Abstract
Protein mannosyltransferases (Pmt proteins) initiate O glycosylation of secreted proteins in fungi. We have characterized PMT6, which encodes the second Pmt protein of the fungal pathogen Candida albicans. The residues of Pmt6p are 21 and 42% identical to those of C. albicans Pmt1p and S. cerevisiae Pmt6p, respectively. Mutants lacking one or two PMT6 alleles grow normally and contain normal Pmt enzymatic activities in cell extracts but show phenotypes including a partial block of hyphal formation (dimorphism) and a supersensitivity to hygromycin B. The morphogenetic defect can be suppressed by overproduction of known components of signaling pathways, including Cek1p, Cph1p, Tpk2p, and Efg1p, suggesting a specific Pmt6p target protein upstream of these components. Mutants lacking both PMT1 and PMT6 are viable and show pmt1 mutant phenotypes and an additional sensitivity to the iron chelator ethylenediamine-di(o-hydroxyphenylacetic acid). The lack of Pmt6p significantly reduces adherence to endothelial cells and overall virulence in a mouse model of systemic infection. The results suggest that Pmt6p regulates a more narrow subclass of proteins in C. albicans than Pmt1p, including secreted proteins responsible for morphogenesis and antifungal sensitivities.
Secreted proteins in fungi can get modified by the attachment of short glycosyl chains consisting of one to seven mannoses to serine or threonine residues (reviewed in reference 38). The first mannosylation step in O glycosylation occurs in the endoplasmic reticulum, presumably cotranslationally, and is mediated by protein mannosyltransferases (Pmt proteins). In the yeast Saccharomyces cerevisiae seven PMT genes are known (20, 24, 28, 37); their paralogous gene products, by their degree of homology, can be grouped in at least two subclasses consisting of either the Pmt1 and Pmt5 proteins or the Pmt2, Pmt3, and Pmt6 proteins (10). We recently isolated and characterized the PMT1 gene of the important human fungal pathogen Candida albicans (41). Pmt homologues in Drosophila melanogaster (25) and humans (21) have also been described, and Pmt homologues deduced from “expressed sequence tags” occur in nematodes, plants, and mammals, although their enzymatic functions as Pmt proteins have not yet been demonstrated. In C. albicans, O-glycosyl chains initiated by Pmt proteins are extended further by mannosyltransferases including Mnt1p (3).
Although the molecular details of target protein recognition by Pmt proteins are unknown, it appears that Pmt proteins can have a preference for certain glycosylation targets. Thus, the lack of Pmt1 and Pmt2 proteins in S. cerevisiae mutants affects O glycosylation of a set of secreted proteins overlapping with, but different from, the set affected in mutants lacking Pmt4p (16), and certain cell wall proteins are affected differently by mutations in PMT genes (28). Recently, the Axl2p protein, involved in axial budding, was recognized as a specific target of mannosylation by Pmt4p (29). Unknown O-glycosylated proteins are needed for general cell viability, since S. cerevisiae pmt1 pmt2 double mutants show reduced growth and some triple pmt mutants are not viable (15). Likewise, the lack of both pmt1 alleles in C. albicans negatively affects growth (41). On the other hand, specific phenotypes have been observed in pmt mutants. S. cerevisiae pmt1 mutants are partially resistant to K1 killer factor (24) and are unable to grow anaerobically in minimal medium (2). C. albicans pmt1/pmt1 homozygous and PMT1/pmt1 heterozygous strains showed an increased sensitivity to aminoglycoside antibiotics and a defect in hyphal morphogenesis (41). A cellular differentiation defect was also observed in Drosophila strains lacking the PMT gene homologue rotated abdomen (25).
Here we describe a second PMT gene of C. albicans, PMT6, which encodes a Pmt protein highly homologous to S. cerevisiae Pmt6p and thus is a representative of the second subgroup of S. cerevisiae proteins. We demonstrate that deletion of PMT6 does not affect growth in general but rather generates specific phenotypes, such as antifungal supersensitivity and defective filamentation. Suppression experiments strongly suggest that O glycosylation by Pmt6p affects a specific component upstream of known signaling cascades, triggering morphogenesis. Defects in properties of adhesion to target cells and the reduced virulence of pmt6 mutants demonstrate the importance of Pmt6p for cellular differentiation and virulence of C. albicans.
MATERIALS AND METHODS
Strains and media.
The C. albicans strains and the plasmids are listed in Table 1. C. albicans strains CAI4 (11) and CAP1-3121 (41) were used for transformations and gene disruptions. Strains were grown in yeast extract-peptone-dextrose (YPD) or SD medium (34), which for Ura− strains was supplemented with 20 μg of uridine/ml. Transformations were performed using the spheroplast method (34). Hyphae were induced on solid “Spider” medium (22) or in liquid using serum (9) or 2.5 mM GlcNAc (18) as the inducer.
TABLE 1.
Strains and plasmids
| C. albicans strain(s) or plasmid(s) | Genotype or description | Reference or source |
|---|---|---|
| Strains | ||
| SC5314 | Prototrophic | 11 |
| CAI4 | ura3Δ::imm34/ura3Δ::imm34 | 11 |
| CAP1-3 | As CAI4, but PMT1/pmt1Δ::hisG-URA3-hisG | 41 |
| CAP1-312 | As CAI4, but pmt1Δ::hisG-URA3-hisG/pmt1Δ::hisG | 41 |
| CAP1-3121 | As CAI4, but pmt1Δ::hisG/pmt1Δ::hisG | 41 |
| SS4 | CAI8 derivative; efg1::ADE2/(URA3-PCK1p)::EFG1 | 36 |
| HLC67 | efg1/efg1 derivative of CAI4 | 23 |
| CAP2-1, CAP2-2, CAP2-3, CAP2-4 | As CAI4, but PMT6/pmt6Δ::hisG-URA3-hisG | This work |
| CAP2-23 | As CAI4, but PMT6/pmt6Δ::hisG | This work |
| CAP2-234, CAP2-239 | As CAI4, but pmt6Δ::hisG/pmt6Δ::hisG-URA3-hisG | This work |
| CAP2-2341, CAP2-2391 | As CAI4, but pmt6Δ::hisG/pmt6Δ::hisG | This work |
| CPP1 | As CAP1-3121, but PMT6/pmt6Δ::hisG-URA3-hisG | This work |
| CPP11 | As CAP1-3121, but PMT6/pmt6Δ::hisG | This work |
| CPP112, CPP117 | As CAP1-3121, but pmt6Δ::hisG/pmt6Δ::hisG-URA3-hisG | This work |
| CPP1123, CPP1171 | As CAP1-3121, but pmt6Δ::hisG/pmt6Δ::hisG | This work |
| Plasmids (transformation vectors) | ||
| pRC18 | URA3-marked CaARS vector | 36 |
| pRC2312 | URA3-marked CaARS vector | 4 |
| pLJ19 (pCPH1) | CPH1 under control of the ADH1 promoter | 7 |
| pCaTPK2 (pTPK2) | URA3-marked CaARS vectors containing TPK2 | 35 |
| pCCa4 (pCEK1) | URA3-marked CaARS vectors containing CEK1 | 7 |
| pBI-HAHYD (pEFG1) | EFG1 fused with the HAa tag under the control of the PCK1 promoter | 35 |
| p99 | pUC18 containing PMT6 | S. Scherer |
| pCT30 | URA3-marked CaARS vector containing PMT1 | 41 |
| pCT34, pCT35 | URA3-marked CaARS vectors containing PMT6 | This work |
| pCT17 | As p99, but without the part of the MCSb between SacI and SphI | This work |
| pCT25 | Plasmid carrying pmt6Δ::hisG-URA3-hisG disruption fragment | This work |
HA, hemagglutinin.
