Abstract
Chitinase A1 from Bacillus circulans WL-12 comprises an N-terminal catalytic domain, two fibronectin type III-like domains, and a C-terminal chitin-binding domain (ChBD). In order to study the biochemical properties and structure of the ChBD, ChBDChiA1 was produced in Escherichia coli using a pET expression system and purified by chitin affinity column chromatography. Purified ChBDChiA1 specifically bound to various forms of insoluble chitin but not to other polysaccharides, including chitosan, cellulose, and starch. Interaction of soluble chitinous substrates with ChBDChiA1 was not detected by means of nuclear magnetic resonance and isothermal titration calorimetry. In addition, the presence of soluble substrates did not interfere with the binding of ChBDChiA1 to regenerated chitin. These observations suggest that ChBDChiA1 recognizes a structure which is present in insoluble or crystalline chitin but not in chito-oligosaccharides or in soluble derivatives of chitin. ChBDChiA1 exhibited binding activity over a wide range of pHs, and the binding activity was enhanced at pHs near its pI and by the presence of NaCl, suggesting that the binding of ChBDChiA1 is mediated mainly by hydrophobic interactions. Hydrolysis of β-chitin microcrystals by intact chitinase A1 and by a deletion derivative lacking the ChBD suggested that the ChBD is not absolutely required for hydrolysis of β-chitin microcrystals but greatly enhances the efficiency of degradation.
Chitin, an insoluble linear β-1,4-linked homopolymer of N-acetylglucosamine, is a common constituent of fungal cell walls, exoskeletons of insects, and shells of crustaceans and is one of the most abundant polysaccharides in nature. Chitinase degrades chitin by hydrolyzing β-1,4-glycosidic-linkages, and the activity has been found in a variety of organisms. Bacteria, which do not contain chitin as a constituent, also produce chitinase, mainly for utilization of chitin as a carbon and energy source. From the ecological point of view, bacterial chitinases play an important role in recycling chitin in nature.
Bacillus circulans WL-12 was isolated as a yeast and fungal cell wall-lytic bacterium from soil (31). When the bacterium was grown in the presence of chitin, more than 10 chitinases were detected in the culture supernatant (2, 39). These chitinases are derived from three genes, chiA, chiC, and chiD (2), and the primary products of these genes are designated chitinases A1, C1, and D1, respectively. All of the three chitinases have multidomain structures, and proteolytic modifications of these chitinases give rise to the large variety of chitinases observed in the culture supernatant. Chitinase A1 binds to insoluble chitin and exhibits the highest degradative activity toward insoluble chitin among the chitinases of this bacterium. The mature form of this enzyme consists of a catalytic domain, two fibronectin type III-like domains (FnIII domains), and a C-terminal domain, in order from its N terminus to its C terminus (38). As described in a previous report, the roles of these domains were studied by using deletion derivatives lacking one or both of the FnIII domains and/or the C-terminal domain (40). The isolated catalytic domain hydrolyzed soluble substrates with an activity level comparable to that of intact chitinase A1. However, loss of the C-terminal domain deprived the enzyme of the ability to bind to insoluble chitin and significantly reduced the ability to hydrolyze colloidal chitin. Therefore, it appeared that the C-terminal domain is the chitin-binding domain (ChBD) and is important for efficient degradation of insoluble chitin. Chitinase D1 of this bacterium also possesses a ChBD that is very similar to that of chitinase A1, but at its N-terminus, and has binding activity to insoluble chitin (2). On the other hand, chitinase C1 has a C-terminal domain with unknown function and lacks significant chitin-binding activity (1).
It has been reported that many insoluble-polysaccharide hydrolases contain discrete substrate-binding domains. Among them, cellulose-binding domains (CBDs) of some cellulases have been studied most extensively. The three-dimensional structures of CBDs of Trichoderma reesei endoglucanase I (CBDEGI), T. reesei cellobiohydrolase I (CBDCBHI), Cellulomonas fimi β-1,4-glucanase Cex (CBDCex), Erwinia chrysanthemi endoglucanase Z (CBDEGZ), C. fimi β-1,4-glucanase CenC (CBDN1), and Clostridium thermocellum cellulosomal scaffolding subunit (Cip-CBD) have been determined (4, 14, 15, 19, 35, 42). From structural analyses, it has been suggested that CBDs which can bind to crystalline cellulose have a hydrophobic face constructed by linearly exposed aromatic amino acid residues, and these residues bind to sugar rings of the crystalline cellulose surface through hydrophobic interactions (3, 6, 17). Although the role of CBDs in the activity of cellulases is not completely clear, two hypotheses have been proposed. One of them is that CBDs simply increase the local enzyme concentration on insoluble substrates (10, 33). The other is that disruption of the structure of the cellulose fiber occurs without hydrolysis for smooth degradation (8).
On the other hand, although the presence of ChBDs and putative ChBDs has been suggested for many chitinases, detailed analyses on the structures and biochemical properties of ChBDs have not yet been reported. In order to understand the mechanisms underlying degradation of insoluble chitin by chitinases, biochemical and structural studies of ChBDs are indispensable. In the present study, we constructed an expression system for ChBD of chitinase A1 (ChBDChiA1), purified the ChBDChiA1, and studied its binding properties without interference from the catalytic domain and FnIII domains.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture medium.
Escherichia coli JM109 was the host strain used throughout the construction of various recombinant plasmids. Chitinase A1 and A1ΔChBD, a deletion derivative of chitinase A1 lacking the ChBD, were produced in E. coli HB101 cells carrying recombinant plasmids pHT012 and pHTX013, respectively (37, 40). ChBDChiA1 was produced in E. coli BL21(DE3) cells carrying pChBD, which was constructed by using expression vector pET3a (Novagen, Madison, Wis.).
E. coli HB101 and BL21(DE3) carrying the recombinant plasmid were generally grown in Luria-Bertani broth (LB) medium containing 100 μg of ampicillin per ml. Bacto Tryptone (Difco, Detroit, Mich.) was used to prepare LB medium for cultivation of E. coli BL21(DE3) cells according to the instructions for the pET expression system. To prepare a uniformly 15N-labeled ChBDChiA1 for nuclear magnetic resonance (NMR) studies, E. coli BL21(DE3) carrying pChBD was grown in modified M9 minimal medium (7.0 g of Na2HPO4 per liter, 3.0 g of KH2PO4 per liter, 0.5 g of NaCl per liter, 20 mg of thymine per liter, 20 mg of adenosine per liter, 20 mg of guanosine per liter, 20 mg of cytidine per liter, 20 mg of biotin per liter, 20 mg of thiamine per liter, 1.0 mM MgSO4, 3.3 μM FeCl3, 50 μM MnCl2, and 100 mM CaCl2) containing 100 μg of ampicillin per ml, 0.1% glycerol, 4.0 g of d-glucose per liter, and 0.5 g of 15NH4Cl per liter as the sole nitrogen source.
Construction of pChBD.
The DNA region encoding the ChBD of chitinase A1 was amplified by PCR with synthetic oligonucleotides 5′-CCGGATCCGCTTGGCAGGTCAAC-3′ and 5′-CCGGATCCCTATTGAAGCTGCCACAA-3′ as the primers and pHT012 carrying the chiA gene as the template. To determine the nucleotide sequence, the amplified fragment was blunt ended by using a DNA blunting kit (Takara Biochemical, Kyoto, Japan), inserted into the SmaI site of pUC119, and sequenced using an automated laser fluorescence DNA sequencer (model 4000L; LI-COR). The inserted fragment was cut out using BamHI and ligated with BamHI-cut pET3a to construct the ChBDChiA1 expression plasmid, pChBD.
