Abstract
Ageing and inflammation strongly drive risk of cardiovascular disease. Work over the last decade has uncovered a common condition characterized by the positive selection of certain somatic mutations in haematopoietic stem cells in ageing humans. This phenomenon, clonal haematopoiesis of indeterminate potential (CHIP), occurs most commonly due to mutations in the transcriptional regulators DNMT3A, TET2 and ASXL1. CHIP associates with a variety of adverse outcomes, including haematological cancer and death. Surprisingly, CHIP also associates with a doubling of the risk of atherosclerotic cardiovascular disease. Studies in mice support the causality of this relationship. Mutations in one gene mutated in CHIP, TET2, lead to increased expression of inflammatory genes in innate immune cells, potentially explaining the link between mutations and increased cardiovascular risk. Therapies targeting the mutant clones or the increased inflammatory mediators might be useful for ameliorating the risk of cardiovascular disease. We propose that the mutations leading to clonal haematopoiesis contribute to the increased inflammation seen in ageing and thereby explain some of the age-related risk of cardiovascular disease.
Introduction
Age is by far the single best predictor for risk of cardiovascular diseases (CVDs), such as coronary artery disease (CAD) and ischaemic stroke1,2. Accordingly, the incidence of atherosclerotic disease increases markedly with age. The effect of ageing has largely been attributed to the duration of exposure to known risk factors, such as high cholesterol levels, hypertension or smoking3. Indeed, middle-aged individuals with an optimal risk-factor profile have considerable protection from developing CAD later in life4.
Nonetheless, there are plausible reasons to suggest that ageing might be more than just a proxy for historical exposure to cardiovascular risk factors. Inflammation contributes causally to atherosclerotic disease5,6. Ageing in humans is associated with a state of chronic, low-grade inflammation characterized by increases in the circulating levels of IL-6 and C-reactive protein (CRP)7,8. A substantial subset of elderly individuals show inflammasome activation and increased IL-1β levels9. Many studies have found that having a high level of these inflammatory molecules associates with risk of chronic diseases of ageing10. Therefore, ‘inflammageing’11 has a robust epidemiological basis, but lacks mechanistic explanations.
Within the last 5 years, work has uncovered a novel link between ageing and inflammation, which might explain this phenomenon. Clonal haematopoiesis, an expansion of blood cell clones due to advantageous somatic mutations, occurs commonly in ageing humans. These clones are rare in those aged <40 years, but can be found in >10% of those aged ≥70 years12-14. Individuals who harbour these clones have an increased risk of haematological cancers, all-cause mortality and, surprisingly, atherosclerotic CVD12,15. The increased cardiovascular risk might occur because many of the same mutations that cause clonal expansion of blood stem cell clones also lead to increased expression of inflammatory genes in innate immune cells15-17. Therefore, clonal haematopoiesis might be a mechanism that links ageing, inflammation and chronic diseases, such as CAD.
In this Review, we discuss the concept of clonal haematopoiesis and the landmark studies in the field. We particularly focus on the link between clonal haematopoiesis and cardiovascular disease, which is likely due to altered function of innate immune cells. We further provide suggestions for clinicians encountering patients with clonal haematopoiesis in their practice, a phenomenon that will become increasingly common over the next few years.
Leukocytes, inflammation and CVD
Leukocytes abound in established human atherosclerotic plaques. Macrophages comprise the most numerous leukocyte type within the atheroma. Macrophages become engorged with lipids, forming foam cells — long considered the hallmark of atherosclerotic lesions5,18. Although some mononuclear phagocytes can reside within the arterial wall from birth, recruitment and proliferation of these cells within lesions contributes to atheroma growth and evolution. Leukocyte recruitment to atherosclerotic plaques starts by binding to adhesion molecules on endothelial cells that line the artery19 (FIG. 1). When adherent to the endothelial surface, leukocytes migrate into the arterial intima in response to a series of chemoattractant cytokines. Pro-inflammatory cytokines induce the expression of the endothelial–leukocyte adhesion molecules and also stimulate the production of chemokines that direct the migration of these cells into the intima. Macrophage foam cells within the plaque undergo death by a variety of mechanisms and give rise to a lipid-rich necrotic core, which his characteristic of advanced atherosclerotic plaques. Within the intima, the macrophages can elaborate mediators that promote smooth muscle cell proliferation and migration. The macrophages also produce proteinases that can degrading the extracellular matrix, a process implicated in weakening of the plaque’s protective fibrous cap that overlies the lipid core. Rupture of a matrix-depleted fibrous cap can provoke thrombosis. The macrophages also produce the major trigger to thrombosis in disrupted plaques, tissue factor pro-coagulant. Therefore, mononuclear phagocytes participate in all phases of atherogenesis from lesion initiation through progression and ultimately the thrombotic complications that cause myocardial infarction (MI) and stroke.
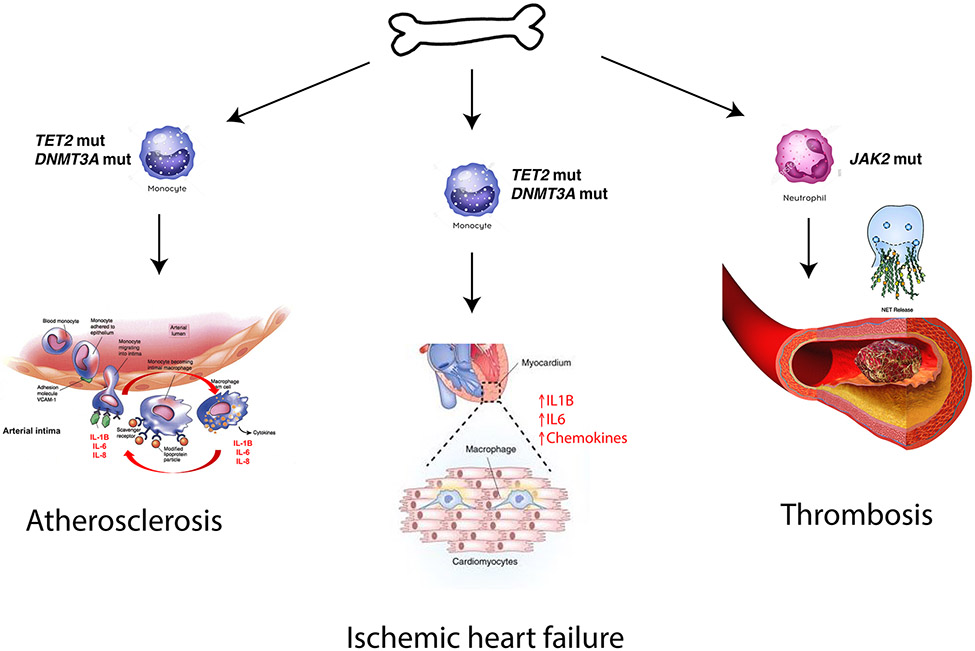
Fig. 1 ∣. Cardiovascular conditions related to CHIP-associated mutations.