MCS, multiple cloning site.
Sequencing of PMT6 and plasmid constructions.
A plasmid containing PMT6 was identified in the C. albicans genome project (p99) (S. Scherer, personal communication; http://www-sequence.stanford.edu/group/candida). Subfragments of p99 were ligated into pUC19 and sequenced from both ends using M13 forward (U-40) and reverse primers or by using insert-specific oligonucleotides. The 5.3-kb HindIII fragment containing PMT6 of p99 was inserted into the HindIII site of pRC18 (36) to generate replicating plasmids (pCT34 and pCT35 with inverse insert orientation).
Disruption of PMT6.
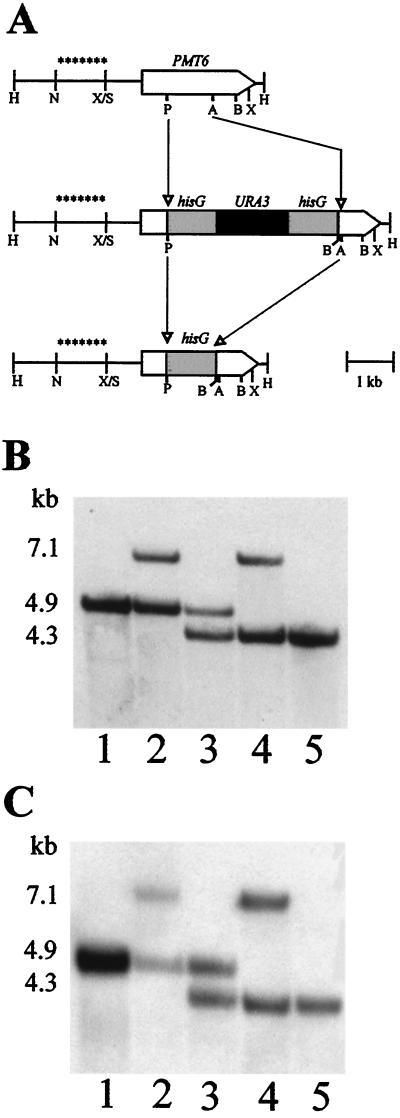
For disruption of the C. albicans PMT6 gene, plasmid pCT17 was cut with Asp718 and PstI and the 4.0-kb “Ura blaster” fragment (cut with Asp718 and PstI) from p5921 (17) was ligated into this vector. From the resulting plasmid, pCT25, a 5.7-kb XhoI fragment (Fig. 1A) was isolated and used for transformation of strains CAI4 and CAP1-3121. Correct insertion of this fragment into one of the two PMT6 alleles was verified by Southern blotting of DNA of transformants, which was cut with BglII and HindIII and probed with a 1.1-kb NcoI-SalI fragment derived from the PMT6 promoter region (Fig. 1A). One of the strains generated, e.g., CAP2-2, with the genotype pmt6Δ::hisG-URA3-hisG/PMT6, was plated out on medium containing 0.02% 5-fluoroorotic acid (5-FOA) (26). Spontaneous 5-FOA-resistant strains were analyzed for loss of the URA3 sequence by Southern blotting. One of the identified strains, CAP2-23, with the genotype pmt6Δ::hisG/PMT6, was used for a second round of gene disruption with the C. albicans pmt6 URA blaster fragment of pCT25. Several transformants had the genotype pmt6Δ::hisG/pmt6Δ::hisG-URA3-hisG, and strain CAP2-239 was chosen to identify strains by 5-FOA resistance. Strain CAP2-2391 is a representative of mutant strains with the genotype pmt6Δ::hisG/pmt6Δ::hisG. The PMT6 gene was reintroduced into CAP2-2391 by transforming this strain with either plasmid pCT34 or pCT35 (Table 1).
FIG. 1.
Sequential disruption of PMT6 alleles. (A) Schematic representation of the construction of the different alleles. The wild-type PMT6 gene and the PMT6 alleles disrupted by the hisG-URA3-hisG cassette or by hisG are shown. H, HindIII; N, NcoI; X, XhoI, S, SalI; P, PstI; B, BglII; A, Asp718. The fragment marked by asterisks was used as a probe for Southern analysis. (B and C) Southern blots of HindIII-BglII-digested chromosomal DNA of the following strains: SC5314 (PMT6/PMT6; lane 1 [B and C]); CAP2-2 (PMT6/pmt6Δ::hisG-URA3-hisG; lane 2 [B]); CAP2-23 (PMT6/pmt6Δ::hisG; lane 3 [B]); CAP2-239 (pmt6Δ::hisG-URA3-hisG/pmt6Δ::hisG; lane 4 [B]); CAP2-2391 (pmt6Δ::hisG/pmt6Δ::hisG; lane 5 [B]); CPP1 (PMT6/pmt6Δ::hisG-URA3-hisG; lane 2 [C]); CPP11 (PMT6/pmt6Δ::hisG; lane 3 [C]); CPP117 (pmt6Δ::hisG-URA3-hisG/pmt6Δ::hisG; lane 4 [C]); CPP1171 (pmt6Δ::hisG/pmt6Δ::hisG; lane 5 [C]).
Adherence to endothelial cells.
Porcine aortic endothelial cells (PAEC) were isolated from aortas of freshly slaughtered pigs, which were obtained from the local slaughterhouse. The lumina of the aortas were washed with phosphate-buffered saline (PBS; 140 mM NaCl, 4 mM KCl, 1 mM Na2HPO4 · 2H2O, 1 mM KH2PO4, 12 mM glucose, pH 7.4) under sterile conditions, and the adventitia was removed (43). The remaining part was immersed completely in 30 ml of dispase solution (0.5 mg/ml; Boehringer, Mannheim, Germany) for 15 min at 37°C in an incubator. Afterwards the aortas were fixed on an aluminum tray and the endothelial cells (EC) were scraped off with a rubber policeman. The cells harvested by each scrape were plated in different wells of a six-well dish (pretreated with 0.2% gelatin solution for 30 min at room temperature). PAEC were cultured at 37°C in the humidified atmosphere of an incubator at 5% CO2–95% air in M199 (Sigma, Munich, Germany) containing 10% fetal calf serum, 30 mM HEPES, 3 mM glutamine, 0.38 g of Dulbecco's PBS/liter, and 4 mg of penicillin and streptomycin/liter. Subcultures were carried out by detaching the cells with trypsin solution (0.5 mg/ml; Sigma), spinning them down at 100 × g for 10 min, and plating them in a dilution of 1:7 (approximately 15,000 cells/cm2). Cells were identified as EC by their cobblestone morphology, the uptake of 1,1′-dioctadecyl-3,3,3′,3′-tetramethyl-indocarbo-cyanine perchlorate-labeled acetylated low-density lipoprotein (Paesel, Frankfurt, Germany), and the immunostaining of factor VIII-related antigen.