Production and purification of chitinase A1, A1ΔChBD, and ChBDChiA1.
Chitinase A1 was produced by E. coli HB101 cells carrying recombinant plasmid pHT012 and purified by chitin affinity column chromatography as described previously (30, 39). A1ΔChBD was produced by E. coli HB101 cells carrying recombinant plasmid pHTX013 and purified by high-performance liquid chromatography as described previously (40).
For production of ChBDChiA1, E. coli BL21(DE3) cells carrying pChBD were grown in 1 liter of LB medium containing 100 μg of ampicillin per ml at 30°C. When the optical density at 600 nm (OD600) reached 0.6 (2.1 × 108 cells/ml), 0.5 mM (final concentration) isopropyl-β-d-thiogalactopyranoside (IPTG) was added to the culture to induce expression of ChBDChiA1, and cultivation was continued for a further 12 h. Then cells were collected by centrifugation, washed once with sonication buffer [10 mM (p-amidinophenyl)methanesulfonyl fluoride and 1 mM EDTA in 100 mM Tris-HCl (pH 8.0)], resuspended in the same buffer, and disrupted by sonication with a Tomy ultrasonic disruptor (model UR-200P). The soluble fraction of disrupted cells was extracted by centrifugation (4°C, 20,000 × g, 15 min) and ultracentrifugation (4°C, 75,000 × g, 2 h). Proteins in the soluble fraction were collected by ammonium sulfate precipitation (60% saturation), dissolved in a small volume of 5 mM sodium phosphate buffer (pH 6.0), dialyzed against the same buffer, and lyophilized. ChBDChiA1 in the collected proteins was purified by chitin affinity column chromatography. Chitin affinity column chromatography was carried out as described previously (30), except that 1 M NaCl was included in the washing solution (20 mM sodium phosphate buffer, pH 6.0). The fractions containing purified ChBDChiA1 eluted with 20 mM acetic acid were collected, dialyzed against 5 mM sodium phosphate buffer (pH 6.0), and lyophilized.
Production and purification of ChBDChiA1 for NMR measurement.
E. coli BL21(DE3) carrying pChBD was grown in LB medium containing 100 μg of ampicillin per ml at 30°C. When the OD600 reached 1.0 (3.5 × 108 cells/ml), 3.0 ml of the culture was withdrawn and centrifuged. The pelleted cells were resuspended in a small volume of modified M9 minimal medium containing 15NH4Cl and other supplements, inoculated into 1 liter of the same medium, and incubated at 30°C with shaking. When the OD600 reached 0.5 (1.8 × 108 cells/ml), IPTG was added to a final concentration of 0.5 mM and cultivation was continued for 24 h. Cells were then collected by centrifugation, and 15N-labeled ChBDChiA1 accumulated in the cytoplasm was extracted and purified as described above.
Chemical shift perturbation experiments.
Samples for NMR measurements consisted of 250 μl of 90% (vol/vol) H2O–10% (vol/vol) 2H2O solution containing 50 μM 15N-labeled ChBDChiA1, 50 mM KH2PO4-K2HPO4 (pH 6.0), and 10 mM deuterated dithiothreitol. The two-dimensional WATERGATE and water-flip-back 15N-1H-HSQC (21) spectra of 15N-labeled ChBDChiA1 in the absence and presence of each of the following soluble chitinous substrates were acquired: hexa-N-acetylchitohexaose [(GlcNAc)6], soluble chitin, carboxymethyl chitin (CM-chitin), and ethylene glycol chitin. The concentration of each added soluble substrate corresponded to 500 μM, assuming the six monosaccharide units to be one molecule. The spectrum in the presence of each substrate was compared with that of the free ChBDChiA1.
NMR experiments were performed using a Bruker DRX500 or DRX800 spectrometer at 310 K. The spectral widths measured with the DRX500 or DRX800 spectrometer were 1,116.1 and 1,785.7 Hz for the 15N dimension and 8,012.8 and 12,019.2 Hz for the 1H dimension, respectively. The 1H carrier was set to the frequency of the water resonance (4.69 ppm), and the 15N carrier was set to 120.0 ppm. NMR data processing and analysis were performed using the nmrPipe and nmrDraw software package (7).
ITC.
Isothermal titration calorimetry (ITC) experiments were carried out using an OMEGA calorimeter (MicroCal Inc., Amherst, Mass.) (41). The reaction cell (ca. 1.4 ml in volume) was filled with 0.1 mM ChBDChiA1 solution, and the injection syringe was filled with 3 mM (GlcNAc)6 or a 2.5-mg/ml soluble chitin solution. Ten-microliter aliquots of the substrate solutions were injected 12 times at 5- to 7-min intervals. Control dilutions of these substrates into buffer were also carried out in order to correct the observed heats of binding. Titration experiments were done at pH 6.0 with 50 mM sodium phosphate buffer for (GlcNAc)6 and 20 mM sodium cacodylate buffer for soluble chitin and also at pH 9.0 with 20 mM pyrophosphate buffer for (GlcNAc)6. The substrates were dissolved in dialysate solutions so that no pH difference existed between the protein and the substrate solutions.
Electron microscopy.
Enzyme-treated β-chitin microcrystals were deposited on carbon-coated grids and allowed to dry. All of the electron micrographs were taken with a JEOL 2000EXII electron microscope operated at 100 kV and recorded on Mitsubishi MEM film. Diffraction contrast imaging in the bright-field mode was used to visualize the sample without further contrast enhancement. The images were taken at magnifications of ×1,000 to ×6,000 under low-dose exposure with the use of a Minimum Dose System (JEOL).
Protein assay.
The protein concentration was estimated from absorbance at 280 nm using the molar extinction coefficients ɛ (chitinase A1) = 153,920, ɛ(A1ΔChBD) = 138,450, and ɛ(ChBDChiA1) = 20,970, which were calculated from the amino acid compositions of each protein (24). To determine relative equilibrium association constants, the protein concentration was estimated by spectrofluorometry (Hitachi F-3010 Spectrofluorometer) at an excitation wavelength of 280 nm and an emission wavelength of 342 nm. A separate standard curve was prepared for each protein.
SDS-polyacrylamide gel electrophoresis.
Sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis in 16.5% polyacrylamide slabs was carried out according to the manufacturer's instructions for SDS molecular weight markers (Sigma Chemical Co., St. Louis, Mo.).
Enzyme assay.
Reducing sugar generated by the degradation of microcrystalline β-chitin from a vestimentiferan tube worm (Lamellibrachia satsuma) was measured by a modification of Schales' procedure using di-N-acetylchitobiose as the standard (12). Each assay mixture (total volume, 300 μl) contained 200 μg (dry weight) of microcrystalline β-chitin and 240 nM enzyme in 0.1 M sodium phosphate buffer (pH 6.0).
Binding assay.
Binding assay mixtures in 1-ml glass microtubes containing various concentrations of protein and 1 mg of binding substrate in 1 ml of 20 mM buffer were incubated on ice with occasional mixing. Each mixture was centrifuged at 4°C for 20 min at 15,000 × g to separate supernatant and substrate with bound protein, the supernatant containing free protein was collected, and the protein concentration was determined. The amount of bound protein was calculated from the difference between the initial protein concentration and the free protein concentration after binding. The relative equilibrium association constants (Kr) was determined from double-reciprocal plots of binding data by the method described by Gilkes et al. (9).
Chemicals.
