Abstract
Histopathological image analysis is the gold standard for pathologists to grade colorectal cancers of different differentiation types. However, the diagnosis by pathologists is highly subjective and prone to misdiagnosis. In this study, we constructed a new attention mechanism named MCCBAM based on channel attention mechanism and spatial attention mechanism, and developed a computer-aided diagnosis (CAD) method based on CNN and MCCBAM, called HCCANet. In this study, 630 histopathology images processed with Gaussian filtering denoising were included and gradient-weighted class activation map (Grad-CAM) was used to visualize regions of interest in HCCANet to improve its interpretability. The experimental results show that the proposed HCCANet model outperforms four advanced deep learning (ResNet50, MobileNetV2, Xception, and DenseNet121) and four classical machine learning (KNN, NB, RF, and SVM) techniques, achieved 90.2%, 85%, and 86.7% classification accuracy for colorectal cancers with high, medium, and low differentiation levels, respectively, with an overall accuracy of 87.3% and an average AUC value of 0.9.In addition, the MCCBAM constructed in this study outperforms several commonly used attention mechanisms SAM, SENet, SKNet, Non_Local, CBAM, and BAM on the backbone network. In conclusion, the HCCANet model proposed in this study is feasible for postoperative adjuvant diagnosis and grading of colorectal cancer.
Subject terms: Colorectal cancer, Image processing, Machine learning
Introduction
Colorectal cancer (CRC), is a highly malignant tumor that forms in the tissues of the colon and rectum1,2. According to the American Cancer Society's Global Cancer Statistics 2021, there will be more than 1.9 million new cases of colorectal cancer and more than 900,000 deaths in 2020, making it the third leading cause of cancer death in the world, after lung cancer and breast cancer3. More than 90% of CRC cases are colorectal adenocarcinoma (CRA), which can be classified into grades I to IV according to Broder's criteria, i.e., highly differentiated (I), moderately differentiated (II), poorly differentiated (III), and undifferentiated (IV), based on the degree of glandular differentiation in histopathological images of colorectal cancer. The histological grading of colorectal cancer is not only a reference basis for assessing its malignancy and staging, but also an important factor affecting its prognosis4. Histopathological image analysis is the gold standard for pathologists to grade colorectal cancers of different differentiation types. In recent years, with the increasing number of colorectal cancer patients1,2,5, the workload of physicians is increasing day by day. In addition, the low differentiation of histopathological images of colorectal cancers of different differentiation types makes the diagnosis complicated and time-consuming6, which may lead to misdiagnosis and missed diagnosis7. Although gastroenterology clinics have a high demand for colon specimens, pathologists have a long training period (> 10 years)8. According to the Chinese Association of Pathologists, China, a country of 1.4 billion people, has only 20,000 professionally accredited pathologists9. Therefore, it is particularly important to build an efficient computerized automatic diagnostic model to effectively identify histopathological images of colorectal cancer at multiple levels, and then assist pathologist in objective diagnosis and grading.
With the rapid development of artificial intelligence technology in the medical field10–12 more and more CAD systems, especially convolutional neural network (CNN)-based CAD systems, are applied to automatic analysis tasks of histopathological images, such as cell nucleus detection and classification13, tumor segmentation14, tumor metastasis detection15,16, and cancer grading17. However, when faced with smaller medical image datasets, CNN models often fail to extract effective information from the dataset. This drawback makes it particularly important to combine CNN models with attention mechanisms18.
In this study, a new convolutional neural network and attention mechanism-based model, HCCANet, was proposed for grading colorectal cancers with different differentiation types. A total of 630 hematoxylin–eosin (H&E) stained histopathology images were included in this study, and the gaussian filtered images were fed into a fine-tuned VGG16 backbone network to extract local features. Then, the MCCBAM module is added in parallel to capture key features that facilitate network classification. Finally, the feature maps of the VGG16 and MCCBAM modules were fused to build the colorectal cancer supplementary diagnosis model HCCANet for the diagnosis of colorectal cancer of three grades: I, II, and III.
In general, the main contributions of this study can be summarized as follows:
This study constructs a new attention mechanism, called MCCBAM, based on multiple channel attention and spatial attention. This attention mechanism outperforms attention modules such as SAM, SENet, SKNet, Non_Local, CBAM, and BAM for classification on the fine-tuned VGG16 model.
In this study, a new automatic colorectal cancer diagnosis model based on a convolutional neural network and MCCBAM, called HCCANet, is proposed. This model enhances feature learning of key regions in histopathological images and outperforms advanced deep learning models and traditional machine learning algorithms in colorectal cancer grading tasks.
In this study, the Grad-CAM visualization method was introduced to convert the model output into a heat map, visualize key regions of interest for the model, enhance the interpretability of the model, and assist pathologists in investigating misdiagnosis cases of false negative and false positive.