Monolayers established in six-well plates were used 2 to 4 days after confluency for adhesion assays. The adhesion of C. albicans cells was determined as described previously (41).
Animal experiments.
Virulence studies were performed as described previously (6). Briefly, strains were harvested from stationary cultures after growth in selective media, adjusted to the desired density in PBS, and injected intravenously into the tail vein in a final volume of 200 μl. The viability of the inoculum was controlled by serial plating of the cells, and the actual infectious load was adjusted accordingly after overnight storage of cells at 4°C. At the time of inoculation viable cell counts were checked again by plating to verify identical loads of infection. After infection, animals were examined for behavioral changes and changes of habit. Survival and mortality were monitored twice a day. Kaplan-Meyer survival graphs were plotted using the GraphPad Prism software, which was also used for log rank test curve comparisons (Mantel-Haenszel test).
Other methods.
For RNA preparation cells were grown in YPD to an optical density at 600 nm between 1.3 and 1.9 or they were induced in 2.5 mM GlcNAc as described by Holmes and Shepherd (18). Total RNA was prepared as described by Schmitt et al. (33). RNA blotting was performed as described previously using the 1.5-kb ClaI-SalI ACT1 fragment (9), a 1.5-kb BamHI-HincII fragment carrying a portion of the PMT1 coding region (41), and the 1.5-kb PstI-XhoI fragment of pCT17 carrying PMT6 as the probes.
The assay for enzymatic Pmt activity was performed as described previously (41). Although measurements of Pmt activity were highly reproducible, the level of residual Pmt activity remaining in pmt1/pmt1 mutants was found to be variable and to depend strongly on the preparations of Dol-P-[14C]Man and of the acceptor peptide.
Nucleotide sequence accession number.
PMT6 was assigned accession no. AF104916 (GenBank/EMBL).
RESULTS
Sequence of PMT6.
A clone containing the whole PMT6 gene (p99) was identified in the C. albicans sequencing project (http://www-sequence.stanford.edu/group/candida). The sequence of PMT6 was determined using M13 standard primers and sequence-specific oligonucleotides. It comprises an open reading frame of 826 codons for a protein with a calculated molecular mass of 94 kDa. Two serine residues at positions 38 and 538 are encoded by nonstandard CUG codons (31). The deduced Pmt6 protein and the S. cerevisiae Pmt6 protein have 42% identical residues, whereas Pmt6p is only 21% identical to Pmt1p (41). The identities to other S. cerevisiae Pmt proteins were much lower; for this reason the gene was designated PMT6. Recently, gene fragments encoding a conceptual Pmt4p homologue of C. albicans were also identified in the C. albicans sequencing project. Sequence comparisons indicated that Pmt6p is clearly different from Pmt4p (22% identity). Computer analysis predicted that Pmt6p is an integral membrane protein, possibly containing 10 transmembrane domains; 7 of these domains coincide with transmembrane regions that have been determined experimentally in Pmt1p of S. cerevisiae (39). Assuming a similar overall structure, the highest degree of identity is present between transmembrane regions I and II and V and VI, respectively, which are the regions constituting both large luminal loops. Pmt6p is also predicted to contain a leucine zipper domain starting at position 723. Four asparagine residues represent potential N glycosylation sites at positions 20, 59, 357, and 453.
Disruption of PMT6 alleles.
The Ura blaster technique (11) was used to disrupt both alleles of PMT6 in the C. albicans PMT1/PMT1 strain CAI4 and the pmt1/pmt1 mutant strain CAP1-3121 (Table 1; Fig. 1A). Chromosomal DNA of transformants was digested with BglII and HindIII and analyzed by Southern hybridization using a specific probe for the PMT6 promoter. The wild-type PMT6 allele displayed a 4.9-kb band (Fig. 1B and C, lane 1), whereas a 7.1-kb band was observed in transformants with the Ura blaster integrated into one allele of PMT6 (Fig. 1B and C, lane 2). After selection on FOA medium, the loss of the URA3 gene and one copy of the hisG element resulted in a 4.3-kb band (Fig. 1B and C, lane 3). The remaining intact PMT6 allele was disrupted similarly, leading to homozygous pmt6::hisG/pmt6Δ::hisG-URA3-hisG strains (Fig. 1B and C, lane 4) and corresponding Ura− derivatives (Fig. 1B and C, lane 5). Thus, these procedures generated strains lacking only PMT6 alleles or doubly mutated strains lacking both PMT6 and PMT1 alleles. Phenotypes reported below were observed in at least two disrupted or reconstituted strains, which were independently isolated.
Supersensitivity of pmt6 mutants.
We previously observed that homozygous pmt1 mutants showed increased sensitivities to various antifungals (G418, hygromycin B, and clotrimazole), calcofluor white, and sodium dodecyl sulfate (SDS) (41). For that reason we also tested the susceptibilities of heterozygous and homozygous pmt6 mutants (Fig. 2A), as well as those of double mutants carrying pmt1 and pmt6 disruptions (Fig. 2B), to various agents.
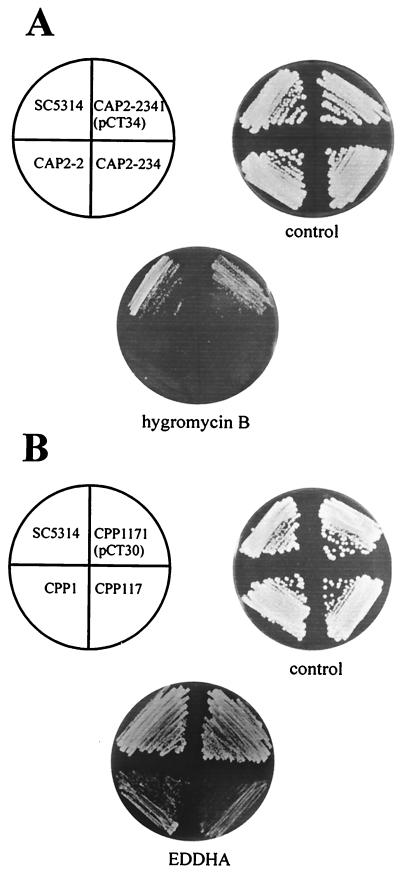
FIG. 2.
Sensitivities of C. albicans strains. The wild-type strain SC5314 (PMT1/PMT1 PMT6/PMT6) was compared with strain CAP2-2 (PMT1/PMT1 PMT6/pmt6), strains CAP2-234 and CAP2-2341 (PMT1/PMT1 pmt6/pmt6), strain CPP1 (pmt1/pmt1 PMT6/pmt6), and strains CPP117 and CPP1171 (pmt1/pmt1 pmt6/pmt6). Plasmid pCT34 carries PMT6, and plasmid pCT30 carries PMT1. Strains were grown on YPD medium without or with hygromycin B (200 μg/ml) or on SD medium without or with EDDHA (300 μM). The plates were incubated for 2 days at 30°C.