Chitin EX (powdered prawn shell chitin), chitosans, and CM-chitin were purchased from Funakoshi Chemical Co. (Tokyo, Japan). (GlcNAc)6 and soluble chitin were obtained from Yaizu Suisan Chemical Co. Ltd. (Shizuoka, Japan). The degree of deacetylation and approximate molecular weight of the soluble chitin were 38.8% and from 200,000 to 300,000, respectively. Avicel and soluble starch were purchased from Asahi Chemical Industry (Osaka, Japan) and Wako Pure Chemical Industries (Osaka, Japan), respectively. Regenerated chitin was prepared from chitosan 8B (approximately 20% acetylated), purchased from Funakoshi Chemical Co., as described by Molano et al. (20). Colloidal chitin and ethylene glycol chitin were prepared from powdered crab shell chitin, purchased from Funakoshi Chemical Co., by the methods described by Jeuniaux (13) and Yamada and Imoto (43), respectively. Microcrystalline β-chitin from vestimentiferan was prepared as described previously (29).
RESULTS
Production and purification of ChBDChiA1.
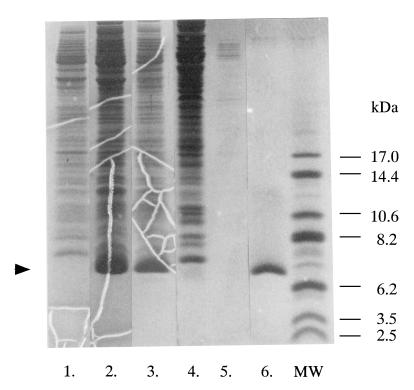
The mature form of chitinase A1 from B. circulans WL-12 is composed of the N-terminal catalytic domain, two FnIII domains and the C-terminal ChBD. In order to study the biochemical properties and structure of ChBD of chitinase A1, a high-level expression system for ChBDChiA1 was constructed by using the pET expression system. The DNA region of the chiA gene corresponding to the ChBD (Ala655 to Gln699 of chitinase A1) was amplified by PCR, and the amplified fragment was first inserted into the SmaI site of pUC119 for sequencing to ensure that the amplified fragment had the expected sequence. The inserted fragment was then cut out using BamHI and reinserted into the BamHI site of pET3a, resulting in the plasmid pChBD. E. coli BL21(DE3) cells carrying pChBD produced ChBDChiA1 in soluble form when induced with IPTG and accumulated it in the cytoplasm. ChBDChiA1 in the cytoplasm was extracted from the cells after 12 h of induction with IPTG, fractionated by ammonium sulfate precipitation, and purified by chitin affinity column chromatography. ChBDChiA1 was eluted from the chitin column with 20 mM acetic acid after the column was washed with 20 mM sodium phosphate buffer (pH 6.0) containing 1 M NaCl. The protein in the peak fraction exhibited a single band on an SDS-polyacrylamide gel, as shown in Fig. 1. From a 1-liter culture of E. coli BL21(DE3) cells carrying pChBD, over 50 mg of ChBDChiA1 was extracted and 20 to 40 mg of purified ChBDChiA1 was obtained. The ChBDChiA1 obtained included a T7 tag at its N terminus consisting of 14 amino acid residues derived from the expression vector.
FIG. 1.
Expression and purification of ChBDChiA1. ChBDChiA1 produced in E. coli BL21 cells was purified by chitin affinity column chromatography from the soluble protein fraction of disrupted cells. Lane 1, soluble protein fraction of E. coli BL21(DE3) cells carrying pET3a (control); lane 2, soluble protein fraction of induced E. coli BL21(DE3) cells carrying pChBD; lane 3, proteins obtained by ammonium sulfate precipitation from the soluble protein fraction; lane 4, flowthrough fraction of chitin affinity column chromatography; lane 5, proteins eluted from the chitin affinity column with 20 mM sodium acetate buffer (pH 5.5); lane 6, purified ChBDChiA1 obtained by elution with 20 mM acetic acid; lane MW, molecular mass standards. The arrowhead indicates the position of the ChBDChiA1 protein band.
Binding activity of purified ChBDChiA1.
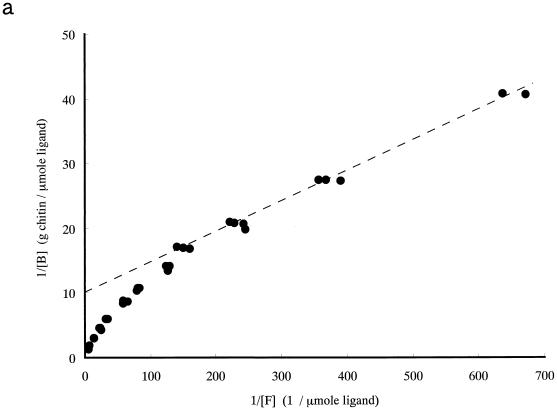
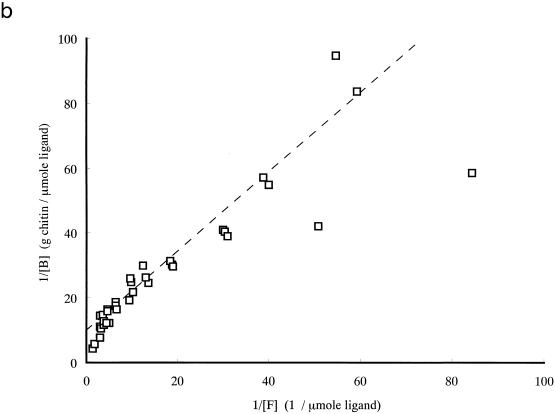
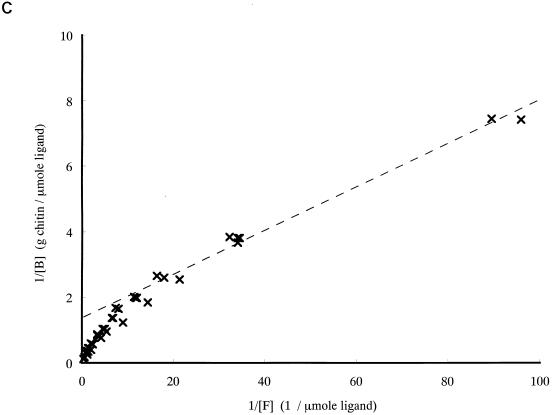
To study the contribution of ChBD to the binding of chitinase A1 to chitin, the binding activity of purified ChBDChiA1 was compared with those of the intact chitinase A1 and A1ΔChBD, a deletion derivative of chitinase A1 lacking ChBD. The relative equilibrium association constants (Kr) of intact chitinase A1, A1ΔChBD, and ChBDChiA1 toward regenerated chitin were determined as shown in Table 1 from the double-reciprocal plots of binding data shown in Fig. 2. The binding assay for these experiments was carried out at pH 6.0, since the pH optimum of chitinase A1 for the hydrolysis reaction is 6.0. The binding reaction times were 3 h for ChBDChiA1 and 1 h for chitinase A1 and A1ΔChBD, since preliminary experiments indicated that the binding of ChBDChiA1 required approximately 3 h and those of chitinase A1 and A1ΔChBD required less than 1 h to reach equilibrium (data not shown). The regenerated chitin used in this experiment was prepared by acetylation of chitosan, and the degree of acetylation was more than 95%. The a/[N0] values, indicating the relative space of substrate surface occupied by a single ligand molecule, are almost consistent with the ligands' molecular weights (a is the number of lattice units occupied by a single ligand molecule, and N0 is the concentration of binding sites on the chitin surface). The Kr of ChBDChiA1 was significantly smaller than that of chitinase A1, and A1ΔChBD exhibited a much smaller Kr than the other two proteins. The Kr of ChBDChiA1 could be an underestimate, since binding assays with ChBDChiA1 in the lower range of protein concentration were difficult to carry out. This small protein is not as sensitive as chitinase A1 and A1ΔChBD in protein concentration measurement. However, the results strongly suggested that the binding activity of intact chitinase A1 depends mostly on the binding activity of ChBDChiA1 and that the catalytic domain is involved in the binding of chitinase A1 to a certain extent. The weak binding activity of A1ΔChBD was considered to be due to the affinity of the catalytic domain for chitin, since the isolated catalytic domain and A1ΔChBD exhibited the same level of affinity toward regenerated chitin, as described previously (40).