Related work
CNN models automatically learn features from input images and build low-level features into high-level features, and have had great success in computer vision fields such as image classification, image segmentation, and object detection tasks19–21. Recently, an increasing number of researchers have used CNNs as an aid in the diagnosis of colorectal cancer. Yoon et al. proposed an improved VGG model for classifying normal and tumor tissue from 10,280 colorectal histological images with an accuracy of 82%22. Ponzio et al. used a pre-trained VGG16 model for migration learning to classify colorectal histopathology images into normal, adenoma, and adenocarcinoma categories and obtained 96% classification accuracy23. Nguyen et al. used a combined model of classical CNN and CapsNet to classify histopathological images of 410 patients into three categories: tumor, normal epithelium, and other tissue types, achieving a multi-classification accuracy of 95.3%24. Zhou et al. proposed a new cell-graph convolutional neural network (CGC-Net) to classify colorectal histopathological images into low-grade cancer (Highly differentiated and moderately differentiated colorectal cancer) and high-grade cancer (Poorly differentiated and undifferentiated colorectal cancer), obtaining 91.60% accuracy on patch images25. Shaban et al. proposed a new context-aware neural network for grading colorectal pathological tissue images (normal, low-grade cancer, high-grade cancer) and obtained an average accuracy of 95.70%26. The above-mentioned studies on colorectal cancer grading have classified colorectal cancer into two grades: high-grade cancer and low-grade cancer25,26, but in the actual treatment process, some studies need to classify colorectal cancer into four grades: I (highly differentiated), II (moderately differentiated), III (undifferentiated), and IV (undifferentiated)4. In addition, some deep learning models based on histopathological images do not perform well on small medical datasets, or even as well as traditional machine learning27,28.
Attention mechanisms (AM) derived from human intuition have been widely used in computer vision, and AM allocates computational resources to the most informative parts of the signal, bringing significant improvements to many visual processing tasks. For example, tasks such as image classification29, object detection30, action recognition31, pose estimation32, and super-resolution33. At present, some investigators have introduced the attention mechanism into the CAD system of colorectal cancer. Pei et al. proposed a model based on a convolutional neural network and attention mechanism to automate colorectal cancer tumor segmentation, which includes a channel attention module and a location attention module to obtain more contextual information in the deeper layers of the network34. Chen et al. proposed a weakly supervised colorectal histopathology image classification model based on interactive learning and multichannel attention mechanisms, which identifies attention regions as accurately as possible in both channel and spatial dimensions by integrating different attention mechanisms35. Although the attentional mechanisms used in the above studies performed well for tasks such as segmentation and classification, they performed poorly for histological grading of colorectal cancer.
In summary, a new attention mechanism called MCCBAM was constructed in this study, and a new model based on convolutional neural network and MCCBAM, HCCANet, was proposed to assist in the diagnosis of histopathological images of colorectal cancer with three different differentiation types: high, medium and low.
Materials and methods
Materials
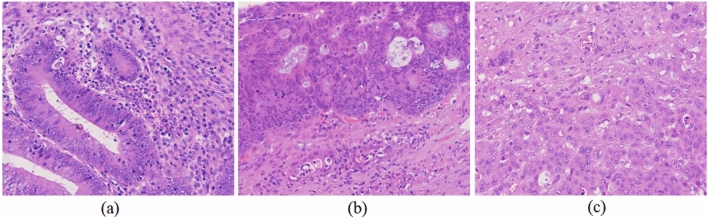
In this study, 105 patients were enrolled in the Cancer Hospital of Xinjiang Medical University between 2012 and 2021, including 35 patients each with colorectal cancer of grades I, II, and III differentiation. The grading of colorectal cancer is based on the degree of glandular differentiation, with grades I, II, and III corresponding to > 95%, 50–95%, and 5–50% of glandular differentiation, respectively. The histological sections of colorectal cancers included in the study were confirmed by postoperative tissue biopsy and were retrieved from the pathology department of the hospital, and two experienced histopathologists labeled the ROI of each patient's tissue section. Sixty of these patients were male (age range, [36–85]) and 45 were female (age range, [23–71]). Histopathological images of colorectal cancers are shown in Fig. 1.
Figure 1.
Different differentiation types of colorectal cancer by digital pathological imager at 40 times magnification. (a) Highly differentiated. (b) Moderately differentiated. (c) Highly differentiated.
The acquisition of the final image used in the experiment consists of two steps. First, the pathological tissue sections were placed horizontally under the digital pathology imager, and two experienced histopathologists selected and labeled ROIs on each patient's tissue section in turn. Second, six non-overlapping images of 1665 × 1393 pixels were extracted from the ROIs at 40 × magnification, and 210 images of each of the three differentiation grades of colorectal cancer, high, medium, and low, were extracted, totaling 630 images. The child images extracted from the histopathological images of the parent images were verified by the pathologist to be of the same differentiation level as the parent image. The details of patients are shown in Table 1.
Table 1.
Patient information sheet.
| Information | Value |
|---|---|
| Gender | |
| Male | 60 (number) |
| Female | 45 (number) |
| Age | |
| Male | 57.97 (average age) |
| Female | 55.23 (average age) |
| Number of patients | |
| High differentiation (I) | 35 |
| Medium differentiation (II) | 35 |
| Low differentiation (III) | 35 |
| Number of images | |
| High differentiation | 210 |
| Medium differentiation | 210 |
| Low differentiation | 210 |
Data augmentation
Due to the limited number of samples in the dataset used in this study, a deep neural network trained with a dataset of this size is risky, and the network is likely to be overfitted due to the small dataset. Therefore, the number of training images is increased using data augmentation methods. First, the data set is divided into a training set, validation set, and test set according to the ratio of 8:1:1. Second, the training set is augmented to 4500 sheets, including rotation, cropping, scaling, etc.