Clonal haematopoiesis of indeterminate potential (CHIP) is associated with atherosclerosis, heart failure and thrombosis. Putative mechanisms for each of these pathologies are shown. Accelerated atherosclerosis in the setting of mutations in TET2 likely occurs due to increased expression of inflammatory molecules in lesional macrophages, such as IL1B, IL6, and IL8, which act to recruit immune cells to vessel walls, thus leading to growth of the plaque. Similarly, risk of heart failure may be due to enhanced inflammation in cardiac macrophages, which leads to progressive dysfunction of the myocardium. Increased thrombotic risk is especially common in those with activating JAK2 mutations, and may be due to enhanced NETosis in mutated neutrophils. NET, neutrophil extracellular trap; VCAM1, vascular cell adhesion protein 1.
Another myeloid cell type, the granulocyte or polymorphonuclear leukocyte (PMN), can also contribute to atherosclerotic events. PMNs do not comprise a major cell type within undisrupted atherosclerotic plaques in humans. However, when a breach in endothelial integrity occurs, granulocytes can congregate at the site of plaque rupture or intimal erosion. These PMNs can elaborate extracellular traps that can amplify thrombi and propagate endothelial injury locally20,21. Therefore, two distinct types of myeloid cells can contribute to various phases of atherosclerosis and its clinical complications. These prominent roles for myeloid cells in atherosclerosis provide a mechanistic framework for understanding how clonal proliferation of myeloid cells that have heightened inflammatory potential can accelerate atherosclerosis and provoke the thrombotic complications that lead to MI and stroke.
A large body of clinical evidence supports the contribution of inflammation to atherosclerotic events. Levels of the acute-phase reactant, CRP, measured using a highly sensitive assay (hsCRP), reliably predict cardiovascular events independently of traditional risk factors22. Inflammatory status assessed by hsCRP can aid the allocation of therapies. Finally, direct inhibition of an inflammatory mediator, IL-1β, by administering a neutralizing antibody, can improve cardiovascular outcomes in individuals with stable CAD but with residual inflammation gauged by above-median hsCRP levels despite all standard therapies, including effective statin treatment6. These data firmly establish the operation of inflammatory pathways in human atherosclerosis and provide a plausible basis by which leukocytes bearing CHIP mutations might aggravate CVD.
Clonal haematopoiesis and ageing
A steady increase in the number of somatic mutations is a hallmark of ageing in several tissues23-25. These mutations arise largely due to processes such as spontaneous deamination of 5-methylcytosine and error-prone repair of double-strand breaks. Humans have an estimated 10,000–200,000 haematopoietic stem cells (HSCs)26,27. Mutational analysis of colonies derived from single HSCs has shown that each cell acquires approximately 170 mutations in the whole genome per decade of life27. Mutations in exons, which account for ~2% of the genome, occur less frequently at ~1 mutation per decade of life24. The overwhelming majority of these random mutations probably do not affect the fitness of the cell. Rarely, a mutation can arise that imparts a selective advantage to a stem cell, allowing it to expand relative to other stem cells. Accordingly, these advantageous mutations could theoretically lead to ‘clonal haematopoiesis’ — an expanded blood cell clone derived from a single mutated ancestor.
Clonal haematopoiesis was first demonstrated in cytogenetic studies of chronic myeloid leukaemia in the 1960s28,29. These studies firmly established that haematological malignancies were clonal in origin. According to the prevailing model, cancers arise due to the acquisition of driver mutations in a single clone that occur sequentially over time30,31. Therefore, clonal expansion due to acquired driver mutations might occur even in healthy individuals with no sign of cancer. Studies of non-random X-chromosome inactivation in healthy women in the 1990s first demonstrated that clonal haematopoiesis could be a prevalent aspect of ageing32,33, although the idea remained controversial because the specific genetic lesions leading to apparent clonal expansion could not be identified in the vast majority of these women34,35.
In 2014, three groups analysing whole-exome sequencing data from tens of thousands of individuals unselected for haematological phenotypes demonstrated that clonal haematopoiesis was common, as evidenced by the presence of specific somatic driver mutations in a large number of people. These mutations occurred primarily in genes that were commonly mutated in myeloid cancers, such as DNMT3A, TET2, ASXL1, TP53, JAK2 and SF3B112-14 (FIG. 2a). The role of these mutations in the development of haematological malignancies and clonal haematopoiesis has been extensively reviewed36-38. These three studies all observed a striking association between clonal haematopoiesis and age. Mutations were rare in those aged <40 years, but were found in ~6% of those aged 60–69 years, ~12% of those aged 70–89 years, and ~20% of those aged ≥90 years36-38. In those with clonal haematopoiesis, the average size of the clone was ~20% of peripheral blood cells12. Therefore, a single mutated stem cell out of ~100,000 could expand enough to contribute to ~20% of the haematopoietic system. These and subsequent studies have established that clonal haematopoiesis commonly accompanies ageing39-45.
