Abstract
Osteopontin (OPN) is a multifunctional glycoprotein that physiologically interacts with different types of integrins. It is considered to be a possible prognostic biomarker in certain tumor types; however, various splicing isoforms exist, which have not been investigated in melanoma. We aimed to define the relative expression pattern of five OPN isoforms and clarify the prognostic significance of the splice variants in melanoma. We also aimed to investigate the expression pattern of eight integrins in the same tumors. Gene expression analyses revealed that the relative expression of OPNa, OPNb, and OPNc is significantly higher in metastatic tumors compared to primary lesions (p < 0.01), whereas the expression of OPN4 and OPN5 was low in both. The more aggressive nodular melanomas had higher expression levels compared to the superficial spreading subtype (p ≤ 0.05). The relative expression of the eight tested integrins was low, with only the expression of ITGB3 being detectable in nodular melanoma (Medianlog2 = 1.274). A positive correlation was found between Breslow thickness and the expression of OPNc variant, whereby thicker tumors (>4 mm) had significantly higher expression (p ≤ 0.05). The Breslow thickness was negatively correlated with the expression of OPN4, and similarly with ITGA2. OPNc also exhibited significant positive correlation with the presence of metastasis. Our data show that high expression of OPNa, OPNb, and especially OPNc and low expression of OPN4 and ITGA2 are associated with an advanced stage of tumor progression and poor prognosis in melanoma.
Keywords: gene expression, melanoma progression, osteopontin, osteopontin splice variants, integrins
Introduction
Osteopontin (OPN or SPP1) is a multifunctional glycoprotein that physiologically interacts with different types of integrins. OPN is considered to be a possible prognostic biomarker in certain tumor types including malignant melanoma (1, 2). Depending on the intracellular or extracellular localization, the expression of OPN is closely related to tumor proliferation, invasion, metastasis, and tumor microenvironment formation (3). These multifunctional biological roles are probably associated with the capacity for OPN to interact with different molecules, including cell surface receptors such as integrin and cluster of differentiation (CD44), intracellular signaling molecules, calcium, and heparin (4). OPN possesses three critical integrin binding sequences: the well conserved RGD domain (arginine–glycine–aspartic acid) that facilitated the interaction of OPN with αv integrins (especially αvβ1, αvβ3, and αvβ5); the SVVYGLR (serine–valine–valine–tyrosine–glutamate–leucine–arginine) domain, which may bind to α4β1, α4β7, and α9β1 integrins; and the ELVTDFP sequence on the N-terminal, which can bind α4β1 (5). The OPN protein is a member of the SIBLING (small integrin-binding ligand N-linked glycoprotein) family, whose members interact with CD44 and integrins through the characteristic domain. OPN is a secreted protein; however, the existence of a nonsecreted intracellular form (iOPN) has also been reported, which can be localized in the cytoplasm and nucleus and has a slightly different function than secreted OPN (sOPN) (6–8). OPN takes part in several normal physiological processes (vascularization, immune responses, inflammation, tissue remodeling, and cell adhesion) but also influences numerous aspects of tumorigenesis and metastasis (cell survival, proliferation, adhesion, migration, and invasion) (9, 10). Through the connection with integrins and CD44, OPN influences the PI3K/AKT signaling pathway, resulting in NF-κB-mediated cell proliferation and survival. Moreover, the interaction of OPN with αvβ3 integrin, in particular, affects the Ras/Raf/MEK/ERK signaling pathway and enhances the metastatic phenotype of several cancer cell types (11).
The primary transcript of the OPN gene is subject to alternative splicing, generating five splice isoforms: OPNa, OPNb, OPNc, OPN4 and OPN5 (3). The splicing variants differ in their gene structure: OPNa can be considered as the full-length isoform containing seven exons, OPNb lacks exon 5, and OPNc lacks exon 4. Both exon 4 and exon 5 are missing from OPN transcript variant 4, and, interestingly OPN5, has all seven exons supplemented with an extra exon derived from the retention of a portion from the intron 3 in the canonical isoform (12). Their translation results proteins called OPNa (314 amino acids), OPNb (300 amino acids), and OPNc (284 amino acids), which are widely studied and functionally well characterized, whereas OPN4 (273 amino acids) and OPN5 (327 amino acids) have only been recently identified (12–14). The splice variants are abnormally expressed in different types of tumors (3, 12). The high expression level of OPNc is indicative of adverse outcomes (nodal involvement, metastasis, and recurrence) in breast cancer, whereas the overexpression of the OPNa splice variant was observed in connection with the tumor growth of lung cancer cells, and OPNa was found to have a key role in thyroid cancer tumor progression (15, 16).
Recently, OPN has received increasing attention, with several studies investigating its role as a potential biomarker in different cancers, including melanoma (17–19). We previously observed, in our high-throughput microarray-based gene expression study, that ulcerated melanomas exhibit 6-fold higher expression of the SPP1/OPN gene compared to non-ulcerated melanomas (20). These results were also validated by qRT-PCR showing that elevated OPN mRNA expression is significantly associated with unfavorable prognostic parameters such as late stages (Clark stages IV–V), elevated Breslow thickness (≥4.00 mm), and ulcerated tumor surface (21). However, the expression patterns and the role of the OPN variants have not yet been described in human malignant melanoma. The expression of OPN splice variants in nonmelanoma skin cancers has only recently been investigated (22).
The aim of our study was to characterize the relative gene expression levels of OPN isoforms and clarify the prognostic significance of the five splice variants in primary and metastatic malignant melanoma. We also aimed to investigate the expression patterns of different integrins (ITGA2, ITGA3, ITGA5, ITGA6, ITGA9, ITGAV, ITGB1, and ITGB3) and assess their potential correlation with the clinicopathological parameters and osteopontin in the same tumors.
Materials and Methods
Melanoma Tissue Samples
Melanoma tissues were obtained from the Department of Dermatology at the University of Debrecen, Hungary. This study was approved by the Regional and Institutional Ethics Committee of the University of Debrecen Clinical Center [ETT TUKEB 26364-1/2012/EKU (449/PI/12)] and was carried out according to all relevant regulations. Written informed consent was obtained from the patients. Lesions were diagnosed on the basis of formalin-fixed paraffin-embedded tissue sections stained with hematoxylin–eosin. A total of 31 primary and 10 metastatic melanoma samples were analyzed. The metastatic samples were derived from different patients other than the primary tumors. The follow up period of patients was 5 years. The clinical–pathological parameters of the melanoma tissue samples are summarized in Table 1.
TABLE 1.
Clinicopathological parameters of melanoma tissue samples.
| Variables | n |
|---|---|
| Primary melanoma samples (n = 31) | |
| SSM a | 21 |
| NM b | 10 |
| Gender | |
| Male | 16 |
| Female | 15 |
| Age (years) | |
| 20–50 | 7 |
| >50 | 24 |
| Breslow thickness (mm) c | |
| <2.00 | 8 |
| 2.01-4.00 | 14 |
| >4.00 | 9 |
| Clark’s level | |
| II–III | 13 |
| IV–V | 17 |
| n.d. | 1 |
| Ulceration | |
| Absent | 18 |
| Present | 13 |
| Localisation | |
| Trunk | 15 |
| Extremities | 13 |
| Head | 3 |
| Metastasis formation d | |
| Non-metastatic | 13 |
| Metastatic | 18 |
| Melanoma metastases (n = 10) | |
| Male | 7 |
| Female | 3 |
| Age (years) | |
| 20–50 | 2 |
| >50 | 8 |
| Localization | |
| Regional lymph node | 4 |
| Regional (sub)cutaneous | 3 |
| Distant | 3 |
SSM, superficial spreading melanoma.
NM, nodular melanoma.
