Abstract
The ςD regulon of Bacillus subtilis is composed of genes encoding proteins for flagellar synthesis, motility, and chemotaxis. Concurrent analyses of ςD protein levels and flagellin mRNA demonstrate that sigD expression and ςD activity are tightly coupled during growth in both complex and minimal media, although they exhibit different patterns of expression. We therefore used the ςD-dependent flagellin gene (hag) as a model gene to study the effects of different nutritional environments on ςD-dependent gene expression. In complex medium, the level of expression of a hag-lacZ fusion increased exponentially during the exponential growth phase and peaked early in the transition state. In contrast, the level of expression of this reporter remained constant and high throughout growth in minimal medium. These results suggest the existence of a nutritional signal(s) that affects sigD expression and/or ςD activity. This signal(s) allows for nutritional repression early in growth and, based on reconstitution studies, resides in the complex components of sporulation medium, as well as in a mixture of mono-amino acids. However, the addition of Casamino Acids to minimal medium results in a dose-dependent decrease in hag-lacZ expression throughout growth and the postexponential growth phase. In work by others, CodY has been implicated in the nutritional repression of several genes. Analysis of a codY mutant bearing a hag-lacZ reporter revealed that flagellin expression is released from nutritional repression in this strain, whereas mutations in the transition state preventor genes abrB, hpr, and sinR failed to elicit a similar effect during growth in complex medium. Therefore, the CodY protein appears to be the physiologically relevant regulator of hag nutritional repression in B. subtilis.
The bacterium Bacillus subtilis is best known for its ability to respond to adverse changes in its environment by developing into a dormant endospore (23, 39). The bacterial cell is capable of sensing when the environment is no longer able to support growth and division. Cells can respond by initiating and undergoing a series of complex changes in gene expression and cell structure that give rise to the spore. As a flagellated, motile bacterium, B. subtilis can also respond to nutrient deprivation by physically moving away from poor conditions and toward better ones.
This physical movement toward more-favorable conditions is mediated by the flagellar organelle in response to chemotactic signals. Early experiments by Nishihara and Freese (30) showed that cells exhibited increased motility (i.e., became hypermotile) as they approached the end of the exponential growth phase. These researchers found by microscopic observation that at the end of the exponential growth phase, when nutrients are scarce, there is both an increased number of motile cells and increased movement by the motile cells. Moreover, it is known that optimal transduction of the flagellum-tropic PBS1 phage of B. subtilis is obtained when phages are added at the end of exponential growth, when the cells are said to be hypermotile (4). Taken together, these studies suggest the occurrence of increased flagellin expression at the end of the exponential growth phase, perhaps triggered by nutrient deprivation, high cell density, and/or the initiation of transition state phenomena (38, 41).
Nutrient deprivation has long been known to be an important signal for the initiation of transition state phenomena and sporulation (34). More recently, researchers have become aware of the important roles of oligopeptides (and perhaps dipeptides) in the initiation of these physiological responses. Specific oligopeptides synthesized as precursors within the growing cell and then secreted, processed, and imported back into the cell have been shown to play an important role in triggering the initiation of sporulation and the development of competence (18). Since these short peptides are secreted from the cell into the culture medium, they can serve as signals for high culture density and have been implicated in a trait common to many bacteria, referred to as quorum sensing (12, 17). Furthermore, the expression of a dipeptide transport system operon (dpp) is induced at the end of the exponential growth phase as nutrients become limiting (27). This pattern of expression is governed by the codY gene product, as it mediates the nutritional repression of the dpp gene during exponential growth in a complex medium (37). In fact, CodY has been implicated in the nutritional repression of several genes during exponential growth in complex medium as well as in minimal medium supplemented with Casamino Acids (CAA) or a mixture of mono-amino acids (10, 35, 37, 46).
In previous work, we demonstrated that the level of B. subtilis flagellin mRNA increases during exponential growth in a complex medium (28). In the present study, we were interested in determining if this increase was due to increased levels of the flagellum-specific alternative sigma factor, ςD, a model which could also account for the hypermotility observed by early investigators. Furthermore, we wished to define the pattern of ςD expression throughout growth and in different media and to compare this pattern to that obtained for hag mRNA. Finally, our goal was to identify environmental signals—and, thus, the signal transduction system(s)—that regulate flagellar gene expression. We speculate that B. subtilis cells possess a regulatory mechanism that senses changes in the culture medium as the cells are growing and increases flagellar gene expression as a result and response.
MATERIALS AND METHODS
Bacterial strains.
The Escherichia coli strains used were TG-2 [Δ(lac pro) supE thi hsdD5 recA EcoK− Mot− F′ traO36 proAB lacIq lacZΔM15] and CSH26 [F− ara thi Δ(lac pro) Mot+]. The Bacillus subtilis strains used are listed in Table 1. All strains were maintained on solid media (Luria-Bertani or tryptose blood agar base [Difco] plates). Antibiotics, when necessary, were used at standard concentrations: ampicillin (Sigma) at 50 μg/ml and chloramphenicol (Sigma) at 5 μg/ml. Growth media and culture conditions for B. subtilis strains are described in detail below.
TABLE 1.
B. subtilis strains used in this study
| Strain | Genotype | Source, or derivation and referencea |
|---|---|---|
| LMB7 | trpC2 pheA1 | J. Hoch, JH642b |
| LMB24 | trpC2 pheA1 hag-lacZ (Cmr) | Transform [LMB7:pDM632, Cmr]; this study |
| LMB25 | trpC2 pheA1 hag-lacZ (Eryr) | Transform [LMB7:pDM632Ery, Eryr]; this study |
| LMB205 | trpC2 pheA1 sinR::cat hag-lacZ (Eryr) | Transform [LMB25:IS432 (from I. Smith), Cmr]; this study |
| LMB206 | trpC2 pheA1 abrB::cat hag-lacZ (Eryr) | Transform [LMB25:JH12-586 (from J. Hoch), Cmr]; this study |
| LMB207 | trpC2 pheA1 codY146::cat hag-lacZ (Eryr) | Transform [LMB25:FJS151 (37), Cmr]; this study |
| LMB253 | trpC2 pheA1 hpr::pJR1 hag-lacZ (Eryr) | Transform [LMB25:pJR1, Cmr]; this study |
| LMB221 | trpC2 pheA1 flgM::mini-Tn10 hag-lacZ (Eryr) | Transform [LMB25:LMB213 (6), Spr]; this study |
Transformation of [recipient strain:with plasmid DNA (pDM632, pJR1) or chromosomal DNA from strain listed, and selecting for resistance given].
Previous name of strain.