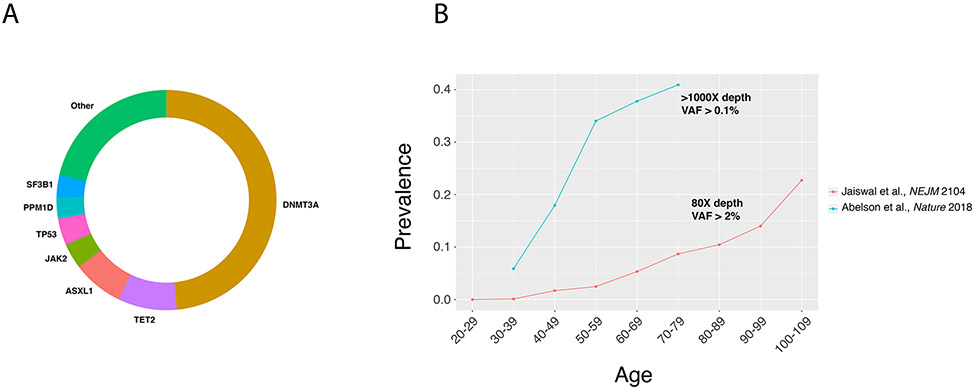
Fig. 2 ∣. Mutational spectrum and prevalence of clonal haematopoiesis.
a ∣ The most commonly mutated genes in clonal haematopoiesis12. The relative number of mutations in each gene is proportional to its representation on the circle’s circumference. b ∣ The prevalence of clonal haematopoiesis according to age12,46.
Of note, the prevalence of clonal haematopoiesis depends strongly on the sensitivity of the sequencing method, which in turn is largely a function of sequencing depth. Whole-exome sequencing is typically performed at 60–80× coverage and can detect mutations down to a variant allele fraction (VAF) of 2% (BOX 1). Studies using deeper sequencing have been able to achieve a lower limit of detection of 0.1% and consequently have detected mutations at a much higher prevalence than studies using exome sequencing46 (FIG. 2b). To add further complexity, apparent clonal haematopoiesis can be detected in the absence of known driver mutations13,44. This situation might result from mutations in genes unknown to be drivers, non-protein-coding mutations or stochastic expansion of clones due to attrition of the stem cell pool.
Box 1 ∣. Terminology related to clonal haematopoiesis.
Variant allele fraction
Variant allele fraction (VAF) is defined as the percentage of reads that support a mutant allele out of the total number of reads. The size of a mutant clone can be inferred from sequence data and is roughly proportional to VAF. Therefore, if sequencing of a blood sample shows 5 mutant reads and 95 wild-type reads, the VAF is calculated to be 5%. Assuming the mutation is heterozygous, this result means that ~10% of the cells from this sample contained the mutation. With increasing sequencing depth, higher levels of sensitivity for detecting mutations can be obtained. Whole-exome sequencing typically achieves an average depth of 60–90 reads and can therefore detect mutations down to ~2–3% VAF. Deeper sequencing can detect clones of <1% VAF, and studies using this level of sensitivity have generally reported higher prevalence of clonal haematopoiesis than studies using whole-genome or whole-exome sequencing.
Clonal haematopoiesis of indeterminate potential
Clonal haematopoiesis is defined by the over-representation of a single clone in the blood or bone marrow. Blood cancers, such as acute myeloid leukaemia, would be considered clonal haematopoiesis, but the condition can also occur in people without cancer. Clonal haematopoiesis most commonly arises due to acquired somatic mutations during ageing, but can also occur in the absence of a known driver mutation or even from non-genetic causes. Furthermore, with very sensitive sequencing methods, a cancer-associated mutation might be found in nearly everyone, rendering the relevance of such a finding moot. For these reasons, clonal haematopoiesis of indeterminate potential (CHIP) was proposed as an entity to distinguish pre-malignant clonal haematopoiesis of clinical importance from other forms of clonal haematopoiesis. CHIP is defined as the presence of cancer-associated mutation in the blood of someone without a blood cancer or other known clonal disorder (such as monoclonal gammopathy). The clone must also be present at a VAF >2%, because clones smaller than this size have uncertain clinical relevance, although this cut-off level might be revised in the future.
Age-related clonal haematopoiesis and pre-leukaemia
Among all patients with clonal haematopoiesis, only a fraction will go on to develop a malignancy, such as acute myeloid leukaemia. Age-related clonal haematopoiesis (ARCH) has been proposed as a term for benign clonal haematopoiesis with lower potential for malignant transformation. By contrast, individuals with certain mutations, such as in TP53 or splicing factors, are ‘pre-leukaemic’ because of a higher risk of developing acute myeloid leukaemia compared with those with mutations in the more common genes DNMT3A, TET2 and ASXL1. Importantly, those with ARCH still have increased risk of death, cardiovascular disease and malignancy relative to those with no mutations, so the condition is not fully benign.
On the basis of these findings, a novel clinical entity, clonal haematopoiesis of indeterminate potential (CHIP), was proposed. CHIP is defined by the presence of a candidate somatic driver mutation from blood cells of individuals without haematological malignancies47 (BOX 1). Because the clinical consequence of very small clones is unknown, the definition of CHIP also requires the VAF to be >2%.
Clinical associations of CHIP
Although most frank malignancies, such as acute myeloid leukaemia (AML) or myelodysplastic syndromes, have several driver mutations, ~90% of CHIP carriers have only one. Therefore, CHIP might represent a pre-malignant state. Indeed, studies have found that individuals with CHIP have an approximately tenfold increased risk of developing haematological cancer in the future, with an absolute risk of transformation of about 1% per year12,13. Most of the malignancies that develop in CHIP carriers are myeloid cancers, such AML or myelodysplastic syndromes, but the risk of lymphoid cancers, especially non-Hodgkin lymphomas, is also increased. Larger studies of AML subsequently corroborated the link between CHIP and haematological cancer, with the risk of transformation most linked to the size of the clone and the number of mutations present46,48,49.
Despite the presence of CHIP-related mutations in a large number of blood cells, CHIP does not associate with substantial alterations in blood counts12,41,44,45. CHIP does, however, associate with increased red blood cell distribution width (RDW) 12, a measure of the variability in the size of red blood cells. Although the majority of individuals with CHIP have a normal RDW, a subset of CHIP carriers have an abnormally high RDW. Epidemiological studies have found that RDW associates with increased mortality, CVD and other adverse outcomes50-52. The factors mediating this link are not known, but a relationship between increased RDW and chronic inflammation provides one possible explanation53.
Several studies have also found an association between CHIP and smoking13,41,44, although whether smoking causally increases the likelihood of developing CHIP is uncertain. CHIP also associates with chronic obstructive pulmonary disease, possibly due to CHIP carriers having a higher rate of smoking44,45.