Thickness categories based on the current melanoma staging system.
Patients with follow-up periods of 5 years were included into the study.
Before RNA isolation all melanoma tissue samples were examined for the content of tumor cells and the adjacent normal tissues were removed to ensure that normal cell contamination will not influence the results. The tumor cell content of tissues analysed were ≥80% for each sample. The total RNA was isolated from frozen melanoma tissues using the RNeasy Plus Mini Kit (Qiagen GmbH, Hilden, Germany) according to the manufacturer’s protocol. The concentration and the quality of the RNA was determined using NanoDrop ND-1000 UV–vis Spectrophotometer V3.3.0 (NanoDrop Technologies, Wilmington, DE, United States). The absorbance ratios of 260 nm/280 nm of all RNA samples were 1.8 or above and the 260 nm/230 nm ratios were 2.0 or above. cDNA synthesis of 600 ng total RNA was performed using a High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, United States) according to the manufacturer’s protocol.
qRT-PCR
Gene expression levels of OPN splice variants (OPNa, OPNb, OPNc, OPN4, and OPN5) and integrins (ITGA2, ITGA3, ITGA5, ITGA6, ITGA9, ITGAV, ITGB1, and ITGB3) were determined by real-time PCR using Xceed qPCR Probe 2x Mix Hi-ROX (Institute of Applied Biotechnologies, Prague, Czech Republic) and a LightCycler® 480 Instrument II (Roche Diagnostics Nederland BV, Almere, Netherlands). Each reaction contained 15 ng cDNA and was run on the LightCycler® 480 instrument. Conditions for real-time PCR included the following steps: preactivation: 95°C 1 min; followed by 45 cycles of the following program: 95°C 5 s (denaturation), annealing 55–62°C for 10 s (specific annealing temperature and the sequence of each primer can be found in Table 2), extension 72°C 15 s, cooling at 40°C for 30 s; finished by melting curve analysis. Primers were obtained from Life Technologies. To analyze qRT-PCR data, cyclophilin A (CYPA) was used as a reference gene and the Livak method (2−ΔΔCT equation) was applied (23). Pooled nevi (n = 8) was used as a normalization control for the melanoma tissue samples. Statistical analysis was performed using IBM SPSS (Statistical Package for Social Sciences) Statistics for Windows, version 25.0 (IBM Corp., Armonk, NY, United States).
TABLE 2.
Primer sequences of OPN splice variants, reference gene and integrins used for qRT-PCR.
| Gene | Nucleotide sequence | T annealing (°C) |
|---|---|---|
| OPNa F | ATCTCCTAGCCCCACAGAAT | 55 |
| OPNa R | CATCAGACTGGTGAGAATCATC | |
| OPNb F | ATCTCCTAGCCCCAGAGAC | 55 |
| OPNb R | AAAATCAGTGACCAGTTCATCAG | |
| OPNc F | TGAGGAAAAGCAGAATGCTG | 57 |
| OPNc R | GTCAATGGAGTCCTGGCTGT | |
| OPN4 F | GGAAAAGCAGACCCTTCC | 55 |
| OPN4 R | CATATGTGTCTACTGTGGGG | |
| OPN5 F | AACAAATGGGCATTGTCCCC | 59 |
| OPN5 R | GCAGTCTAATTGCAGTGACCC | |
| CYPA F | CTCGAATAAGTTTGACTTG | 60 |
| CYPA R | CTAGGCATGGGAGGGAACA | |
| ITGA2 F | CACAAAGACACAGGTGGGGT | 62 |
| ITGA2 R | TGGGATGTCTGGGATGTTGC | |
| ITGA3 F | GCCCACAAGGATGACTGTG | 60 |
| ITGA3 R | GCTGGTCTTCTGACCCTGA | |
| ITGA5 F | GAGCAAGAGCCGGATAGAGG | 55 |
| ITGA5 R | CTGCTCCCCAAACACTTCCA | |
| ITGA6 F | AAACTGCGTCCCATTCCCA | 60 |
| ITGA6 R | TGTCGTCTCCACATCCCTC | |
| ITGA9 F | CGGTACACCTACCTGGGCTA | 58 |
| ITGA9 R | AAACCTTGCCGATGCCTTTG | |
| ITGAV F | AATGTTGTGCCGGATGTTTCTT | 58 |
| ITGAV R | CGGGTAGAAGACCAGTCACAT | |
| ITGB1 F | CCAAATGGGACACGGGTGAA | 58 |
| ITGB1 R | GTGTTGTGGGATTTGCACGG | |
| ITGB3 F | CCTCATCACCATCCACGACC | 62 |
| ITGB3 R | GTTGTTGGCTGTGTCCCATT |
Statistical Analysis
The expression data were analyzed in the melanoma tissue samples classified by histological tissue subtype, Breslow thickness of the primary tumors, and Clark staging. The following Breslow thickness groups were applied based on the current melanoma staging system: tissues from tumors with less than 2 mm thickness (n = 8), between 2 and 4 mm (n = 14), and tumors with more than 4 mm thickness (n = 9) (24). The Kruskal–Wallis H test was used to determine the significant differences of expression data between more than two groups. A two-sided Mann–Whitney U test was applied to reveal significant differences between the expression data of two certain tissue sample groups. Stepwise regression analysis was applied to select those OPN variants and integrins whose expression demonstrate a relationship with the Breslow thickness (as a continuous variant) independently of each other and without collinearity. Linear regression (adjusted for age and sex) was carried out to determine which of these variants were significantly associated with Breslow thickness. Primary melanomas were also grouped according to whether the patient was diagnosed with metastasis or did not develop metastasis during the follow-up (5 years) period. The expression data were analyzed by logistic regression. p < 0.05 was considered to indicate statistically significant differences in each case.
Results
Relative Gene Expression of OPN Splicing Isoforms and Their Comparison in Melanoma Tissue Subtypes
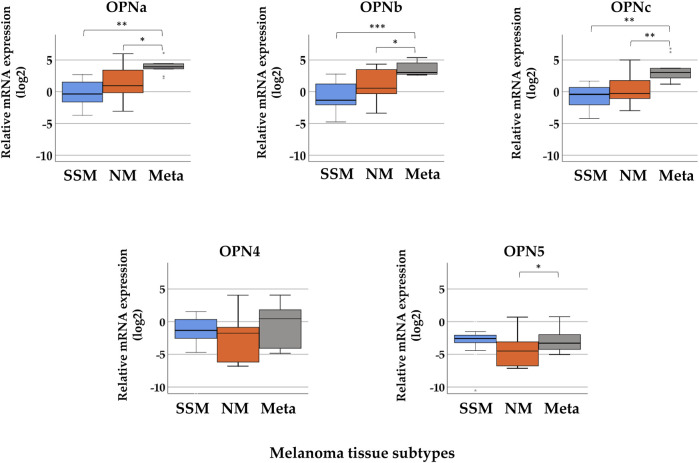
The median gene expression values determined for OPN splicing isoforms in SSM, NM, and melanoma metastasis tissue samples are summarized in Supplementary Table S1. In Figure 1, boxplots show the relative expression of the five splice variants (OPNa, OPNb, OPNc, OPN4, and OPN5) in superficial spreading melanomas, nodular melanomas, and melanoma metastasis. The highest expression of OPNa (median = 3.925), OPNb (median = 3.043), and OPNc (median = 3.060) was detected in melanoma metastasis, which significantly differed when compared to the nodular and superficial tissue samples (p ≤ 0.05). The relative expression levels of OPN4 and OPN5 were very low in all pathological subtypes, and they were downregulated relative to nevi (except in the case of OPN4 in metastasis). A significant difference in the expression of OPN5 was observed only between nodular and melanoma metastasis tissues (p ≤ 0.05).