B. subtilis liquid culture media. (i) Complex medium.
2XSG medium (45) was used as the standard complex medium for the growth of B. subtilis. 2XSG base is an autoclaved solution containing (per liter) 16 g of nutrient broth (Difco), 0.5 g of MgSO4 · 7H2O, and 2 g of KCl; it was stored in the dark and used within 2 weeks of preparation. For use as a culture medium, the following supplements were added: 1 mM Ca(NO3)2, 0.1 mM MnSO4, 1 μM FeSO4, 0.1% glucose, and (when appropriate) 5 μg of chloramphenicol/ml.
(ii) Minimal medium.
S7 minimal medium was employed for growth of B. subtilis LMB7 and LMB24 in a defined synthetic medium (44) containing trace metals as previously described (4). The base consists of 100 mM morpholinepropanesulfonic acid (MOPS) (adjusted to pH 7.0 with KOH), 10 mM (NH4)2SO4, 5 mM potassium phosphate (pH 7.0), 2 mM MgCl2, 0.9 mM CaCl2, 50 μM MnCl2, 5 μM FeCl3, 10 μM ZnCl2, and 2 μM thiamine-hydrochloride. Sodium glutamate (20 mM) and d-glucose (2 to 5 mM) were added aseptically to the autoclaved base (glucose at 5 mM was used in the fermentor run). The medium was further supplemented with phenylalanine and tryptophan to 0.1 mg/ml each, and chloramphenicol was added to 5 μg/ml when appropriate.
(iii) Conditioned and concentrated media.
Preparation of conditioned medium was accomplished by collecting the bacterial suspension from a growing culture and filtering it through a 0.2-μm-pore-size filtration apparatus (Millipore). The resulting medium, in addition to being free from cells, did not possess any β-galactosidase activity, indicating that the hag-lacZ fusion protein was not secreted. Concentration of conditioned and fresh 2XSG media was accomplished by rotary evaporation under a vacuum at 60°C, and the concentrates were stored at 4°C until used.
(iv) Complex, amino acid, and vitamin supplements.
Nutrient broth, Bacto Peptone, Bacto Tryptone, and CAA (all from Difco) were prepared as autoclaved 8 to 32% solutions and stored in the dark. Amino acids and vitamins (Sigma, Aldrich, Calbiochem, National Biochemical Corp., or Eastman Organic) were prepared as filter-sterilized solutions and stored away from light when appropriate.
Growth of LMB7 for protein and RNA extraction. (i) Complex medium.
B. subtilis LMB7 prepared for protein extraction was grown in 2XSG medium in a New Brunswick fermentor (model SF-116) at 37°C with an impeller speed of 350 rpm. When the culture reached an A600 of 0.2 (T0), a sample was harvested and stored on ice; this was repeated every 15 min until T1 and then every hour through T5. LMB7 for RNA extraction was grown in 500 ml of 2XSG in a 2-liter Fernbach flask on a rotary shaker in a 37°C warm room. A 100-ml volume of 2XSG was inoculated with a plate colony and incubated for 2 h before being added to 400 ml of additional medium. When the culture reached an A600 of 0.4 (T0), a cell sample was collected and placed on ice; this was repeated every 15 min through T1 and then every hour through T5.
(ii) Minimal medium.
LMB7 prepared for protein and RNA extractions was grown in 12 liters of S7 medium in the fermentor as described above. A 120-ml volume of this medium was inoculated with a colony, and the bacteria were grown to mid-exponential phase in a shaker culture and then added to the fermentor. When the A600 of the culture reached 0.2 (T0), a cell sample was obtained and placed on ice; this was repeated approximately every 30 min through T6.
Quantitative hag mRNA primer extension analysis.
Extraction of total RNA from frozen cell pellets and measurement of levels of hag mRNA by primer extension were performed essentially as described by Mirel and Chamberlin (28).
Quantitative ςD protein immunoblot analysis.
Rabbit antiserum to the B. subtilis ςD protein was prepared previously (15). The amount of ςD protein present was determined by Western analysis using iodinated protein A. 125I-labeled Staphylococcus aureus protein A (70 to 100 mCi/mg) was purchased from New England Nuclear. Nitrocellulose membranes were obtained from Schleicher and Schuell. Frozen cell pellets, each representing 10 to 250 ml of cell culture, were treated as described previously in order to extract and quantitate the total protein (26). Protein (50 μg) from each lysate was resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis using 12.5% acrylamide gels and then electrophoretically transferred to a nitrocellulose membrane. The membrane containing the transferred protein was treated as previously described (26), except that a 1:100 dilution of ςD antiserum and a 1:1,000 dilution of iodinated protein A were used. To decrease nonspecific binding, the iodinated protein A was used within 3 weeks of its date of synthesis.
Quantitation of the amount of ςD protein in a sample was accomplished by excision of the reactive protein from the nitrocellulose (after autoradiography) and subsequent counting of the gamma particles in an LKB 1272 Clinigamma Counter. The background radioactivity was determined by measuring the radioactivity in a piece of nitrocellulose located immediately above the reactive protein on the filter and having the same dimensions as that bearing the ςD protein. The number of counts obtained for the background sample was subtracted from the number obtained for the reactive protein to obtain net counts.
Construction of hag-lacZ integrational vectors.
An in-frame translational fusion containing 180 bp of DNA upstream of the hag transcriptional start site and encoding a fusion protein comprising the first 71 amino acids of B. subtilis flagellin fused to the sixth amino acid (Gly) of the E. coli β-galactosidase enzyme was constructed and confirmed as follows. The 2.38-kb ClaI-PstI fragment of plasmid pDM67 (28), containing most of the coding region for hag, was replaced with the 3.1-kb SmaI-PstI fragment of pMC1871 that contains the lacZ coding region (3). To create an in-frame fusion, it was necessary to fill in the ClaI end of the digested pDM67 fragment. ClaI leaves a 5′ overhang (PstI does not), allowing for the fill-in reaction by the Klenow fragment of DNA polymerase (Boehringer Mannheim) and dCTP (Pharmacia) solely at this end. Any resulting C-tail overhang was removed by the addition of S1 nuclease (Boehringer Mannheim). After these manipulations, the 5.1-kb pDM67 fragment containing the transcriptional and translational start site information for hag in pJM102 (32) was isolated from the above-described 2.38-kb fragment by gel purification (Geneclean kit; BIO 101, Inc.). This 5.1-kb blunt-end PstI fragment was then ligated to the similarly purified 3.1-kb SmaI-PstI fragment of pMC1871 bearing lacZ.