Strains carrying one or two pmt6 mutant alleles grew well in the presence of 100 μg of hygromycin B/ml, whereas they failed to grow at 200 μg/ml (Fig. 2A). Remarkably, the heterozygous PMT6/pmt6 and the homozygous pmt6/pmt6 mutant showed the same phenotypes. No increased sensitivities to nystatin (10 to 15 μg/ml), amphotericin B (0.5 to 1.5 μg/ml), clotrimazole (1 to 2 μg/ml), fluconazole (5 μg/ml), fluphenazine (50 μg/ml), SDS (0.06%), calcofluor white (10 to 25 μg/ml), G418 (0.8 to 1.2 mg/ml), and sodium orthovanadate (10 to 20 mM) were detected in pmt6 disruptants. C. albicans pmt1/pmt1 strains with disruptions in at least one PMT6 allele showed the same phenotype with regard to antifungals as pmt1 mutants: supersensitivity to G418, SDS, calcofluor white, clotrimazole, and low concentrations of hygromycin B (41). Furthermore, we could detect an increased sensitivity of pmt1 or pmt1 pmt6 mutants to Congo red (200 μg/ml), which was not observed in pmt6 disruptants (data not shown).
Interestingly, the pmt1 pmt6 mutants also showed new phenotypes that were not detected in strains carrying homozygous single mutations. pmt1 and pmt6 single disruptants were resistant to a 300 μM concentration of the iron chelating agent ethylenediamine-di(o-hydroxyphenylacetic acid) (EDDHA), as was the wild-type strain (SC5314). However, the pmt1/pmt1 pmt6/pmt6 double mutant (CPP117) and the pmt1/pmt1 PMT6/pmt6 heterozygous strain (CPP1) were no longer able to grow in the presence of 300 μM EDDHA (Fig. 2B). This new phenotype was completely suppressed by overexpression of PMT1 (plasmid pCT30). In addition to EDDHA sensitivity, slightly reduced growth of strains CPP1 and CPP117 compared to that of singly mutated strains was also observed on YPD medium containing 20 mM caffeine (data not shown).
PMT6 is required for hyphal morphogenesis.
Hypha formation is induced in certain media including Spider medium (22) or in the presence of positive stimuli including serum or N-acetylglucosamine (GlcNAc) (5). We reported previously that pmt1 mutants are unable to develop hyphae on Spider medium, while they still form hyphae if induced by serum or GlcNAc (41). Therefore, the ability of pmt6 mutants to form hyphae was tested.
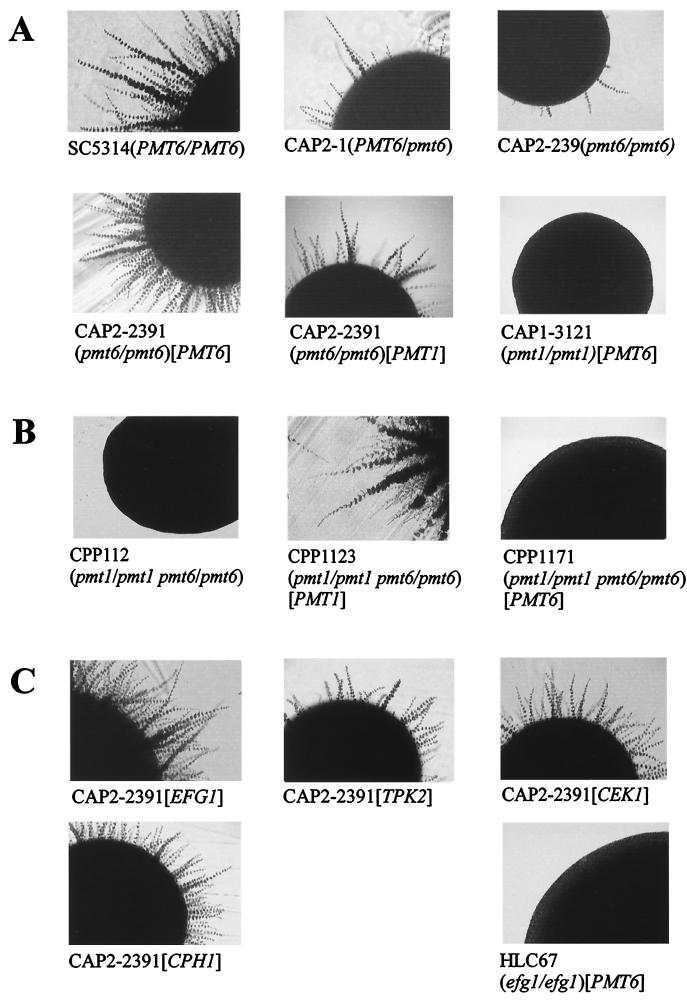
The heterozygous PMT6/pmt6 strains, as well as the homozygous pmt6/pmt6 disruptants showed normal hyphal formation during induction by serum and GlcNAc (data not shown) but a decreased ability to form hyphae on Spider medium (Fig. 3A). Again, as for hygromycin B sensitivity (Fig. 2A), the heterozygous and the homozygous pmt6 mutants showed identical defective phenotypes. A doubly mutated strain lacking both PMT1 and PMT6 alleles was completely morphogenesis negative on Spider medium (Fig. 3B), as were pmt1 single mutants (41). Reintroduction of PMT6 into the pmt6/pmt6 mutant restored morphogenesis as expected; in addition, PMT1 expression complemented the pmt6 mutant phenotype, suggesting common functions of Pmt6p and Pmt1p. Pmt6p may have a more narrow substrate specificity than Pmt1p since PMT6 overexpression did not complement the pmt1 phenotype (Fig. 3A).
FIG. 3.
Hypha formation of C. albicans strains. Shown are sections of colonies grown for 3 days on Spider medium at 37°C. (A) Phenotypes of pmt single mutants. The indicated pmt6/pmt6 strains were complemented with a plasmid carrying PMT6 (pCT34) or PMT1 (pCT30); as controls a pmt1/pmt1 strain complemented by PMT6 (pCT35) and a wild-type strain (SC5314) were analyzed. (B) Phenotypes of pmt double mutants. The indicated pmt1/pmt1 pmt6/pmt6 double mutants were transformed with plasmid pCT30 (PMT1) or pCT35 (PMT6). (C) Suppression of the pmt6 phenotype by genes encoding signaling components. Strain CAP2-2391 (pmt6/pmt6) was transformed with plasmids carrying EFG1, CEK1, CPH1, or TPK2 genes (Table 1). As a control, an efg1/efg1 mutant strain transformed with pCT35 (PMT6) was analyzed.
Because strains lacking one or two PMT6 alleles had the same defective phenotype, we speculated that only one allele of PMT6 was functional, as in the natural heterozygosity of some C. albicans strains (32), or that pairing of alleles was required for expression (1). Alternatively, a threshold level of Pmt6p was presumed to be necessary for cells to generate the wild-type morphogenetic phenotype. To determine if PMT6 is expressed in an allele-specific manner, Northern analyses were performed. All four independent heterozygous PMT6/pmt6 transformants contained a 3-kb PMT6 transcript (data not shown), suggesting that PMT6 is not expressed in such a manner.