TABLE 1.
Adsorption parameters
| Protein or domain | Kr (liters g−1) | a/[N0] (g μmol−1) |
|---|---|---|
| Chitinase A1 | 21.1 | 10.1 |
| A1ΔChBD | 0.8 | 9.9 |
| ChBDChiA1 | 14.9 | 1.3 |
FIG. 2.
Double-reciprocal plots of binding data for chitinase A1 (a), A1ΔChBD (b), and ChBDChiA1 (c). The binding assay mixture contained 1 mg (dry weight) of regenerated chitin and from 1 to 70 μg of each protein (14 to 1,000 pmol of chitinase A1, 15 to 1,050 pmol of A1ΔChBD, and 154 to 10,770 pmol of ChBDChiA1) in 1 ml of 20 mM sodium phosphate buffer (pH 6.0). The binding assay was performed by keeping the binding assay mixtures on ice for 1 h for chitinase A1 and A1ΔChBD and 3 h for ChBDChiA1. [B], bound protein concentration; [F], free protein concentration.
Effect of pH and salt concentration on binding.
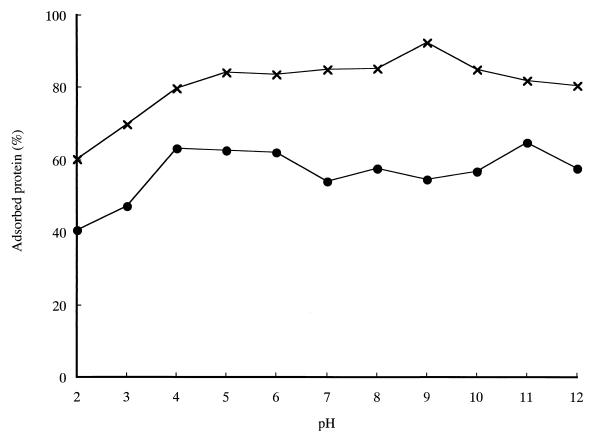
The effect of pH on the binding of chitinase A1 and ChBDChiA1 to regenerated chitin was investigated. As shown in Fig. 3, both chitinase A1 and ChBDChiA1 showed binding activity over a wide range of pH. The highest binding activity of ChBDChiA1 was observed at pH 9.0. The binding of both chitinase A1 and ChBDChiA1 significantly decreased at pHs below 3. This may explain the observation that these proteins were eluted from a chitin affinity column by 20 mM acetic acid. ChBDChiA1 exhibited higher percentages of bound protein than chitinase A1 did at all tested pHs under this reaction condition, in contrast to the lower Kr value compared to chitinase A1. The conflict may be explained by the possible underestimate of the Kr of ChBDChiA1 described above and the fact that the relative binding of chitinase A1 decreases much faster than that of ChBDChiA1 with the increase in the amount of added proteins in the binding assay mixture, probably due to its higher a/[N0] value.
FIG. 3.
Effect of pH on the binding of chitinase A1 and ChBDChiA1 to regenerated chitin. Assay mixtures contained 1 mg (dry weight) of regenerated chitin and 1 nmol of either chitinase A1 (70 μg) or ChBDChiA1 (6.5 μg) in 1 ml of buffers with various pHs. The buffers used in this experiment were 20 mM sodium citrate (pH 2.0 to 6.0), sodium phosphate (pH 7.0), Tris-HCl (pH 8.0), glycine-NaOH (pH 9.0 and 10.0), and disodium hydrogen phosphate-NaOH (pH 11 and 12). Assay mixtures were incubated on ice for 1 h for chitinase A1 and 3 h for ChBDChiA1. ●, chitinase A1; ×, ChBDChiA1.
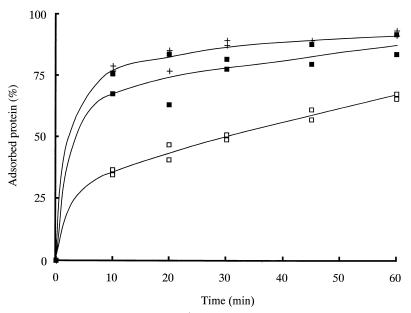
Figure 4 shows the kinetics of binding of ChBDChiA1 over 60 min of incubation at pH 9.0 and in the presence of 0.5 M NaCl at pH 6.0. ChBDChiA1 bound better at pH 9.0 than that at pH 6.0. The isoelectric points (pIs) calculated from the amino acid sequences of ChBDChiA1 with and without a T7 tag are 9.3 and 8.8, respectively. Therefore, the electric charge of ChBDChiA1 was almost minimized at pH 9.0. In the presence of 0.5 M NaCl at pH 6.0, ChBDChiA1 also bound better than it did without NaCl. These results suggest that the binding of ChBDChiA1 to chitin is mediated mainly by hydrophobic interactions.
FIG. 4.
Time courses of binding of ChBDChiA1 at pH 9.0 and in the presence of NaCl. Assay mixtures contained 6 mg (dry weight) of regenerated chitin and 2.3 nmol (15 μg) of ChBDChiA1 in 1 ml of 20 mM sodium phosphate buffer (pH 6.0) (□), 20 mM sodium phosphate buffer (pH 6.0) containing 0.5 M NaCl (■), or 20 mM Tris-HCl buffer (pH 9.0) (+).
Specificity of binding to insoluble polysaccharides.
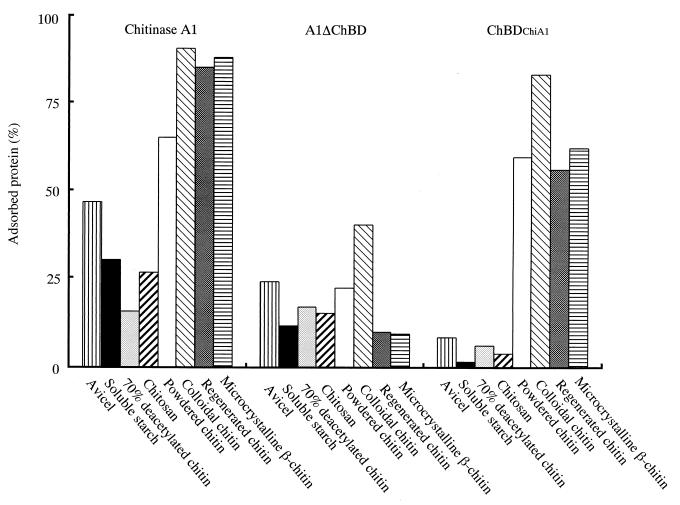
Figure 5 shows the specificities of binding of intact chitinase A1, A1ΔChBD, and ChBDChiA1 to various insoluble polysaccharides. Chitinase A1 preferentially bound to various forms of insoluble chitin, but it also bound, although weakly, to other insoluble polysaccharides such as avicel and soluble starch. On the other hand, ChBDChiA1 bound highly specifically to chitin. It bound to colloidal chitin, regenerated chitin, chitin powder (chitin EX), and microcrystalline β-chitin from L. satsuma and did not show significant binding to chitosan, starch, or avicel. A1ΔChBD exhibited weak affinity to all insoluble β-1,4-polysaccharides examined in this experiment, and thus, the affinity was not restricted to chitin, although the binding activity to colloidal chitin was the highest among the tested polysaccharides. Therefore, the weak binding activity of the intact chitinase A1 observed with polysaccharides other than chitin is suggested to be due to the affinity of the catalytic domain for these polysaccharides.
