Abstract
Our understanding of the importance of microbiomes on large aquatic animals—such as whales, sea turtles and manatees—has advanced considerably in recent years. The latest observations indicate that epibiotic diatom communities constitute diverse, polyphyletic, and compositionally stable assemblages that include both putatively obligate epizoic and generalist species. Here, we outline a successful approach to culture putatively obligate epizoic diatoms without their hosts. That some taxa can be cultured independently from their epizoic habitat raises several questions about the nature of the interaction between these animals and their epibionts. This insight allows us to propose further applications and research avenues in this growing area of study. Analyzing the DNA sequences of these cultured strains, we found that several unique diatom taxa have evolved independently to occupy epibiotic habitats. We created a library of reference sequence data for use in metabarcoding surveys of sea turtle and manatee microbiomes that will further facilitate the use of environmental DNA for studying host specificity in epizoic diatoms and the utility of diatoms as indicators of host ecology and health. We encourage the interdisciplinary community working with marine megafauna to consider including diatom sampling and diatom analysis into their routine practices.
Subject terms: Speciation, Biodiversity, Microbial ecology
Introduction
Common health indicators currently used to monitor cetaceans, sirenians and sea turtles include mortality rates, demographics, disease prevalence and frequency of stranding events. Since animal-associated microbiota may affect and be affected by their host, both internal and external microbiome composition at any given time could also reflect mid- and longer-term effects of disturbances or stressors experienced by the animal1. New health and fitness indices based on compositional changes in the native microbiomes could be a valuable addition to comprehensive health assessments for aquatic vertebrates2.
Studies on the external microbiome of large aquatic vertebrates have typically focused on the bacterial and/or viral components. In contrast, epizoic microeukaryotes remain poorly explored despite the observation of diatoms on whales over a century ago3,4. Diatoms (Bacillariophyta) are a diverse group of largely photosynthetic microalgae characterized by their uniquely shaped siliceous thecae (frustules) and are commonly found in the plankton and benthos of many different aquatic habitats. Recent studies have expanded the known diversity of epizoic diatoms through increased sampling of hosts to include sea turtles5–22, sea snakes23 and manatees24,25.
Competition for limited resources among diatoms has led to niche partitioning and significant habitat specificity in some taxa. The epizoic diatom communities growing on aquatic vertebrates appear to be formed by a combination of opportunistic surface-attached taxa and putatively obligate epizoic (POE) taxa. While the opportunistic taxa are shared across the benthic habitats of the local environment, the POE taxa thus far have only been observed in the epizoic microbiome7,21,26,27. This mixture of opportunistic and POE taxa is an intriguing assemblage, as it is potentially influenced by the host’s biology (e.g. physiology, anatomy and host-specific prokaryotic microbiome) and behavior (e.g. long-distance migrations, diving, basking, and terrestrial nesting which expose epibionts to extremes in temperature, pressure, irradiance, nutrient concentration and desiccation) as well as the environment (e.g. mean temperature, salinity, nutrient load, local biocenoses). Moreover, the unique and highly specific diatom flora composition can be documented long past the death of the diatom cells by the weathering-resistant inorganic frustules. This has resulted in diatoms being utilized extensively for paleoecological reconstructions and bioindication in freshwater environments; for multiple reviews, see28. Similar diatom-based health indices may be developed for the marine animals and their habitats.
However, before this can happen, at least two issues must be addressed:
1) We must expand upon our knowledge of the specific molecular, genomic and ecological nature of the interactions between POE diatoms and their host and environment.
2) We need to simplify the identification of epizoic diatoms, which currently requires specialized equipment (such as electron microscopy) and literature that can be highly fragmented and incomplete, particularly in the case of marine diatoms.
Both of these issues could be addressed by metagenomic and metabarcoding techniques, respectively. Currently, however, the dearth of reference data—both in annotated genome and transcriptomes as well as vouchered DNA barcodes for diatoms—would limit the effectiveness of either effort. For example, a metabarcoding attempt on sea turtle epiflora29 failed to recover some of the diatom taxa identified in microscopical surveys, including the dominant POE taxon Labellicula lecohuiana Majewska, De Stefano & Van de Vijver. The authors acknowledged that this failure was likely due to the lack of any relevant reference sequences for the genus Labellicula. Further, the position of Labellicula in the molecular phylogeny of diatoms is unknown. This uncertainty significantly hinders any bioinformatic efforts to find sequence data even closely related to Labellicula among both the metabarcoding reads and the reference databases. Many other POE taxa have uncertain phylogenetic affinities within the raphid diatoms, including Tursiocola Holmes, Nagasawa & Takano, Epiphalaina Holmes, Nagasawa & Takano and the “Tripterion complex”. This latter assemblage of diatom genera (Tripterion Holmes, Nagasawa & Takano, Chelonicola Majewska, De Stefano & Van de Vijver, Poulinea Majewska, De Stefano & Van de Vijver and Medlinella Frankovich, Ashworth & M.J.Sullivan) is of particular taxonomic interest as they represent a radiation of exclusively epizoic diatom taxa. Their current taxonomy is not universally accepted15, and distinguishing the genera can be difficult without the use of electron microscopy due to a similar overall frustule morphology (heteropolar, stalked and septate or pseudoseptate) and relatively small size (< 20 um).
To address the aforementioned issues, we have cultured and sequenced DNA data from POE diatom taxa. These were isolated from sea turtles and manatees from the wild, rehabilitation and rescue centers as well as aquaria from the United States of America, The Bahamas, Croatia, Italy and South Africa. While DNA sequence data from vouchered specimens alone would be useful for molecular identification, the ability to maintain these diatoms away from their hosts facilitates the formulation of hypotheses and laboratory experiments to test the molecular nature of the relationship between the diatom and host.
Results
Culture success
We successfully cultured > 600 strains, both POE and opportunistic diatoms on the epizoic habitat. This manuscript focuses on 76 of these sequenced strains (Table 1) and the sequences from the single-cell DNA extractions of the non-photosynthetic Tursiocola spp. (Figs. 1, 2). Sequence data from 21 additional diatoms are included (Figs. S1, S2). While these additional sequenced diatom taxa were isolated from epizoic collections, they are known opportunistic taxa, occur in non-epizoic habitats, or their habitat preferences are unclear.
Table 1.