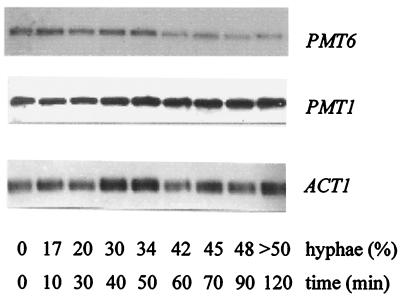
Because of the effects of PMT6 on morphogenesis, we tested if PMT6 expression depends on hyphal induction. For this experiment the PMT strain CAI4(pRC2312) was induced by 2.5 mM GlcNAc according to standard procedures (9); at different time points after induction the percentage of hyphae was determined and RNA was isolated for Northern analysis (Fig. 4). The results demonstrate that the PMT6 transcript level is lowered slightly during the first 50 min of induction, but decreases severalfold after this time; thus, hyphal morphogenesis and PMT6 transcript levels are not closely correlated. Reprobing the same Northern blot with PMT1 revealed that the PMT1 transcript is also not correlated closely to the degree of hyphal development, since it remains relatively constant during the first 50 min of induction and increases only slightly after this time. Figure 4 allows a rough estimation of the relative amounts of both PMT transcripts, because probes of similar lengths and specific activities were used. The autoradiographic exposure times were 4 days for PMT1 and 9 days for PMT6. Considering that the PMT1 signal has about twice the strength of that of PMT6 at 0 min, we estimate that the level of the PMT6 transcript is about 25% relative to the level of the PMT1 transcript; this ratio changes during hyphal formation further in favor of the PMT1 transcript. The downregulation of the PMT6 transcript had a different kinetics from that of the downregulation of the EFG1 transcript described previously (36), because the latter transcript disappeared much more rapidly during hyphal induction. Furthermore, we could show by Northern blottings that high and low EFG1 expression levels in strain SS4 grown in different media (36) did not alter PMT6 transcript levels, thus arguing against a close expressional correlation of both genes (data not shown).
FIG. 4.
PMT6 and PMT1 transcripts during hyphal induction. The wild-type strain CAI4(pRC2312) was induced to form hyphae in the presence of 2.5 mM GlcNAc. At the indicated times the percentages of hypha-forming cells were determined and total RNA was prepared and analyzed by Northern blotting using probes for the indicated genes.
Suppression of pmt6 morphogenetic phenotypes.
Conceivably, the inability of pmt6/pmt6 and PMT6/pmt6 strains to form hyphae was due to one of several possibilities, including (i) defects in structural components required for hyphal morphogenesis and (ii) defects in a component of signaling pathways that translate environmental stimuli into alterations of the cell form (e.g., an O-glycosylated membrane “sensor”). Because in pmt6 mutants hyphal morphogenesis occurred in the presence of serum or GlcNAc, the former hypothesis appeared unlikely. The alternative hypothesis predicts that stimulation of components situated downstream of Pmt6p targets would suppress the morphogenetic defects of pmt6 mutants. To test this possibility, we transformed the pmt6/pmt6 strain CAP2-2391 with plasmids allowing overexpression of the mitogen-activated protein (MAP) kinase Cek1p and its downstream transcription factor, Cph1p (7, 22), or with plasmids allowing overexpression of the catalytic subunit of the protein kinase A isoform (Tpk2p) (35) and of the morphogenetic regulator Efg1p (36).
As shown in Fig. 3C overproduction of all tested components of signaling pathways was able to relieve the morphogenetic block of the pmt6/pmt6 mutant, although to various degrees. Overexpression of genes encoding transcription factors Efg1p and Cph1p showed a strong complementation, similar to complementation by PMT6 itself, while overexpression of the genes encoding the kinases Cek1p and Tpk2p resulted in weaker restoration. The different degrees of suppression may be related to promoter strengths, because in the plasmids used EFG1 and CPH1 are controlled by the strong PCK1 and ADH1 promoters, while the CEK1 and TPK2 genes are transcribed by their natural promoters. On the other hand, the complete block of hyphal formation in a pmt1/pmt1 mutant or the partial block of morphogenesis in a PMT1/pmt1 mutant could not be altered by overexpression of the above signaling components (data not shown), suggesting (i) that pmt1 mutants contain defects unrelated to signaling and (ii) that residual morphogenesis was not responsible for suppression of the pmt6 morphogenetic defect. We point out that the antifungal sensitivities of the pmt6 mutants described above could not be suppressed by any of the signaling components (data not shown), indicating that the functions of Pmt6p in morphogenesis and antifungal sensitivities are separable. These results are compatible with the hypothesis that the pmt6 morphogenetic phenotype is caused by defects in one or more specific components which function upstream of known signaling components, leading to hyphal morphogenesis; however, more-complicated mechanisms cannot be excluded.
Other phenotypes of pmt6 and pmt6 pmt1 mutants.
The heterozygous PMT6/pmt6 mutant CAP2-2 and the homozygous pmt6 disruptant CAP2-234 showed the same generation times in SD medium as the wild-type SC5314 or the PMT6-reconstituted strain CAP2-2341(pCT34). Furthermore, the homozygous pmt6/pmt6 mutant strains did not aggregate, unlike the pmt1/pmt1 mutants (41), and grew as regular yeast cells. On the other hand, doubly mutated strains lacking both PMT6 and PMT1 (strain CPP117) essentially showed the phenotypes of pmt1 mutants, including slower growth, aggregation, antifungal sensitivity, and complete loss of hyphal morphogenesis on Spider medium (but retained the ability for hyphal morphogenesis in serum media) (41); a few additional sensitivities not present in strains containing single mutations were also observed (see above).
To test if known secreted proteins are modified by Pmt6p, we compared the electrophoretic mobilities in SDS-polyacrylamide gel electrophoresis of several known secreted proteins in pmt6/pmt6 mutants and wild-type cells. Immunoblottings were performed using antibodies to Als1p (19) Int1p (14), Cdr1p (30), and Pma1p (27), as described previously (41). None of the tested proteins showed a different electrophoretic migration in pmt6/pmt6 mutants, indicating that these proteins are not extensively modified by Pmt6p. Furthermore, we did not observe any different activities and intra- or extracellular distributions of chitinase activities in pmt6 mutants compared to those in PMT6 strains. Different results were obtained previously for pmt1 mutants, which showed altered migration of Als1p and altered activities and distributions of chitinase (41).
An analysis of the enzymatic Pmt activity was performed by an in vitro assay measuring the transfer of [14C]mannose residues from Dol-P-[14C]mannose to the acceptor peptide acetyl-YATAV-NH2 (40–42). Wild-type strain SC5314 showed high levels of Pmt activity, while in the homozygous pmt1 mutants the enzymatic activity was decreased to 27% of wild-type activity (Table 2). In a homozygous pmt1 pmt6 knockout mutant the Pmt activity was nearly identical (26% compared to that of the homozygous pmt1 disruptant). Thus, Pmt6p is not detectably active under standard assay conditions and contributes little to the overall Pmt activity of cells. Interestingly, we observed that in a strain with only PMT6 deleted Pmt activity was increased, rising to 129% of wild-type activity (Table 2). Possibly, in strains lacking Pmt6p a compensatory increase in activities of Pmt1p and/or other Pmt proteins occurs.
TABLE 2.
Pmt enzymatic activity in strains
| C. albicans strain | Genotype | Amt of [14C]mannose transferred (cpm/min/mg of protein) | % Activity |
|---|---|---|---|
| SC5314 | PMT1/PMT1 PMT6/PMT6 | 7,870 ± 116 | 100 |
| CAP1-312 | pmt1/pmt1 PMT6/PMT6 | 2,100 ± 206 | 27 |
| CAP2-239 | PMT1/PMT1 pmt6/pmt6 | 10,170 ± 223 | 129 |
| CPP117 | pmt1/pmt1 pmt6/pmt6 | 2,000 ± 302 | 26 |
Pmt6p is required for adherence and virulence of C. albicans.