FIG. 5.
Binding specificities of chitinase A1, A1ΔChBD, and ChBDChiA1. Binding assay mixtures contained 1 mg (dry weight) of various insoluble polysaccharides and 25 μg of protein in 1 ml of 20 mM Tris-HCl (pH 9.0). Assay mixtures were kept on ice for 24 h for binding.
Interaction with soluble substrates.
Interactions of ChBDChiA1 with soluble substrates, including (GlcNAc)6, ethylene glycol chitin, CM-chitin, and soluble chitin, were examined by NMR and ITC.
The simplest approach for detecting interactions between proteins and substrates by NMR is the chemical shift perturbation experiment (25), where changes of amide chemical shifts of a protein are monitored by 15N-1H-HSQC spectra upon the addition of a substrate. The spectra of the 15N-labeled ChBDChiA1 in the presence and absence of one of the four soluble substrates were compared. These comparisons revealed no changes of the chemical shifts of the NMR peaks, although slight overall increases in the peak widths were observed, probably due to the increase in the solution viscosity caused by the additional oligosaccharide (data not shown). Thus, we concluded that ChBDChiA1 does not interact specifically with any of the four soluble substrates.
Interaction between ChBDChiA1 and either (GlcNAc)6 or soluble chitin was also studied by means of ITC. Titration of 0.1 mM ChBDChiA1 solution with either 2.5 mg of soluble chitin per ml or 3 mM (GlcNAc)6 was carried out, but no heat change was observed for either titration with the two substrates within the sensitivity range of the calorimeter. These results indicated that no specific interaction occurred between the protein and the substrates. This was also confirmed by differential scanning calorimetry measurements conducted on the protein solutions in both the presence and absence of each of three substrates, (GlcNAc)6, soluble chitin, and CM-chitin, at pH 6.0. The denaturation temperature of ChBDChiA1 was not shifted to a higher temperature range by the presence of these substrates, indicating that there was no complex formation, as judged using the Le Chaterier principle (28).
In addition, the effect of the presence of (GlcNAc)6 and CM-chitin on the binding of ChBDChiA1 to regenerated chitin was examined. Equal amounts of either (GlcNAc)6 or CM-chitin and regenerated chitin, the binding substrate, were included in the assay mixture, and binding of ChBDChiA1 was carried out for 24 h at 4°C. The protein concentration in the supernatant was measured after centrifugation, and the amount of bound protein was estimated. However, no significant decrease in the amount of bound protein was observed even in the presence of soluble substrates, and therefore, these soluble substrates do not interfere with the binding of ChBDChiA1 to regenerated chitin.
From all of these data, it was concluded that ChBDChiA1 does not interact with either chito-oligosaccharide or soluble derivatives of chitin.
Degradation of microcrystalline β-chitin.
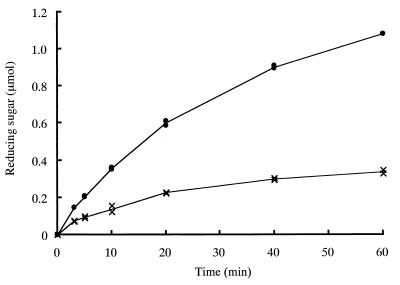
Since ChBDChiA1 was shown not to interact with either soluble chitin or chito-oligosaccharide, the effect of the absence of the ChBD from chitinase A1 on the hydrolysis of highly crystalline β-chitin was examined. β-Chitin microcrystals isolated from protective tubes of L. satsuma were treated with intact chitinase A1 and A1ΔChBD, and the amount of reducing sugar generated was measured. As shown in Fig. 6, degradation of microcrystalline β-chitin by A1ΔChBD was much less efficient than degradation by intact chitinase A1. Hydrolysis by chitinase A1 continued and reducing sugar increased during 60 min of incubation, although the rate of hydrolysis decreased gradually with incubation time. Since approximately 30% of the substrate was hydrolyzed at the end of the incubation period, the decrease in the rate of hydrolysis was probably due to the substrate shortage and/or product inhibition of the hydrolysis reaction. On the other hand, significant degradation of microcrystalline β-chitin by A1ΔChBD was observed only at the beginning of the incubation period, and soon hydrolysis slowed down and nearly stopped. The effects of substrate consumption and/or product inhibition must therefore be much smaller than in the case of chitinase A1. Therefore, it is clear that ChBD plays a vital role in the degradation of β-chitin microcrystals.
FIG. 6.
Hydrolysis of β-chitin microfibrils by intact chitinase A1 and A1ΔChBD. Reaction mixtures contained 200 μg (dry weight) of microcrystalline β-chitin from L. satsuma and 71 pmol of each chitinase. Reactions were performed at 37°C, and the amount of reducing sugar generated was monitored by the modified Schales procedure. ●, chitinase A1; ×, A1ΔChBD.
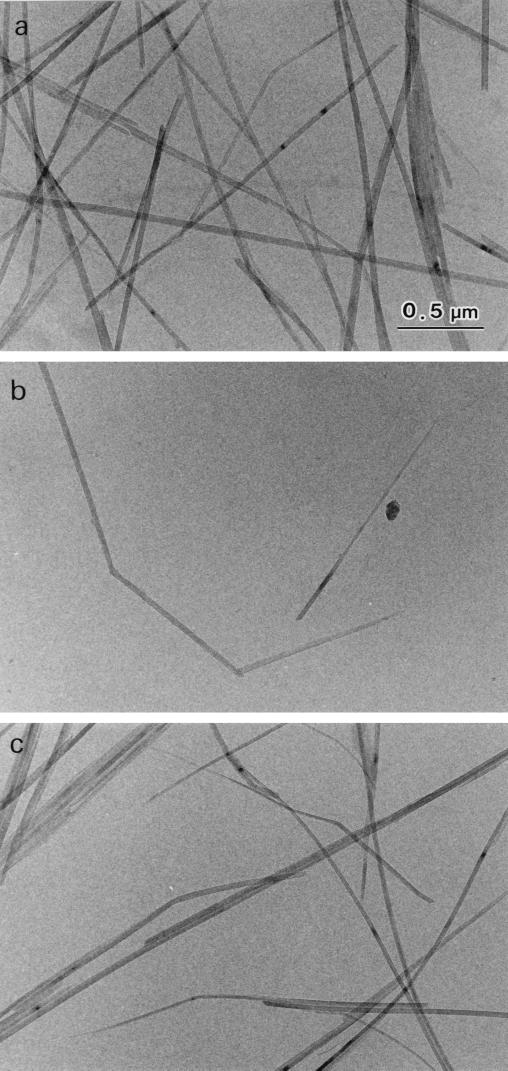
However, since significant degradation by A1ΔChBD was observed only at the beginning of incubation, it is possible that degradation by A1ΔChBD occurred at less-crystalline regions of the substrate. To see whether A1ΔChBD can truly hydrolyze β-chitin microcrystals, β-chitin microfibrils were treated with intact chitinase A1 and A1ΔChBD and then examined by electron microscopy. As demonstrated previously, intact chitinase A1 shortened β-chitin microcrystals and formed a pointed tip at one end of the microcrystal (29). As shown in Fig. 7c, A1ΔChBD also shortened β-chitin microcrystals and formed a pointed tip at one end of the microcrystal, just as chitinase A1 did. The shape of the β-chitin microcrystals treated with A1ΔChBD was indistinguishable from the shape of those treated with chitinase A1. The means that without ChBD, the enzyme is still able to hydrolyze the crystalline part of the substrate. Therefore, it appeared that ChBDChiA1 is not absolutely required for hydrolysis of β-chitin microcrystals but that it greatly improves the efficiency of degradation.