FIGURE 1.
Relative mRNA expression levels of osteopontin isoforms (OPNa, OPNb, OPNc, OPN4, and OPN5) in distinct pathological subgroups of malignant melanoma: superficial spreading melanoma (SSM, n = 21), nodular melanoma (NM, n = 10), and melanoma metastasis (Meta, n = 10) tissue samples. Asterisks indicate significant differences: *p < 0.05; **p < 0.01; ***p < 0.001.
Comparison of the Relative Gene Expression of OPN Splice Variants in Tissues Classified by Breslow Thickness and Clark Stages in Primary Tumor Samples
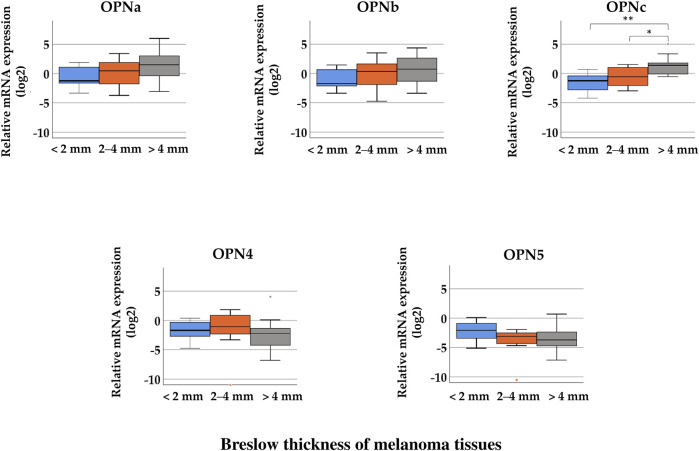
Investigating the relative gene expression of the OPN isoforms in the three thickness groups, the Kruskal–Wallis H test showed significant differences in the relative expression of OPNc between the groups (<2 mm (median = −1.24), 2–4 mm (median = −0.57), and >4 mm (median = 1.40); p = 0.008). When comparing two groups with each other, tissues samples with >4 mm Breslow thickness showed significantly higher relative expression of OPNc than samples with 2–4 mm thickness (Mann–Whitney U test p = 0.023), and tissues with less than 2 mm exhibited the lowest expression (Figure 2). This trend of elevation can also be observed in the expression of OPNa and OPNb variants as the thickness increases; however, these differences were not considered significant. The mRNA expression of OPN4 and OPN5 did not show significant association with Breslow thickness. Supplementary Table S2 summarizes the median values of each group. We did not find significant association between the relative gene expression levels of the different OPN splice variants and Clark stages.
FIGURE 2.
Comparison of the relative mRNA expression of osteopontin isoforms (OPNa, OPNb, OPNc, OPN4, and OPN5) in malignant melanoma tissue samples from tumors with different Breslow thicknesses: < 2 mm (n = 8), 2–4 mm (n = 14), and >4 mm (n = 9). Significant differences (*p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001 Mann–Whitney test, Kruskal–Wallis test) are indicated by asterisks.
Relative Gene Expression of Integrins and Their Comparison in Pathological Subgroups of Melanoma Tissues
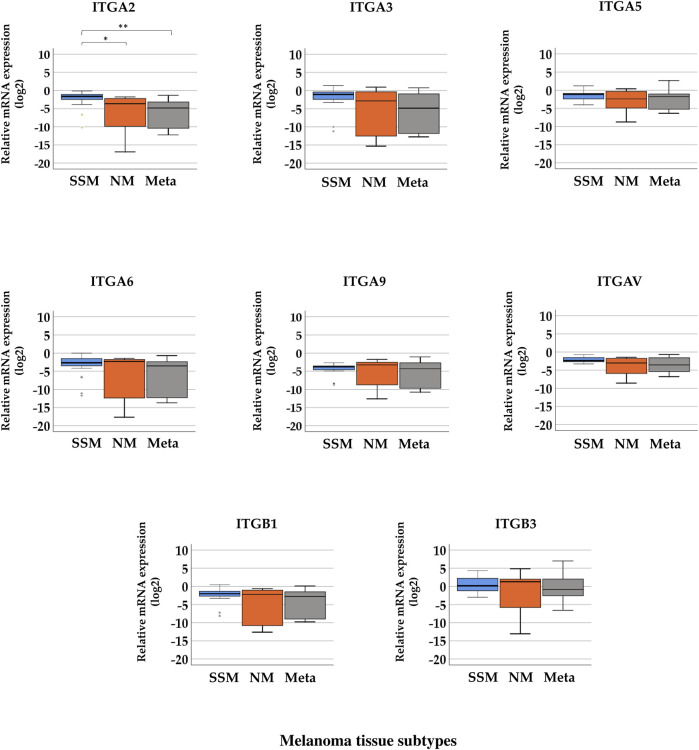
Boxplots of the expression levels of the eight integrin genes in subgroups of melanoma tissue samples are presented in Figure 3. Excluding ITGA2, seven integrins exhibited no significant differences in gene expression between different tissue subtypes. A significant difference in the gene expression level of ITGA2 was found in SSM tissue samples (median = −1.622) compared with NM (median = −3.630) and melanoma metastasis (median = −4.807) tissue samples. (Kruskal–Wallis H test, p = 0.001; Mann–Whitney U test, p = 0.002). However, the expression of the eight tested integrin genes was, overall, extremely low (with negative values for medians indicating downregulation) except ITGB3, which showed measurable values without significant differences in expression (median values: MSSM = 0.168, MNM = 1.274, and MMetastasis = −0.863). Supplementary Table S3 summarizes the median values of relative gene expression data in each melanoma tissue subgroup.
FIGURE 3.
Relative mRNA expression levels of integrin (ITGA2, ITGA3, ITGA5, ITGA6, ITGA9, ITGAV, ITGB1, and ITGB3) genes in distinct pathological subgroups of malignant melanoma: superficial spreading melanoma (SSM, n = 20), nodular melanoma (NM, n = 9), and melanoma metastasis (Meta, n = 10) tissue samples. Asterisks indicate significant differences (*p < 0.05; **p < 0.01; ***p < 0.001).
Relationship Between the Relative Gene Expression of Integrins and the Clinicopathological Data (Breslow Thickness and Clark Staging) of Primary Melanoma Tissues
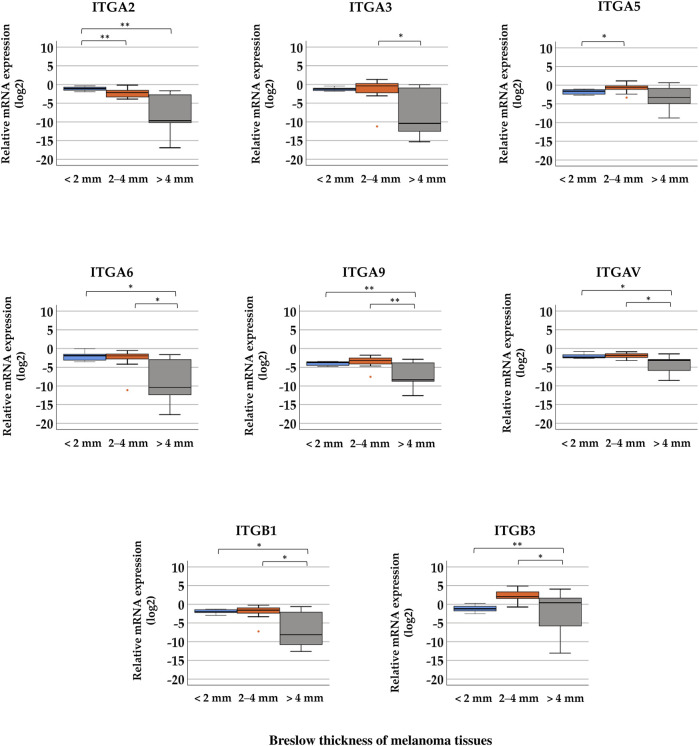
In the cases of ITGA2, ITGA6, ITGA9, ITGAV, ITGB1, and ITGB3, significant differences were observed between the three Breslow thickness groups (Figure 4, Kruskal–Wallis test, p < 0.05). Tumor samples with more than 4 mm thickness exhibited significantly lower relative expression levels of ITGA3, ITGA6, ITGA9, ITGAV, and ITGB1 than samples belonging to the 2–4 mm Breslow thickness category (Mann–Whitney test, p < 0.05). ITGB3 exhibited significantly higher relative expression in tissues with 2–4 mm and >4 mm Breslow thickness, and was the highest in the group with 2–4 mm thickness. Median values can be found in Supplementary Table S4.