Several studies have found that CHIP also associates with increased all-cause mortality12,13,44. In one study, those with CHIP and elevated RDW were particularly at risk, with a 3.7-fold increase in mortality compared with those without CHIP and normal RDW12. Surprisingly, the cause of increased death in CHIP carriers was not an increase in haematological malignancies, but an increased rate of fatal strokes and heart attacks. A subsequent exploratory analysis found that those with CHIP were approximately twice as likely to develop CAD and ischaemic stroke, even after adjusting for known risk factors, such as smoking and elevated cholesterol levels12. In this study, the cumulative incidence of CAD in those without CHIP was ~7% at 10 years compared with ~14% for those with CHIP12.
CHIP and CVD in humans
Since the initial finding of an association between CHIP and cardiovascular outcomes came from an unplanned secondary analysis, replication of the finding in additional cohorts was necessary. Additional case–control cohorts for CAD were selected for this purpose15. In two cohorts of older individuals without a history of heart disease at baseline, the risk of incident CAD was found to be increased 1.9-fold in those with CHIP. In two cohorts of younger individuals with early-onset MI and age-matched controls, the prevalence of CHIP was approximately fourfold higher in those with early MI. The magnitude of risk conferred by the presence of CHIP was as great or greater than many commonly assessed risk factors for CVD (TABLE 1).
Table 1 ∣.
Regression model of risk factors for cardiovascular disease
| Risk factor | HR (95% CI) |
|---|---|
| Age 50–59 years | 2.20 (1.32–3.69) |
| Age 60–69 years | 2.41 (1.44–4.02) |
| Age ≥70 years | 6.27 (3.77–10.42) |
| Female sex | 0.68 (0.50–0.93) |
| Has type 2 diabetes mellitus | 2.18 (1.62–2.94) |
| Former or current smoker | 1.40 (1.04–1.90) |
| Hypertension stage II–IV | 1.20 (0.89–1.62) |
| Total-cholesterol level >200 mg/dl | 1.40 (1.04–1.88) |
| HDL-cholesterol level <35 mg/dl | 1.46 (0.98–2.18) |
| HDL-cholesterol level >60 mg/dl | 0.77 (0.52–1.13) |
| CHIP present | 1.82 (1.15–2.89) |
Hazard ratios and 95% confidence intervals associated with Framingham risk factors and CHIP in longitudinal population-based cohorts. Data obtained from a Cox regression using 3,661 individuals from Jackson Heart Study, Framingham Heart Study and Finland–United States Investigation of NIDDM Genetics in REF.15. CHIP, clonal haematopoiesis of indeterminate potential; NIDDM, non-insulin-dependent diabetes mellitus.
To test the link between CHIP and vascular wall inflammation, coronary artery calcium (CAC) scores — a radiological measure of atherosclerosis — were assessed in the same cohorts15. Those with CHIP had substantially greater CAC, regardless of whether they developed CAD or not, supporting the hypothesis that CHIP could increase the burden of atherosclerosis. In further support of this model, those harbouring mutations in TET2 have increased levels of circulating IL-8, an atherogenic chemokine15,54.
Heart failure (HF) commonly complicates ischaemic heart disease and also strongly links to ageing. In a study of patients with a history of MI and stable HF, the presence of mutations in DNMT3A or TET2 in bone marrow cells was associated with a hazard ratio for all-cause mortality of 3.25, with most deaths occurring due to complications of HF55. The risk of death was strongly linked to the size of the mutant clone, as was also shown for CAD15. This association could not be explained by CHIP carriers having worse baseline HF as assessed by alterations in left ventricular ejection fraction, serum levels of N-terminal pro-B-type natriuretic peptide or HF risk scores. Therefore, HF is another cardiovascular complication of CHIP.
The effect of CHIP on arterial or venous thrombosis is less well studied. Whereas the risk of CAD was increased approximately twofold for those with mutations in DNMT3A, TET2 or ASXL1, the risk of CAD was approximately tenfold higher in those with activating mutations in JAK215. JAK2 is a non-receptor tyrosine kinase that is frequently mutated in myeloproliferative neoplasms56. Individuals with JAK2-mutated myeloproliferative neoplasms have a markedly increased risk of both arterial and venous thrombosis57,58. The risk of thrombosis correlates most strongly with increased leukocyte, but not platelet, count, suggesting leukocytosis or altered leukocyte function as the culprit59,60. An increased propensity to the formation of neutrophil extracellular traps in cells with the JAK2 mutation might contribute to this thrombotic diathesis61. Those with non-JAK2-mutated CHIP also had a doubling of the risk of venous thrombosis, although the mechanism is unknown61.
CHIP and CVD in mice
The robust human genetic association between CHIP and heart disease alone cannot prove causality, because the link could be purely correlative if CHIP were simply a marker of ageing. For example, those individuals who are more biologically aged due to genetic predisposition might have a higher rate of developing both mutant stem cell clones and worse vascular health, independently. Alternatively, the association might reflect confounding by unmeasured environmental or lifestyle factors, such as exposure to pollutants or underlying medications. However, it is biologically plausible that mutated blood cells relate causally to CVD because the most common mutations in CHIP occur in epigenetic regulators and, therefore, could lead to altered gene expression in blood cells. Theoretically, the mutations could either alter thrombotic risk via greater propensity to platelet and leukocyte activation, or influence atherosclerosis via alteration of innate immune cell function.
The availability of several mouse models for genes commonly mutated in CHIP enabled the testing of causality in mice with experimental atherosclerosis. In 2017, two groups reported experiments in which Ldlr−/− mice were transplanted with bone marrow deficient for Tet2, the second most commonly mutated gene in CHIP, and quantitatively assessed for atherosclerotic burden15,17. These studies yielded highly concordant findings. Mice with either heterozygous or homozygous loss of Tet2 developed aortic root lesions that were ~50–70% larger than those in control mice at early time points, while lesional area was also approximately twofold larger in the descending aorta at later time points. Neither group found quantitative differences in peripheral blood counts in the mice, arguing against leukocytosis as the primary driver of accelerated atherosclerosis. This finding is also true in humans, given that those with CHIP mutations typically have normal blood counts. Instead, both studies found that knocking out Tet2 in only the myeloid compartment was sufficient to lead to increased lesional area, suggesting a qualitative change in myeloid cell function.