Image processing for model training
Image pre-processing work can improve the performance of the model to some extent36. In this study, the image pre-processing work includes three points, first, the image noise processing: this study uses four filtering techniques: mean filtering, median filtering, gaussian filtering, and bilateral filtering to de-noise the image respectively. The best filtering technique was selected by comparing the performance of the images on HCCANet after different filtering techniques. Second, image resizing: the original pixel size of 1665 × 1393 is scaled to 224 × 224 to better fit the backbone network in the HCCANet model7,37,38. Third, image normalization: The image is normalized by calling the image preprocessing method scale() of the Sklearn.preprocessing module. The scale() method subtracts the data by its attributes from its mean value and divides it by its variance so that all data for each attribute are clustered around 0 and the variance value is 1.
Methods
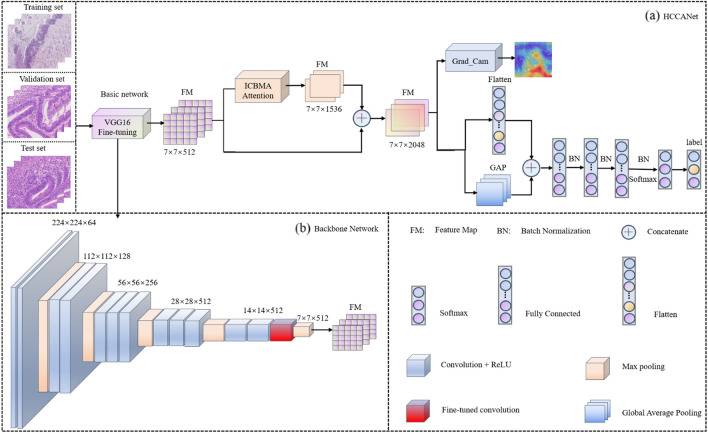
In this study, a cleverly designed network structure named HCCANet is proposed to perform the task of grading histopathological images of colorectal cancer of different differentiation types. Figure 2a shows the overall architecture of HCCANet. Figure 2b shows the visual attention mechanism module MCCBAM constructed in this paper.
Figure 2.
The framework of HCCANet. (a) The overall architecture of HCCANet. (b) The backbone of HCCANet.
HCCANet consists of two parts, the backbone network VGG16 and the multichannel converged attention mechanism MCCBAM. In the medical field, one of the main challenges in adopting deep learning models is the lack of training data due to the difficulty in collecting and labeling data39. To make the VGG16 model more applicable to the dataset in this study and reduce the risk of overfitting the model on small datasets, we migrate the weights of the VGG16 model trained on the ImageNet dataset and fine-tune the Block_Conv3 layer of the VGG16 model (the module marked in red in Fig. 2b).
MCCBAM attention mechanism
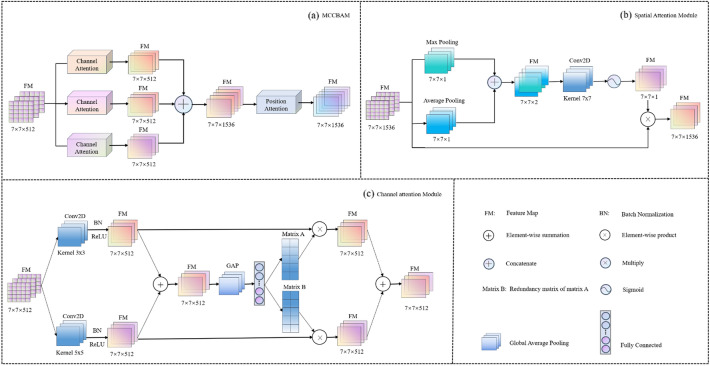
AM has become one of the most essential concepts in the field of deep learning40. However, traditional AMs have some drawbacks. For example, individual AMs may have difficulty in capturing useful features, AMs may capture redundant information, etc. To reduce the drawbacks of AMs, we construct a new attention module called MCCBAM. The module consists of three parallel SKNets41 and Spatial Attention Mechanism (SAM)29, and Fig. 3a shows the overall architecture of MCCBAM.
Figure 3.
The framework of MCCBAM. (a) The overall architecture of MCCBAM. (b) Components of the Spatial Attention Block. (c) Components of the Channel Attention Block.
The MCCBAM module consists of two parts, namely SKNet and SAM. The processing of features in the MCCBAM module consists of two steps: First, the features are processed by SKNet with three reduction ratios of 4, 8, and 16, and the processed features are fused by concatenating. Second, the fused features are fed into the spatial attention mechanism with a kernel size of 7 for further processing. As shown in Fig. 3a, given an intermediate feature map as input, MCCBAM sequentially derives three-channel attention maps and one spatial attention map , and the whole attention process can be summarized as follows:
| 1 |
where is the final output feature map, the superscript r in represents the magnitude of the reduction ratio, and means the element multiplication.
The essence of the channel attention module comes from the squeeze and excitation network42. Its essence is to allow the network to use global information to selectively enhance beneficial feature channels and suppress useless ones, thus enabling adaptive channel selection. SKNet is a new channel attention mechanism in which each neuron in the module adapts the size of its receptive field to capture target objects at different scales according to the multiple scales of the input information41. As shown in Fig. 3c, given an intermediate feature map as input, channel attention derives a channel attention map , and the whole attention process can be summarized as:
| 2 |
| 3 |
where denotes Global Average Pooling, denotes Fully Connected, ⊕ means element summing, and represent matrices.
Unlike channel attention, spatial attention adopts a global perspective to learn the connections between voxels and tasks, focusing on the spatial location information of key features by establishing rich contextual relationships between local features and assigning different weights to them43. As shown in Fig. 3b, given an intermediate feature map as input, the spatial attention is derived as a spatial attention map , and the whole attention process can be summarized as:
| 4 |
where denotes sigmoid function, represents a convolution operation with the filter size of 7 × 7, and represent the average pooling and maximum pooling operations, respectively.