POE diatoms cultured in this study, sorted by host species.
| Host species | Location | Host status | POE Diatoms cultured (# of strains) [total cultures] |
|---|---|---|---|
| Chelonia mydas (Green Sea Turtle) | Bahamas | Wild animal: “turtle1” | Cca (2), Td (2), Pv (1) 9 |
| Chelonia mydas (Green Sea Turtle) | Durban, South Africa | Aquarium resident: “Calypso” | Cco (1), Cm (3), Pl (2) 14 |
| Chelonia mydas (Green Sea Turtle) | Durban, South Africa | Aquarium resident: “Wasabi” | Ma (1), Pl (4) 12 |
| Chelonia mydas (Green Sea Turtle) | Florida, USA | Wild animal: “FL noname” | Ae (2), Tg (1) 6 |
| Chelonia mydas (Green Sea Turtle) | Florida, USA | Rehabilitation animal: “Fleming” | Ae (5), Pl (3), Pv (3) 22 |
| Eretmochelys imbricata (Hawksbill Sea Turtle) | Texas, USA | Aquarium resident: “Einstein” | Ca (2) 4 |
| Eretmochelys imbricata (Hawksbill Sea Turtle) | Durban, South Africa | Aquarium resident: “Tripod” | Ma (3) 11 |
| Lepidochelys kempii (Kemp's Ridley Sea Turtle) | Georgia, USA | Wild animal: “Z6” | Ae (3) 12 |
| Dermochelys coriacea (Leatherback Sea Turtle) | Kosi Bay, South Africa | Wild animal: “ZA0019A/ZA1824E” | Cd (1) 7 |
| Caretta caretta (Loggerhead Sea Turtle) | Durban, South Africa | Aquarium resident: “Shiv” | Pl (3) 6 |
| Caretta caretta (Loggerhead Sea Turtle) | Kosi Bay, South Africa | Wild animal: “ZA00940/ZA10860” | Ma (1) 4 |
| Caretta caretta (Loggerhead Sea Turtle) | Kosi Bay, South Africa | Wild animal: “ZA1595E/ZA1826E” | Pl (1) 10 |
| Caretta caretta (Loggerhead Sea Turtle) | Kosi Bay, South Africa | Wild animal | Csp (2) 10 |
| Caretta caretta (Loggerhead Sea Turtle) | Florida, USA | Wild animal: “A2” | Ae (7) 8 |
| Caretta caretta (Loggerhead Sea Turtle) | Florida, USA | Wild animal: “CC032217a” | Cca (2), Pv (2) 19 |
| Caretta caretta (Loggerhead Sea Turtle) | Florida, USA | Wild animal: “FL Christine” | Cca (2) 6 |
| Caretta caretta (Loggerhead Sea Turtle) | Brijuni Islands, Croatia | Aquarium resident: “Lunga” | Ps (1) 3 |
| Caretta caretta (Loggerhead Sea Turtle) | Bisceglie, Italy | Rehabilitation animal: “Iracus” | Pl (3) 38 |
| Lepidochelys olivacea (Olive Ridley Sea Turtle) | Long Beach, California | Aquarium resident: “LoMain” | Pl (1) 14 |
| Lepidochelys olivacea (Olive Ridley Sea Turtle) | Florida, USA | Rehabilitation animal: “Harry” | Ae (2), Pl (3) 10 |
| Trichechus manatus latirostris (West Indian Manatee) | Florida, USA | Wild animal: “FLMan40” | Ae (2) 11 |
| Trichechus manatus latirostris (West Indian Manatee) | Georgia, USA | Wild animal: “CGA1605” | Ae (1) 23 |
POE diatoms are abbreviated and followed by the number of strains cultured from the indicated host: Ae = Achnanthes elongata, Ca = Craspedostauros alatus, Cd = Craspedostauros danayanus, Cm = Craspedostauros macewanii, Cca = Chelonicola caribeana, Cco = Chelonicola costaricensis, Csp = Chelonicola sp., Ma = Medlinella amphoroidea, Pl = Poulinea lepidochelicola, Ps = Proschkinia sulcata, Pv = Proschkinia vergostriata, Td = Tursiocola denysii, Tg = Tursiocola guyanensis.
Figure 1.
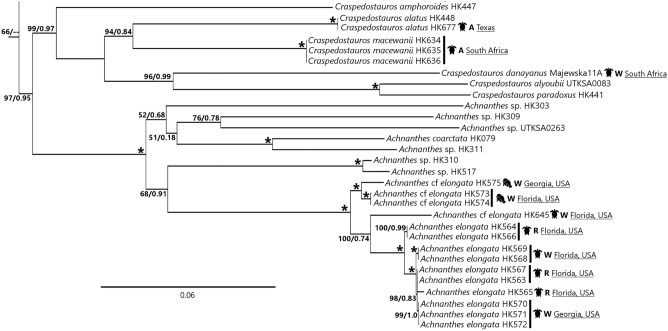
Maximum likelihood phylogenetic tree derived from a concatenated 3-gene DNA sequence dataset, representing the Achnanthes, Craspedostauros and Staurotropis clades (complete tree shown in Fig. S1). Support values (ML bootstrap support/BI posterior probability) shown above nodes; “*” = nodes with 100%/1.0 values. Taxon name followed by DNA extraction voucher number or strain ID. Taxa isolated from epizoic habitats followed by a diagrammatic representation of the host from which the strain was isolated, and metadata on the location and setting in which the host was sampled (A = aquarium, R = rehabilitation facility, W = wild).
Figure 2.
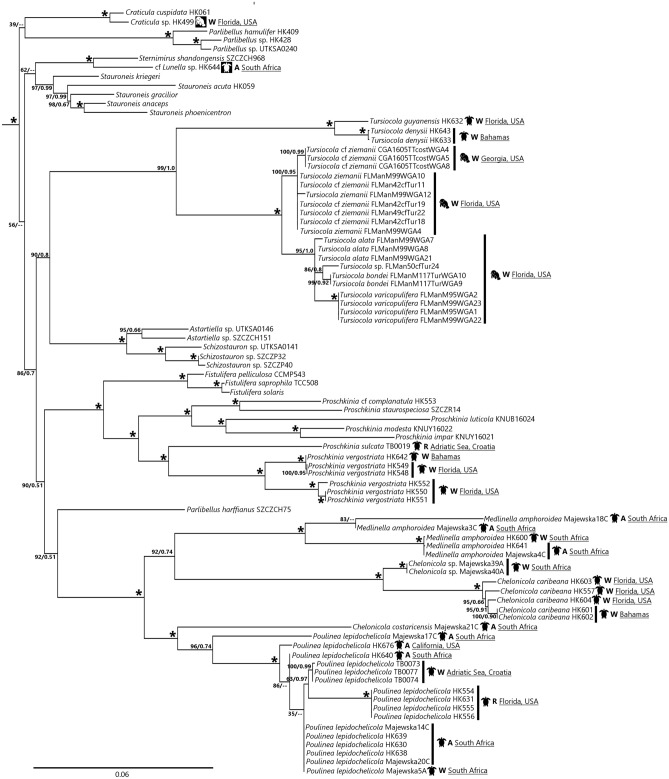
Maximum likelihood phylogenetic tree derived from a concatenated 3-gene DNA sequence dataset, representing the clade containing the Tripterion complex, Tursiocola and Proschkinia clades (complete tree shown in Fig. S1). Support values (ML bootstrap support/BI posterior probability) shown above nodes; “*” = nodes with 100%/1.0 values. Taxon name followed by DNA extraction voucher number or strain ID. Taxa isolated from epizoic habitats followed by a diagrammatic representation of the host from which the strain was isolated, and metadata on the location and setting in which the host was sampled (A = aquarium, R = rehabilitation facility, W = wild). Black host icon = POE taxon; white host icon = unclear habitat preference.