Sequencing across the junctions of a number of candidate constructs demonstrated that the intended junction site was not created. Instead, junctions at a variety of sites further 5′ of the ClaI site were found, suggesting that gratuitous exonucleolytic activity had occurred. The hag-lacZ fusion construct identified and subsequently used consisted of an in-frame junction of amino acid residue 71 (Ser) of B. subtilis hag with amino acid residue 6 (Gly) of the E. coli lacZ gene found on pMC1871. The fusion gene was subcloned as a 3.7-kb HindIII fragment into pJH101 (a pBR322-based integrational vector) (7). The occurrence of the appropriate junction event was reconfirmed by sequence analysis, and the resultant plasmid was named pDM632. Additionally, a hag-lacZ integrational vector that confers erythromycin rather than chloramphenicol resistance on the transformed B. subtilis cell was prepared by replacing the cat gene on pDM632 with an erythromycin resistance cassette, yielding pDM632Ery.
Construction of hpr disruptional vector and null mutant.
To create a null mutation in the hpr gene of B. subtilis, a disruptional vector was first prepared. A region of DNA internal to the hpr coding sequence was amplified by PCR using primers OJW HPR 5′ (AAAAGAATTCCTTAGCAAGGCTCTTTGG) and OJW HPR 3′ (AAAACTGCAGTTCCGTTTACGCTTTCA). The PCR conditions used were as follows. The PCR buffer was 1× Taq buffer containing 1.5 mM MgCl2, 0.2 mM deoxynucleoside triphosphates, 10 μM each primer, 0.2 ng of LMB7 chromosomal DNA, and 5 U of Taq polymerase. PCR involved 30 cycles each of 1 min at 94°C, 1 min at 55°C, and 2 min at 72°C, preceded by a denaturing step of 3 min at 94°C and followed by a prolonged extension step of 10 min at 72°C. The resulting 500-bp fragment was digested with EcoRI and PstI and cloned into integrational vector pJM102 (32) which had been similarly digested. The identity of the resultant hpr-disrupting plasmid, pJR1, was confirmed by restriction map analysis. pJR1 was transformed into B. subtilis LMB25 (4), and disruption of the hpr gene was verified by monitoring protease production. Several transformants that produced a large halo on skim-milk plates, indicating hyperprotease production, were obtained. One of these transformants, designated LMB253, was used to monitor hag-lacZ expression in the absence of the Hpr transition state preventor.
Construction of reporter strains.
Transformation of pDM632 or pDM632Ery into B. subtilis LMB7 (4) resulted in reporter strains LMB24 and LMB24, respectively. The occurrence of the appropriate integration events was confirmed by Southern blot analysis (25).
Growth of reporter strains for determination of β-galactosidase assay.
Batch cultures of LMB24 were grown with good aeration (high rotation speed and flask not more than one-fifth filled) in a gyratory water bath shaker at 37°C. Cultures were started by loop inoculation of two to three isolated colonies from a plate into prewarmed medium. To inoculate sterile medium with cells from an already growing culture, two methods were used, as described below. Culture density was determined with a spectrophotometer at time intervals that were generally less than the doubling time of the culture. Furthermore, culture samples were diluted appropriately with the same medium to ensure that readings were within the linear range of the spectrophotometer. To stop growth before a reading, a culture sample was first kept on ice for 5 min in an Eppendorf tube. After the optical density had been determined, the cells were returned to the Eppendorf tube and kept on ice until assayed for β-galactosidase activity (see below). It was found that samples could be stored on ice for at least 5 h without any loss of β-galactosidase enzymatic activity.
For experiments in which cultures were grown in the same medium throughout the experiment, a sample of one culture was simply introduced directly into the recipient medium. Inoculum cultures (2 ml), started from single colonies, were grown at 37°C until turbid (ca. 2 h; A600 ≥ 1.2), then diluted 1:10 in 5 ml of prewarmed medium. Late in the exponential growth phase (at an A600 of 0.8), the culture was diluted to an A600 of 0.1 into 25 ml of prewarmed medium in a 125-ml flask. Samples were withdrawn from this culture for analysis of hag-lacZ expression as described below. The regimen outlined allowed the cells to undergo several doublings during exponential growth to ensure that the cells were fully adapted to the medium used.
For experiments in which cells were first cultured in one medium and then switched to another, a method involving removal of the source medium and resuspension of cells in the recipient medium was employed. Although this method requires more time and greater manipulation of the cells, it had been found that the carryover of medium from the source culture, such as from fresh medium into conditioned medium, can perceptibly alter the recipient medium conditions. The cells to be inoculated were collected by centrifugation in a microcentrifuge. The supernatant was aspirated, the recipient medium was added (in the same volume as the inoculum), and the pelleted cells were resuspended and then inoculated into the recipient culture. This entire procedure could be performed in 4 min or less.
β-Galactosidase assay.
Expression of the hag-lacZ reporter construct in LMB24 cells growing in different media was measured by β-galactosidase assay as previously described (6).
RESULTS
ςD protein and hag mRNA levels in 2XSG complex medium.
We have previously observed that flagellin mRNA levels in a wild-type strain of B. subtilis increase during exponential growth in complex sporulation medium (2XSG), peak at or near the end of the exponential growth phase, and then decrease during the postexponential growth period (28). Similar patterns of expression have been found for all ςD-dependent genes studied in this way (13, 28, 36). We therefore performed studies to determine the role of ςD protein levels in regulating ςD-dependent gene expression.
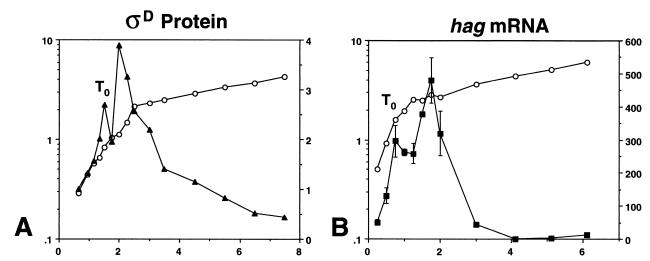
The amount of ςD protein present per cell in a wild-type strain (LMB7) growing in 2XSG sporulation medium was determined by quantitative immunoblot analysis. The growth of this culture and ςD protein levels are shown as functions of time in Fig. 1A. ςD protein levels increased exponentially during exponential growth in the complex medium 2XSG. At the end of the exponential growth phase (T0, as defined by the time of the break from the maximal doubling rate), ςD protein levels continued to increase (until T0.5) and then began to decay. Between T0.5 and T1, ςD protein levels decreased slightly and then continued increasing. After reaching a maximum amount per cell, ςD protein levels declined slowly; however, even at T6 there was still a substantial amount of this sigma factor present (Fig. 1A).
FIG. 1.
ςD protein and hag mRNA levels during growth of wild-type strain LMB7 in a complex medium. For each panel, the left y axis is the absorbance at 600 nm, the right y axis is the expression level in arbitrary units, and the x axis is time expressed in hours. Symbols: open circles, growth; closed triangles, ςD protein; closed squares, hag mRNA. (A) Typical results of quantitative immunoblot analysis of ςD protein during growth. Standard error levels ranged from 5 to 9% of the value indicated. (B) Results of a primer extension analysis using a primer specific to hag mRNA. Data presented are the means of values from two experiments. The break from exponential growth is indicated in each panel as T0.