FIG. 7.
Bright-field diffraction contrast micrographs of L. satsuma β-chitin microfibrils. (a) no enzyme treatment (control); (b) after treatment with chitinase A1; (c) after treatment with A1ΔChBD.
DISCUSSION
ChBDChiA1 was successfully expressed separately from the catalytic domain and FnIII domains by using a pET expression system. This made it possible to study the properties of ChBD without influence from the other domains, and we found two important and unexpected features of the ChBD of chitinase A1. One is that the ChBDChiA1 interacted only with insoluble chitin, and the other is that binding of ChBDChiA1 was highly specific for chitin. As mentioned above, no interactions between ChBDChiA1 and a chito-oligosaccharide, (GlcNAc)6, was detected. Interactions between ChBDChiA1 and soluble substrates were also studied with ITC. Titration of ChBDChiA1 solution with either (GlcNAc)6 or soluble derivatives of chitin did not give significant reaction heat, which indicates a lack of interaction between ChBDChiA1 and the tested substrates. In addition, (GlcNAc)6 and CM-chitin did not interfere with the binding of ChBDChiA1 to regenerated chitin. Thus, no interaction was observed using three different approaches attempting to detect interaction with soluble chitinous substrates.
Since chitinase A1 exhibited weak but significant binding to cellulose in addition to various forms of insoluble chitin, we first expected that ChBD would also have some binding activity to cellulose. However, it appeared that ChBDChiA1 bound only to chitin, and there was no significant binding to cellulose or other polysaccharides. These observations suggest that ChBDChiA1 recognizes a structure that is present only in insoluble or crystalline chitin and not in chito-oligosaccharides, soluble derivatives of chitin, or other insoluble polysaccharides.
As far as we know, the three-dimensional structures of six CBDs from five families, five from cellulases and one from cellulosome, have been reported (4, 14, 15, 19, 35, 42). Among them, the CBD of CenC from C. fimi binds to amorphous cellulose but not to crystalline cellulose, and the others can bind both amorphous and crystalline celluloses. CBDs that can bind to crystalline cellulose share the common surface feature of a planar array of three aromatic residues. The three aromatic residues have been proposed to make a major contribution to the binding of CBDs to the cellulose surface (3, 17). Interactions of CBDs and cello-oligosaccharides have been demonstrated with CBDs of exoglucanase from C. fimi, endoglucanase I from T. reesei, and xylanase A from Pseudomonas fluorescens subsp. cellulosa by NMR spectroscopy (18, 23, 42) and titration calorimetry (6). Binding to chitin in addition to cellulose has been demonstrated with CBDCBHII, CBDCex, CBDCenA, and Cip-CBD, while CBDCBHI did not bind to chitin (11, 16, 32, 34). On the other hand, ChBD bound specifically to chitin and not to cellulose and did not interact with chito-oligosaccharide, as mentioned above. These observations suggest that there must be significant differences in the mechanisms of binding between ChBDChiA1 and these CBDs. In fact, the solution structure determined very recently revealed that ChBDChiA1 does not have a planar array of aromatic residues on its surface (unpublished data).
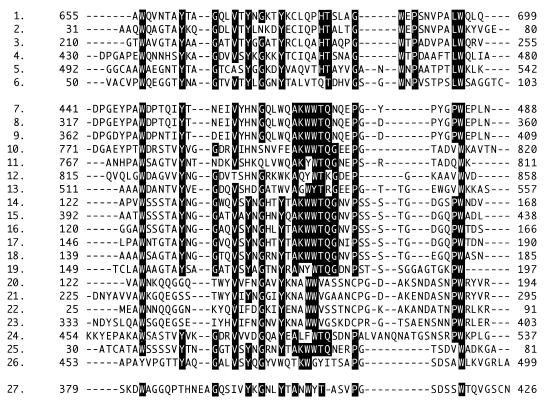
The ChBD of chitinase A1 exhibits amino acid sequence similarity with many sequence segments in other chitinases, cellulases, and a protease, as shown in Fig. 8. These sequences can be divided into two groups: one is represented by the ChBD of chitinase A1 (upper group in Fig. 8) and the other is represented by a family V CBD from E. chrysanthemi endoglucanase (CBDZEGZ). As seen in the alignment, the N-terminal halves of the sequences are relatively conserved between the two groups of sequences, but the C-terminal halves are very different between the two groups of sequences. It is most striking that the upper group does not have the AKWWTQG motif, which is well conserved in the lower-group sequences, and the upper group contains solely known or proposed ChBDs. The C-terminal domain of protease C from Streptomyces griseus is a member of the upper group. The homology of this domain with ChBDs of B. circulans chitinases A1 and D1 was described by Sidhu et al. (27), and they suggested that the enzyme is specialized for the degradation of chitin-linked proteins. On the other hand, the lower group contains both CBDs and ChBDs, including those of chitinase B from Clostridium paraputrificum (22), chitinase 85 from Altermonas sp. strain O-7 (36), and chitinases from Aeromonas sp. strain 10S-24 (26). CBDEGZ exhibits a ski-boot shape, and three aromatic residues (Trp382, Trp407, and Tyr408) are localized on one face (4). These aromatic residues are proposed to play a major role in the binding of this CBD to the cellulose surface, as mentioned above. Trp407 and Tyr408 of CBDEGZ correspond to the two aromatic residues in this motif.
FIG. 8.
Alignment of amino acid sequences that have similarity to ChBD of chitinase A1. Rows: 1, ChBD of B. circulans WL-12 chitinase A1; 2, N-terminal domain (ChBD) of B. circulans WL-12 chitinase D1; 3, C-terminal region of S. griseus proteinase C; 4, C-terminal region of Serratia marcescens 2170 chitinase CI; 5, C-terminal region of Aeromonas sp. strain 10S chitinase II; 6, C-terminal region of Janthinobacterium lividum chitinase 69a; 7 to 9, Bacillus sp. (strain N-4) cellulase A, cellulase B1, and cellulase B, respectively; 10, Alteromonas sp. (strain O-7) chitinase 85; 11-12, Aeromonas caviae chitinase A; 13, Vibrio harveyi chitinase A; 14 to 18, Aeromonas sp. strain 10S ORF1, chitinase II, ORF3, ORF4, and ORF2, respectively; 19, J. lividum chitinase 69b; 20 to 24, repeating units found in E. coli hypothetical 97.1-kDa protein YheB; 25, S. griseus chitinase C; 26, S. marcescens 2170 chitinase B; 27, E. chrysanthemi endglucasase Z. Highly conserved amino acid sequences in the upper and lower groups are indicated by a black background. The starting and ending amino acid positions, based on the translational start methionine being residue 1, are shown on both sides of each protein segment. The CLUSTAL X program was used to make the alignment.