FIGURE 4.
Comparison of relative mRNA expression levels of integrin (ITGA2, ITGA3, ITGA5, ITGA6, ITGA9, ITGAV, ITGB1, and ITGB3) genes in melanoma tissue samples grouped by distinct Breslow thickness: <2 mm (n = 8), 2–4 mm (n = 12), and >4 mm (n = 9). Significant differences (*p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001 Mann–Whitney test, Kruskal–Wallis test) are indicated by asterisks.
Tissue samples of primary tumors were also differentiated according to Clark stages: earlier stages (II–III, n = 13), and later stages (IV–V, n = 17) Supplementary Figure S1 shows that a significant difference was observed only in the case of ITGA2: tissues with a later Clark stage (IV–V) exhibited lower mRNA expression (mean = −1.35) than samples with earlier stage (II–III) (mean = −3.57) (Mann–Whitney U test, p < 0.005).
Correlation of the Relative Expression of Osteopontin Variants and Integrins
The gene expression data of OPN variants and integrins analyzed with Spearman’s correlation revealed positive correlations of expression between OPN4 and most of the integrins: ITGB3 (r = 0.604), ITGA5 (r = 0.530), ITGA9 (r = 0.530), ITGAV (r = 0.520), ITGB1 (r = 0.590), ITGA3 (r = 0.585), and ITGA6 (r = 0.500) (p < 0.01). Positive correlations of expression were also observed between OPN5 and ITGA2 (r = 0.447), ITGAV (r = 0.504) (p < 0.01), ITGB1 (r = 0.348), ITGA3 (r = 0.361), and ITGA6 (r = 0.406) (p < 0.05). Negative correlations of expression were observed between ITGA2 and OPNa (r = −0.480), OPNb (r = −0.416), and OPNc (r = −0.540) (p < 0.01) and between ITGA6 and OPNc (r = −0.392) (p < 0.05). A table with the results of the Spearman’s rho correlation can be found in Supplementary Table S5 and graphs of significantly correlating variables in Supplementary Figures S2–S5.
Correlation of OPN Variants and ITGs With Breslow Thickness (As a Continuous Outcome in mm) Based on the Results of Stepwise and Linear Regression Analysis
As a result of stepwise regression analysis, the expression of two osteopontin variants, OPNc and OPN4, and two integrins, ITGA5 and ITGA2, was determined to be linked with Breslow thickness independently. The linear regression analysis of these variants revealed that their relationship with Breslow thickness was significant; however, while the beta value of OPNc was 0.217, indicating a positive correlation, the negative beta value of OPN4, −0.762, indicates a negative correlation with Breslow thickness. In the case of integrins, ITGA2 had a significant negative correlation (β value = −23.061), but ITGA5 showed a significant positive correlation (β value = 2.697) with Breslow thickness. A 0.1 unit (2−ΔΔCt) decrease in ITGA2 expression is associated with 2.3 mm increase of Breslow thickness. See Table 3 for more details.
TABLE 3.
Linear regression analyses of the gene expression of OPN splice variants and integrins with the Breslow thickness of primary melanomas.
| Osteopontin isoforms | β | 95% CI | p-value |
|---|---|---|---|
| OPNa | 0.115 | −0.011–0.242 | 0.071 |
| OPNb | 0.172 | −0.069–0.414 | 0.155 |
| OPNc | 0.217 | 0.112–0.322 | <0.001 |
| OPN4 | −0.762 | −1.343–(−0.181) | 0.012 |
| OPN5 | −3.030 | −8.237–2,176 | 0.242 |
| Integrins | β | 95% CI | p-value |
| ITGA2 | −23.061 | −30.158–(−15.964) | <0.001 |
| ITGA3 | −2.338 | −6.861–2.185 | 0.296 |
| ITGA5 | 2.697 | 0.956–4.438 | 0.004 |
| ITGA6 | −9.079 | −20.317–2.158 | 0.108 |
| ITGA9 | −14.345 | −41.876–13.185 | 0.292 |
| ITGAV | −15.465 | −30.958–0.046 | 0.051 |
| ITGB1 | −5.086 | −13.612–3.440 | 0.229 |
| ITGB3 | 0.023 | −0.099–0.146 | 0.7 |
β is a regression coefficients which indicates the direction and size of effect between variables. CI: confidence interval. Bold values indicates significant associations.
Logistic Regression: Relative Expression of OPN Variants and Presence of Metastasis
The results of logistic regression show that OPNc expression (log2 transformed data) is significantly positively correlated with the presence of metastasis (OR = 1.931, p = 0.044) (Table 4), whereas OPN4 was not significantly correlated (OR = 0.962, p = 0.773) with the presence of metastasis . Integrins did not show significant correlation with the presence of metastasis.
TABLE 4.
Logistic regression analyses of the gene expressions OPN splice variants and integrins in relation of metastasis formation of malignant melanoma.
| Osteopontin isoforms | OR | 95% CI | p-value |
|---|---|---|---|
| OPNa | 1.223 | 0.855–1.749 | 0.270 |
| OPNb | 1.070 | 0.759–1.507 | 0.699 |
| OPNc | 1.931 | 1.018–3.661 | 0.044 |
| OPN4 | 0.962 | 0.743–1.248 | 0.773 |
| OPN5 | 1.144 | 0.867–1.509 | 0.341 |
| Integrins | OR | 95% CI | p-value |
| ITGA2 | 0.685 | 0.348–1.349 | 0.274 |
| ITGA3 | 0.411 | 0.135–1.250 | 0.117 |
| ITGA5 | 0.889 | 0.396–1.996 | 0.776 |
| ITGA6 | 0.560 | 0.234–1.337 | 0.191 |
| ITGA9 | 0.675 | 0.407–1.120 | 0.129 |
| ITGAV | 0.280 | 0.065–1.207 | 0.088 |
| ITGB1 | 0.333 | 0.085–1.305 | 0.115 |
| ITGB3 | 0.836 | 0.642–1.088 | 0.182 |
OR, odds ratio; CI, confidence interval. Bold values indicates significant associations.
Discussion
Osteopontin (OPN), a multifunctional protein, has been widely studied as a promising biomarker in various types of tumors for monitoring tumor progression, invasion, metastasis formation and drug resistance (13, 25). The association between the aberrant expression of osteopontin and melanoma invasion, metastasis formation, and radio/drug resistance has been recently described (1, 3, 25–27). The biological functions of tumor-associated gene products are extensively regulated via pre- and posttranscriptional modifications, resulting in alternative splicing of OPN. Alternative splicing of various mRNA products of a single gene is a critical mechanism for generating proteomic diversity. OPNa, OPNb, and OPNc variants were first described in glioma, and the two additional splice variants (OPN4 and OPN5) were found in esophageal adenocarcinomas and glioblastomas (28). Because the different splice variants of OPN are associated with different types of cancers, it is assumed that the isotypes may have different functions. Osteopontin isoforms display functional heterogeneity and cell and tissue specificity, which still poses challenges while providing opportunities for novel diagnostic, prognostic, and therapeutic strategies (13).