The steady-state levels of B. subtilis flagellin message present in cells grown in complex medium were measured by quantitative primer extension analysis. These studies extended the original hag mRNA work presented by Mirel and Chamberlin (28). Flagellin gene mRNA levels increased exponentially during exponential-phase growth of LMB7 in 2XSG (Fig. 1B). At the end of the exponential growth phase (T0), hag mRNA levels generally continued to increase for 1 h (until T1) and then began to decay (Fig. 1B). Between T0 and T1, hag mRNA levels decreased slightly and then continued to increase, demonstrating a tight coupling between sigD expression and ςD activity during the exponential growth and early postexponential growth phases. After reaching a maximum amount per cell at T1, flagellin message levels decayed to zero by T4, in agreement with data presented previously (28). Thus, ςD protein persists in the cell longer than hag mRNA; this suggests that the rapid decay of hag mRNA levels after T0 is a result of the inactivation of the ςD protein or its RNA polymerase form. Preliminary data suggest that this inactivation may be mediated by FlgM, the anti-sigma factor specific for ςD (see section on genetic regulation below).
ςD protein and hag mRNA levels in S7 minimal medium.
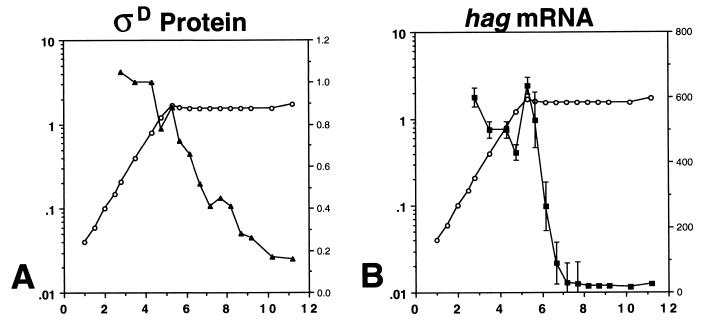
To determine if ςD expression and/or activity is influenced by different nutrient conditions, ςD and hag mRNA levels in cells growing in the synthetic minimal medium S7 were measured (44) (Fig. 2). In this medium, ςD protein and hag mRNA were present at sustained, higher levels than those found at time points measured during exponential growth in complex medium. In S7, the end of the exponential growth phase is extremely well defined, and the decay of ςD protein and hag mRNA can be observed to occur more rapidly than in complex medium. Furthermore, as was also seen in complex medium, ςD protein persists in the cell longer than does hag mRNA. In S7, flagellin gene expression, as measured by primer extension, apparently is entirely an exponential-phase process.
FIG. 2.
ςD protein and hag mRNA levels during growth of wild-type strain LMB7 in synthetic minimal medium S7. For each panel, the left y axis is the absorbance at 600 nm, the right y axis is the expression level in arbitrary units, and the x axis is time expressed in hours. Symbols: open circles, growth; closed triangles, ςD protein; closed squares, hag mRNA. (A) Typical results of quantitative immunoblot analysis of ςD protein during growth. Standard error levels ranged from 0.8 to 1% of the value indicated. (B) Results of a primer extension analysis using a primer specific to hag mRNA. Data presented are the means of values from two experiments.
hag-lacZ expression in complex and minimal media.
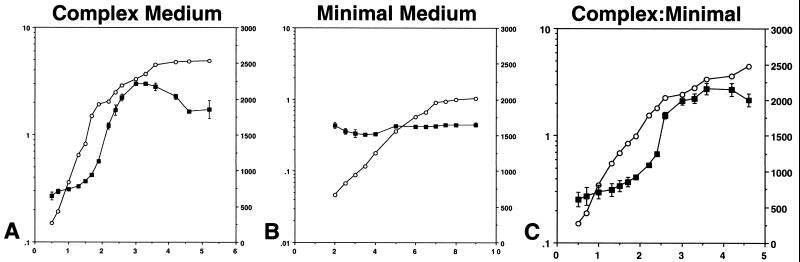
The observation that ςD-dependent gene expression increases steadily during exponential growth in complex medium while remaining high and constant in minimal medium was especially intriguing and elicited further study. To allow the rapid (and nearly real-time) assessment of flagellin gene expression during growth and in different nutritional media, a hag-lacZ translational fusion was constructed (see Materials and Methods). The hag-lacZ-bearing plasmid pDM632 was integrated into the genome of a wild-type strain (LMB7) to create strain LMB24, and flagellin gene expression was monitored by β-galactosidase assay. We found that expression of hag-lacZ in complex and minimal media replicated the flagellin gene expression patterns obtained by primer extension analysis during and shortly after the exponential growth phase (Fig. 3A and B). In complex medium, hag-lacZ expression increased exponentially during the exponential growth phase, peaked early in the postexponential growth phase, and declined thereafter (Fig. 3A). In minimal medium, hag-lacZ expression remained high and constant during exponential growth (Fig. 3B).
FIG. 3.
hag-lacZ levels during growth of reporter strain LMB24 in complex and minimal media. For each panel, the left y axis is the absorbance at 600 nm, the right y axis is β-galactosidase activity expressed in Miller units, and the x axis is time expressed in hours. Symbols: open circles, growth; closed squares, β-galactosidase activity. (A) Complex sporulation medium; (B) minimal medium; (C) 1:1 mixture of complex and minimal media. Data presented are the means and standard errors of values from two experiments.
The data in Fig. 3 demonstrate that the hag-lacZ reporter accurately reflected the behavior of hag mRNA during the exponential and early postexponential phases of growth. Thereafter, the expression level of the hag-lacZ reporter remained high in the postexponential growth phase (Fig. 3B) while hag mRNA levels rapidly decayed after T1 (Fig. 2B). This observation suggests that the hag-lacZ fusion protein is turned over much more slowly than hag mRNA in the postexponential growth phase. Since this study focused on the regulation of expression during the exponential and early postexponential growth phases, we can confidently employ the hag-lacZ construct as an accurate reporter of ςD-dependent gene expression during these two growth phases.
Evidence for nutritional control of hag-lacZ expression.
Flagellin gene expression per cell increases with time during exponential growth in the complex medium 2XSG, while in S7 minimal medium it remains constant. Furthermore, it appears that the expression level observed in minimal medium corresponds to the maximum level of expression seen in cells growing in 2XSG at and around T0 (compare Fig. 3A and B). Several hypotheses can be advanced to explain the different patterns of hag-lacZ expression in complex and minimal media. We proposed that 2XSG medium was being altered during growth and that this change was responsible for the pattern of hag-lacZ expression in this medium. The change in 2XSG could be due to production by the cells of an activating signal for hag expression that accumulates with time or to the depletion, during growth, of a repressing signal (perhaps a nutrient) that is present in complex medium and is entirely absent from minimal medium.