Target POE taxa
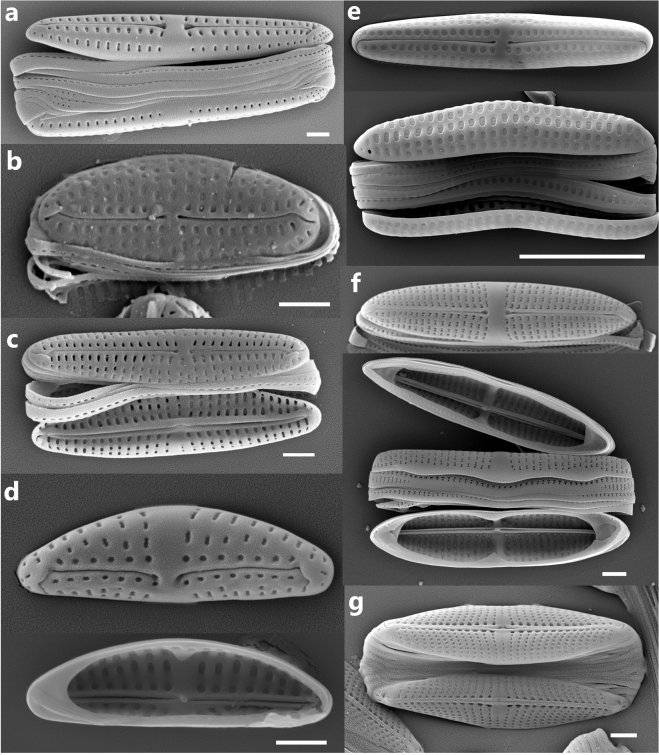
POE taxa were identified based on the available literature and included diatom species that have only ever been observed in association with the epizoic habit being found on multiple animal specimens6,8,10,11,14–16,24,25,30. Among these were epizoic taxa typically reaching high relative abundances (> 25%)—Achnanthes elongata Majewska & Van de Vijver, Chelonicola costaricensis Majewska, De Stefano & Van de Vijver, C. caribeana Riaux-Gobin, Witkowski, Ector & Chevallier, Craspedostauros danayanus Majewska & Ashworth, Medlinella amphoroidea Frankovich, Ashworth & M.J.Sullivan, Poulinea lepidochelicola Majewska, De Stefano & Van de Vijver, Tursiocola spp., as well as species often present on animals but never exceeding 10% of the diatom relative abundance—Craspedostauros alatus Majewska & Ashworth, C. macewanii Majewska & Ashworth, Proschkinia sulcata Majewska, Van de Vijver & Bosak and P. vergostriata Frankovich, Ashworth & M.J Sullivan. SEM images of some of these taxa sampled for DNA can be found in Fig. 3. This list of POE taxa is not exhaustive as the full diversity of POE diatoms remains to be documented. Moreover, it does not include several probable POE species (e.g. Achnanthes squaliformis Majewska & Van de Vijver, Navicula dermochelycola Riaux-Gobin, Witkowski, Kociolek & Chevallier), which have not yet been isolated and cultured.
Figure 3.
Scanning electron micrographs of some of the POE diatom taxa successfully cultured and sampled for DNA. a = Poulinea lepidochelicola HK630, complete frustule. b = Chelonicola cf costaricensis Majewska 21C, valve exterior. c = Chelonicola sp. Majewska 40A, complete frustule. d = Medlinella amphoroidea HK600 (valve exterior above, interior below). e = Achnanthes elongata HK563 (valve exterior above, complete frustule below). f = Tursiocola denysii HK633 (valve exterior above, complete frustule below). g = Proschkinia vergostriata HK552, complete frustule. All scale bars = 1 µm.
Molecular phylogenetic results
The currently recognized POE strains were predominantly located in two clades in the molecular phylogeny—Achnanthes sensu stricto + Craspedostauros (Fig. 1) and the clade containing the Tripterion complex, Tursiocola and Proschkinia (Fig. 2). With regards to Achnanthes, most of the sampled diversity comes from three species of sea turtles (green, Kemp’s ridley and loggerhead) and West Indian manatees sampled in the southeastern US. These strains formed a well-supported clade (ML bootstrap support [bs] = 100%, BI posterior probability [pp] = 1.0) sister to the rest of the sequenced Achnanthes spp. The POE Achnanthes clade also sorted by host, with strains collected from manatee (100%/1.0 bs/pp) and sea turtle (100%/0.74 bs/pp) hosts in their own clades. The POE Craspedostauros taxa showed a different pattern to the rest of the POE diatoms. Their clade included both POE and non-POE species, with POE taxon C. danayanus sister to C. alyoubii and C. paradoxus (96%/0.99 bs/pp) rather than to the POE C. macewanii and C. alatus.
The “Tripterion complex + ” clade (strains illustrated in Fig. 3a–d) was resolved with strong support (100%/1.0 bs/pp). While we were able to sample taxa from the Chelonicola, Poulinea and Medlinella genera in this complex, we were unable to observe any taxa within Tripterion sensu stricto in our collections. The “Tripterion complex + ” clade also contained the POE genus Tursiocola and Proschkinia Karayeva, which has both POE and non-POE species, as well as the non-epizoic genera Stauroneis Ehrenberg, Craticula Grunow, Parlibellus E.J.Cox, Fistulifera Lange-Bertalot and some monoraphid genera such as Schizostauron Grunow and Astartiella Witkowski, Lange-Bertalot & Metzeltin. The molecular data suggested no common origin for the POE clades; Tursiocola and the Tripterion complex are sister to non-POE taxa rather than each other, and the POE Proschkinia (P. vergostriata and P. sulcata) formed a clade sister (100%/1.0 bs/pp) to the rest of the Proschkinia spp.
Within Tursiocola, both nutritional types appear monophyletic, with the non-photosynthetic manatee-associated taxa (T. alata, T. bondei, T. varicopulifera and T. ziemanii) and the photosynthetic sea turtle-associated taxa (T. denysii and T. guyanensis) in their own clades (100%/1.0 bs/pp for both clades). It should be noted, however, that there were only two photosynthetic Tursiocola taxa sampled. Tree topology in the Tripterion complex remained the same regardless of analysis, with Chelonicola costaricensis “Majewska21C” + Poulinea lepidochelicola (100%/1.0 bs/pp) sister to Medlinella amphoroidea + Chelonicola sp. “Majewska39A/40A” + C. caribeana (92%/0.74 bs/pp).