Most of the studies investigating the role of OPN splice variants have focused on the expression of OPNa, OPNb, and OPNc; however, the data in distinct tumor types are conflicting, and the functional heterogeneity of the variants serves as motivation for researchers to define the role of OPN splice variants in each type of cancer. In addition, at present, no data on the isoform expression patterns in malignant melanoma are available. Therefore, in this study, our primary aim was to study the expression profile of five OPN splice variants (OPNa, OPNb, OPNc, OPN4, and OPN5) in different types of malignant melanoma tissues and investigate the association with the clinicopathological features of tumor tissues. In addition, because OPN signaling occurs through different integrin receptors, we also aimed to examine the relative mRNA expression profile of eight integrins (ITGA2, ITGA3, ITGA5, ITGA6, ITGA9, ITGAV, ITGB1, and ITGB3) and analyze the correlation of the relative expression of osteopontin variants and integrins in the same melanoma samples.
We found elevated relative mRNA expression of OPN variants (OPNa, OPNb, and OPNc) in nodular melanomas and melanoma metastasis compared to samples of superficial spreading melanoma. The significant increase of these isoforms in the more advanced stages indicates that they may contribute to tumor progression and worse outcome, since lower survival rates were observed in NM and metastatic melanoma patients (29). The significant elevation of OPNc expression in thicker melanoma tissue samples suggests that it is associated with increasing Breslow thickness. The elevating tendencies of the expression levels of the other variants were similar; however, none were found to be significant in the comparison. Our further statistical analysis also confirmed the significant positive association of OPNc with Breslow thickness: a one-unit increase in OPNc expression is associated with a 0.217 increase in Breslow thickness. Based on our observations, it is possible that OPNc expression has a crucial role in melanoma tumor progression, and that elevated levels of this variant can contribute to progression toward advanced stages of disease and even the induction of metastasis formation, and it can thus serve as an indicator of an aggressive phenotype.
The expression of OPN splice variants has already been investigated in hematological malignancies, thyroid tumors, and gastric cancers (18, 30). Wide variations were observed in the expression patterns and the predominantly expressed variant(s) depending on the tumor type; therefore, it is difficult to establish a possible universal nature or profile. It could be convenient to establish the expression profile of each splice variant in each distinct cancer type. To date, it has been observed that the OPNa mRNA levels were significantly associated with high TNM staging and unfavorable clinical outcomes in gastric cancer; moreover, OPNa and OPNb are correlated with short overall and disease-free survival of patients (30). It was pointed out that OPNc expression is also associated with advanced stage, tumor recurrence, and metastasis formation; thus, OPNc is considered to be a promising prognostic factor in breast cancer (15). In our results, the elevated expression of the OPNc variant in an advanced stage of primary melanoma and melanoma metastasis samples in addition to its significant correlation with the presence of metastasis indicates the importance of OPNc in melanoma progression, metastasis formation, and the relationship with the aggressive phenotype, which justifies its further investigation at multiple levels as a promising prognostic biomarker in malignant melanoma. Since no specific antibodies against these splice variants are currently available (except for OPNc from Gallus Immunotech Inc. which is a polyclonal antibody), the future development of the OPN isotype antibodies could also be an important step forward in the characterization of these variants. Moreover, investigating the specific role of each splice variant in melanoma progression could bring us closer to developing potential targetable molecules in melanoma therapy.
While the exact role of OPNc overexpression on tumor progression is unknown, there are several possible explanations for its association with metastasis. Since OPNc variant lacks exon 4, which contains the target sequence for transglutaminase, it lacks an important domain for calcium induced aggregation and transglutamination (31). OPNc, unlike the other isoforms, cannot form polymeric complexes. This might be the essential reason for the pathological role of OPNc and it may not be cross-linked with extracellular matrix and thus it results cell migration. The full length OPN aggregates and enhances cell adhesion and therefore reduces dissemination of tumor cells, whereas OPNc promotes tumor invasion and metastasis formation because of its lack of aggregation (32). The non-aggregative nature of OPNc is in concert with the relative resistance to polymerization (33). On the other hand, OPNc can stimulate cell proliferation rates independently of growth factors, a feature of proteins typically involved in tumor progression (34). While numerous studies suggest that OPN plays a key role in mediating tumor progression and metastasis by regulating various pathways, very few data are available for the role of different OPN splice variants, the expression patterns of the OPN isoforms in malignant melanoma was first described in the present study. In order to discover the detailed functional role of OPNc, the 3D structure of the variant might be useful. It is an important step that the tertiary structure of OPNc was successfully predicted by Sivakumar and this predicted structure might be used for computational drug design of OPNc with respect to cancer prevention (31).
Aside from the three most frequently studied OPN variants, we also investigated the expression of OPN4 and OPN5 isoforms that were recently described (4, 35). Surprisingly, relative expression of OPN4 and OPN5 was low in primary as well as metastatic tissues. Though statistical analysis showed a significant difference in OPN5 expression between nodular melanoma and melanoma metastasis histological subtypes, OPN4 and OPN5 were downregulated, and the median expression values did not exceed zero on the logarithmic scale (except in the case of OPN4 in melanoma metastasis). Although significant differences were not found when comparing their expression between the sample groups for different Breslow thicknesses, logistic regression demonstrated a significant negative correlation between OPN4 and Breslow thickness and conversely in the case of OPNc, which was positive correlated. Moreover, the analysis of the Spearman’s correlation between osteopontin splice variants and integrins in expression data revealed the positive correlation of OPN4 expression with that of most of the integrins and the negative correlation of ITGA2 expression with that of most of the osteopontins, suggesting that OPN4 may have an expression profile more similar to that of the integrins than that of the osteopontins.
The expression of the OPN4 and OPN5 variants was previously investigated in esophageal adenocarcinoma tissue samples (4), where, unlike in our study, it was found that the expression of OPN4 and OPN5 was elevated in primary tumors when compared to normal and Barrett’s samples, and the isoforms were co-overexpressed. In another study, the expression of these two isoforms was found to be variable in most of the tested 7 cell lines (prostate tumor, ovarian cancer, B-cell precursor acute lymphoid leukemia, breast cancer, colorectal cancer, and thyroid and lung tumors) (31). Except for in the two breast cancer cell lines, OPN4 and OPN5 were found to be co-expressed in the other 5 cell lines, but the expression patterns differed from those of the previously characterized OPNa, OPNb, and OPNc variants (31). According to the study of Chou et al., besides the predominant expression of OPNa variant, OPN4 was found to be minimally expressed in normal skin and nonmelanoma skin cancer, but OPN5 exhibited higher expression in normal skin than OPNb and OPNc, and OPN5 was more highly expressed in nonmelanoma skin cancer than OPNc (22). Taken together, the expression of OPN4 and OPN5 splice variants appears to vary widely in distinct tumor types, but in melanoma, they slightly have a different expression profile than the three predominant splice variants.
In connection with the expression profile of integrins, our results show that the relative mRNA expression levels of the investigated integrin genes are extremely low except in one case. Median values of the integrin expression levels in SSM, NM, and melanoma metastasis were equally below zero, which means they are downregulated compared to the nevus control. As the exception, ITGB3 appeared to be upregulated in SSM and NM tissue samples. Moreover, when comparing integrin gene expression in melanoma tissues with distinct Breslow thickness, a significantly higher ITGB3 expression was observed in thicker tissue samples (2–4 mm and > 4 mm) compared to tissues with lower Breslow thickness (<2 mm) (p = 0.004). This result is in accordance with the relevant literature: in malignant melanoma, upregulated expression of subunit β3 was found in the vertical growth phase, which was linked with disease progression and correlated with poor survival and lymph node and lung metastasis formation (36, 37).