To test for the presence of an activator of expression that accumulates with time or culture density, we grew LMB24 in 2XSG to which conditioned medium was added (see Materials and Methods). The pattern of hag-lacZ expression was indistinguishable from that seen in 2XSG alone (data not shown); this observation provided evidence against the presence of an activating environmental signal.
Several experiments did support a model in which a component present in the complex medium causes repression. The addition of twofold-concentrated 2XSG to twofold-concentrated S7 minimal medium at a 1:1 ratio supported both a growth rate similar to and the exponential increase in hag-lacZ specific activity observed for growth in 2XSG alone (Fig. 3C). This was also true when nutrient broth alone was added to S7 at its concentration in 2XSG (data not shown). When S7 was supplemented with the remaining components found in 2XSG (not including nutrient broth), the LMB24 hag-lacZ expression patterns obtained were equivalent to those seen for S7 alone (data not shown). These experiments demonstrate that one can identify a repressing signal on the basis of its ability, when added to S7 minimal medium, to promote the hag-lacZ pattern of expression found in cells growing in complex medium. We term this pattern of low-level expression early in growth, followed by an exponential increase in expression as the cells approach T0, nutritional repression release.
CAA and mono-amino acids control hag-lacZ expression.
The experiments described above suggested that we might be able to identify a repressing signal if hag-lacZ expression in LMB24, when added to S7, displayed the exponential increase seen in 2XSG. The repressing signal might be lifted by its depletion during growth and then allow hag-lacZ expression to increase exponentially as growth proceeded toward T0.
It seemed plausible that one or more amino acids were acting as a repressing signal, and therefore hag-lacZ expression was monitored in cells growing in medium consisting of S7 and various concentrations of CAA. To determine if changes in CAA concentration (e.g., its depletion caused by consumption during growth) were influencing hag-lacZ expression, conditions under which CAA was nearly limiting for growth but glucose was still utilized as the energy source were found. This goal was achieved by using S7 minimal medium containing 2 mM glucose; growth stopped at an A600 of around 0.85 when glucose was depleted. hag-lacZ expression was repressed throughout growth in a concentration-dependent manner, with the maximal effect occurring at 0.32% (Fig. 4A). Under these conditions, the level of hag-lacZ expression did not display the nutritional repression release pattern found for cells growing in 2XSG. Thus, the values given in Fig. 4A are for the constant β-galactosidase specific activity measured for all samples obtained during growth.
FIG. 4.
hag-lacZ levels during growth of reporter strain LMB24 in supplemented minimal media. For each panel, the y axis is β-galactosidase activity expressed in Miller units. The x axis is percent CAA in panel A and absorbance at 600 nm in panel B. (A) β-Galactosidase specific activity during growth in S7 minimal medium supplemented with increasing concentrations of CAA; (B) hag-lacZ expression in S7 (open squares), S7 plus 1.6% nutrient broth (closed triangles), S7 plus 0.32% CAA (closed squares), and S7 plus 0.32% amino acids (closed circles). The results of a typical experiment for S7, S7 plus 1.6% nutrient broth, and S7 plus 0.32% CAA are given, and for S7 plus 0.32% amino acids the data presented are the means and standard errors of values from two experiments.
CAA is a complete hydrolysate of casein composed of mono-amino acids, although di- and oligopeptides may also exist in the mixture. While mono-amino acids play a role in governing the chemotaxis response in B. subtilis (31), di- and oligopeptides have been implicated in the control of gene expression (18–20, 33, 37). We therefore supplemented S7 with purified mono-amino acids in the same ratios as were listed in the specification sheet for CAA from Difco. S7 media containing amino acid mixtures equivalent to those found in 0.32 and 0.64% CAA were analyzed for their effect on hag-lacZ expression. The results for medium with amino acids equivalent to 0.32% CAA are presented (Fig. 4B); identical results were obtained for medium with amino acids equivalent to 0.64% CAA, demonstrating that the maximal response occurs at the lower amino acid concentration. Surprisingly, the purified amino acids were able to reconstitute the nutritional repression release pattern for hag-lacZ expression found in complex medium and did not result in the uniform expression found in S7 plus CAA (Fig. 4B).
Genetic regulation.
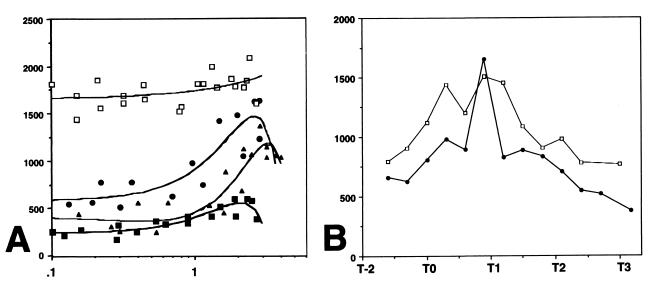
Having determined that hag-lacZ is subject to nutritional repression in complex medium or medium supplemented with a mixture of amino acids early in growth, we sought to identify the physiologically relevant regulator molecule. The codY gene product has been implicated in the nutritional repression of several genes during exponential growth in complex medium as well as in minimal medium supplemented with CAA or a mixture of mono-amino acids (10, 35, 37, 46). Therefore, hag-lacZ expression was monitored in strain LMB207, which bears a null mutation in codY. The level of flagellin expression in this strain is high and constant throughout growth (Fig. 5A), with the pattern of expression being similar to that found for cells growing in minimal medium (Fig. 3B).
FIG. 5.
hag-lacZ expression in strains bearing null mutations in codY, the genes encoding the transition state preventors, and flgM during growth in complex medium. For each panel, the y axis is β-galactosidase activity expressed in Miller units. The x axis is absorbance at 600 nm in panel A and time relative to the break from exponential growth (T0) in panel B. (A) β-Galactosidase specific activity in strains bearing null mutations as follows: in codY, LMB207 (open squares); in abrB, LMB206 (closed circles); in hpr, LMB253 (closed triangles); and in sinR, LMB205 (closed squares). (B) hag-lacZ expression in wild-type strain LMB24 (closed circles) and in flgM-null mutant LMB221 (open squares). The results presented for each strain are the averages of values from two experiments.
We also measured expression in strains bearing null mutations in the transition state preventor genes abrB, hpr, and sinR, since their gene products have been implicated in the repression of postexponential-phase functions during growth (40, 41). In all cases, the level of hag-lacZ expression was lower early in growth and increased throughout growth as cells approached T0, indicative of nutritional repression release. It appears that although the transition state preventor gene products are involved in maximal expression of the flagellin gene at the end of the logarithmic growth phase, they are not required for nutritional repression early in growth. While the abrB and hpr products may play a modest role in allowing maximal flagellin gene expression at the end of the exponential growth phase, the sinR gene product is apparently required in all growth phases. Flagellin expression in the sinR mutant is decreased to about 20% of the wild-type level, in agreement with data obtained by others for a single time point at the end of growth (11).