Mannoproteins are necessary for adhesion of C. albicans to a number of surfaces (13). Recently we demonstrated that PMT1 is required for adhesion of C. albicans to EC (41). To explore the role of Pmt6p in C. albicans adhesion, we tested the ability of heterozygous and homozygous pmt6 strains to adhere to PAEC. In comparison to wild-type cells (strain SC5314), strains bearing disruptions in both C. albicans PMT6 alleles adhered less to a monolayer of PAEC (Table 3). While 35% of wild-type cells and 35 to 39% of heterozygous PMT6/pmt6 mutants adhered to the PAEC monolayer in 45 min, the adherence of homozygous pmt6 disruptants CAP2-234 and CAP2-239 was reduced to 13 and 25%, respectively. Reintroduction of the PMT6 gene into one pmt6/pmt6 mutant increased adherence, as expected.
TABLE 3.
Adhesion of C. albicans strains to PAEC
| Strain (genotype) | Adhesion (%)a |
|---|---|
| SC5314 (PMT6/PMT6) | 34.6 ± 3.3 |
| CAP2-2 (PMT6/pmt6) | 34.9 ± 3.9 |
| CAP2-3 (PMT6/pmt6) | 39.0 ± 6.9 |
| CAP2-234 (pmt6/pmt6) | 13.1 ± 0.9 |
| CAP2-239 (pmt6/pmt6) | 24.8 ± 6.9 |
| CAP2391(pCT34) (pmt6/pmt6 [PMT6]) | 41.6 ± 5.0 |
The percentages of 200 C. albicans cells adhering to a monolayer of PAEC in 45 min were determined. Values represent the means of three independent measurements ± standard deviations.
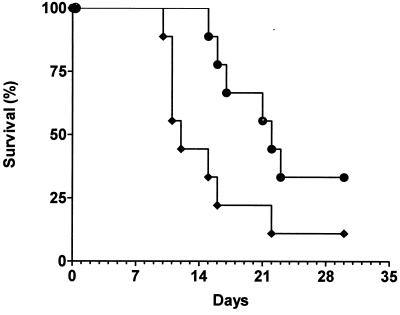
To test whether Pmt6p influences the virulence of C. albicans in a mouse model of systemic infection, 105 cells of the homozygous disruptant CAP2-239 and the reconstituted strain CAP2-2391[pCT35] were injected into the tail vein of immunocompetent mice. The mice were observed for 30 days. The data of Fig. 5 are representative of a total of four independent infections with similar results. Mice infected with the reconstituted wild-type strain (pmt6/pmt6 [PMT6]) showed a mean survival time (MST) of 12 days (Fig. 5), while animals treated with the homozygous pmt6-disrupted strain survived significantly longer (MST, 22 days). Thus, we could prove that PMT6 is involved in the virulence of C. albicans.
FIG. 5.
Virulence of C. albicans strains. Strains CAP2-239 (●; pmt6/pmt6) and the PMT6-reconstituted strain CAP2-2391(pCT35) (⧫; pmt6/pmt6 [PMT6]) were compared. The survival of mice (n = 12) injected with 105 C. albicans cells in the tail vein was determined.
DISCUSSION
The C. albicans Pmt6 protein corresponds to one subclass of Pmt proteins in S. cerevisiae comprising the Pmt2, Pmt3, and Pmt6 proteins. We previously described the C. albicans Pmt1 protein, which corresponds to a second subclass comprising the S. cerevisiae Pmt1 and Pmt5 proteins (41). By nonstringent hybridization using S. cerevisiae PMT1 to PMT7 probes we only detected two PMT genes in C. albicans DNA by Southern blotting (41); these genes correspond to PMT1 and PMT6, which are described here. In the C. albicans genome sequencing project (http://www-sequence.stanford.edu/group/candida/), which at present covers approximately 95% of the genome (S. Scherer, personal communication), gene fragments designated PMT1 to PMT5 have been discovered. Computer comparisons revealed that the genes designated PMT1 and PMT5 correspond to the PMT1 sequence described by us (41), while the gene fragments designated PMT2 and PMT3 are identical to PMT6, which is described here. Recently, other gene fragments, named PMT4, were identified by the C. albicans genome project; PMT4 is different from PMT1 and PMT6. Thus, the present evidence suggests that the set of PMT genes in C. albicans comprises PMT1, PMT4, and PMT6. The existence of only three PMT genes in C. albicans differs from what is found for S. cerevisiae, which harbors seven paralogous PMT genes (38). Deletion of three or four of the seven PMT genes is lethal in S. cerevisiae (15), while we show here that in C. albicans two of the three PMT genes can be deleted without a loss in viability. This finding raises the intriguing question of whether proteins other than Pmt proteins could mediate O glycosylation in fungi.
The structure of Pmt6p corresponds to that of the S. cerevisiae Pmt proteins, which presumably includes a seven-transmembrane helical configuration in endoplasmic reticulum membranes (39). Nevertheless, although Northern blotting demonstrated that PMT6 is expressed (although at a lower level than PMT1), no effect of PMT6 deletion on in vitro Pmt enzymatic activity was observed. It is possible that the standard acceptor peptide used in the enzymatic assay does not correspond to the substrate specificity of Pmt6p or that assay conditions able to detect Pmt1p activity fail to reveal Pmt6p. Several of our findings support the hypothesis that Pmt6p does not have a general role in O glycosylation but rather modifies and/or regulates a relatively narrow set of target proteins. First, in vitro Pmt enzymatic activities are identical in extracts of the wild-type and pmt6 disruptants. Second, we did not observe altered electrophoretic migrations of several secreted and cell wall proteins, including Als1p, Int1p, Cdr1p, and Pma1p, as detected by immunoblotting. The Als1 protein was of special interest because its overproduction in S. cerevisiae induced adhesion to host cells (12) and because the electrophoretic migration of Als1p was altered in pmt1 mutants (41). Third, activity and intra- and extracellular distributions of chitinase were not affected by deletion of PMT6, while these parameters were altered in pmt1 mutants (41). Fourth, the generation of new phenotypes in pmt6 pmt1 double mutants compared to that in pmt6 and pmt1 single mutants, i.e., supersensitivity to the iron chelator EDDHA and to caffeine, suggests that Pmt1p and Pmt6p proteins mannosylate an overlapping, but different, set of target proteins. EDDHA sensitivity has not yet been described in connection with alterations in the cell surface structure of C. albicans. In S. cerevisiae some combinations of pmt mutations are known to cause increased sensitivities to caffeine (15).
An unexpected phenotype of C. albicans pmt mutants was their inability to form hyphae in certain inducing conditions, although growth of the yeast form was unaffected. The pmt1 mutation led to a complete block of morphogenesis, while the pmt6 mutant, although severely compromised, still formed short hyphal extensions on solid Spider medium. Current models of dimorphism in C. albicans comprise two parallel signaling pathways consisting of a protein kinase (Cek1 MAP kinase or Tpk2 protein kinase A [PKA]) and a transcription factor that are regulated by these kinases (Cph1p or Efg1p) (23, 35, 36). We do not favor the hypothesis that lack of O glycosylation led to defects in, e.g., structural components which are needed for hyphal formation, because hyphae developed in both pmt1 and pmt6 mutants in the presence of serum or GlcNAc. Instead, we speculate that O-glycosylated components situated functionally upstream of the Cek1 MAP kinase and/or the Tpk2 PKA were compromised in pmt mutants, since we could restore filamentation by overexpression of both kinases and the associated transcription factors. Suppression of only the morphogenetic phenotype, not the antifungal sensitivity phenotype, of pmt6 strains was observed, indicating different functions of Pmt6p in both processes. Hypothetical components upstream of signaling pathways could, for example, be O-glycosylated sensor proteins located in the cytoplasmic membrane that mediate external signals. To our knowledge this is the first report describing suppression of a specific glycosylation defect by an elevated level of signaling components.