Only two clades in the Tripterion complex had any geographic variation: the Poulinea clade and Chelonicola caribeana clade. For Poulinea, strains collected in South Africa were not monophyletic, with “Majewska 17C” sister to the rest of the clade, which included strains isolated from the Adriatic, Florida, California and South Africa. It should be noted that the Florida clade represented strains collected from a single location—a rehabilitation facility—while the South African strains were isolated from collections of both wild and captive host animals. The C. caribeana clade, on the other hand, contained strains isolated exclusively from wild host animals in South Africa, Florida and the Bahamas, with the South African strains (“Majewska39A/40A”) sister to the rest.
Discussion
Based on our molecular phylogeny, it appears that the epizoic habit has evolved several times and in several different raphid diatom morphotypes: elongate biraphid (Tursiocola and Proschkinia , Fig. 3f,g, respectively) and monoraphid frustules (Achnanthes, Fig. 3e), asymmetric, clavate biraphid frustules (Tripterion complex, Fig. 3a) and thin oval monoraphid frustules (Bennettella, Epipellis31). These independent gains of the epizoic habit could be driven by the host biology and evolution. The various epizoic diatom lineages, if eventually resolved to be closely linked to a specific type of host animal, might have diverged from non-epizoic taxa under different ecological and evolutionary constraints and at different times corresponding to the emergence of various groups of marine megafauna.
Among others, the eco-physiological constraints shaping epizoic diatom speciation through adaptive radiation would include the nature and character of the animal substrate. Variations of the dermal layer of sirenians and sea turtles including the ultrastructure, topology, physiology (e.g. shedding patterns), and biochemistry (e.g. enzymatic activity) would require different attachment and colonization (and re-colonization) strategies, thus encouraging the development of specific adaptations. Such a specific adaptation is evidenced by Melanothamnus maniticola Woodworth, Frankovich & Freshwater, an epizoic red alga on manatees that has unique skin penetrating rhizoids that anchor the thallus to the deeper epidermis and permit the alga to persist as the host surface skin cells are shed32. In marine reptiles, the carapace scutes are often shed periodically, while the skin scales are either shed continuously (sea turtles) or the epidermis is renewed completely in a process called ecdysis (sea snakes33). These patterns differ from those observed in marine mammals in which skin shedding may be regulated by external factors such as temperature34. Similarly, animals with different diving regimes may host diatoms with different physiological and metabolic adaptations as various stages of photosynthesis will be differently affected by changes in hydrostatic pressure related to the depth, duration, and frequency of dives35.
Moreover, the diversification dynamics in POE diatoms may be linked to the host animal behavior and lifestyle. The niche heterogeneity, biodiversity, productivity, and nutrient concentrations typical of shallow-water habitats occupied by sirenians and some sea turtles may increase colonization rates by new species and favor benthic diatom immigration to the epizoic community, thus spurring the observed diversity of diatom forms associated with manatees24,25 or sea turtles using neritic foraging habitats (e.g. loggerheads;21). The opposite phenomenon could explain low epizoic diatom diversity on leatherback sea turtles5,30, and pelagic sea snakes23 that spend significant time feeding in the pelagic zone rather than on benthic organisms36. This follows the general pattern of low macro-epibiotic diversity on leatherbacks37. Epizoic diatom diversity might also be driven by intrinsic biotic factors, such as gregariousness and range of the host species as both factors may affect the new species encounter and colonization rates. However, in these systems in which epizoic diatom species richness is driven mainly by speciation rates as opposed to benthic species immigration, the total epizoic diatom diversity may remain low. The higher number of diatom taxa observed on neritic megafauna species as compared to open-water animals seem to support this hypothesis20.
Currently, taxon sampling is still scattered, and while strains were isolated from multiple geographic localities, much of the strain diversity in species-level clades come from a single collection. The Florida Poulinea lepidochelicola clade, for example, represents strains isolated exclusively from the Turtle Hospital rehabilitation facility in Marathon, Florida. Among the South African P. lepidochelicola strains, six strains (Majewksa 14C, Majewska 20C, HK630, HK638, HK639 and HK640) came from collections from three turtles at the uShaka Sea World facility in Durban, and likely represent one population. However, a morphological difference does exist between the sequenced Medlinella amphoroidea strains from South Africa and the type population of Florida Bay. The valve areolae of the former appear to be occluded by hymenes (Fig. 3d) as opposed to the volae of the type population14. Whether this corresponds to a genetic, and perhaps species differentiation remains to be seen, once the Florida Bay population is sequenced.
While we do not yet have enough information to assign any sort of host specificity to certain POE diatom taxa, we have enough DNA sequence data to suggest that some genetic differentiation among POE diatoms is occurring. While we do not know if the genetic distance between the Florida, Mediterranean and South African Poulinea strains is driven by speciation or intraspecific biogeography, they are genetically distinct. Data collected from loggerheads suggests little mixing between sea turtle individuals across ocean basins38, with the Mediterranean population being distinct from the northeast Atlantic one, which is then distinct from northwest Atlantic (including the Gulf of Mexico) population. Even within closer geographic boundaries, such as the western Atlantic, there is demonstrated genetic distance between POE strains (C. caribeana of Florida and the Bahamas; Achnanthes elongata of Florida and Georgia) in DNA sequence markers which are generally considered too conserved to show intraspecific variation in diatoms39,40.
The collection of molecular information from a larger number of POE diatom strains may reveal whether genetic diversity in epizoic diatoms reflects biogeographic, ecological, and behavioral patterns observed in the host animal populations. For example, it was demonstrated that sea turtle phylogeography is shaped by the sea turtle species thermal regime and habitat preference41. Provided the close relationship between epizoic diatoms and sea turtles holds up under the scrutiny of increased data sampling, it may be expected that POE diatoms associated with the cold-tolerant leatherbacks, which are able to use the southwestern corridors to migrate across the oceans, will be characterized by lower genetic diversity than diatom taxa growing on tropical species such as green turtles, hawksbills, and olive ridley sea turtles, whose Atlantic and Indo-Pacific populations appear to be genetically distinct42. This knowledge may significantly advance our understanding about evolutionary relationships between diatoms and their animal hosts as well as shed more light on the mechanistic processes of divergence and adaptive evolution of diatoms and other marine microbes.