Finally, flagellin expression was determined in a strain lacking the FlgM anti-sigma factor, a known inhibitor of ςD activity (2, 29). Whereas the patterns of flagellin expression in the wild type and the flgM-null mutant were nearly identical, levels were slightly elevated throughout growth, in agreement with previous results (29). More significantly, these levels persisted longer in the cell during the postexponential phase. These preliminary data suggest that FlgM may be involved in the postexponential-phase inhibition of ςD activity found in complex and minimal media (Fig. 1 and 2).
DISCUSSION
In this paper we have outlined an experimental approach to gaining a better understanding of how B. subtilis ςD-dependent gene expression is influenced by the cells' environment. We have used the flagellin gene (hag) as a model ςD-dependent gene. Singer first described the general phenomenon of increased expression of four ςD-dependent genes during growth in a complex medium (36). Subsequent studies of other ςD-dependent genes (13, 28) demonstrated a similar pattern of increased expression during growth in a complex medium. These results suggest that the members of the sigD regulon are regulated by a common mechanism. We therefore sought to determine if ςD activity was directly related to sigD expression by concurrently analyzing ςD protein levels and hag mRNA in complex and minimal media.
Our results demonstrate that ςD protein and hag mRNA exhibit virtually identical patterns of expression during both the exponential and the early postexponential growth phases in complex and minimal media (Fig. 1 and 2). These observations support the notions that ςD activity is directly related to sigD expression during these growth phases and that ςD protein drives hag mRNA expression. In the postexponential phase, ςD protein levels persist longer than hag mRNA, leading to the prediction that ςD activity is inhibited during this growth phase (see below).
In the experiments involving S7-glucose minimal medium, flagellin gene expression paralleled growth. In other words, hag gene transcription is entirely an exponential-phase phenomenon. Transcription occurs while glucose is present and being utilized, and it ceases when this substance is exhausted. This suggests that either the presence or the utilization of the primary energy or carbon source for growth is somehow a requirement for ςD-dependent gene expression. The expression of hag in the presence of glucose also implies that flagellin synthesis in B. subtilis is not subject to glucose catabolite repression as it is in E. coli (24).
When one compares ςD-dependent hag expression in a complex medium to that in a minimal medium (Fig. 1B and 2B), a significant difference in the patterns of expression is apparent. Whereas the level of hag expression is low early in growth and then increases exponentially during exponential growth in a complex medium, it remains high and constant in a minimal medium. These patterns of gene expression are accurately replicated during the growth and early postexponential phases by using a hag-lacZ fusion reporter construct (Fig. 3A and B). Therefore, the reporter construct was determined to be an appropriate tool for rapidly monitoring hag expression during these growth phases. We have termed the pattern of hag-lacZ expression found in complex medium (Fig. 3A) nutritional repression release since hag-lacZ activity is low early in growth and increases exponentially once the nutritional repression is relieved. This pattern is missing in strains grown in minimal medium (Fig. 3B) but can be reconstituted by adding a mixture of amino acids to S7 (Fig. 4B). In contrast, the addition of a CAA mixture to S7 prevents this release at any time during the exponential growth phase (Fig. 4).
We speculate that early in growth a substance in complex medium mediates a repression of hag-lacZ that is released as the culture proceeds toward T0. The substance, which we have termed a nutritional repressing signal, appears to be either a single or a mixture of amino acids. Work exploring the possibility that this signal is monitored or mediated through the same mechanism as that involved in chemotaxis is in progress. Interestingly, chemotaxis in B. subtilis is regulated by the interaction of all 20 amino acids with membrane-bound proteins, the methyl-accepting chemotaxis proteins (31). These proteins are responsible for monitoring the external environment and influencing chemotaxis. They serve as amino acid receptors that interact with a signal transduction cascade, leading to the activation of a two-component regulatory system that governs the rotation of the flagellar motor and thereby chemotaxis (31). Our results suggest that there may be a common mechanism for monitoring the environment and controlling both the physiological response of chemotaxis and expression of ςD-dependent genes required for motility during the exponential growth phase.
Once the nutritional repressing signal is detected across the cellular membrane, it must be transduced to the transcriptional machinery; therefore, we sought to identify the intracellular regulator for nutritional repression of hag gene expression. The codY gene product has been implicated in the nutritional repression of several genes involved in nitrogen metabolism, including the hut, dpp, bkd, and ure operons (5, 8, 10, 37, 46), as well as in the expression of comK and srfA, which are required for competence development (9, 35). In a strain lacking the CodY regulator, hag-lacZ expression appears to be derepressed (Fig. 5A), suggesting that this protein is the physiologically relevant regulator of nutritional repression of the hag gene in B. subtilis. We are investigating the molecular mechanism by which CodY appears to exert its effect on flagellin expression. Our preliminary results suggest that purified CodY protein binds to both the hag and fla/che promoter regions (F. Vergara, J. Iwamasa, J. C. Patarroyo, S. Santa Anna-Arriola, and L. M. Márquez-Magaña, submitted for publication). Binding at the hag promoter appears to be a direct effect of CodY activity on hag gene expression. However, binding to the fla/che promoter fragment is likely an indirect effect since expression of the sigD gene, which encodes ςD, is dependent on the fla/che dual promoters (6).
Although CodY appears to be responsible for nutritional repression of hag gene expression early in growth, the rapid decrease in hag mRNA relative to ςD protein in postexponential growth in both complex and minimal media is probably regulated by other means and by different factors (Fig. 1 and 2). During the E. coli heat shock response, induction of expression of heat shock genes is dependent on the concentration of the heat shock sigma factor, ς32 (14, 43), just as ςD-dependent gene expression appears to be dependent on ςD protein levels (Fig. 1 and 2). The activity of ς32, however, is inhibited posttranscriptionally by the action of heat shock proteins encoded by ς32-dependent genes (21, 42). Inhibition of ςD activity in the postexponential growth phase may occur similarly via the product of a ςD-dependent gene, flgM, a known negative regulator of this sigma factor (29).