CNN-based classifiers for comparison
This study used four advanced deep learning models ResNet50, MobileNetV2, Xception, and DenseNet121 to build the classifier. Inspired by Tajbakhsh44, four CNN models were fine-tuned using weights pre-trained on ImageNet. Araújo27 and Yan28 showed that features extracted with a pre-trained CNN were able to achieve better performance than some end-to-end CNN classifiers on SVM classifiers. Therefore, this study uses a pre-trained VGG16 network to extract features and uses the extracted features to train classifiers such as KNN, RF, NB, and SVM. The classification performance of the above models is compared with that of HCCANet, and the specific experimental configuration is shown in Table 2.
Table 2.
Hyperparameter settings for each classifier.
| Model name | Hyper-parameters |
|---|---|
| Deep learning algorithms | |
| ResNet50 |
Input size: (224, 224, 3), Learning rate: 0.005, Epochs: 100 Optimizer: Adam ( 9), Batch size: 32 Loss function: Categorical Cross-Entropy |
| MobileNetV2 | |
| Xception | |
| DenseNet121 | |
| Traditional machine learning algorithms | |
| KNN | Neighbors: 5 |
| RF | Estimators: 850, Random state: 0, Bootstrap: True |
| NB | Alpha: 1.0 |
| SVM | Kernel: RBF, C: 1.0, Gamma: 0.005 |
Performance evaluation
In this study, receiver operating characteristic (ROC) curves and confusion matrices were plotted to assess the performance of HCCANet in terms of accuracy and reliability. Area under curve (AUC) is a quantitative measure of the model's performance, and the closer the value of AUC is to 1, the better the model performs. In addition, we also calculate the precision, recall, F1-score, and accuracy of the model when predicting samples to evaluate the model, and these metrics are calculated as shown in Table 3.
Table 3.
Performance metric calculation formulas.
| Performance metric | Precision | Recall | F1-score | Accuracy |
|---|---|---|---|---|
| Formula |
Informed consent
This study has been approved by the Cancer Affiliated Hospital of Xinjiang Medical University (in these studies). Informed consent was obtained from all participants before participating in the interview study. All methods were carried out in accordance with relevant guidelines and regulations (e.g. Helsinki guidelines). This article is based on the project "PI3K/AKT and MEK/ERK signaling pathways in microRNA-106b-induced epithelial transformation process in colorectal cancer cells the process of mesothelial transformation in colorectal cancer cells", which was approved by the ethics committee of the Cancer Hospital of Xinjiang Medical University, so the article does not require a separate ethics report.
Results
All experiments in this study are based on the Python programming language, using TensorFlow-GPU deep learning framework to build the deep learning models needed during the experiments, and using GeForce RTX 1080ti for training. The Sklearn machine learning library was used to build the machine learners needed for the experiments. All classifiers were trained using a five-fold cross-validation method.
Selection of filters or image denoising
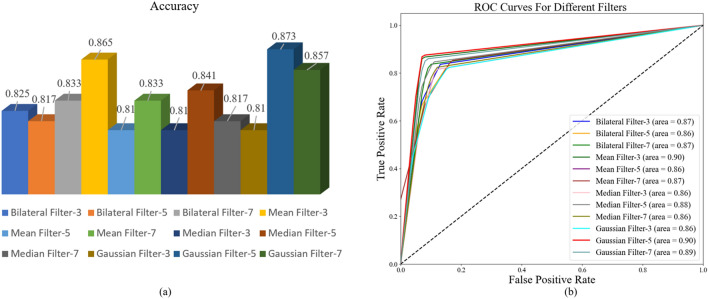
Medical images usually have a noise component, and the removal of this noise is essential for medical diagnosis45. To reduce the impact of the noise present on medical images on the classification performance of the model, four filtering techniques, namely, mean filter, median filter, Gaussian filter, and bilateral filter, are used in this study to de-noise the images respectively. A cross-sectional comparison of the precision, recall, F1-score, and accuracy of HCCANet based on different filtering techniques on histopathological images of colorectal cancer with different differentiation types was performed to select the optimal filter. From Table 4, it can be seen that choosing a Gaussian filter with a kernel size of 5 can improve the ability of HCCANet for automatic diagnosis of colorectal cancer histopathology images. Figure 4a,b shows the performance of HCCANet based on different filters in terms of accuracy and AUC values, respectively. (See Supplementary Material for the confusion matrix).
Table 4.
Comparison of the denoising effect of different filtering techniques.