Brun et al. suggested that the sequence regions shown in the lower group form individual functional domains structurally related to CBDEGZ, and they proposed the possibility that CBDEGZ and some ChBDs might form a new family of functionally and structurally related protein modules (4). They also noted that the ChBDs of B. circulans chitinases A1 and D1 are presumably not included in this family, since they do not exhibit the highly conserved stWWst motif (AKWWTQG motif), where s and t represent small residues and turn-like residues, respectively. Unique features of ChBDChiA1 with respect to its binding properties and striking differences in the surface structure between ChBDChiA1 and CBDEGZ may indicate the relevance of their suggestion. Therefore, presumably, the upper group forms a new family of functionally and structurally related protein modules distinct from the family of the lower group. The family may share common features with the ChBD of chitinase A1 characterized by chitin-specific binding activity and interaction only with insoluble chitin.
Isolated ChBDChiA1 has a tightly packed structure and remarkable stability in spite of its small size (45 amino acids) and the absence of disulfide bonds, as demonstrated by NMR and differential scanning calorimetry (unpublished data). The binding activity of ChBDChiA1 is highly specific for insoluble chitin, and it can be controlled by changing the pH, as shown above. The ChBD of chitinase A1 has been successfully used for purification of target proteins in foreign protein expression systems as described by Chong et al. (5). In addition to the remarkable stability and small size of ChBDChiA1, its specific binding to insoluble chitin and easy desorption by controlling the pH demonstrated in this study may open new aspects regarding applications of the ChBD of chitinase A1.
ACKNOWLEDGMENTS
We thank Kazuo Sakai, Yaizu Suisan Chemical Co., Ltd., for supplying soluble chitin. We thank Jun Hashimoto, Japan Marine Science and Technology Center, Yokosuka, Japan, for the kind gift of tubes from the vestimentiferan tubeworm, Lamellibrachia satsuma.
This work was partly supported by a grant-in-aid for scientific research (10660077) from the Ministry of Education, Science and Culture of Japan.
REFERENCES
- 1.Alam M M, Nikaidou N, Tanaka H, Watanabe T. Cloning and sequencing of chiC gene of Bacillus circulans WL-12 and relationship of its product to some other chitinases and chitinase-like proteins. J Ferment Bioeng. 1995;80:454–461. [Google Scholar]
- 2.Alam M M, Mizutani T, Isono M, Nikaidou N, Watanabe T. Three chitinase genes (chiA, chiC, and chiD) comprise the chitinase system of Bacillus circulans WL-12. J Ferment Bioeng. 1996;82:28–36. [Google Scholar]
- 3.Bray M R, Johnson P E, Gilkes N R, McIntosh L P, Kilburn D G, Warren R A. Probing the role of tryptophan residues in a cellulose-binding domain by chemical modification. Protein Sci. 1996;5:2311–2318. doi: 10.1002/pro.5560051117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brun E, Moriaud F, Gans P, Blackledge M J, Barras F, Marion D. Solution structure of the cellulose-binding domain of the endoglucanase Z secreted by Erwinia chrysanthemi. Biochemistry. 1997;36:16074–16086. doi: 10.1021/bi9718494. [DOI] [PubMed] [Google Scholar]
- 5.Chong S, Mersha F B, Comb D G, Scott M E, Landry D, Vence L M, Perler F B, Benner J, Kucera R B, Hirvonen C A, Pelletier J J, Paulus H, Xu M Q. Single-column purification of free recombinant proteins using a self-cleavable affinity tag derived from a protein splicing element. Gene. 1997;192:271–281. doi: 10.1016/s0378-1119(97)00105-4. [DOI] [PubMed] [Google Scholar]
- 6.Creagh A L, Ong E, Jervis E, Kilburn D G, Haynes C A. Binding of the cellulose-binding domain of exoglucanase Cex from Cellulomonas fimi to insoluble microcrystalline cellulose is entropically driven. Proc Natl Acad Sci USA. 1996;93:12229–12234. doi: 10.1073/pnas.93.22.12229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Delaglio F, Grzesiek S, Vuister G W, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 8.Din N, Gilkes N R, Tekant B, Miller R C, Warren R A J, Kilburn D G. Non-hydrolytic disruption of cellulose fibers by the binding domain of a bacterial cellulase. Bio/Technology. 1991;9:1096–1099. [Google Scholar]
- 9.Gilkes N R, Jervis E, Henrissat B, Tekant B, Miller R C, Jr, Warren R A, Kilburn D G. The adsorption of a bacterial cellulase and its two isolated domains to crystalline cellulose. J Biol Chem. 1992;267:6743–6749. [PubMed] [Google Scholar]
- 10.Gilkes N R, Warren R A, Miller R C, Jr, Kilburn D G. Precise excision of the cellulose binding domains from two Cellulomonas fimi cellulases by a homologous protease and the effect on catalysis. J Biol Chem. 1988;263:10401–10407. [PubMed] [Google Scholar]
- 11.Goldstein M A, Takagi M, Hashida S, Shoseyov O, Doi R H, Segel I H. Characterization of the cellulose-binding domain of the Clostridium cellulovorans cellulose-binding protein A. J Bacteriol. 1993;175:5762–5768. doi: 10.1128/jb.175.18.5762-5768.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Imoto T, Yagishita K. A simple activity measurement of lysozyme. Agric Biol Chem. 1971;35:1154–1156. [Google Scholar]
- 13.Jeuniaux C. Chitinases. Methods Enzymol. 1966;8:644–650. [Google Scholar]
- 14.Johnson P E, Joshi M D, Tomme P, Kilburn D G, McIntosh L P. Structure of the N-terminal cellulose-binding domain of Cellulomonas fimi CenC determined by nuclear magnetic resonance spectroscopy. Biochemistry. 1996;35:14381–14394. doi: 10.1021/bi961612s. [DOI] [PubMed] [Google Scholar]
- 15.Kraulis J, Clore G M, Nilges M, Jones T A, Pettersson G, Knowles J, Gronenborn A M. Determination of the three-dimensional solution structure of the C-terminal domain of cellobiohydrolase I from Trichoderma reesei. A study using nuclear magnetic resonance and hybrid distance geometry-dynamical simulated annealing. Biochemistry. 1989;28:7241–7257. doi: 10.1021/bi00444a016. [DOI] [PubMed] [Google Scholar]
- 16.Linder M, Salovuori I, Ruohonen L, Teeri T T. Characterization of a double cellulose-binding domain. Synergistic high affinity binding to crystalline cellulose. J Biol Chem. 1996;271:21268–21272. doi: 10.1074/jbc.271.35.21268. [DOI] [PubMed] [Google Scholar]
- 17.Mattinen M L, Kontteli M, Kerovuo J, Linder M, Annila A, Lindeberg G, Reinikainen T, Drakenberg T. Three-dimensional structures of three engineered cellulose-binding domains of cellobiohydrolase I from Trichoderma reesei. Protein Sci. 1997;6:294–303. doi: 10.1002/pro.5560060204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mattinen M L, Linder M, Teleman A, Annila A. Interaction between cellohexaose and cellulose binding domains from Trichoderma reesei cellulases. FEBS Lett. 1997;407:291–296. doi: 10.1016/s0014-5793(97)00356-6. [DOI] [PubMed] [Google Scholar]