Recent biochemical studies of FlgM function have demonstrated that it binds specifically to ςD protein, thereby inhibiting its activity (1). Although primarily transcribed from a ςD-dependent promoter, the gene for the FlgM anti-sigma factor is also expressed as a result of read-through from the ςA-dependent promoter for comF (22). It has been postulated that production of FlgM via a non-ςD-dependent pathway is part of a molecular switch responsible for the increased development of competence and the decreased motility found in cells growing postexponentially (22). In this study, the decreased level of hag mRNA production relative to ςD protein found during this phase is consistent with the non-ςD-dependent expression of FlgM, as demonstrated in Fig. 5B. hag-lacZ expression in a flgM-null mutant is of a higher level and persists longer in the postexponential growth phase than in the wild-type strain (Fig. 5B). However, the increased stability of the hag-lacZ fusion protein at this time makes interpretation of these data problematic. We plan to perform primer extension analyses of hag mRNA in the flgM mutant in order to more accurately assess the importance of FlgM in the postexponential-phase inhibition of ςD activity. It is also possible that the postexponential-phase decrease in ςD activity is due to increased competition for the holoenzyme by consecutive sporulation ς factors (16); validation of this hypothesis will require more-complex genetic and biochemical studies.
ACKNOWLEDGMENTS
We are grateful to Michael Chamberlin for providing the foundation on which this work was initiated, to Joyce West for critical readings of early versions of the manuscript, and to Robert Ramirez for input on the final version.
This work was supported by NIH-AREA grant GM54342-02 and by DOE-GAANN support (P200A80128) to E.O., as well as by NIH-MARC support to J.R. (5 T34-GM08574-01).
REFERENCES
- 1.Bertero M G, Gonzales B, Tarricone C, Ceciliani F, Galizzi A. Overproduction and characterization of the Bacillus subtilis anti-sigma factor FlgM. J Biol Chem. 1999;274:12103–12107. doi: 10.1074/jbc.274.17.12103. [DOI] [PubMed] [Google Scholar]
- 2.Caramori T, Barillà D, Nessi C, Sacchi L, Galizzi A. Role of FlgM in ςD-dependent gene expression in Bacillus subtilis. J Bacteriol. 1996;178:3113–3118. doi: 10.1128/jb.178.11.3113-3118.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Casadaban M, Martinez-Arias J A, Shapira S K, Chou J. Beta-galactosidase gene fusions for analyzing gene expression in Escherichia coli and yeast. Methods Enzymol. 1983;100:293–308. doi: 10.1016/0076-6879(83)00063-4. [DOI] [PubMed] [Google Scholar]
- 4.Cutting S M, Vander Horn P B. Genetic analysis. In: Harwood C R, Cutting S M, editors. Molecular biological methods for Bacillus. West Essex, England: John Wiley and Sons, Ltd.; 1990. pp. 27–74. [Google Scholar]
- 5.Debarbouille M, Gardan R, Arnaud M, Rapoport G. Role of BkdR, a transcriptional activator of the SigL-dependent isoleucine and valine degradation pathway in Bacillus subtilis. J Bacteriol. 1999;181:2059–2066. doi: 10.1128/jb.181.7.2059-2066.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Estacio W, Santa Anna-Arriola S, Adedipe M, Márquez- Magaña L M. Dual promoters are responsible for transcription initiation of the fla/che operon in Bacillus subtilis. J Bacteriol. 1998;180:3548–3555. doi: 10.1128/jb.180.14.3548-3555.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferrari F A, Nguyen A, Lang D, Hoch J A. Construction and properties of an integrable plasmid for Bacillus subtilis. J Bacteriol. 1983;154:1513–1515. doi: 10.1128/jb.154.3.1513-1515.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferson A E, Wray L V, Jr, Fisher S H. Expression of the Bacillus subtilis gabP gene is regulated independently in response to nitrogen and amino acid availability. Mol Microbiol. 1996;22:693–701. doi: 10.1046/j.1365-2958.1996.d01-1720.x. [DOI] [PubMed] [Google Scholar]
- 9.Fisher S. Regulation of nitrogen metabolism in Bacillus subtilis: vive la différence! Mol Microbiol. 1999;32:223–232. doi: 10.1046/j.1365-2958.1999.01333.x. [DOI] [PubMed] [Google Scholar]
- 10.Fisher S H, Rohrer K, Ferson A E. Role of CodY in regulation of the Bacillus subtilis hut operon. J Bacteriol. 1996;178:3779–3784. doi: 10.1128/jb.178.13.3779-3784.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fredrick K, Helmann J D. FlgM is a primary regulator of ςD activity, and its absence restores motility to a sinR mutant. J Bacteriol. 1996;178:7010–7013. doi: 10.1128/jb.178.23.7010-7013.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fuqua C, Winans S C, Greenberg E P. Census and consensus in bacterial ecosystems: the LuxR and LuxI family of quorum-sensing transcriptional regulators. Annu Rev Microbiol. 1996;50:727–751. doi: 10.1146/annurev.micro.50.1.727. [DOI] [PubMed] [Google Scholar]
- 13.Gilman M, Chamberlin M J. Development and genetic regulation of Bacillus subtilis genes transcribed by ς28 RNA polymerase. Cell. 1983;35:285–293. doi: 10.1016/0092-8674(83)90231-3. [DOI] [PubMed] [Google Scholar]
- 14.Grossman A D, Erickson J W, Gross C A. The htpR gene product of Escherichia coli is a sigma factor for heat shock promoters. Cell. 1984;38:383–390. doi: 10.1016/0092-8674(84)90493-8. [DOI] [PubMed] [Google Scholar]
- 15.Helmann J D, Márquez L M, Chamberlin M J. Cloning, sequencing, and disruption of the Bacillus subtilis ς28 gene. J Bacteriol. 1988;170:1568–1574. doi: 10.1128/jb.170.4.1568-1574.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ju J, Mitchell T, Peters III H, Haldenwang W G. Sigma factor displacement from RNA polymerase during Bacillus subtilis sporulation. J Bacteriol. 1999;181:4969–4977. doi: 10.1128/jb.181.16.4969-4977.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kleerebezem M, Quadri L E, Kuipers O P, de Vos W M. Quorum sensing by peptide pheromones and two-component signal-transduction systems in gram-positive bacteria. Mol Microbiol. 1997;24:895–904. doi: 10.1046/j.1365-2958.1997.4251782.x. [DOI] [PubMed] [Google Scholar]
- 18.Koide A, Perego M, Hoch J A. ScoC regulates peptide transport and sporulation initiation in Bacillus subtilis. J Bacteriol. 1999;181:4114–4117. doi: 10.1128/jb.181.13.4114-4117.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lazazzera B A, Kurtser I G, McQuade R S, Grossman A D. An autoregulatory circuit affecting peptide signaling in Bacillus subtilis. J Bacteriol. 1999;181:5193–5200. doi: 10.1128/jb.181.17.5193-5200.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lazazzera B A, Solomon J M, Grossman A D. An exported peptide functions intracellularly to contribute to cell density signaling in Bacillus subtilis. Cell. 1997;89:917–925. doi: 10.1016/s0092-8674(00)80277-9. [DOI] [PubMed] [Google Scholar]