| Filter type/kernel size | Grading | Precision | Recall | F1-score | Accuracy |
|---|---|---|---|---|---|
| Mean filtering/3 | I | 0.87 | 0.952 | 0.91 | 0.865 |
| II | 0.872 | 0.81 | 0.84 | ||
| III | 0.854 | 0.833 | 0.843 | ||
| Mean filtering/5 | I | 0.868 | 0.786 | 0.825 | 0.810 |
| II | 0.775 | 0.738 | 0.756 | ||
| III | 0.792 | 0.905 | 0.844 | ||
| Mean filtering/7 | I | 1.00 | 0.810 | 0.895 | 0.833 |
| II | 0.816 | 0.738 | 0.775 | ||
| III | 0.741 | 0.952 | 0.833 | ||
| Median filtering/3 | I | 0.804 | 0.881 | 0.841 | 0.810 |
| II | 0.767 | 0.786 | 0.776 | ||
| III | 0.865 | 0.762 | 0.810 | ||
| Median filtering/5 | I | 0.860 | 0.881 | 0.871 | 0.841 |
| II | 0.795 | 0.833 | 0.814 | ||
| III | 0.872 | 0.810 | 0.840 | ||
| Median filtering/7 | I | 0.826 | 0.905 | 0.864 | 0.817 |
| II | 0.861 | 0.738 | 0.795 | ||
| III | 0.773 | 0.810 | 0.791 | ||
| Bilateral filtering/3 | I | 0.860 | 0.881 | 0.871 | 0.825 |
| II | 0.848 | 0.667 | 0.747 | ||
| III | 0.780 | 0.929 | 0.848 | ||
| Bilateral Filtering / 5 | I | 0.854 | 0.833 | 0.843 | 0.817 |
| II | 0.729 | 0.833 | 0.778 | ||
| III | 0.892 | 0.786 | 0.835 | ||
| Gaussian filtering/3 | I | 0.917 | 0.786 | 0.846 | 0.810 |
| II | 0.723 | 0.810 | 0.764 | ||
| III | 0.814 | 0.833 | 0.824 | ||
| Gaussian filtering/5 | I | 0.902 | 0.881 | 0.892 | 0.873 |
| II | 0.850 | 0.810 | 0.829 | ||
| III | 0.867 | 0.929 | 0.897 | ||
| Gaussian filtering/7 | I | 0.860 | 0.881 | 0.871 | 0.857 |
| II | 0.833 | 0.833 | 0.833 | ||
| III | 0.878 | 0.857 | 0.867 |
Figure 4.
(a) Accuracy of HCCANet based on different filters. (b) AUC values for HCCANet based on different filters.
Comparison of MCCBAM with other attention mechanisms
To evaluate the performance of the MCCBAM attention mechanism constructed in this study on the histopathological image grading of colorectal carcinoma, we incorporated six commonly used attention mechanisms, SAM, SENet42, SKNet, Non_Local31, CBAM29, and BAM to construct a comparison experiment. All attention mechanisms are added to the tail of the same backbone network in a parallel manner. The experimental results showed that the model based on the MCCBAM attention mechanism outperformed other attention mechanisms in terms of precision, recall, F1-score, and accuracy, demonstrating the superiority and usability of the MCCBAM attention mechanism constructed in this study for histopathological image grading of colorectal carcinoma. The experimental results are shown in Table 5. Figure 5a,b shows the performance of the VGG16 backbone network based on different attention mechanisms in terms of accuracy and AUC values, respectively. (See Supplementary Material for the confusion matrix).
Table 5.
Comparison of MCCBAM with other attention mechanisms.
| Attention mechanism | Grading | Precision | Recall | F1-score | Accuracy |
|---|---|---|---|---|---|
| SAM | I | 0.769 | 0.714 | 0.741 | 0.754 |
| II | 0.698 | 0.714 | 0.706 | ||
| III | 0.795 | 0.833 | 0.814 | ||
| SENet | I | 0.696 | 0.929 | 0.796 | 0.762 |
| II | 0.806 | 0.595 | 0.685 | ||
| III | 0.821 | 0.762 | 0.790 | ||
| SKNet | I | 0.878 | 0.857 | 0.867 | 0.833 |
| II | 0.761 | 0.833 | 0.795 | ||
| III | 0.872 | 0.810 | 0.840 | ||
| Non_Local | I | 0.857 | 0.857 | 0.857 | 0.841 |
| II | 0.846 | 0.786 | 0.815 | ||
| III | 0,0.822 | 0.881 | 0.851 | ||
| CBAM | I | 0.850 | 0.810 | 0.829 | 0.817 |
| II | 0.786 | 0.786 | 0.786 | ||
| III | 0.818 | 0.857 | 0.837 | ||
| BAM | I | 0.923 | 0.857 | 0.889 | 0.833 |
| II | 0.767 | 0.786 | 0.776 | ||
| III | 0.818 | 0.857 | 0.837 | ||
| MCCBAM | I | 0.902 | 0.881 | 0.892 | 0.873 |
| II | 0.850 | 0.810 | 0.829 | ||
| III | 0.867 | 0.929 | 0.897 |
Figure 5.
(a) Accuracy of VGG16 based on different attention mechanisms. (b) AUC values of VGG16 based on different attention mechanisms.
CNN-based classifiers for comparison
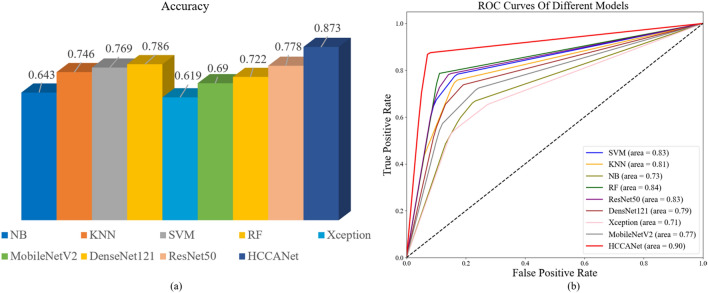
This study uses four advanced deep learning models ResNet50, MobileNetV2, Xception, and DenseNet121, and four commonly used machine learning models KNN, RF, NB, and SVM to build classifiers for training and compare them with HCCANet. From Table 6, the average classification accuracy of HCCANet is 9.5% higher than that of ResNet50, 18.3% higher than that of MobileNetV2, 25.4% higher than that of Xception, 15.1% higher than that of DenseNet121, 12.7% higher than that of KNN, 8.7% higher than that of RF, 23% higher than that of NB, and 10.4% higher than that of SVM 10.4%. The experimental results show that HCCANet outperforms other classifiers in terms of precision, recall, F1-score, and accuracy, proving the superiority and usability of the HCCANet model for histopathological image grading of colorectal cancer. Figure 6a,b shows the accuracy and AUC values of different models for grading histopathological images, respectively. (See Supplementary Material for the confusion matrix).