Mouse experiments have also assessed the effect of mutations in Tet2 or Dnmt3a on other cardiovascular phenotypes. In two experimental models of HF, loss of Tet2 in bone marrow cells led to lower ejection fraction and increased cardiac fibrosis and remodelling. Tet2 loss restricted to the myeloid cells caused similar findings, suggesting that macrophages or monocytes were responsible for the exacerbation of experimental HF62. In another study, CRISPR-mediated deletion of either Tet2 or Dnmt3a in bone marrow sufficed to cause increased cardiac hypertrophy, a reduction in cardiac function and increased cardiac fibrosis in mice with HF produced by angiotensin II infusion. These results suggest that the reduced survival seen in humans with TET2 or DNMT3A mutations relate causally to altered immune cell function in the myocardium63 (FIG. 1).
Other studies have explored the role of JAK2 mutations in atherothrombosis. Mice harbouring the JAK2-V617F mutation can develop spontaneous pulmonary venous thrombosis, which could be blocked by administration of the JAK2 inhibitor ruxolitinib61. The mechanism of increased thrombosis might relate to the elevated levels of neutrophil extracellular trap formation in neutrophils expressing JAK2-V617F (FIG. 1). Another study examined the role of Jak2 mutations in atherosclerotic mice64. Bone marrow with JAK2-V617F transplanted into Ldlr−/− recipients led to larger lesions in the aortic root, despite reduced serum cholesterol levels in the mutant mice. The accelerated atherosclerosis might be due to increased inflammation and erythrophagocytosis by macrophages, although the marked leukocytosis and erythrocytosis in these mice could confound interpretation of these studies. Interestingly, humans with JAK2-mutated CHIP also have lower circulating cholesterol levels, possibly due to sequestration in plasma membranes from increased erythropoiesis65. The fact that the risk of CAD remains elevated despite the lower cholesterol levels suggests this mutation is a particularly potent driver.
Mutations in the DNA-damage response genes TP53 and PPM1D are also common in CHIP. Some studies have found that loss of Tp53 in myeloid cells increased plaque size in mice66,67, but the role of PPM1D is atherosclerosis is unstudied. The mechanism of accelerated atherosclerosis in the setting of macrophage TP53 deficiency is unknown, but could relate to increased proliferation of lesional macrophages67.
Mechanisms of atherosclerosis due to CHIP
Atherosclerosis is a disease of chronic inflammation that accompanies cholesterol accumulation in the arterial wall. Therefore, factors that either increase underlying inflammation or diminish the capacity of cholesterol to efflux from lesions can result in worsening atherosclerosis. Most studies to date have indicated increased inflammation as the outcome when mutating either TET2 or DNMT3A.
In murine macrophages and dendritic cells, Tet2 represses the transcription of pro-inflammatory molecules, such as IL-6, a known pro-atherogenic mediator. Surprisingly, this effect is reported to be independent of the known role for Tet2 in catalysing the oxidation of 5-methylcytosine, a chemical modification of DNA well known to regulate transcription. Other studies have found that loss of Tet2 in murine macrophages exposed to either LDL or lipopolysaccharide leads to increased expression of a variety of inflammatory mediators, such as IL-1β, IL-6, CXCL1, CXCL2 and CXCL3. Increased expression of these molecules in the setting of CHIP might lead to further leukocyte recruitment to plaques, initiating a feed-forward loop that results in accelerated atherosclerosis (FIG. 1). Targeting these molecules might provide a strategy to lower the risk of cardiovascular morbidity in those with CHIP. Blockade of inflammasome activation and of the subsequent activation of IL-1β sufficed to reverse the accelerated atherosclerosis in mice with Tet2-deficient bone marrow17. IL-1β blockade was also sufficient to ameliorate the diminished cardiac function in the Tet2-deficient model of HF63. The CANTOS trial6 reported a reduction in major adverse cardiovascular events in individuals given a monoclonal antibody to IL-1β. A subgroup analysis of whether those with CHIP had better responses to the drug will be of interest68.
The role of DNMT3A in regulating innate immune function has not undergone as thorough study as that of TET2 or JAK2, but most reports have found evidence of increased inflammation when the gene is perturbed. In one study, Dnmt3a-deficient mast cells produced higher levels of IL-6, tumour necrosis factor and IL-13 in response to stimulation with immunoglobulin E in vitro, and mast cell activity was also increased in vivo in an allergic model69. In another study, mutation of Dnmt3a with CRISPR in mouse RAW 264.7 macrophages resulted in increased expression of Cxcl1, Cxcl2 and Il6 in response to bacterial lipopolysaccharide63. However, the mechanisms of these specific gene-expression changes remain poorly understood. Virtually nothing is known about the effect of mutations in ASXL1, SF3B1, and SRSF2 on innate immune function.
CHIP for cardiologists
Practitioners of cardiovascular medicine might encounter individuals with CHIP in several contexts. When cardiologists collaborate in the care of patients with cancer, they will encounter patients in whom sequencing of the tumour and/or blood reveals the presence of CHIP70. Oncologists look to cardiovascular specialists for guidance in how to manage cardiovascular risk in patients who have presented through their portal. Increasingly, cardiovascular specialists will encounter individuals without cancer who have undergone DNA sequencing for other reasons. The incidental finding of CHIP might emerge from DNA sequencing undertaken by personal curiosity or by liquid biopsy screening for solid tumours through analysis of cell-free DNA in the blood. These individuals who have sought sequencing because of concerns regarding cardiovascular risk or because of premature or otherwise unexplained cardiovascular events have begun to present to practitioners.
The current lack of an evidence base to inform the management of cardiovascular risk in patients with CHIP discourages the screening of unselected individuals. Requests from patients for screening for CHIP should stimulate a shared decision-making conversation with explicit explanation that we currently lack evidence-based, actionable responses to the finding of CHIP. In the absence of such evidence, intensification of lifestyle measures to control cardiovascular risk seems appropriate. Institution of pharmacological therapies, such as statin treatment, again requires a clear discussion with patients that we lack evidence or advice from guidelines in this regard. On an individual basis, seeking CHIP mutations might warranted in some patients. For example, an individual with an unexplained increase in RDW might warrant a discussion regarding the advisability of seeking a CHIP mutation to inform the intensity of cardiovascular risk-factor management.