- 19.Mattinen M L, Linder M, Drakenberg T, Annila A. Solution structure of the cellulose-binding domain of endoglucanase I from Trichoderma reesei and its interaction with cello-oligosaccharides. Eur J Biochem. 1998;256:279–286. doi: 10.1046/j.1432-1327.1998.2560279.x. [DOI] [PubMed] [Google Scholar]
- 20.Molano J, Duran A, Cabib E. A rapid and sensitive assay for chitinase using tritiated chitin. Anal Biochem. 1977;83:648–656. doi: 10.1016/0003-2697(77)90069-0. [DOI] [PubMed] [Google Scholar]
- 21.Mori S, Abeygunawardana C, Johnson M O, van Zijl P C. Improved sensitivity of HSQC spectra of exchanging protons at short interscan delays using a new fast HSQC (FHSQC) detection scheme that avoids water saturation. J Magn Reson B. 1995;108:94–98. doi: 10.1006/jmrb.1995.1109. . (Erratum, 110:321, 1996.) [DOI] [PubMed] [Google Scholar]
- 22.Morimoto K, Karita S, Kimura T, Sakka K, Ohmiya K. Cloning, sequencing, and expression of the gene encoding Clostridium paraputrificum chitinase ChiB and analysis of the functions of novel cadherin-like domains and a chitin-binding domain. J Bacteriol. 1997;179:7306–7314. doi: 10.1128/jb.179.23.7306-7314.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nagy T, Simpson P, Williamson M P, Hazlewood G P, Gilbert H J, Orosz L. All three surface tryptophans in Type IIa cellulose binding domains play a pivotal role in binding both soluble and insoluble ligands. FEBS Lett. 1998;429:312–316. doi: 10.1016/s0014-5793(98)00625-5. [DOI] [PubMed] [Google Scholar]
- 24.Pace C N, Vajdos F, Fee L, Grimsley G, Gray T. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 1995;4:2411–2423. doi: 10.1002/pro.5560041120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rajagopal P, Waygood E B, Reizer J, Saier M H, Jr, Klevit R E. Demonstration of protein-protein interaction specificity by NMR chemical shift mapping. Protein Sci. 1997;6:2624–2627. doi: 10.1002/pro.5560061214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shiro M, Ueda M, Kawaguchi T, Arai M. Cloning of a cluster of chitinase genes from Aeromonas sp. no. 10S-24. Biochim Biophys Acta. 1996;1305:44–48. doi: 10.1016/0167-4781(95)00213-8. [DOI] [PubMed] [Google Scholar]
- 27.Sidhu S S, Kalmar G B, Willis L G, Borgford T J. Streptomyces griseus protease C. A novel enzyme of the chymotrypsin superfamily. J Biol Chem. 1994;269:20167–20171. [PubMed] [Google Scholar]
- 28.Sturtevant J M. Biochemical applications of differential scanning calorimetry. Ann Rev Phys Chem. 1987;38:463–488. [Google Scholar]
- 29.Sugiyama J, Boisset C, Hashimoto M, Watanabe T. Molecular directionality of β-chitin biosynthesis. J Mol Biol. 1999;286:247–255. doi: 10.1006/jmbi.1998.2458. [DOI] [PubMed] [Google Scholar]
- 30.Suzuki K, Suzuki M, Taiyoji M, Nikaidou N, Watanabe T. Chitin binding protein (CBP21) in the culture supernatant of Serratia marcescens 2170. Biosci Biotechnol Biochem. 1998;62:128–135. doi: 10.1271/bbb.62.128. [DOI] [PubMed] [Google Scholar]
- 31.Tanaka H, Phaff H J. Enzymatic hydrolysis of yeast cell walls. J Bacteriol. 1976;89:1570–1580. doi: 10.1128/jb.89.6.1570-1580.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tomme P, Driver D P, Amandoron E A, Miller R C, Jr, Antony R, Warren J, Kilburn D G. Comparison of a fungal (family I) and bacterial (family II) cellulose-binding domain. J Bacteriol. 1995;177:4356–4363. doi: 10.1128/jb.177.15.4356-4363.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tomme P, Van Tilbeurgh H, Pettersson G, Van Damme J, Vandekerckhove J, Knowles J, Teeri T, Claeyssens M. Studies of the cellulolytic system of Trichoderma reesei QM 9414. Analysis of domain function in two cellobiohydrolases by limited proteolysis. Eur J Biochem. 1988;170:575–581. doi: 10.1111/j.1432-1033.1988.tb13736.x. [DOI] [PubMed] [Google Scholar]
- 34.Tomme P, Gilkes N R, Miller R C, Jr, Warren A J, Kilburn D G. An internal cellulose-binding domain mediates adsorption of an engineered bifunctional xylanase/cellulase. Protein Eng. 1994;7:117–123. doi: 10.1093/protein/7.1.117. [DOI] [PubMed] [Google Scholar]
- 35.Tormo J, Lamed R, Chirino A J, Morag E, Bayer E A, Shoham Y, Steitz T A. Crystal structure of a bacterial family-III cellulose-binding domain: a general mechanism for attachment to cellulose. EMBO J. 1996;15:5739–5751. [PMC free article] [PubMed] [Google Scholar]
- 36.Tsujibo H, Orikoshi H, Shiotani K, Hayashi M, Umeda J, Miyamoto K, Imada C, Okami Y, Inamori Y. Characterization of chitinase C from a marine bacterium, Alteromonas sp. strain O-7, and its corresponding gene and domain structure. Appl Environ Microbiol. 1998;64:472–478. doi: 10.1128/aem.64.2.472-478.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Watanabe T, Kobori K, Miyashita K, Fujii T, Sakai H, Uchida M, Tanaka H. Identification of glutamic acid 204 and aspartic acid 200 in chitinase A1 of Bacillus circulans WL-12 as essential residues for chitinase activity. J Biol Chem. 1993;268:18567–18572. [PubMed] [Google Scholar]
- 38.Watanabe T, Suzuki K, Oyanagi W, Ohnishi K, Tanaka H. Gene cloning of chitinase A1 from Bacillus circulans WL-12 revealed its evolutionary relationship to Serratia chitinase and to the type III homology units of fibronectin. J Biol Chem. 1990;265:15659–15665. [PubMed] [Google Scholar]
- 39.Watanabe T, Oyanagi W, Suzuki K, Tanaka H. Chitinase system of Bacillus circulans WL-12 and importance of chitinase A1 in chitin degradation. J Bacteriol. 1990;172:4017–4022. doi: 10.1128/jb.172.7.4017-4022.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Watanabe T, Ito Y, Yamada T, Hashimoto M, Sekine S, Tanaka H. The roles of the C-terminal domain and type III domains of chitinase A1 from Bacillus circulans WL-12 in chitin degradation. J Bacteriol. 1994;176:4465–4472. doi: 10.1128/jb.176.15.4465-4472.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wiseman T, Williston S, Brandts J F, Lin L-N. Rapid measurement of binding constants and heats of binding using a new titration calorimeter. Anal Biochem. 1989;179:131–137. doi: 10.1016/0003-2697(89)90213-3. [DOI] [PubMed] [Google Scholar]
- 42.Xu G Y, Ong E, Gilkes N R, Kilburn D G, Muhandiram D R, Harris-Brandts M, Carver J P, Kay L E, Harvey T S. Solution structure of a cellulose-binding domain from Cellulomonas fimi by nuclear magnetic resonance spectroscopy. Biochemistry. 1995;34:6993–7009. [PubMed] [Google Scholar]
- 43.Yamada H, Imoto T. A convenient synthesis of glycolchitin, a substrate of lysozyme. Carbohydr Res. 1981;92:160–162. doi: 10.1016/s0008-6215(00)85993-5. [DOI] [PubMed] [Google Scholar]