Much of the recent interest in C. albicans biology is due to the need to develop new and effective antifungals. In this respect it is of interest that C. albicans cells lacking Pmt6p were supersensitive to hygromycin B, a phenotype that in S. cerevisiae emerges only if at least two pmt mutations are combined or in mutants defective in N glycosylation (8). Because sensitivities to other agents were not observed, it appears that the pmt6 mutation causes less-drastic sensitivity phenotypes than the pmt1 mutation. The molecular mechanisms by which O or N glycosylation modify sensitivity characteristics of fungi in general and which contribute to the relatively high intrinsic resistance of C. albicans to antifungals and other toxic agents remain to be established. The function of Pmt6p in antifungal resistance is not related to its function in morphogenesis, because overexpression of signaling components (see above) did not alter the antifungal sensitivities of pmt6 mutants. Conceivably, agents lowering levels of resistance to antifungals may be of great interest to complement current antifungal therapies. Furthermore, fungal Pmt proteins are potential selective targets for novel antifungals, because the bulk of O glycosylation in mammals occurs via a biosynthetic pathway different from that in fungi. We have shown here that deletion of pmt6 generates a significant drop in virulence in a system model of mouse infection, while the pmt1 mutation produces absolute avirulence (41). The reduced virulence observed for pmt6 mutants is not caused by a general effect on growth but may involve the block in morphogenesis as described above and/or the defective adhesion to endothelial cells.
ACKNOWLEDGMENTS
Plasmid p99 was generously supplied by S. Scherer. We thank A. Goffeau, M. Hostetter, L. Hoyer, and D. Sanglard for contributing antisera. We thank A. Sonneborn for help with Northern blottings.
REFERENCES
- 1.Aramayo R, Metzenberg R L. Meiotic transvection in fungi. Cell. 1996;86:103–113. doi: 10.1016/s0092-8674(00)80081-1. [DOI] [PubMed] [Google Scholar]
- 2.Bourdineaud J-P, van der Vaart J M, Donzeau M, de Sampaio G, Verrips C T, Lauquin G J-M. Pmt1 mannosyltransferase is involved in cell wall incorporation of several proteins in Saccharomyces cerevisiae. Mol Microbiol. 1998;27:85–98. doi: 10.1046/j.1365-2958.1998.00660.x. [DOI] [PubMed] [Google Scholar]
- 3.Buurman E T, Westwater C, Hube B, Brown A J P, Odds F C, Gow N A R. Molecular analysis of CaMnt1p, a mannosyltransferase important for adhesion and virulence of Candida albicans. Proc Natl Acad Sci USA. 1998;95:7670–7675. doi: 10.1073/pnas.95.13.7670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cannon R D, Jenkinson H F, Shepherd M G. Cloning and expression of Candida albicans ADE2 and proteinase genes on a replicative plasmid in C. albicans and in Saccharomyces cerevisiae. Mol Gen Genet. 1992;235:453–457. doi: 10.1007/BF00279393. [DOI] [PubMed] [Google Scholar]
- 5.Cassone A, Sullivan P A, Shepherd M G. N-acetyl-D-glucosamine-induced morphogenesis in Candida albicans. Microbiologica. 1985;8:85–99. [PubMed] [Google Scholar]
- 6.Csank C, Makris C, Meloche S, Schröppel K, Röllinghoff M, Dignard D, Thomas D Y, Whiteway M. Derepressed hyphal growth and reduced virulence in a VH1 family-related protein phosphatase mutant of the human pathogen Candida albicans. Mol Biol Cell. 1997;8:2539–2551. doi: 10.1091/mbc.8.12.2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Csank C, Schröppel K, Leberer E, Harcus D, Mohamed O, Meloche S, Thomas D Y, Whiteway M. Roles of the Candida albicans mitogen-activated protein kinase homolog, Cek1p, in hyphal development and systemic candidiasis. Infect Immun. 1998;66:2713–2721. doi: 10.1128/iai.66.6.2713-2721.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dean N. Yeast glycosylation mutants are sensitive to aminoglycosides. Proc Natl Acad Sci USA. 1995;92:1287–1291. doi: 10.1073/pnas.92.5.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Delbrück S, Ernst J F. Morphogenesis-independent regulation of actin transcript levels in the pathogenic yeast Candida albicans. Mol Microbiol. 1993;10:859–866. doi: 10.1111/j.1365-2958.1993.tb00956.x. [DOI] [PubMed] [Google Scholar]
- 10.Finck M, Bergmann N, Jansson B, Ernst J F. Defective threonine-linked glycosylation of human insulin-like growth factor in mutants of the yeast Saccharomyces cerevisiae. Glycobiology. 1996;6:313–320. doi: 10.1093/glycob/6.3.313. [DOI] [PubMed] [Google Scholar]
- 11.Fonzi W A, Irwin M Y. Isogenic strain construction and gene mapping in Candida albicans. Genetics. 1993;134:717–728. doi: 10.1093/genetics/134.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fu Y, Rieg G, Fonzi W A, Belanger P H, Edwards J E, Jr, Filler S G. Expression of the Candida albicans gene ALS1 in Saccharomyces cerevisiae induces adherence to endothelial and epithelial cells. Infect Immun. 1998;66:1783–1786. doi: 10.1128/iai.66.4.1783-1786.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fukazawa Y, Kagaya K. Molecular bases of adhesion of Candida albicans. J Med Vet Mycol. 1997;35:87–99. doi: 10.1080/02681219780000971. [DOI] [PubMed] [Google Scholar]
- 14.Gale C A, Bendel C M, McClellan M, Hauser M, Becker J M, Berman J, Hostetter M K. Linkage of adhesion, filamentous growth, and virulence in Candida albicans to a single gene, INT1. Science. 1998;279:1355–1358. doi: 10.1126/science.279.5355.1355. [DOI] [PubMed] [Google Scholar]
- 15.Gentzsch M, Tanner W. The PMT gene family: protein O-glycosylation in Saccharomyces cerevisiae is vital. EMBO J. 1996;15:5752–5759. [PMC free article] [PubMed] [Google Scholar]
- 16.Gentzsch M, Tanner W. Protein-O-glycosylation in yeast: protein-specific mannosyltransferases. Glycobiology. 1997;7:481–486. doi: 10.1093/glycob/7.4.481. [DOI] [PubMed] [Google Scholar]
- 17.Gow N A R, Robbins P W, Lester J W, Brown A J, Fonzi W A, Chapman T, Kinsman O S. A hyphal-specific chitin synthase gene (CHS2) is not essential for growth, dimorphism, or virulence of Candida albicans. Proc Natl Acad Sci USA. 1994;91:6216–6220. doi: 10.1073/pnas.91.13.6216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holmes A R, Shepherd M G. Nutritional factors determine germ tube formation in Candida albicans. J Med Vet Mycol. 1988;26:127–131. [PubMed] [Google Scholar]