The very uncertainty regarding the management of individuals found to have CHIP, either with or without known cancer, highlights an urgent need for further clinical investigation to furnish a database. Particularly with the ageing of the population and the high prevalence of CHIP in the aged, we urgently need information to guide our clinical management. The heterogeneity in the mutations and the VAF will render large-scale clinical trials challenging. Certainly, assembling registries of individuals with CHIP to follow their natural history and to mine for management strategies seem highly desirable.
CHIP and CVD: unanswered questions
Although our understanding of CHIP has burgeoned over the last five years, research into this area is still new and evolving. In this section, we highlight several areas related to CHIP and CVD that, in our view, merit further study in the coming years.
Understanding whether the other commonly mutated genes in CHIP, in addition to TET2 and JAK2, causally link to atherosclerosis is of central importance. Common mechanisms might promote cardiovascular risk in each of the CHIP-associated mutations in many cases, but important gene-specific differences could also exist. Mice with Dnmt3a mutations faithfully recapitulate the clonal advantage seen in humans and should, therefore, provide a tractable model for studies of atherosclerosis71,72. HSCs bearing mutations in Asxl173 and the splicing factors74-76, however, actually show a competitive disadvantage in vivo compared with wild-type stem cells. Whether this finding results from the mutations not having the same effect in mice, or whether the mutations exert a selective advantage only in certain scenarios, is uncertain. Therefore, CVD experiments in these mice require cautious interpretation. Moreover, some mutations might conceivably have a beneficial effect on the risk of CVD. For example, loss of PPM1D results in hyper-inflammation77, so gain-of-function PPM1D mutations seen in humans might dampen inflammatory responses in innate immune cells.
Although CHIP is a potent risk factor for CVD, only a minority of individuals with CHIP develop the complications of atherosclerotic disease in their lifetime. Studies of biomarkers or environmental exposures from large epidemiological cohorts might shed light on other factors that influence outcomes in those with CHIP, such as markers of inflammation or dietary factors. Whether CHIP has synergistic interactions with known cardiovascular risk factors is also unknown. Furthermore, the interaction with traditional cardiovascular risk factors might differ depending on the mutated gene or genes. Again, large cohorts with genetic sequencing and animal experiments might help to answer this question.
Most importantly, we need to find ways to reduce the CVD burden in those with CHIP. Targeting the inflammatory molecules associated with CHIP, such as IL-1β, IL-6 or IL-8, is an attractive option. Indeed, studies to determine whether CHIP carriers respond better to drugs such as canakinumab are ongoing68. Although inflammatory blockade seems to be closest on the horizon, drugs will ideally be found that can directly suppress the growth of mutant CHIP clones in the bone marrow, which might reduce the risk of both cancer and CVD.
Conclusions
CHIP is a common, and perhaps inevitable, consequence of ageing. Individuals who harbour these mutated clones are at increased risk of haematological cancer, but also several adverse cardiovascular outcomes, such as MI, stroke, thrombosis and HF. Our understanding of the link between CHIP and CVD is incomplete, but early results suggest that the heightened risk might be due to increased inflammation in innate immune cells bearing these mutations. Currently, no therapies are available for those with CHIP that can reduce the risk of cancer or CVD. As our understanding of CHIP grows, so too should our armamentarium of therapies that can prevent its detrimental effects.
Key points.
Clonal haematopoiesis of indeterminate potential (CHIP) is a common age-related condition characterized by the clonal expansion of haematopoietic stem cells bearing mutations in certain genes, especially DNMT3A, TET2 and ASXL1.
CHIP is associated with increased risk of haematological malignancies and all-cause mortality, but also increased risk of atherosclerotic cardiovascular disease, venous thrombosis and worse outcomes in heart failure.
Mutations associated with CHIP seem to have effects on immune effector cells, such as macrophages and neutrophils, which might account for the increased risk of cardiovascular complications in individuals with CHIP.
No treatments are currently available to lower the risk of cardiovascular disease in those with CHIP, but blockade of inflammatory molecules is a potential strategy to mitigate the effects of CHIP.
Individuals incidentally found to have CHIP should undergo evaluation for lifestyle modifications to reduce the risk of cardiovascular disease.
Acknowledgements
The authors were supported by Burroughs Wellcome Fund Career Award for Medical Scientists and Fondation Leducq Transatlantic Network of Excellence (S.J.) and by the National Heart, Lung, and Blood Institute (R01HL080472), AHA (18CSA34080399) and the RRM Charitable Fund (P.L.).
Footnotes
Competing interests
S.J. has filed patents related to the topic of this Review and is a consultant for GRAIL. P.L. is an unpaid consultant to, or is involved in clinical trials for, Amgen, AstraZeneca, Esperion Therapeutics, Ionis Pharmaceuticals, Kowa Pharmaceuticals, Novartis, Pfizer, Sanofi-Regeneron and XBiotech. P.L. is a member of the scientific advisory board for Amgen, Corvidia Therapeutics, DalCor Pharmaceuticals, IFM Therapeutics, Kowa Pharmaceuticals, Olatec Therapeutics, Medimmune, Novartis and XBiotech.