Other previous in vivo and in vitro studies have also investigated altered integrin expression, summarized in detail by Arias-Mejias et al. (38). It was found that elevated expression of integrin β3 protein (the dimerized form is αvβ3 or αIIbβ3) in human melanoma cells and tissues was associated with tumor progression, organ-specific metastasis formation, disease recurrence, and decreased long-term survival (39, 40). In our earlier study we identified metastasis correlated genes, including many genes involved in signaling in the immune system (HLA antigens), cell adhesion and cell motility networks (40, 41). These networks involve genes such as that of integrins (ITGA2, ITGA3, ITGA4, ITGA9, ITGB5 or ITGB8). Investigating the expression of these genes in metastatic primary melanomas and metastases, we found that ITGA3 was downregulated in both regional and distant organ metastases compared to the metastatic primary lesions. In the present study, even the direction of the mRNA expression was similar for ITGA3, we observed significant decrease only for the ITGA2 gene in the metastatic tumors. The inconsistency between the two investigations can be explained by the fact that the composition of primary tumor groups was different between the two studies.
In our case, interestingly, a significant difference between the histological subtypes was observed only in the expression of ITGA2; however, it displays a decreasing tendency in NM and metastasis samples compared to SSM. Relative expression of integrins in melanoma tissue samples with distinct Breslow thicknesses varies, six integrins (ITGA3, ITGA6, ITGA9, ITGAV, ITGB1, and ITGB3) out of the eight showed a significant difference between the 2–4 mm and the > 4 mm group (lowest expression in the thickest group), and again, expression of ITGA2 showed a significant decrease as the thickness increased. Linear regression analysis indicated that while ITGA5 had positive correlation with Breslow thickness (as a continuous variant) ITGA2 was in strong significant inverse correlation with the Breslow thickness of the tumor. These results suggest that downregulation of ITGA2 may be linked with tumor progression in malignant melanoma. This hypothesis seems to be supported by the results of in vitro studies with breast cancer mouse models, which suggest that integrin α2 might function as a metastasis suppressor (42). Moreover, decreased expression of the α2 subunit was found to be associated with more advanced status, such as higher tumor nodal status or presence of metastasis (41). Madamanchi et al. describe that certain other cancer types (prostate, colon, and lung cancer) also seem to be associated with reduced integrin α2β1, which is associated with tumor progression and metastasis. However, they also note that other cancer types were associated with high α2β1 integrin expression levels; hence, the exact biological role of this integrin is being heavily debated (43). Indeed, the relevance of the α2β1 integrin as a main regulator of metastasis in tumor cells was discovered only in recent years, and these findings appear to be controversial compared with the results of the previously mentioned studies. The crucial role of the α2β1 integrin has been determined in cancer types including melanoma, as it is responsible for regulating cell migration, survival, proliferation, and metastasis formation in the lung and liver (44, 45). Upregulation of α2β1 was found in highly metastatic melanoma compared to nonmetastatic or poorly metastatic cell lines, where it was associated with enhanced cell migration (38). Increased expression of α2β1 in malignant melanoma compared to benign tumors was found to stimulate angiogenesis and facilitate tumor growth (46).
Though acquiring a comprehensive understanding of integrin signaling is challenging, some possible reasons may explain the differences in the expression of the various integrins and their role in tumor progression. Single nucleotide polymorphisms may change the affinity of ITGA2 for transcription factors, which can alter the transcription rate (47). Enhancing transcription–coactivator complex binding can increase ITGA2 transcription. In addition, different posttranslational modifications, such as sialylation and glycosylation, can modify the role of integrins in tumor progression (48, 49).
The data of the current study are the first to describe the relative mRNA expression of five osteopontin splice variants in primary and metastatic melanoma tissue samples. We found that the expression levels of OPNa, OPNb, and OPNc were significantly higher in the metastatic lesions compared to the primary tumors, and OPNc was significantly positively correlated with increasing Breslow thickness in the primary tumors. The expression of the recently described OPN4 and OPN5 isoforms was shown to be downregulated in the evaluated melanoma subtypes, and OPN4 exhibited a significant negative correlation with Breslow thickness. The relative expression of eight integrins was very low; only ITGB3 showed detectable expression in metastatic tumors compared to the primary lesions; moreover, ITGA2 showed significant negative correlation with the Breslow thickness of the primary tumors. Our data show that high expression of OPNa, OPNb, and OPNc is associated with poor prognosis, and OPN4 and ITGA2 may have an opposite role in melanoma progression. Nevertheless, further studies are needed to more specifically characterize the involvement of osteopontin splice variants in malignant melanoma progression and their interaction with integrins in cancer.
Data Availability Statement
The original data presented in this study are included in the article as well as in the Supplementary Material, further inquiries can be directed to the corresponding author.
Ethics Statement
The study was conducted according to the guidelines of the Declaration of Helsinki and approved by the Regional and Institutional Ethics Committee of the University of Debrecen [Document No.: 25364-1/2012/EKU (449/PI/12. and DE RKEB/IKEB: 4820-2017)] and by the Ethics Committee of the Hungarian Scientific Council on Health (Reference No.: 6674/2014 EKU and 17876/218). Informed consent was obtained from all subjects involved in the study. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
MB, KJ, and TK conceived and designed the study. KJ and TK performed the experiments. KJ, TK, and PP analyzed the data. KJ, IS, PP, and VK performed statistical analyses and designed the figures and tables. KJ wrote the first draft of the manuscript. VK, IS, and KJ were involved to write the final version of the manuscript. PP, TK, KJ, VK, and IS approved the manuscript. MB finalized the manuscript. MB were responsible for supervision and funding acquisition.
Funding
This research was co-financed by the National Research Development and Innovation Fund (grant number K-112327 and K-135752), the European Regional Development Fund (GINOP-2.3.2-15-2016-00005), and the Hungarian Academy of Sciences (TK2016-78). New National Excellence Program of the Ministry for Innovation and Technology from the Source of National research, Development and Innovation Fund (UNKP-21-4-II-DE-361, UNKP-21-4-II-DE-136, and UNKP-21-4-II-DE-363).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.por-journal.com/articles/10.3389/pore.2022.1610608/full#supplementary-material
Relative mRNA expression of ITGA2 gene in malignant melanoma tissue samples from tumors with different Clark stages: earlier stages (II–III. n = 13) and later stages (IV–V. n = 17).
Graphs illustrating the significant positive correlations (Spearman’s rho) between expression of OPN4 and integrin genes (ITGB3, ITGA5, ITGA9, ITGAV, ITGB1, ITGA3, and ITGA6).
Graphs illustrating the significant positive correlations (Spearman’s rho) between the expression of OPN5 and integrin genes (ITGA2, ITGAV, ITGB1, ITGA3, and ITGA6).
Graphs illustrating the significant negative correlations (Spearman’s rho) between the gene expression of ITGA2 and osteopontin splice variants (OPNa, OPNb, OPNc).
Graphs illustrating the significant negative correlation (Spearman’s rho) between the expression of ITGA6 and OPNc.