Table 6.
CNN-based classifiers for comparison.
| Classifier | Grading | Precision | Recall | F1-Score | Accuracy |
|---|---|---|---|---|---|
| ResNet50 | I | 0.786 | 0.786 | 0.786 | 0.778 |
| II | 0.714 | 0.714 | 0.714 | ||
| III | 0.833 | 0.833 | 0.833 | ||
| MobileNetV2 | I | 0.727 | 0.571 | 0.640 | 0.690 |
| II | 0.608 | 0.738 | 0.667 | ||
| III | 0.762 | 0.762 | 0.762 | ||
| Xception | I | 0.676 | 0.595 | 0.632 | 0.619 |
| II | 0.511 | 0.571 | 0.539 | ||
| III | 0.690 | 0.690 | 0.690 | ||
| DenseNet121 | I | 0.789 | 0.714 | 0.750 | 0.722 |
| II | 0.644 | 0.690 | 0.667 | ||
| III | 0.744 | 0.762 | 0.753 | ||
| KNN | I | 0.865 | 0.762 | 0.810 | 0.746 |
| II | 0.675 | 0.643 | 0.659 | ||
| III | 0.714 | 0.833 | 0.769 | ||
| RF | I | 0.791 | 0.810 | 0.800 | 0.786 |
| II | 0.757 | 0.667 | 0.709 | ||
| III | 0.804 | 0.881 | 0.841 | ||
| NB | I | 0.703 | 0.619 | 0.658 | 0.643 |
| II | 0.583 | 0.500 | 0.583 | ||
| III | 0.642 | 0.810 | 0.716 | ||
| SVM | I | 0.805 | 0.786 | 0.795 | 0.769 |
| II | 0.689 | 0.738 | 0.713 | ||
| III | 0.825 | 0.786 | 0.805 | ||
| HCCANet | I | 0.902 | 0.881 | 0.892 | 0.873 |
| II | 0.850 | 0.810 | 0.829 | ||
| III | 0.867 | 0.929 | 0.897 |
Figure 6.
(a) Accuracy of different models in grading histopathological images. (b) AUC values for different models on histopathological image grading.
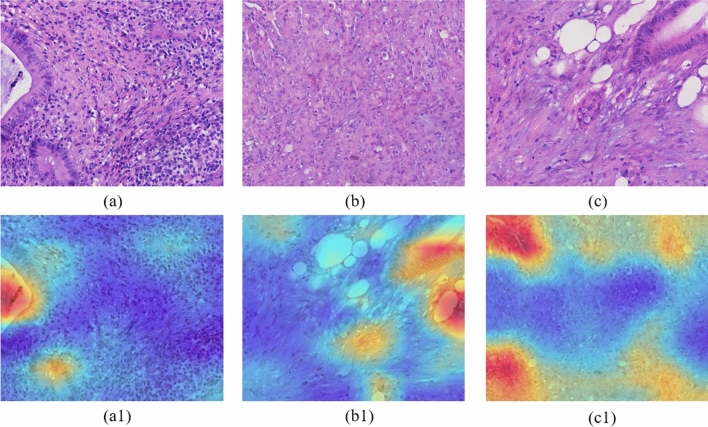
Grad-CAM visual analysis
Grad_CAM is a widely used method for visualizing feature maps that uses gradients to calculate the importance of spatial locations in a convolutional layer46. In the heat map generated by Grad_CAM, the blue color is the unimportant region, while the red color is the critical region associated with that category. The classifier makes judgments based on these local pixel-level features in red. As shown in Fig. 7, the upper part is the histopathological image of colorectal cancer stained by H&E, and the lower part is the Grad-CAMs generated by HCCANet after extracting relevant features. Figure 7a–c shows the images of colorectal cancer at grade I, grade II, and grade III stages (i.e., highly differentiated, moderately differentiated, and poorly differentiated stages, respectively), Fig. 7(a1), (b1) and (c1) are their corresponding Grad-CAMs.
Figure 7.
Histopathological images of colorectal cancer and its corresponding CAMs.
Discussion
In this study, we propose a new computer-aided diagnostic model that can use histopathological images to distinguish three differentiation grades of colorectal cancer: high, intermediate, and low. Due to the scarce and precious nature of medical images, the model often fails to perform as expected in solving actual disease diagnosis using advanced deep learning models. To address these issues, we use image denoising, image enhancement, weight migration and model fine-tuning to make the deep learning models perform better on the dataset in this study. In addition, this study constructs a new attention mechanism, called MCCBAM, based on channel attention and spatial attention, which outperforms multiple current state-of-the-art attention mechanisms in the colorectal cancer 3-level grading task in this study, resulting in a large improvement in the discriminative power of the model.