References
- 1.Wang TJ et al. Multiple biomarkers for the prediction of first major cardiovascular events and death. N Engl J Med 355, 2631–2639 (2006). [DOI] [PubMed] [Google Scholar]
- 2.Pencina MJ, D’Agostino RB Sr, Larson MG, Massaro JM & Vasan RS Predicting the 30-year risk of cardiovascular disease: the Framingham Heart Study. Circulation 119, 3078–3084 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sniderman AD & Furberg CD Age as a modifiable risk factor for cardiovascular disease. Lancet 371, 1547–1549 (2008). [DOI] [PubMed] [Google Scholar]
- 4.Lloyd-Jones DM et al. Prediction of lifetime risk for cardiovascular disease by risk factor burden at 50 years of age. Circulation 113, 791–798 (2006). [DOI] [PubMed] [Google Scholar]
- 5.Libby P, Nahrendorf M & Swirski FK Leukocytes link local and systemic inflammation in ischemic cardiovascular disease: an expanded “cardiovascular continuum”. J Am Coll Cardiol 67, 1091–1103 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ridker PM et al. Antiinflammatory therapy with canakinumab for atherosclerotic disease. N Engl J Med 377, 1119–1131 (2017). [DOI] [PubMed] [Google Scholar]
- 7.Wikby A et al. The immune risk phenotype is associated with IL-6 in the terminal decline stage: findings from the Swedish NONA immune longitudinal study of very late life functioning. Mech Ageing Dev 127, 695–704 (2006). [DOI] [PubMed] [Google Scholar]
- 8.Ferrucci L et al. The origins of age-related proinflammatory state. Blood 105, 2294–2299 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Furman D et al. Expression of specific inflammasome gene modules stratifies older individuals into two extreme clinical and immunological states. Nat Med 23, 174–184 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Franceschi C & Campisi J Chronic inflammation (inflammaging) and its potential contribution to age-associated diseases. J Gerontol A Biol Sci Med Sci 69 (Suppl. 1), S4–S9 (2014). [DOI] [PubMed] [Google Scholar]
- 11.Franceschi C et al. Inflamm-aging. An evolutionary perspective on immunosenescence. Ann N Y Acad Sci 908, 244–254 (2000). [DOI] [PubMed] [Google Scholar]
- 12.Jaiswal S et al. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med 371, 2488–2498 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Genovese G et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med 371, 2477–2487 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xie M et al. Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat Med 20, 1472–1478 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jaiswal S et al. Clonal hematopoiesis and risk of atherosclerotic cardiovascular disease. N Engl J Med 377, 111–121 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Q et al. Tet2 is required to resolve inflammation by recruiting Hdac2 to specifically repress IL-6. Nature 525, 389–393 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fuster JJ et al. Clonal hematopoiesis associated with Tet2 deficiency accelerates atherosclerosis development in mice. Science 355, 842–847 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moore KJ et al. Macrophage trafficking, inflammatory resolution, and genomics in atherosclerosis: JACC Macrophage in CVD Series (Part 2). J Am Coll Cardiol 72, 2181–2197 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schumski A, Winter C, Doring Y & Soehnlein O The ins and outs of myeloid cells in atherosclerosis. J Innate Immun 10, 479–486 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Doring Y, Soehnlein O & Weber C Neutrophil extracellular traps in atherosclerosis and atherothrombosis. Circ Res 120, 736–743 (2017). [DOI] [PubMed] [Google Scholar]
- 21.Franck G et al. Roles of PAD4 and NETosis in experimental atherosclerosis and arterial injury: implications for superficial erosion. Circ Res 123, 33–42 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ridker PM A test in context: high-sensitivity C-reactive protein. J Am Coll Cardiol 67, 712–723 (2016). [DOI] [PubMed] [Google Scholar]
- 23.Armitage P & Doll R A two-stage theory of carcinogenesis in relation to the age distribution of human cancer. Br J Cancer 11, 161–169 (1957). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Welch JS et al. The origin and evolution of mutations in acute myeloid leukemia. Cell 150, 264–278 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martincorena I & Campbell PJ Somatic mutation in cancer and normal cells. Science 349, 1483–1489 (2015). [DOI] [PubMed] [Google Scholar]
- 26.Abkowitz JL, Catlin SN, McCallie MT & Guttorp P Evidence that the number of hematopoietic stem cells per animal is conserved in mammals. Blood 100, 2665–2667 (2002). [DOI] [PubMed] [Google Scholar]
- 27.Lee-Six H et al. Population dynamics of normal human blood inferred from somatic mutations. Nature 561, 473–478 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nowell PC The minute chromosome (Phl) in chronic granulocytic leukemia. Blut 8, 65–66 (1962). [DOI] [PubMed] [Google Scholar]
- 29.Rowley JD Letter: A new consistent chromosomal abnormality in chronic myelogenous leukaemia identified by quinacrine fluorescence and Giemsa staining. Nature 243, 290–293 (1973). [DOI] [PubMed] [Google Scholar]
- 30.Nowell PC The clonal evolution of tumor cell populations. Science 194, 23–28 (1976). [DOI] [PubMed] [Google Scholar]
- 31.Reya T, Morrison SJ, Clarke MF & Weissman IL Stem cells, cancer, and cancer stem cells. Nature 414, 105–111 (2001). [DOI] [PubMed] [Google Scholar]
- 32.Fey MF et al. Clonality and X-inactivation patterns in hematopoietic cell populations detected by the highly informative M27 beta DNA probe. Blood 83, 931–938 (1994). [PubMed] [Google Scholar]
- 33.Busque L et al. Nonrandom X-inactivation patterns in normal females: lyonization ratios vary with age. Blood 88, 59–65 (1996). [PubMed] [Google Scholar]
- 34.Champion KM, Gilbert JG, Asimakopoulos FA, Hinshelwood S & Green AR Clonal haemopoiesis in normal elderly women: implications for the myeloproliferative disorders and myelodysplastic syndromes. Br J Haematol 97, 920–926 (1997). [DOI] [PubMed] [Google Scholar]
- 35.Busque L et al. Recurrent somatic TET2 mutations in normal elderly individuals with clonal hematopoiesis. Nat Genet 44, 1179–1181 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sperling AS, Gibson CJ & Ebert BL The genetics of myelodysplastic syndrome: from clonal haematopoiesis to secondary leukaemia. Nat Rev Cancer 17, 5–19 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jan M, Ebert BL & Jaiswal S Clonal hematopoiesis. Semin Hematol 54, 43–50 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bowman RL, Busque L & Levine RL Clonal hematopoiesis and evolution to hematopoietic malignancies. Cell Stem Cell 22, 157–170 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McKerrell T et al. Leukemia-associated somatic mutations drive distinct patterns of age-related clonal hemopoiesis. Cell Rep 10, 1239–1245 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Acuna-Hidalgo R et al. Ultra-sensitive sequencing identifies high prevalence of clonal hematopoiesis-associated mutations throughout adult life. Am J Hum Genet 101, 50–64 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Coombs CC et al. Therapy-related clonal hematopoiesis in patients with non-hematologic cancers is common and associated with adverse clinical outcomes. Cell Stem Cell 21, 374–382.e4 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Young AL, Challen GA, Birmann BM & Druley TE Clonal haematopoiesis harbouring AML-associated mutations is ubiquitous in healthy adults. Nat Commun 7, 12484 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gibson CJ et al. Clonal hematopoiesis associated with adverse outcomes after autologous stem-cell transplantation for lymphoma. J Clin Oncol 35, 1598–1605 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zink F et al. Clonal hematopoiesis, with and without candidate driver mutations, is common in the elderly. Blood 130, 742–752 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Buscarlet M et al. DNMT3A and TET2 dominate clonal hematopoiesis and demonstrate benign phenotypes and different genetic predispositions. Blood 130, 753–762 (2017). [DOI] [PubMed] [Google Scholar]