This study lays the groundwork for biodiversity and biogeographical work in marine epibioses by starting the development of a database of DNA sequence data from 16 of the known POE diatom species for sea turtles and manatees. These sequences will also be useful in not only identifying more POE taxa, but searching for potential refugia of these taxa in non-epizoic habitats. Large areas of the world’s marine shallow benthic environment are poorly studied for diatoms, and therefore we cannot exclude the possibility that the POE taxa do exist outside of epizoic habitats. Even in localities that are relatively well-studied for benthic diatoms, variation in the composition and relative abundance in an assemblage due to substrate specificity and seasonality make the assembly of an exhaustive diatom flora extremely difficult. Environmental DNA surveys, such as metabarcoding, have an advantage over microscope-based surveys with regards to relatively small-sized taxa. Based on the molecular phylogeny of the Tripterion complex, it is easy to see how these taxa might have remained undetected in a bioinformatic summary of OTUs by sequence similarity, as there is significant genetic difference between the Tripterion complex and the only other sequenced representatives of the Rhoicospheniaceae—the freshwater taxon Rhoicosphenia abbreviata (C.Agardh) Lange-Bertalot. In fact, there are no morphological characters exclusive to the taxa in the molecular clade containing Tursiocola and the Tripterion complex that would cause a diatomist to expect a close match in sequence identity to the POE taxa. With curated sequence data now available for the most common POE taxa, we may find evidence for their occurrence in non-epizoic habitats through eDNA studies.
One of the stated goals of this study was to generate additional DNA sequence data from POE diatom taxa on sea turtles and sirenians. This goal was greatly aided by our ability to culture many of these POE diatoms away from their hosts, which raises several questions about the ecological requirements and adaptations of epizoic diatoms. The isolated strains of POE diatoms, which can be maintained in artificial conditions and without the animal hosts, provide opportunities to further study the molecular, genomic and physiological nature of the unique relationship between the diatoms and marine megafauna in a laboratory setting. For example, we can examine how different species may be affected by different conditions or possess specific adaptations to epizoic lifestyle. It is possible that some trade-off in obtaining those adaptations makes the POE taxa less competitive in non-epizoic benthic environments. We know little about the extent to which the microbes associated with the diatom (“phycosphere”) might affect the competitive ability of diatoms, and/or whether the phycosphere may itself manufacture some critical compound only in an epizoic community. Since all cultured POE diatoms were maintained as non-axenic cultures, it is yet unclear what role the bacterial strains played in the development and survival of the targeted diatom species and whether the long-term maintenance of axenic POE strains would be feasible. Future studies may also determine the number of evolutionary leaps to the epizoic habitat and the number of host switches, shedding more light on the co-evolution of diatom-animal relationships.
Methods
Cultures and microscopy
Diatoms were collected from the skin of West Indian manatees and the skin and carapace of six species of sea turtles (see Table 1 for details). These collections were made following the protocol outlined by Pinou et al.43. Wild sea turtles were either sampled on nesting beaches after oviposition (as to not disturb the nesting process) or from turtles captured in water via a rodeo method44. The seven sea turtles resident at the uShaka Sea World in Durban (South Africa) were sampled during feeding. The Adriatic Sea turtles were sampled upon arrival to the rescue center after being caught accidentally during trawling (Iracus) or during rehabilitation in an outdoor pool with freely circulating seawater (Lunga). Manatees were sampled during annual health assessments conducted by the USGS Sirenia Project.
Individual diatom cells were isolated by micropipette into sterile f/2 culture medium45 with a salinity matching that of the collection area. Strains isolated from the Bahamas, and the US were maintained under natural light in a north-facing window at UT Austin at room temperature (between 20 and 24 °C). South African strains were lit by natural light from a south-facing window and maintained at a temperature of 20–24 °C at the Unit of Environmental Sciences and Management in Potchefstroom. The strains isolated from the Adriatic were grown at 18–20 °C at 7–10 μmol m2 s−1, 12:12 (light:dark) cycle .In the case of non-photosynthetic taxa (like some Tursiocola species), individual cells were documented by light micrograph (“photovouchered”) and isolated into WGA whole-genome amplification cocktail25.
Cultures were harvested into separate pellets for microscopy preparation and DNA sequencing. Pellets for microscopy were cleaned with hydrogen peroxide and nitric acid, rinsed to neutral pH and dried onto 22 × 22 mm and 12 mm coverslips for light microscopy (LM) and scanning electron microscopy (SEM), respectively. Permanent mounts for the LM slides were made with Naphrax® mounting medium (Brunel Microscopes, www.brunelmicroscopessecure.co.uk) and micrographs were taken with a Zeiss Axioskop. Coverslips for SEM were coated with iridium by a Cressington 208 Bench Top Sputter Coater (Cressington Scientific Instruments, Watford, UK) and micrographs taken with a Zeiss SUPRA 40 VP scanning electron microscope (Carl Zeiss Microscopy, Thornwood, NY, USA). Additional micrographs of the strains are available from the authors.
DNA isolation, amplification and sequencing
Pellets for DNA sequencing were extracted using the DNeasy Plant Minikit, with an extra 45 s incubation in a Beadbeater (Biospec Products, Bartlesville, OK, USA) with 1.0 mm glass pellets for colony and frustule disruption. The nuclear-encoded ribosomal SSU and chloroplast-encoded rbcL and psbC markers were amplified by PCR using the primers outlined in Theriot et al.46 in 25 µL reactions with 1–3 µL of template DNA, 0.5 µL of each primer, 0.25 µL of Taq polymerase, 12.5 µL of pre-mixed FailSafe Buffer E (Lucigen Corporation) and 8.25–10.25 µL of sterile water. PCR conditions were identical for rbcL and psbC: 94 °C for 3.5 min., 35 cycles of (94 °C for 30 s, 48 °C for 60 s., 72 °C for 2 min.), and a final extension at 72 °C for 15 min. PCR conditions for SSU were: 94 °C for 3.5 min., 35 cycles of (94 °C for 30 s., 51 °C for 60 s., 72 °C for 3 min.), and a final extension at 72 °C for 15 min. The amplicons were purified using an EXO-SAP protocol: a 3 µL of an EXO-SAP solution containing 0.5 µL of shrimp alkaline phosphatase, 0.25 µL of exonuclease I and 2.25 µL of sterile water were added to the PCR products and incubated at 37 °C for 30 min. followed by 80 °C for 15 min. Purified products were then sequenced on an ABI 3730 DNA Analyzers using BigDye Terminator v3.1 chemistry.
Sequence data were added to a dataset of raphid and araphid pennate diatoms, with Asterionellopsis glacialis used as an outgroup (see Table S1 for GenBank accession numbers). SSU data were aligned by the SSUalign program, using the covariance model outlined in Lobban et al.47. Data were initially partitioned by gene, by paired and unpaired sites in SSU secondary structure and codon position in rbcL and psbC. Model testing and grouping of partitions were performed by PartitionFinder 248 using all nucleotide substitution models, linked branches, and rcluster search49 settings for trees inferred by RAxML 850. The best model was chosen using the corrected Akaike information criterion (AICc). Maximum Likelihood and Bayesian Inference based phylogenies were inferred using IQ-TREE version 1.6.12 for Linux51 with partitioned models52 and multi-threaded MPI hybrid variant of ExaBayes version 1.553, respectively. Nodal support for the maximum likelihood phylogeny was assessed using 1000 bootstrap replicates via IQ-TREE. ExaBayes analyses included four independent runs with two coupled chains where branch lengths were linked. Convergence parameters included an average deviation of split frequencies (ASDSF) of less than or equal to 5% with a minimum of 10,000,000 generations. Bayesian nodal support was assessed using posterior probabilities, with the first 25% of the trees removed as “burn-in”.