This study combines MCCBAM and a fine-tuned VGG16 network architecture to construct a new model for histological grading of colorectal cancer, called HCCANet. To our knowledge, this is the first study that combines deep learning and attentional mechanisms to grade colorectal cancer of different differentiation types. The model has a good performance on the dataset used in this study, and the classification performance is better than existing deep learning models and classical machine learning models, which has some practical value in solving the realistic problems of manual grading of colorectal cancer after surgery to some extent. In addition, the gradient-weighted class activation map (Grad-CAM) visualization method is used to display the fused feature maps, which can improve the interpretability of the model and better help pathologists understand the output feature maps of HCCANet.
In the future, we will continue to collect more samples from different center institutions to build a companion diagnostic model with better performance and higher generalization ability, so as to make greater use of the clinical significance of postoperative diagnosis of colorectal cancer. In addition, we plan to combine histopathological images with clinical data to build a complementary diagnostic model based on multimodal information.
Supplementary Information
Author contributions
P.Z. developed the computer-aided diagnosis system, trained and evaluated the convolutional neural network model. P.Z., M.L. and Y.C. drafted the manuscript. Y.C. collected and labeled the dataset. X.G., Y.M., J.W. and C.C. participated in the study design and revised the manuscript. X.L., M.L. and C.C. provided guidance and reviewed the experiments and manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the Key Laboratory of Clinical Gene Detection and Biomedical Information of Xinjiang; the Xinjiang Uygur Autonomous Region Science Foundation for Distinguished Young Scholars [grant number 2022D01E11&2022D01E27]; the National Natural Science Foundation of China [grant number 81860430]; the Xinjiang Autonomous Region Science and Technology Plan Project [grant number 2018D01C257]; the Karamay Central Hospital Project: Research on Molecular Mechanism and Application of DNA Methylation Liquid Biopsy in the "Prevention, Diagnosis and Treatment" of Malignant Tumors.
Data availability
The datasets generated and analyzed during the current study are not publicly available due to data privacy laws, but are available from the corresponding author on reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Panyun Zhou and Yanzhen Cao.
Contributor Information
Xiaoyi Lv, Email: xjuwawj01@163.com.
Cheng Chen, Email: chenchengoptics@gmail.com.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-022-18879-1.
References
- 1.Sung H, et al. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Mattiuzzi C, Sanchis-Gomar F, Lippi G. Concise update on colorectal cancer epidemiology. Ann. Transl. Med. 2019;7:609. doi: 10.21037/atm.2019.07.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sung H, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 4.Chen S, et al. Automatic tumor grading on colorectal cancer whole-slide images: Semi-quantitative gland formation percentage and new indicator exploration. Front. Oncol. 2022 doi: 10.3389/fonc.2022.833978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Freddie, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 6.Komura D, Ishikawa S. Machine learning methods for histopathological image analysis. Comput. Struct. Biotechnol. J. 2018;16:34–42. doi: 10.1016/j.csbj.2018.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim S-H, Koh HM, Lee B-D. Classification of colorectal cancer in histological images using deep neural networks: An investigation. Multimed. Tools Appl. 2021 doi: 10.1007/s11042-021-10551-6. [DOI] [Google Scholar]
- 8.Black-Schaffer WS, Morrow JS, Prystowsky MB, Steinberg JJ. Training pathology residents to practice 21st century medicine: A proposal. Acad. Pathol. 2016;3:2374289516665393. doi: 10.1177/2374289516665393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun H, Zeng X, Xu T, Peng G, Ma Y. Computer-aided diagnosis in histopathological images of the endometrium using a convolutional neural network and attention mechanisms. IEEE J. Biomed. Health Inform. 2019;24:1664–1676. doi: 10.1109/JBHI.2019.2944977. [DOI] [PubMed] [Google Scholar]
- 10.Chen C, et al. Raman spectroscopy combined with multiple algorithms for analysis and rapid screening of chronic renal failure. Photodiagn. Photodyn. Ther. 2020;30:101792. doi: 10.1016/j.pdpdt.2020.101792. [DOI] [PubMed] [Google Scholar]
- 11.Yue F, et al. Fourier transform infrared spectroscopy combined with deep learning and data enhancement for quick diagnosis of abnormal thyroid function. Photodiagn. Photodyn. Ther. 2020;32:101923. doi: 10.1016/j.pdpdt.2020.101923. [DOI] [PubMed] [Google Scholar]
- 12.Chen C, et al. Urine Raman spectroscopy for rapid and inexpensive diagnosis of chronic renal failure (CRF) using multiple classification algorithms. Optik. 2020;203:164043. doi: 10.1016/j.ijleo.2019.164043. [DOI] [Google Scholar]
- 13.Song TH, Sanchez V, Eidaly H, Rajpoot NM. Simultaneous cell detection and classification in bone marrow histology images. IEEE J. Biomed. Health Inf. 2019;23:1469–1476. doi: 10.1109/JBHI.2018.2878945. [DOI] [PubMed] [Google Scholar]
- 14.Shirazi AZ, et al. A deep convolutional neural network for segmentation of whole-slide pathology images identifies novel tumour cell-perivascular niche interactions that are associated with poor survival in glioblastoma. Br. J. Cancer. 2021;125:337–350. doi: 10.1038/s41416-021-01394-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koohbanani NA, Qaisar T, Shaban M, Gamper J, Rajpoot N. Significance of Hyperparameter Optimization for Metastasis Detection in Breast Histology Images. Springer; 2018. [Google Scholar]
- 16.Lin H, et al. Fast ScanNet: Fast and dense analysis of multi-gigapixel whole-slide images for cancer metastasis detection. IEEE Trans. Med. Imaging. 2019;38:1948–12958. doi: 10.1109/TMI.2019.2891305. [DOI] [PubMed] [Google Scholar]
- 17.Arvaniti E, et al. Automated Gleason grading of prostate cancer tissue microarrays via deep learning. Eur. Urol. Suppl. 2018;17:e3020–e3021. doi: 10.1016/S1569-9056(18)33852-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen H, et al. IL-MCAM: An interactive learning and multi-channel attention mechanism-based weakly supervised colorectal histopathology image classification approach. Comput. Biol. Med. 2022;143:105265. doi: 10.1016/j.compbiomed.2022.105265. [DOI] [PubMed] [Google Scholar]
- 19.Lei H, et al. A deeply supervised residual network for HEp-2 cell classification via cross-modal transfer learning. Pattern Recogn. 2018;79:290–302. doi: 10.1016/j.patcog.2018.02.006. [DOI] [Google Scholar]
- 20.Cai, Z. & Vasconcelos, N. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition 6154–6162.
