the discovery and development of 3-chymotrypsin-like cysteine protease inhibitors targeting SARS-CoV-2 infection. Curr Opin Virol. 2021;49:36–40. doi: 10.1016/j.coviro.2021.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Joshi T., Sharma P., Mathpal S., Joshi T., Maiti P., Nand M., et al. Computational investigation of drug bank compounds against 3C-like protease (3CL(pro)) of SARS-CoV-2 using deep learning and molecular dynamics simulation. Mol Divers. 2021:1–14. doi: 10.1007/s11030-021-10330-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Arshia A.H., Shadravan S., Solhjoo A., Sakhteman A., Sami A. De novo design of novel protease inhibitor candidates in the treatment of SARS-CoV-2 using deep learning, docking, and molecular dynamic simulations. Comput Biol Med. 2021;139 doi: 10.1016/j.compbiomed.2021.104967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zhang H., Liao L., Cai Y., Hu Y., Wang H. IVS2vec: a tool of inverse virtual screening based on word2vec and deep learning techniques. Methods. 2019;166:57–65. doi: 10.1016/j.ymeth.2019.03.012. [DOI] [PubMed] [Google Scholar]
- 114.Zhang H, Zhang T, Saravanan KM, Liao L, Wu H, Zhang H, Zhang H, Pan Y, Wu X, Wei Y. A novel virtual drug screening pipeline with deep-leaning as core component identifies inhibitor of pancreatic alpha-amylase, 2021 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Houston, TX, USA., 2021, pp. 104–111.
- 115.Trott O., Olson A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading. J Comput Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kirchdoerfer R.N., Ward A.B. Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors. Nat Commun. 2019;10(1) doi: 10.1038/s41467-019-10280-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wu C., Liu Y., Yang Y., Zhang P., Zhong W., Wang Y., et al. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm Sin B. 2020;10(5):766–788. doi: 10.1016/j.apsb.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Gao Y., Yan L., Huang Y., Liu F., Zhao Y., Cao L., et al. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science. 2020;368(6492):779–782. doi: 10.1126/science.abb7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zhang H., Yang Y., Li J., Wang M., Saravanan K.M., Wei J., et al. A novel virtual screening procedure identifies Pralatrexate as inhibitor of SARS-CoV-2 RdRp and it reduces viral replication in vitro. PLoS Comput Biol. 2021;16(12):e1008489. doi: 10.1371/journal.pcbi.1008489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Casalino L., Dommer A.C., Gaieb Z., Barros E.P., Sztain T., Ahn S.-H., et al. AI-driven multiscale simulations illuminate mechanisms of SARS-CoV-2 spike dynamics. Int J High Perform Comput Appl. 2021;35(5):432–451. doi: 10.1177/10943420211006452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lee H., Turilli M., Jha S., Bhowmik D., Ma H., Ramanathan A., et al. IEEE/ACM third workshop on deep learning on supercomputers (DLS) Denver, CO, USA. 2019;2019 doi: 10.1109/DLS49591.2019.00007. [DOI] [Google Scholar]
- 122.van der Kamp M.W., Shaw K.E., Woods C.J., Mulholland A.J. Biomolecular simulation and modelling: status, progress and prospects. J R Soc Interface. 2008;5(suppl_3):173–190. doi: 10.1098/rsif.2008.0105.focus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Gershenson A., Gosavi S., Faccioli P., Wintrode P.L. Successes and challenges in simulating the folding of large proteins. J Biol Chem. 2020;295(1):15–33. doi: 10.1074/jbc.REV119.006794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Freddolino P.L., Harrison C.B., Liu Y., Schulten K. Challenges in protein-folding simulations. Nat Phys. 2010;6(10):751–758. doi: 10.1038/nphys1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Mottaqi M.S., Mohammadipanah F., Sajedi H. Contribution of machine learning approaches in response to SARS-CoV-2 infection. Inform Med Unlocked. 2021;23 doi: 10.1016/j.imu.2021.100526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Keshavarzi Arshadi A, Webb J, Salem M, Cruz E, Calad-Thomson S, Ghadirian N, Collins J, Diez-Cecilia E, Kelly B, H. Goodarzi, J. S. Yuan, Artificial intelligence for COVID-19 drug discovery and vaccine development, Front Artif Intell, 3 (2020), 10.3389/frai.2020.00065. [DOI] [PMC free article] [PubMed]
- 127.Torrens-Fontanals M., Stepniewski T.M., Aranda-García D., Morales-Pastor A., Medel-Lacruz B., Selent J. How do molecular dynamics data complement static structural data of GPCRs. Int J Mol Sci. 2020;21(16) doi: 10.3390/ijms21165933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Lazim R., Suh D., Choi S. Advances in molecular dynamics simulations and enhanced sampling methods for the study of protein systems. Int J Mol Sci. 2020;21(17) doi: 10.3390/ijms21176339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Yu W., MacKerell A.D., Jr. Computer-aided drug design methods. Methods Mol Biol. 2017;1520:85–106. doi: 10.1007/978-1-4939-6634-9_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Schenone M., Dančík V., Wagner B.K., Clemons P.A. Target identification and mechanism of action in chemical biology and drug discovery. Nat Chem Biol. 2013;9(4):232–240. doi: 10.1038/nchembio.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.G. Arora, J. Joshi, R. S. Mandal, N. Shrivastava, R. Virmani, T. Sethi, Artificial intelligence in surveillance, diagnosis, drug discovery and vaccine development against COVID-19, Pathogens, 10 (8) (2021), 10.3390/pathogens10081048. [DOI] [PMC free article] [PubMed]
- 132.Reis G., dos Santos Moreira-Silva E.A., Silva D.C.M., Thabane L., Milagres A.C., Ferreira T.S., et al. Effect of early treatment with fluvoxamine on risk of emergency care and hospitalisation among patients with COVID-19: the TOGETHER randomised, platform clinical trial. Lancet Glob Health. 2022;10(1):e42–e51. doi: 10.1016/S2214-109X(21)00448-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Piccialli F., di Cola V.S., Giampaolo F., Cuomo S. The role of artificial intelligence in fighting the COVID-19 pandemic. Inf Syst Front. 2021;23(6):1467–1497. doi: 10.1007/s10796-021-10131-x. [DOI] [PMC free article] [PubMed] [Google Scholar]