Supplementary Information
Acknowledgements
Financial support for sequencing and SEM comes from the Jane and the Roland Blumberg Centennial Professorship in Molecular Evolution at UT Austin and the US Department of Defense (grant number W911NF-17-2-0091). Sampling in South Africa was done with partial financial support from The Systematics Association (UK) through the Systematics Research Fund Award granted to RM (2017 and 2020). Work in the Adriatic Sea was supported by Croatian Science Foundation, project UIP-05-2017-5635 (TurtleBIOME). KF has been fully supported by the “Young researchers' career development project – training of doctoral students” of the CSF funded by the EU from the European Social Fund. NJR was funded by the Spanish government (AEI) through the ‘Severo Ochoa Centre of Excellence’ accreditation (CEX2019-000928-S). The authors would like to thank Bob and Cathy Bonde, Martine DeWit and Jim Powell as well as the UF Aquatic Animal Health Program, Sea to Shore Alliance, and the entire manatee capture team. We also thank Richie Moretti and Bette Zirkelbach of the Turtle Hospital in Marathon, Florida for sampling access. We thank Jack Cuffley at the Cape Eleuthera Institute in The Bahamas for assistance with epizoic collection. We thank Sandy Trautwein and Janet Monday for collections at the Aquarium of the Pacific in Long Beach, California and Taylor Yaw and Catherina Razal for collections at the Texas State Aquarium in Corpus Christi, Texas. We thank Diane Z. M. Le Gouvello du Timat (Nelson Mandela University, Port Elizabeth, South Africa) and Tony McEwan, Leanna Botha, Simon Chater at the uShaka Sea World staff and members of the South African Association for Marine Biological Research (SAAMBR), Durban, South Africa for collection support and advice. We would also like to thank Melanie Parker of California Department of Fish and Game, for advice and support. Phylogenetic analyses were performed on the University of Alabama High Performance Computer Cluster (UAHPC). Many thanks are also due to Adriana Trotta, Marialaura Corrente and the staff from Sea Turtle Clinic (Bari, Italy), as well as Milena Mičić and Karin Gobić Medica along with the rest of staff from Marine Turtle Rescue Centre, Aquarium Pula (Croatia). This is contribution #1472 from the Coastlines and Oceans Division of the Institute of Environment at Florida International University.
Abbreviations
- POE
Putatively obligate epizoic
- SEM
Scanning electron microscope
- bs
Bootstrap support
- pp
Posterior probability
Author contributions
M.P.A. and R.M. contributed equally to the study design, culture isolation and manuscript writing and editing. M.P.A. also extracted and sequenced DNA and constructed the phylogenetic datasets, while R.M. collected samples in South Africa and the Bahamas and managed South African collections. T.F. contributed to managing collections in the US and study design and interpretation, as well as manuscript editing. M.S. contributed to data interpretation and manuscript editing. S.B. and K.F. contributed to collecting samples in Croatia, extracting and sequencing DNA as well as study design, interpretation and manuscript editing. B.V. contributed to study design, data interpretation and manuscript editing. M.A., J.S., N.I.S., J.R.P. and C.A.M. collected samples in the US and contributed to data interpretation and manuscript editing. R.N. obtained permits for sample collection in South Africa and contributed to manuscript editing. N.J.R. and M.G. obtained permits and coordinated sample collection in the Bahamas and contributed to data interpretation and manuscript editing. E.C.T. and D.W.L. contributed to the phylogenetic analysis of DNA data, data interpretation and manuscript editing. S.R.M. contributed to culture maintenance, DNA work in the US and manuscript editing.
Data availability
DNA sequence data generated for this study are published on the NCBI GenBank online sequence depository under the accession numbers listed in Table S1. Additional micrographs and cleaned voucher material from the sequenced cultures are available from lead author MPA.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Matt P. Ashworth and Roksana Majewska.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-022-19064-0.
References
- 1.Zaneveld JR, McMinds R, Thurber RV. Stress and stability: Applying the Anna Karenina principle to animal microbiomes. Nat. Microbiol. 2017;2:1–8. doi: 10.1038/nmicrobiol.2017.121. [DOI] [PubMed] [Google Scholar]
- 2.Trevelline BK, Fontaine SS, Hartup BK, Kohl KD. Conservation biology needs a microbial renaissance: A call for the consideration of host-associated microbiota in wildlife management practices. Proc. R. Soc. B. 2019;286:2018–2448. doi: 10.1098/rspb.2018.2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bennett AG. On the occurrence of diatoms on the skin of whales. Proc. R. Soc. Lond. B. 1920;91:352–357. doi: 10.1098/rspb.1920.0021. [DOI] [Google Scholar]
- 4.Denys L. Morphology and taxonomy of epizoic diatoms (Epiphalaina and Tursiocola) on a sperm whale (Physeter macrocephalus) stranded on the coast of Belgium. Diatom. Res. 1997;12:1–18. doi: 10.1080/0269249X.1997.9705398. [DOI] [Google Scholar]
- 5.Majewska R. Tursiocola neliana sp. nov (Bacillariophyceae) epizoic on South African leatherback sea turtles (Dermochelys coriacea) and new observations on the genus Tursiocola. Phytotaxa. 2020;453:1–15. doi: 10.11646/phytotaxa.453.1.1. [DOI] [Google Scholar]
- 6.Majewska R, Kociolek J, Thomas E, De Stefano M, Santoro M, Bolanos F, Van de Vijver B. Chelonicola and Poulinea, two new gomphonemoid genera living on marine turtles from Costa Rica. Phytotaxa. 2015;233:236–250. doi: 10.11646/phytotaxa.233.3.2. [DOI] [Google Scholar]