References
- 1. Zhao Y, Huang C. The Role of Osteopontin in the Development and Metastasis of Melanoma. Melanoma Res (2021) 31(4):283–9. 10.1097/CMR.0000000000000753 [DOI] [PubMed] [Google Scholar]
- 2. Rangel J, Nosrati M, Torabian S, Shaikh L, Leong SP, Haqq C, et al. Osteopontin as a Molecular Prognostic Marker for Melanoma. Cancer (2008) 112(1):144–50. 10.1002/cncr.23147 [DOI] [PubMed] [Google Scholar]
- 3. Gimba ERP, Brum MCM, De Moraes GN. Full-length Osteopontin and its Splice Variants as Modulators of Chemoresistance and Radioresistance (Review). Int J Oncol (2019) 54(2):420–30. 10.3892/ijo.2018.4656 [DOI] [PubMed] [Google Scholar]
- 4. Lin J, Myers AL, Wang Z, Nancarrow DJ, Ferrer-Torres D, Handlogten A, et al. Osteopontin (OPN/SPP1) Isoforms Collectively Enhance Tumor Cell Invasion and Dissemination in Esophageal Adenocarcinoma. Oncotarget (2015) 6(26):22239–57. 10.18632/oncotarget.4161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yokosaki Y, Tanaka K, Higashikawa F, Yamashita K, Eboshida A. Distinct Structural Requirements for Binding of the Integrins αvβ6, αvβ3, αvβ5, α5β1 and α9β1 to Osteopontin. Matrix Biol (2005) 24(6):418–27. 10.1016/j.matbio.2005.05.005 [DOI] [PubMed] [Google Scholar]
- 6. Inoue M, Shinohara ML. Intracellular Osteopontin (iOPN) and Immunity. Immunol Res (2011) 49(1-3):160–72. 10.1007/s12026-010-8179-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Del Prete A, Scutera S, Sozzani S, Musso T. Role of Osteopontin in Dendritic Cell Shaping of Immune Responses. Cytokine Growth Factor Rev (2019) 50:19–28. 10.1016/j.cytogfr.2019.05.004 [DOI] [PubMed] [Google Scholar]
- 8. Leavenworth JW, Verbinnen B, Wang Q, Shen E, Cantor H. Intracellular Osteopontin Regulates Homeostasis and Function of Natural Killer Cells. Proc Natl Acad Sci U S A (2015) 112(2):494–9. 10.1073/pnas.1423011112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Anborgh PH, Mutrie JC, Tuck AB, Chambers AF. Role of the Metastasis-Promoting Protein Osteopontin in the Tumour Microenvironment. J Cel Mol Med (2010) 14(8):2037–44. 10.1111/j.1582-4934.2010.01115.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cho HJ, Cho HJ, Kim HS. Osteopontin: A Multifunctional Protein at the Crossroads of Inflammation, Atherosclerosis, and Vascular Calcification. Curr Atheroscler Rep (2009) 11(3):206–13. 10.1007/s11883-009-0032-8 [DOI] [PubMed] [Google Scholar]
- 11. Pang X, Gong K, Zhang X, Wu S, Cui Y, Qian BZ, et al. Osteopontin as a Multifaceted Driver of Bone Metastasis and Drug Resistance. Pharmacol Res (2019) 144:235–44. 10.1016/j.phrs.2019.04.030 [DOI] [PubMed] [Google Scholar]
- 12. Briones-Orta MA, Avendano-Vazquez SE, Aparicio-Bautista DI, Coombes JD, Weber GF, Syn WK, et al. Osteopontin Splice Variants and Polymorphisms in Cancer Progression and Prognosis. Biochim Biophys Acta Rev Cancer (2017) 1868(1):93–108. 10.1016/j.bbcan.2017.02.005 [DOI] [PubMed] [Google Scholar]
- 13. Hao C, Cui Y, Owen S, Li W, Cheng S, Jiang WG, et al. Human Osteopontin: Potential Clinical Applications in Cancer (Review). Int J Mol Med (2017) 39(6):1327–37. 10.3892/ijmm.2017.2964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lamort AS, Giopanou I, Psallidas I, Stathopoulos GT. Osteopontin as a Link between Inflammation and Cancer: The Thorax in the Spotlight. Cells (2019) 8(8):E815. 10.3390/cells8080815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pang H, Lu H, Song H, Meng Q, Zhao Y, Liu N, et al. Prognostic Values of Osteopontin-C, E-Cadherin and Beta-Catenin in Breast Cancer. Cancer Epidemiol (2013) 37(6):985–92. 10.1016/j.canep.2013.08.005 [DOI] [PubMed] [Google Scholar]
- 16. Sun SJ, Wu CC, Sheu GT, Chang HY, Chen MY, Lin YY, et al. Integrin β3 and CD44 Levels Determine the Effects of the OPN-A Splicing Variant on Lung Cancer Cell Growth. Oncotarget (2016) 7(34):55572–84. 10.18632/oncotarget.10865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Han X, Wang W, He J, Jiang L, Li X. Osteopontin as a Biomarker for Osteosarcoma Therapy and Prognosis. Oncol Lett (2019) 17(3):2592–8. 10.3892/ol.2019.9905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Viana B, Gomes AVP, Gimba ERP, Ferreira LB. Osteopontin Expression in Thyroid Cancer: Deciphering EMT-Related Molecular Mechanisms. Biomedicines (2021) 9(10):1372. 10.3390/biomedicines9101372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Amilca-Seba K, Sabbah M, Larsen AK, Denis JA. Osteopontin as a Regulator of Colorectal Cancer Progression and its Clinical Applications. Cancers (Basel) (2021) 13(15):3793. 10.3390/cancers13153793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rakosy Z, Ecsedi S, Toth R, Vizkeleti L, Hernandez-Vargas H, Lazar V, et al. Integrative Genomics Identifies Gene Signature Associated with Melanoma Ulceration. PLoS One (2013) 8(1):e54958. 10.1371/journal.pone.0054958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kiss T, Ecsedi S, Vizkeleti L, Koroknai V, Emri G, Kovacs N, et al. The Role of Osteopontin Expression in Melanoma Progression. Tumour Biol (2015) 36(10):7841–7. 10.1007/s13277-015-3495-y [DOI] [PubMed] [Google Scholar]
- 22. Chou CF, Huang CC, Bin Dabil N, Chang PL. Assessing SPP1/Osteopontin (OPN) Splice Variants and Their Association to Nonmelanoma Skin Cancer by Absolute Quantification: Identification of OPN-5 Subvariants and Their Protein Coding Potential. Cancer Invest (2021) 39(6-7):559–70. 10.1080/07357907.2021.1933015 [DOI] [PubMed] [Google Scholar]
- 23. Livak KJ, Schmittgen TD. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods (2001) 25(4):402–8. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 24. Scolyer RA, Rawson RV, Gershenwald JE, Ferguson PM, Prieto VG. Melanoma Pathology Reporting and Staging. Mod Pathol (2020) 33(1):15–24. 10.1038/s41379-019-0402-x [DOI] [PubMed] [Google Scholar]
- 25. Patel V, Szasz I, Koroknai V, Kiss T, Balazs M. Molecular Alterations Associated with Acquired Drug Resistance during Combined Treatment with Encorafenib and Binimetinib in Melanoma Cell Lines. Cancers (2021) 13(23):6058. 10.3390/cancers13236058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kiss T, Jambor K, Koroknai V, Szasz I, Bardos H, Mokanszki A, et al. Silencing Osteopontin Expression Inhibits Proliferation, Invasion and Induce Altered Protein Expression in Melanoma Cells. Pathol Oncol Res (2021) 27:581395. 10.3389/pore.2021.581395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Szasz I, Koroknai V, Kiss T, Vizkeleti L, Adany R, Balazs M, et al. Molecular Alterations Associated with Acquired Resistance to BRAFV600E Targeted Therapy in Melanoma Cells. Melanoma Res (2019) 29(4):390–400. 10.1097/CMR.0000000000000588 [DOI] [PubMed] [Google Scholar]