- 21.Tang, P. et al. In Proceedings of the European Conference on Computer Vision (ECCV) 352–368.
- 22.Yoon H, et al. Tumor identification in colorectal histology images using a convolutional neural network. J. Dig. Imaging. 2019;32:131–140. doi: 10.1007/s10278-018-0112-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ponzio, F., Macii, E., Ficarra, E. & Cataldo, S. D. In 5th International Conference on Bioimaging.
- 24.Nguyen, H.-G., Blank, A., Lugli, A. & Zlobec, I. In 2020 IEEE 17th International Symposium on Biomedical Imaging (ISBI) 1271–1274 (IEEE).
- 25.Zhou, Y. et al. CGC-Net: Cell graph convolutional network for grading of colorectal cancer histology images. IEEE (2020). [DOI] [PubMed]
- 26.Shaban M, et al. Context-aware convolutional neural network for grading of colorectal cancer histology images. IEEE Trans. Med. Imaging. 2020;39:2395–2405. doi: 10.1109/TMI.2020.2971006. [DOI] [PubMed] [Google Scholar]
- 27.Araújo T, et al. Classification of breast cancer histology images using convolutional neural networks. PLoS ONE. 2017;12:e0177544. doi: 10.1371/journal.pone.0177544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu Y, et al. Large scale tissue histopathology image classification, segmentation, and visualization via deep convolutional activation features. BMC Bioinform. 2017;18:1–17. doi: 10.1186/s12859-017-1685-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Woo, S., Park, J., Lee, J.-Y. & Kweon, I. S. In Proceedings of the European Conference on Computer Vision (ECCV) 3–19.
- 30.Carion, N. et al. In European conference on computer vision 213–229 (Springer).
- 31.Wang, X., Girshick, R., Gupta, A. & He, K. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition 7794–7803.
- 32.Chu, X. et al. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition 1831–1840.
- 33.Dai, T., Cai, J., Zhang, Y., Xia, S.-T. & Zhang, L. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition 11065–11074.
- 34.Pei Y, et al. Colorectal tumor segmentation of CT scans based on a convolutional neural network with an attention mechanism. IEEE ACCESS. 2020;8:64131–64138. doi: 10.1109/ACCESS.2020.2982543. [DOI] [Google Scholar]
- 35.Chen H, et al. IL-MCAM: An interactive learning and multi-channel attention mechanism-based weakly supervised colorectal histopathology image classification approach. Comput. Biol. Med. 2022;143:105265. doi: 10.1016/j.compbiomed.2022.105265. [DOI] [PubMed] [Google Scholar]
- 36.Vasuki, P., Kanimozhi, J. & Devi, M. B. In 2017 IEEE International Conference on Electrical, Instrumentation and Communication Engineering (ICEICE) 1–6 (IEEE).
- 37.Ds A, Rhp A, Ab A, Pa B. Deep learning in image classification using residual network (ResNet) variants for detection of colorectal cancer. Procedia Comput. Sci. 2021;179:423–431. doi: 10.1016/j.procs.2021.01.025. [DOI] [Google Scholar]
- 38.Sarwinda, D., Bustamam, A., Paradisa, R. H., Argyadiva, T. & Mangunwardoyo, W. In 2020 4th International Conference on Informatics and Computational Sciences (ICICoS).
- 39.Alzubaidi L, et al. Towards a better understanding of transfer learning for medical imaging: A case study. Appl. Sci. 2020;10:4523. doi: 10.3390/app10134523. [DOI] [Google Scholar]
- 40.Niu Z, Zhong G, Yu H. A review on the attention mechanism of deep learning. Neurocomputing. 2021;452:48–62. doi: 10.1016/j.neucom.2021.03.091. [DOI] [Google Scholar]
- 41.Li, X., Wang, W., Hu, X. & Yang, J. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition 510–519.
- 42.Hu, J., Shen, L. & Sun, G. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition 7132–7141.
- 43.Chen, L. et al. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition 5659–5667.
- 44.Tajbakhsh N, et al. Convolutional neural networks for medical image analysis: Full training or fine tuning? IEEE Trans. Med. Imaging. 2016;35:1299–1312. doi: 10.1109/TMI.2016.2535302. [DOI] [PubMed] [Google Scholar]
- 45.Ravishankar, A. et al. In 2017 International conference of Electronics, Communication and Aerospace Technology (ICECA) 385–389 (IEEE).
- 46.Hou, Q., Zhou, D. & Feng, J. In 2021 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR) 13708–13717.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and analyzed during the current study are not publicly available due to data privacy laws, but are available from the corresponding author on reasonable request.