- 7.Majewska R, Van de Vijver B, Nasrolahi A, Ehsanpour M, Afkhami M, Bolaños F, Iamunno F, Santoro M, De Stefano M. Shared epizoic taxa and differences in diatom community structure between green turtles (Chelonia mydas) from distant habitats. Microb Ecol. 2017;74:969–978. doi: 10.1007/s00248-017-0987-x. [DOI] [PubMed] [Google Scholar]
- 8.Majewska R, De Stefano M, Ector L, Bolaños F, Frankovich TA, Sullivan MJ, Ashworth MP, Van de Vijver B. Two new epizoic Achnanthes species (Bacillariophyta) living on marine turtles from Costa Rica. Bot. Mar. 2017;60:303–318. doi: 10.1515/bot-2016-0114. [DOI] [Google Scholar]
- 9.Majewska R, De Stefano M, Van de Vijver B. Labellicula lecohuiana, a new epizoic diatom species living on green turtles in Costa Rica. Nova Hedwig Beih. 2018;146:23–31. doi: 10.1127/1438-9134/2017/023. [DOI] [Google Scholar]
- 10.Majewska R, Ashworth MP, Lazo-Wasem E, Robinson NJ, Rojas L, Van de Vijver B, Pinou T. Craspedostauros alatus sp. nov., a new diatom (Bacillariophyta) species found on museum sea turtle specimens. Diatom Res. 2018;33:229–240. doi: 10.1080/0269249X.2018.1491426. [DOI] [Google Scholar]
- 11.Majewska R, Bosak S, Frankovich TA, Ashworth MP, Sullivan MJ, Robinson NJ, Lazo-Wasem EA, Pinou T, Nel R, Manning SR, Van de Vijver B. Six new epibiotic Proschkinia (Bacillariophyta) species and new insights into the genus phylogeny. Eur. J. Phycol. 2019;54:609–631. doi: 10.1080/09670262.2019.1628307. [DOI] [Google Scholar]
- 12.Majewska R, Robert K, Van de Vijver B, Nel R. A new species of Lucanicum (Cyclophorales, Bacillariophyta) associated with loggerhead sea turtles from South Africa. Bot. Lett. 2020;167:7–14. doi: 10.1080/23818107.2019.1691648. [DOI] [Google Scholar]
- 13.Frankovich TA, Sullivan MJ, Stacy NI. Tursiocola denysii sp. Nov. (Bacillariophyta) from the neck skin of Loggerhead sea turtles (Caretta caretta) Phytotaxa. 2015;234:227–236. doi: 10.11646/phytotaxa.234.3.3. [DOI] [Google Scholar]
- 14.Frankovich TA, Ashworth MP, Sullivan MJ, Vesela J, Stacy NI. Medlinella amphoroidea gen. et sp. Nov. (Bacillariophyta) from the neck skin of Loggerhead sea turtles (Caretta caretta) Phytotaxa. 2016;272:101–114. doi: 10.11646/phytotaxa.272.2.1. [DOI] [Google Scholar]
- 15.Riaux-Gobin C, Witkowski A, Kociolek JP, Ector L, Chevallier D, Compère P. New epizoic diatom (Bacillariophyta) species from sea turtles in the Eastern Caribbean and South Pacific. Diatom Res. 2017;32:109–125. doi: 10.1080/0269249X.2017.1299042. [DOI] [Google Scholar]
- 16.Riaux-Gobin C, Witkowski A, Chevallier D, Daniszewska-Kowalczyk G. Two new Tursiocola species (Bacillariophyta) epizoic on green turtles (Chelonia mydas) in French Guiana and Eastern Caribbean. Fottea Olomouc. 2017;17:150–163. doi: 10.5507/fot.2017.007. [DOI] [Google Scholar]
- 17.Riaux-Gobin C, Witkowski A, Kociolek JP, Chevallier D. Navicula dermochelycola sp. Nov., presumably an exclusively epizoic diatom on sea turtles Dermochelys coriacea and Lepidochelys olivacea from French Guiana. Oceanol. Hydrobiol. Stud. 2020;49:132–139. doi: 10.1515/ohs-2020-0012. [DOI] [Google Scholar]
- 18.Robert K, Bosak S, Van de Vijver B. Catenula exigua sp. Nov., a new marine diatom (Bacillariophyta) species from the Adriatic Sea. Phytotaxa. 2019;414:113–118. doi: 10.11646/phytotaxa.414.2.3. [DOI] [Google Scholar]
- 19.Van de Vijver B, Bosak S. Planothidium kaetherobertianum, a new marine diatom (Bacillariophyta) species from the Adriatic Sea. Phytotaxa. 2019;425:105–112. doi: 10.11646/phytotaxa.425.2.5. [DOI] [Google Scholar]
- 20.Robinson NJ, Majewska R, Lazo-Wasem EA, Nel R, Paladino FV, Rojas L, Zardus JD, Pinou T. Epibiotic diatoms are universally present on all sea turtle species. PLoS ONE. 2016;11:e0157011. doi: 10.1371/journal.pone.0157011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Van de Vijver B, Robert K, Majewska R, Frankovich TA, Panagopolou A, Bosak S. Diversity of diatom communities (Bacillariophyta) associated with loggerhead sea turtles. PLoS ONE. 2020;15:e0236513. doi: 10.1371/journal.pone.0236513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van de Vijver B, Robert K, Witkowski A, Bosak S. Majewskaea gen. nov. (Bacillariophyta), a new marine benthic diatom genus from the Adriatic Sea. Fottea. 2020;20:112–120. doi: 10.5507/fot.2020.001. [DOI] [Google Scholar]
- 23.Majewska R. Nagumoea hydrophicola sp. Nov. (Bacillariophyta), the first diatom species described from sea snakes. Diatom Res. 2021;36:49–59. doi: 10.1080/0269249X.2020.1870159. [DOI] [Google Scholar]
- 24.Frankovich TA, Sullivan MJ, Stacey NI. Three new species of Tursiocola (Bacillariophyta) from the skin of the West Indian manatee (Trichechus manatus) Phytotaxa. 2015;204:33–48. doi: 10.11646/phytotaxa.204.1.3. [DOI] [Google Scholar]
- 25.Frankovich TA, Ashworth MP, Sullivan MJ, Theriot EC, Stacy NI. Epizoic and apochlorotic Tursiocola species (Bacillariophyta) from the skin of Florida manatees (Trichechus manatus latirostris) Protist. 2018;169:539–568. doi: 10.1016/j.protis.2018.04.002. [DOI] [PubMed] [Google Scholar]
- 26.Azari M, Farjad Y, Nasrolahi A, De Stefano M, Ehsanpour M, Dobrestov S, Majewska R. Diatoms on sea turtles and floating debris in the Persian Gulf (Western Asia) Phycologia. 2020;59:292–304. doi: 10.1080/00318884.2020.1752533. [DOI] [Google Scholar]
- 27.Majewska R, Goosen WE. For better, for worse: Manatee-associated Tursiocola (Bacillariophyta) remain faithful to their host. J. Phycol. 2020;56:1019–1027. doi: 10.1111/jpy.12993. [DOI] [PubMed] [Google Scholar]
- 28.Smol JP, Stoermer EF. The Diatoms: Applications for the Environmental and Earth Sciences. Cambridge University Press; 2010. [Google Scholar]