- 21.Liberek K, Georgopoulos C. Autoregulation of the Escherichia coli heat shock response by the DnaK and DnaJ heat shock proteins. Proc Natl Acad Sci USA. 1993;90:11019–11023. doi: 10.1073/pnas.90.23.11019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu J, Zuber P. A molecular switch controlling competence and motility: competence regulatory factors ComS, MecA, and ComK control ςD-dependent gene expression in Bacillus subtilis. J Bacteriol. 1998;180:4243–4251. doi: 10.1128/jb.180.16.4243-4251.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Losick R, Youngman P, Piggot P J. Genetics of endospore formation in Bacillus subtilis. Annu Rev Genet. 1986;20:625–669. doi: 10.1146/annurev.ge.20.120186.003205. [DOI] [PubMed] [Google Scholar]
- 24.Macnab R. Genetics and biogenesis of bacterial flagella. Annu Rev Genet. 1992;26:131–158. doi: 10.1146/annurev.ge.26.120192.001023. [DOI] [PubMed] [Google Scholar]
- 25.Maniatis T, Fritsch E, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 26.Márquez L M, Helmann J D, Ferrari E, Parker H M, Ordal G W, Chamberlin M J. Studies of ςD-dependent functions in Bacillus subtilis. J Bacteriol. 1990;172:3435–3443. doi: 10.1128/jb.172.6.3435-3443.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mathiopoulos C, Mueller J P, Slack F J, Murphy C G, Patankar S, Bukusoglu G, Sonenshein A L. A Bacillus subtilis dipeptide transport system expressed early during sporulation. Mol Microbiol. 1991;5:1903–1913. doi: 10.1111/j.1365-2958.1991.tb00814.x. [DOI] [PubMed] [Google Scholar]
- 28.Mirel D B, Chamberlin M J. The Bacillus subtilis flagellin gene (hag) is transcribed by the ς28 form of RNA polymerase. J Bacteriol. 1989;171:3095–3101. doi: 10.1128/jb.171.6.3095-3101.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mirel D B, Lauer P, Chamberlin M J. Identification of flagellar synthesis regulatory and structural genes in a ςD-dependent operon of Bacillus subtilis. J Bacteriol. 1994;176:4492–4500. doi: 10.1128/jb.176.15.4492-4500.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nishihara T, Freese E. Motility of Bacillus subtilis during growth and sporulation. J Bacteriol. 1975;123:366–371. doi: 10.1128/jb.123.1.366-371.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ordal G W, Márquez-Magaña L, Chamberlin M J. Motility and chemotaxis. In: Sonenshein A L, Hoch J A, Losick R, editors. Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. Washington, D.C.: American Society for Microbiology; 1993. pp. 765–784. [Google Scholar]
- 32.Perego M. Integrational vectors for genetic manipulation in Bacillus subtilis. In: Sonenshein A L, Hoch J A, Losick R, editors. Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. Washington, D.C.: American Society for Microbiology; 1993. pp. 615–624. [Google Scholar]
- 33.Perego M. A peptide export-import control circuit modulating bacterial development regulates protein phosphatases of the phosphorelay. Proc Natl Acad Sci USA. 1997;94:8612–8617. doi: 10.1073/pnas.94.16.8612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Piggot P J, Coote J G. Genetic aspects of bacterial endospore formation. Bacteriol Rev. 1976;40:908–962. doi: 10.1128/br.40.4.908-962.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Serror P, Sonenshein A L. CodY is required for nutritional repression of Bacillus subtilis genetic competence. J Bacteriol. 1996;178:5910–5915. doi: 10.1128/jb.178.20.5910-5915.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Singer V L. Characterization of promoters and genes controlled by Bacillus subtilis ς28 RNA polymerase. Ph.D. dissertation. Berkeley: University of California; 1987. [Google Scholar]
- 37.Slack F J, Serror P, Joyce E, Sonenshein A L. A gene required for nutritional repression of the Bacillus subtilis dipeptide permease operon. Mol Microbiol. 1995;15:689–702. doi: 10.1111/j.1365-2958.1995.tb02378.x. [DOI] [PubMed] [Google Scholar]
- 38.Sonenshein A L. Metabolic regulation of sporulation and other stationary-phase phenomena. In: Smith I, Slepecky R A, Setlow P, editors. Regulation of procaryotic development: structural and functional analysis of bacterial sporulation and germination. Washington, D.C.: American Society for Microbiology; 1989. pp. 109–130. [Google Scholar]
- 39.Stragier P, Losick R. Molecular genetics of sporulation in Bacillus subtilis. Annu Rev Genet. 1996;30:297–341. doi: 10.1146/annurev.genet.30.1.297. [DOI] [PubMed] [Google Scholar]
- 40.Strauch M A, Hoch J A. Transition-state regulators: sentinels of Bacillus subtilis post-exponential gene expression. Mol Microbiol. 1993;7:337–342. doi: 10.1111/j.1365-2958.1993.tb01125.x. [DOI] [PubMed] [Google Scholar]
- 41.Strauch M A. Regulation of Bacillus subtilis gene expression during the transition from exponential to stationary phase. Prog Nucleic Acid Res Mol Biol. 1993;46:121–153. doi: 10.1016/s0079-6603(08)61020-x. [DOI] [PubMed] [Google Scholar]
- 42.Straus D, Walter W, Gross C A. DnaK, DnaJ, and GrpE heat shock proteins negatively regulate heat shock gene expression by controlling the synthesis and stability of ς32. Genes Dev. 1990;4:2202–2209. doi: 10.1101/gad.4.12a.2202. [DOI] [PubMed] [Google Scholar]
- 43.Straus D B, Walter W, Gross C A. The activity of ς32 is reduced under conditions of excess heat shock protein production in Escherichia coli. Genes Dev. 1989;3:2003–2010. doi: 10.1101/gad.3.12a.2003. [DOI] [PubMed] [Google Scholar]
- 44.Vasantha N, Freese E. Enzyme changes during Bacillus subtilis sporulation caused by deprivation of guanine nucleotides. J Bacteriol. 1980;144:1119–1125. doi: 10.1128/jb.144.3.1119-1125.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang P Z, Doi R H. Overlapping promoters transcribed by Bacillus subtilis ς55 and ς37 RNA polymerase holoenzymes during growth and stationary phases. J Biol Chem. 1984;259:8619–8625. [PubMed] [Google Scholar]
- 46.Wray L V, Jr, Ferson A E, Fisher S H. Expression of the Bacillus subtilis ureABC operon is controlled by multiple regulatory factors including CodY, GlnR, TnrA, and Spo0H. J Bacteriol. 1997;179:5494–5501. doi: 10.1128/jb.179.17.5494-5501.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]