- 28. Kijewska M, Kocyk M, Kloss M, Stepniak K, Korwek Z, Polakowska R, et al. The Embryonic Type of SPP1 Transcriptional Regulation is Re-activated in Glioblastoma. Oncotarget (2017) 8(10):16340–55. 10.18632/oncotarget.14092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. El Sharouni MA, van Diest PJ, Witkamp AJ, Sigurdsson V, van Gils CH. Subtyping Cutaneous Melanoma Matters. JNCI Cancer Spectr (2020) 4(6):pkaa097. 10.1093/jncics/pkaa097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hao CC, Cui YX, Lane J, Jia SQ, Ji JF, Jiang WG, et al. Distinctive Prognostic Value and Cellular Functions of Osteopontin Splice Variants in Human Gastric Cancer. Cells (2021) 10(7):1820. 10.3390/cells10071820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sivakumar S, Niranjali Devaraj S. Tertiary Structure Prediction and Identification of Druggable Pocket in the Cancer Biomarker - Osteopontin-C. J Diabetes Metab Disord (2014) 13(1):13. 10.1186/2251-6581-13-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. He B, Mirza M, Weber GF. An Osteopontin Splice Variant Induces Anchorage Independence in Human Breast Cancer Cells. Oncogene (2006) 25(15):2192–202. 10.1038/sj.onc.1209248 [DOI] [PubMed] [Google Scholar]
- 33. Nishimichi N, Hayashita-Kinoh H, Chen C, Matsuda H, Sheppard D, Yokosaki Y, et al. Osteopontin Undergoes Polymerization In Vivo and Gains Chemotactic Activity for Neutrophils Mediated by Integrin Alpha9beta1. J Biol Chem (2011) 286(13):11170–8. 10.1074/jbc.M110.189258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tilli TM, Franco VF, Robbs BK, Wanderley JL, da Silva FR, de Mello KD, et al. Osteopontin-c Splicing Isoform Contributes to Ovarian Cancer Progression. Mol Cancer Res (2011) 9(3):280–93. 10.1158/1541-7786.MCR-10-0463 [DOI] [PubMed] [Google Scholar]
- 35. Silva GR, Mattos DS, Bastos ACF, Viana B, Brum MCM, Ferreira LB, et al. Osteopontin-4 and Osteopontin-5 Splice Variants are Expressed in Several Tumor Cell Lines. Mol Biol Rep (2020) 47(10):8339–45. 10.1007/s11033-020-05867-9 [DOI] [PubMed] [Google Scholar]
- 36. Danen EH, Ten Berge PJ, Van Muijen GN, Van 't Hof-Grootenboer AE, Brocker EB, Ruiter DJ, et al. Emergence of α5β1 Fibronectin- and αvβ3 Vitronectin-Receptor Expression in Melanocytic Tumour Progression. Histopathology (1994) 24(3):249–56. 10.1111/j.1365-2559.1994.tb00517.x [DOI] [PubMed] [Google Scholar]
- 37. Hieken TJ, Ronan SG, Farolan M, Shilkaitis AL, Das Gupta TK. Molecular Prognostic Markers in Intermediate-Thickness Cutaneous Malignant Melanoma. Cancer (1999) 85(2):375–82. [DOI] [PubMed] [Google Scholar]
- 38. Arias-Mejias SM, Warda KY, Quattrocchi E, Alonso-Quinones H, Sominidi-Damodaran S, Meves A, et al. The Role of Integrins in Melanoma: A Review. Int J Dermatol (2020) 59(5):525–34. 10.1111/ijd.14850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Huang R, Rofstad EK. Integrins as Therapeutic Targets in the Organ-specific Metastasis of Human Malignant Melanoma. J Exp Clin Cancer Res (2018) 37(1):92. 10.1186/s13046-018-0763-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Vizkeleti L, Kiss T, Koroknai V, Ecsedi S, Papp O, Szasz I, et al. Altered Integrin Expression Patterns Shown by Microarray in Human Cutaneous Melanoma. Melanoma Res (2017) 27(3):180–8. 10.1097/Cmr.0000000000000322 [DOI] [PubMed] [Google Scholar]
- 41. Martin TA, Jiang WG. Evaluation of the Expression of Stem Cell Markers in Human Breast Cancer Reveals a Correlation with Clinical Progression and Metastatic Disease in Ductal Carcinoma. Oncol Rep (2014) 31(1):262–72. 10.3892/or.2013.2813 [DOI] [PubMed] [Google Scholar]
- 42. Ramirez NE, Zhang Z, Madamanchi A, Boyd KL, O'Rear LD, Nashabi A, et al. The αβ Integrin is a Metastasis Suppressor in Mouse Models and Human Cancer. J Clin Invest (2011) 121(1):226–37. 10.1172/JCI42328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Madamanchi A, Santoro SA, Zutter MM. α2β1 Integrin. Adv Exp Med Biol (2014) 819:41–60. 10.1007/978-94-017-9153-3_3 [DOI] [PubMed] [Google Scholar]
- 44. Bartolome RA, Torres S, Isern de Val S, Escudero-Paniagua B, Calvino E, Teixido J, et al. VE-Cadherin RGD Motifs Promote Metastasis and Constitute a Potential Therapeutic Target in Melanoma and Breast Cancers. Oncotarget (2017) 8(1):215–27. 10.18632/oncotarget.13832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yoshimura K, Meckel KF, Laird LS, Chia CY, Park JJ, Olino KL, et al. Integrin Alpha2 Mediates Selective Metastasis to the Liver. Cancer Res (2009) 69(18):7320–8. 10.1158/0008-5472.CAN-09-0315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Naci D, Vuori K, Aoudjit F. Alpha2beta1 Integrin in Cancer Development and Chemoresistance. Semin Cancer Biol (2015) 35:145–53. 10.1016/j.semcancer.2015.08.004 [DOI] [PubMed] [Google Scholar]
- 47. Di Paola J, Jugessur A, Goldman T, Reiland J, Tallman D, Sayago C, et al. Platelet Glycoprotein I(b)alpha and Integrin Alpha2 Beta1 Polymorphisms: Gene Frequencies and Linkage Disequilibrium in a Population Diversity Panel. J Thromb Haemost (2005) 3(7):1511–21. 10.1111/j.1538-7836.2005.01273.x [DOI] [PubMed] [Google Scholar]
- 48. Cheli Y, Williams SA, Ballotti R, Nugent DJ, Kunicki TJ. Enhanced Binding of poly(ADP-Ribose)polymerase-1 and Ku80/70 to the ITGA2 Promoter via an Extended Cytosine-Adenosine Repeat. PLoS One (2010) 5(1):e8743. 10.1371/journal.pone.0008743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Adorno-Cruz V, Liu H. Regulation and Functions of Integrin α2 in Cell Adhesion and Disease. Genes Dis (2019) 6(1):16–24. 10.1016/j.gendis.2018.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Relative mRNA expression of ITGA2 gene in malignant melanoma tissue samples from tumors with different Clark stages: earlier stages (II–III. n = 13) and later stages (IV–V. n = 17).
Graphs illustrating the significant positive correlations (Spearman’s rho) between expression of OPN4 and integrin genes (ITGB3, ITGA5, ITGA9, ITGAV, ITGB1, ITGA3, and ITGA6).
Graphs illustrating the significant positive correlations (Spearman’s rho) between the expression of OPN5 and integrin genes (ITGA2, ITGAV, ITGB1, ITGA3, and ITGA6).
Graphs illustrating the significant negative correlations (Spearman’s rho) between the gene expression of ITGA2 and osteopontin splice variants (OPNa, OPNb, OPNc).
Graphs illustrating the significant negative correlation (Spearman’s rho) between the expression of ITGA6 and OPNc.
Data Availability Statement
The original data presented in this study are included in the article as well as in the Supplementary Material, further inquiries can be directed to the corresponding author.