- 29.Rivera SF, Vasselon V, Ballorain K, Carpentier A, Wetzel CE, Ector L, Bouchez A, Rimet F. DNA metabarcoding and microscopic analyses of sea turtles biofilms: Complementary to understand turtle behavior. PLoS ONE. 2018;13:e0195770. doi: 10.1371/journal.pone.0195770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Majewska R, Ashworth MP, Bosak S, Goosen WE, Nolte C, Filek K, Van de Vijver B, Taylor JC, Manning SR, Nel R. On sea turtle-associated Craspedostauros with description of three novel species. J Phycol. 2021;57:199–208. doi: 10.1111/jpy.13086. [DOI] [PubMed] [Google Scholar]
- 31.Holmes RW. The morphology of diatoms epizoic on cetaceans and their transfer from Cocconeis to two new genera, Bennettella and Epipellis. Br. Phycol. J. 1985;20:43–57. doi: 10.1080/00071618500650061. [DOI] [Google Scholar]
- 32.Woodworth KA, Frankovich TA, Freshwater DW. Melanothamnus maniticola (Ceramiales, Rhodophyta): An epizoic species evolved for life on the West Indian Manatee. J. Phycol. 2019;55:1239–1245. doi: 10.1111/jpy.12912. [DOI] [PubMed] [Google Scholar]
- 33.Vitt LJ, Caldwell JP. Herpetology: An Introductory Biology of Amphibians and Reptiles. Academic Press; 2013. [Google Scholar]
- 34.Pitman LR, Durban JW, Joyce T, Fearnbach H, Panigada S, Lauriano G. Skin in the game: Epidermal molt as a driver of long-distance migration in whales. Mar. Mamm. Sci. 2020;36:565–594. doi: 10.1111/mms.12661. [DOI] [Google Scholar]
- 35.Pope DH, Berger LR. Algal photosynthesis at increased hydrostatic pressure and constant pO2. Arch. Microbiol. 1973;89:321–325. [Google Scholar]
- 36.Calcagno V, Jarne P, Loreau M, Mouquet N, David P. Diversity spurs diversification in ecological communities. Nat. Commun. 2017;8:15810. doi: 10.1038/ncomms15810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Robinson NJ, Pfaller JB. Sea turtle epibiosis: Global patterns and knowledge gaps. Trends Evol. Ecol. 2021;10:844021. [Google Scholar]
- 38.Conant, T. A., Dutton, P. H., Eguchi, T., Epperly, S. P., Fahy, C. C., Godfrey, M. H., MacPherson, S. L., Possardt, E. E., Schroeder, B. A., Seminoff, J. A., Snover, M. L. Loggerhead sea turtle (Caretta caretta) 2009 status review under the US Endangered Species Act. In Report of the loggerhead biological review Team to the National Marine Fisheries Service. 222, 1–230 (2009).
- 39.Evans KM, Wortley AH, Mann DG. An assessment of potential diatom ‘‘barcode’’ genes (cox1, rbcL, 18S and ITS rDNA) and their effectiveness in determining relationships in Sellaphora (Bacillariophyta) Protist. 2007;158:349–364. doi: 10.1016/j.protis.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 40.Hamsher SE, Evans KM, Mann DG, Poulíčková A, Saunders GW. Barcoding diatoms: Exploring alternatives to COI-5P. Protist. 2011;162:405–422. doi: 10.1016/j.protis.2010.09.005. [DOI] [PubMed] [Google Scholar]
- 41.Bowen BW, Karl SA. Population genetics and phylogeography of sea turtles. Mol Ecol. 2007;16:4886–4907. doi: 10.1111/j.1365-294X.2007.03542.x. [DOI] [PubMed] [Google Scholar]
- 42.Shanker K, Ramadevi J, Choudhury BC, Singh L, Aggarwal RK. Phylogeography of olive ridley turtles (Lepidochelys olivacea) on the east coast of India: implications for conservation theory. Mol. Ecol. 2004;13:1899–1909. doi: 10.1111/j.1365-294X.2004.02195.x. [DOI] [PubMed] [Google Scholar]
- 43.Pinou T, Domenech F, Lazo-Wasem EA, Majewska R, Pfaller JB, Zardus JD, Robinson NJ. Standardizing sea turtle epibiont sampling: Outcomes of the epibiont workshop at the 37th International Sea Turtle Symposium. Mar. Turt. Newsl. 2019;157:22–32. [Google Scholar]
- 44.Ehrhert L., Ogren L. H. Studies in foraging habitats: capturing and handling turtles. In Research and management techniques for the conservation of sea turtles (eds. Eckert, K. L., Bjorndal, K. A., Abreu-Grobois, F. A., Donnelly, M.). IUCN/SSC Marine Turtle Specialist Group. Publication No. 4. (1999).
- 45.Guillard, R. R. Culture of phytoplankton for feeding marine invertebrates. In Culture of Marine Invertebrate Animals 29–60 (Springer, 1975).
- 46.Theriot EC, Ashworth MP, Nakov T, Ruck E, Jansen RK. Dissecting signal and noise in diatom chloroplast protein encoding genes with phylogenetic information profiling. Mol. Phylogenet. Evol. 2015;89:28–36. doi: 10.1016/j.ympev.2015.03.012. [DOI] [PubMed] [Google Scholar]
- 47.Lobban CS, Ashworth MP, Calaor JJ, Theriot EC. Extreme diversity in fine-grained morphology reveals fourteen new species of conopeate Nitzschia (Bacillariophyta: Bacillariales) Phytotaxa. 2019;401:199–238. doi: 10.11646/phytotaxa.401.4.1. [DOI] [Google Scholar]
- 48.Lanfear R, Frandsen PB, Wright AM, Senfeld T, Calcott B. PartitionFinder 2: New methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol. Biol. Evol. 2017;34:772–773. doi: 10.1093/molbev/msw260. [DOI] [PubMed] [Google Scholar]
- 49.Lanfear R, Calcott B, Kainer D, Mayer C, Stamatakis A. Selecting optimal partitioning schemes for phylogenomic datasets. BMC Evol. Biol. 2014;14:1–14. doi: 10.1186/1471-2148-14-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stamatakis A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum likelihood phylogenies. Mol. Biol. Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chernomor O, Von Haeseler A, Minh BQ. Terrace aware data structure for phylogenomic inference from supermatrices. Syst. Biol. 2016;65:997–1008. doi: 10.1093/sysbio/syw037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Aberer AJ, Kobert K, Stamatakis A. ExaBayes: Massively parallel bayesian tree inference for the whole-genome Era. Mol. Biol. Evol. 2014;31:2553–2556. doi: 10.1093/molbev/msu236. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
DNA sequence data generated for this study are published on the NCBI GenBank online sequence depository under the accession numbers listed in Table S1. Additional micrographs and cleaned voucher material from the sequenced cultures are available from lead author MPA.