- 46.Abelson S et al. Prediction of acute myeloid leukaemia risk in healthy individuals. Nature 559, 400–404 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Steensma DP et al. Clonal hematopoiesis of indeterminate potential and its distinction from myelodysplastic syndromes. Blood 126, 9–16 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Desai P et al. Somatic mutations precede acute myeloid leukemia years before diagnosis. Nat Med 24, 1015–1023 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sellar RS, Jaiswal S & Ebert BL Predicting progression to AML. Nat Med 24, 904–906 (2018). [DOI] [PubMed] [Google Scholar]
- 50.Tonelli M et al. Relation between red blood cell distribution width and cardiovascular event rate in people with coronary disease. Circulation 117, 163–168 (2008). [DOI] [PubMed] [Google Scholar]
- 51.Patel KV, Ferrucci L, Ershler WB, Longo DL & Guralnik JM Red blood cell distribution width and the risk of death in middle-aged and older adults. Arch Intern Med 169, 515–523 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Perlstein TS, Weuve J, Pfeffer MA & Beckman JA Red blood cell distribution width and mortality risk in a community-based prospective cohort. Arch Intern Med 169, 588–594 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lippi G et al. Relation between red blood cell distribution width and inflammatory biomarkers in a large cohort of unselected outpatients. Arch Pathol Lab Med 133, 628–632 (2009). [DOI] [PubMed] [Google Scholar]
- 54.Gerszten RE et al. MCP-1 and IL-8 trigger firm adhesion of monocytes to vascular endothelium under flow conditions. Nature 398, 718–723 (1999). [DOI] [PubMed] [Google Scholar]
- 55.Dorsheimer L et al. Association of mutations contributing to clonal hematopoiesis with prognosis in chronic ischemic heart failure. JAMA Cardiol 4, 25–33 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Morgan KJ & Gilliland DG A role for JAK2 mutations in myeloproliferative diseases. Annu Rev Med 59, 213–222 (2008). [DOI] [PubMed] [Google Scholar]
- 57.Marchioli R et al. Vascular and neoplastic risk in a large cohort of patients with polycythemia vera. J Clin Oncol 23, 2224–2232 (2005). [DOI] [PubMed] [Google Scholar]
- 58.Hinds DA et al. Germ line variants predispose to both JAK2 V617F clonal hematopoiesis and myeloproliferative neoplasms. Blood 128, 1121–1128 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Carobbio A et al. Leukocytosis is a risk factor for thrombosis in essential thrombocythemia: interaction with treatment, standard risk factors, and Jak2 mutation status. Blood 109, 2310–2313 (2007). [DOI] [PubMed] [Google Scholar]
- 60.Landolfi R et al. Leukocytosis as a major thrombotic risk factor in patients with polycythemia vera. Blood 109, 2446–2452 (2007). [DOI] [PubMed] [Google Scholar]
- 61.Wolach O et al. Increased neutrophil extracellular trap formation promotes thrombosis in myeloproliferative neoplasms. Sci Transl Med 10, eaan8292 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sano S et al. Tet2-mediated clonal hematopoiesis accelerates heart failure through a mechanism involving the IL-1β/NLRP3 inflammasome. J Am Coll Cardiol 71, 875–886 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sano S et al. CRISPR-mediated gene editing to assess the roles of Tet2 and Dnmt3a in clonal hematopoiesis and cardiovascular disease. Circ Res 123, 335–341 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang W et al. Macrophage inflammation, erythrophagocytosis, and accelerated atherosclerosis in Jak2 (V617F) mice. Circ Res 123, e35–e47 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu DJ et al. Exome-wide association study of plasma lipids in >300,000 individuals. Nat Genet 49, 1758–1766 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.van Vlijmen BJ et al. Macrophage p53 deficiency leads to enhanced atherosclerosis in APOE*3-Leiden transgenic mice. Circ Res 88, 780–786 (2001). [DOI] [PubMed] [Google Scholar]
- 67.Merched AJ, Williams E & Chan L Macrophage-specific p53 expression plays a crucial role in atherosclerosis development and plaque remodeling. Arterioscler Thromb Vasc Biol 23, 1608–1614 (2003). [DOI] [PubMed] [Google Scholar]
- 68.Svensson EC et al. Abstract 15111: TET2-driven clonal hematopoiesis predicts enhanced response to canakinumab in the CANTOS trial: an exploratory analysis [abstract]. Circulation 138, A15111–A15111 (2018). [Google Scholar]
- 69.Leoni C et al. Dnmt3a restrains mast cell inflammatory responses. Proc Natl Acad Sci USA 114, E1490–E1499 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Coombs CC et al. Identification of clonal hematopoiesis mutations in solid tumor patients undergoing unpaired next-generation sequencing assays. Clin Cancer Res 24, 5918–5924 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Guryanova OA et al. Dnmt3a regulates myeloproliferation and liver-specific expansion of hematopoietic stem and progenitor cells. Leukemia 30, 1133–1142 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cole CB et al. Haploinsufficiency for DNA methyltransferase 3A predisposes hematopoietic cells to myeloid malignancies. J Clin Invest 127, 3657–3674 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Abdel-Wahab O et al. Deletion of Asxl1 results in myelodysplasia and severe developmental defects in vivo. J Exp Med 210, 2641–2659 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Obeng EA et al. Physiologic expression of Sf3b1(K700E) causes impaired erythropoiesis, aberrant splicing, and sensitivity to therapeutic spliceosome modulation. Cancer Cell 30, 404–417 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shirai CL et al. Mutant U2AF1 expression alters hematopoiesis and pre-mRNA splicing in vivo. Cancer Cell 27, 631–643 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kim E et al. SRSF2 mutations contribute to myelodysplasia by mutant-specific effects on exon recognition. Cancer cell 27, 617–630 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sun B et al. Phosphatase Wip1 negatively regulates neutrophil migration and inflammation. J Immunol 192, 1184–1195 (2014). [DOI] [PubMed] [Google Scholar]