- 19.Hoyer L L, Clevenger J, Hecht J E, Ehrhart E J, Poulet F M. Detection of Als proteins on the cell wall of Candida albicans in murine tissues. Infect Immun. 1999;67:4251–4255. doi: 10.1128/iai.67.8.4251-4255.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Immervoll T, Gentzsch M, Tanner W. PMT3 and PMT4, two new members of the protein-O-mannosyltransferase gene family of Saccharomyces cerevisiae. Yeast. 1995;11:1345–1351. doi: 10.1002/yea.320111403. [DOI] [PubMed] [Google Scholar]
- 21.Jurado L A, Coloma A, Cruces J. Identification of a human homolog of the drosophila rotated abdomen gene (POMT1) encoding a putative protein O-mannosyltransferase, and assignment to human chromosome 9q34.1. Genomics. 1999;58:171–180. doi: 10.1006/geno.1999.5819. [DOI] [PubMed] [Google Scholar]
- 22.Liu H, Köhler J R, Fink G R. Suppression of hyphal formation in Candida albicans by mutation of a STE12 homolog. Science. 1994;266:1723–1726. doi: 10.1126/science.7992058. [DOI] [PubMed] [Google Scholar]
- 23.Lo H-J, Köhler J R, Didomenico B, Loebenberg D, Cacciapuoti A, Fink G R. Nonfilamentous C. albicans mutants are avirulent. Cell. 1997;90:939–949. doi: 10.1016/s0092-8674(00)80358-x. [DOI] [PubMed] [Google Scholar]
- 24.Lussier M, Gentzsch M, Sdicu A-M, Bussey H, Tanner W. Protein O-glycosylation in yeast: the PMT2 gene specifies a second protein O-mannosyltransferase that functions in addition to the PMT1-encoded activity. J Biol Chem. 1995;270:2770–2775. doi: 10.1074/jbc.270.6.2770. [DOI] [PubMed] [Google Scholar]
- 25.Martín-Blanco E, García-Bellido A. Mutations in the rotated abdomen locus affect muscle development and reveal intrinsic asymmetry in Drosophila. Proc Natl Acad Sci USA. 1996;93:6048–6052. doi: 10.1073/pnas.93.12.6048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McCusker J H, Davis R W. The use of proline as a nitrogen source causes hypersensitivity to, and allows more economical use of 5-FOA in Saccharomyces cerevisiae. Yeast. 1991;7:607–608. doi: 10.1002/yea.320070608. [DOI] [PubMed] [Google Scholar]
- 27.Monk B C, Kurtz M B, Marrinan J A, Perlin D S. Cloning and characterization of the plasma membrane H+-ATPase from Candida albicans. J Bacteriol. 1991;173:6826–6836. doi: 10.1128/jb.173.21.6826-6836.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mrsa V, Seidl T, Gentzsch M, Tanner W. Specific labelling of cell wall proteins by biotinylation. Identification of four covalently linked O-mannosylated proteins of Saccharomyces cerevisiae. Yeast. 1997;13:1145–1154. doi: 10.1002/(SICI)1097-0061(19970930)13:12<1145::AID-YEA163>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 29.Sanders S L, Gentzsch M, Tanner W, Herskowitz I. O-glycosylation of Axl2/Bud10p by Pmt4p is required for its stability, localization, and function in daughter cells. J Cell Biol. 1999;145:1177–1188. doi: 10.1083/jcb.145.6.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sanglard D, Kuchler K, Ischer F, Pagani J L, Monod M, Bille J. Mechanisms of resistance to azole antifungal agents in Candida albicans isolates from AIDS patients involve specific multidrug transporters. Antimicrob Agents Chemother. 1995;39:2378–2386. doi: 10.1128/aac.39.11.2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Santos M A S, Tuite M F. The CUG codon is decoded in vivo as serine and not leucine in Candida albicans. Nucleic Acids Res. 1995;23:1481–1486. doi: 10.1093/nar/23.9.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scherer S, Magee P T. Genetics of Candida albicans. Microbiol Rev. 1990;54:226–241. doi: 10.1128/mr.54.3.226-241.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schmitt M E, Brown T A, Trumpower B L. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 1990;18:3091. doi: 10.1093/nar/18.10.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sherman F, Fink G R, Hicks J B. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1986. [Google Scholar]
- 35.Sonneborn A, Bockmühl D P, Gerads M, Kurpanek K, Sanglard D, Ernst J F. Protein kinase A encoded by TPK2 regulates dimorphism of Candida albicans. Mol Microbiol. 2000;35:386–396. doi: 10.1046/j.1365-2958.2000.01705.x. [DOI] [PubMed] [Google Scholar]
- 36.Stoldt V R, Sonneborn A, Leuker C E, Ernst J F. Efg1p, an essential regulator of morphogenesis of the human pathogen Candida albicans, is a member of a conserved class of bHLH proteins regulating morphogenetic processes in fungi. EMBO J. 1997;16:1982–1991. doi: 10.1093/emboj/16.8.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Strahl-Bolsinger S, Immervoll T, Deutzmann R, Tanner W. PMT1, the gene for a key enzyme of protein O-glycosylation in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1993;90:8164–8168. doi: 10.1073/pnas.90.17.8164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Strahl-Bolsinger S, Gentzsch M, Tanner W. Protein O-mannosylation. Biochim Biophys Acta. 1999;1426:297–307. doi: 10.1016/s0304-4165(98)00131-7. [DOI] [PubMed] [Google Scholar]
- 39.Strahl-Bolsinger S, Scheinost A. Transmembrane topology of Pmt1p, a member of an evolutionarily conserved family of protein O-mannosyltransferases. J Biol Chem. 1999;274:9068–9075. doi: 10.1074/jbc.274.13.9068. [DOI] [PubMed] [Google Scholar]
- 40.Strahl-Bolsinger S, Tanner W. Protein O-glycosylation in Saccharomyces cerevisiae. Purification and characterization of the dolichyl-phosphate-D-mannose-protein O-D-mannosyltransferase. Eur J Biochem. 1991;196:185–190. doi: 10.1111/j.1432-1033.1991.tb15802.x. [DOI] [PubMed] [Google Scholar]
- 41.Timpel C, Strahl-Bolsinger S, Ziegelbauer K, Ernst J F. Multiple functions of Pmt1p-mediated protein O-mannosylation in the fungal pathogen Candida albicans. J Biol Chem. 1998;273:20837–20846. doi: 10.1074/jbc.273.33.20837. [DOI] [PubMed] [Google Scholar]
- 42.Weston A, Nassau P M, Henly C, Marriott M S. Protein O-mannosylation in Candida albicans. Determination of the amino acid sequences of peptide acceptors for protein O-mannosyltransferase. Eur J Biochem. 1993;215:845–849. doi: 10.1111/j.1432-1033.1993.tb18101.x. [DOI] [PubMed] [Google Scholar]
- 43.Zink S, Naß T, Rösen P, Ernst J F. Migration of the fungal pathogen Candida albicans across endothelial monolayers. Infect Immun. 1996;64:5085–5091. doi: 10.1128/iai.64.12.5085-5091.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]