Abstract
Bacillus subtilis grows in the absence of oxygen using nitrate ammonification and various fermentation processes. Lactate, acetate, and 2,3-butanediol were identified in the growth medium as the major anaerobic fermentation products by using high-performance liquid chromatography. Lactate formation was found to be dependent on the lctEP locus, encoding lactate dehydrogenase and a putative lactate permease. Mutation of lctE results in drastically reduced anaerobic growth independent of the presence of alternative electron acceptors, indicating the importance of NADH reoxidation by lactate dehydrogenase for the overall anaerobic energy metabolism. Anaerobic formation of 2,3-butanediol via acetoin involves acetolactate synthase and decarboxylase encoded by the alsSD operon. Mutation of alsSD has no significant effect on anaerobic growth. Anaerobic acetate synthesis from acetyl coenzyme A requires phosphotransacetylase encoded by pta. Similar to the case for lctEP, mutation of pta significantly reduces anaerobic fermentative and respiratory growth. The expression of both lctEP and alsSD is strongly induced under anaerobic conditions. Anaerobic lctEP and alsSD induction was found to be partially dependent on the gene encoding the redox regulator Fnr. The observed fnr dependence might be the result of Fnr-induced arfM (ywiD) transcription and subsequent lctEP and alsSD activation by the regulator ArfM (YwiD). The two-component regulatory system encoded by resDE is also involved in anaerobic lctEP induction. No direct resDE influence on the redox regulation of alsSD was observed. The alternative electron acceptor nitrate represses anaerobic lctEP and alsSD transcription. Nitrate repression requires resDE- and fnr-dependent expression of narGHJI, encoding respiratory nitrate reductase. The gene alsR, encoding a regulator potentially responding to changes of the intracellular pH and to acetate, is essential for anaerobic lctEP and alsSD expression. In agreement with its known aerobic function, no obvious oxygen- or nitrate-dependent pta regulation was observed. A model for the regulation of the anaerobic fermentation genes in B. subtilis is proposed.
Bacillus subtilis was long considered to be unable to grow in the absence of molecular oxygen as a terminal electron acceptor. However, as in the case of other members of the genus Bacillus, the ability of B. subtilis to utilize nitrate as an alternative electron acceptor has been described by several groups (5, 9, 14, 31). During the process of anaerobic nitrate ammonification, nitrate is reduced by a respiratory nitrate reductase (NarGHI) to nitrite, which is subsequently reduced further to ammonia by a general cellular nitrite reductase (NasDE) (4, 8, 9, 22). The nar locus, consisting of the narGHJI operon (encoding respiratory nitrate reductase), narK (for a potential nitrite extrusion protein), and the open reading frames ywiC and ywiD (of unknown function) also contains the regulatory gene fnr (4). The nitrite reductase genes nasDE were found downstream of the nasAB operon, which encodes assimilatory nitrate reductase (24). Two regulatory systems for the transition from aerobic to anaerobic nitrate respiratory conditions have already been identified (4, 21). First, Fnr, a member of Escherichia coli Crp-Fnr regulatory protein family, acts directly on the expression of several anaerobic transcriptional units (i.e., narK and narGHJI), most likely via interaction with a conserved DNA binding site similar to the E. coli Crp binding site, which is usually centered 41.5 nucleotides (nt) upstream of the transcriptional start point (4). Second, ResD-ResE, the pleiotropic two-component response regulator system encoded by the last two genes of the resABCDE operon, regulates, directly or indirectly, aerobic and anaerobic respiration (23, 33). The binding site for the phosphorylated form of ResD, the active form of the regulator, is still unknown. B. subtilis fnr is strongly induced in the absence of oxygen in a Fnr-independent manner (4). This induction is abolished in a resDE background (23). It is not known, however, if ResD∼P acts directly to activate fnr transcription or if an unknown intermediary regulator is required. This contrasts with mainly autonomous fnr function in E. coli (34, 35).
Several groups demonstrated that B. subtilis is able to grow anaerobically on minimal media in the absence of terminal electron acceptors (8, 19). E. coli and other bacteria use a mixed acid fermentation for glucose metabolism to form the end products ethanol, succinate, lactate, acetate, formate, hydrogen, and carbon dioxide (2). Typical indicators of this process arise from the activity of its key enzyme, pyruvate formate-lyase (Pfl), which leads to massive excretion of formate and acetate as fermentative by-products. For B. subtilis, Nakano and coworkers (19) have identified, via nuclear magnetic resonance analysis, lactate, acetate, acetoin, ethanol, and succinate as main fermentation products (Fig. 1). No significant amounts of formate were detected (19), although the gas phase was not investigated. This observation is supported by the absence of any obvious counterpart to E. coli Pfl among the protein sequences deduced from the complete B. subtilis genome sequence (12).
FIG. 1.
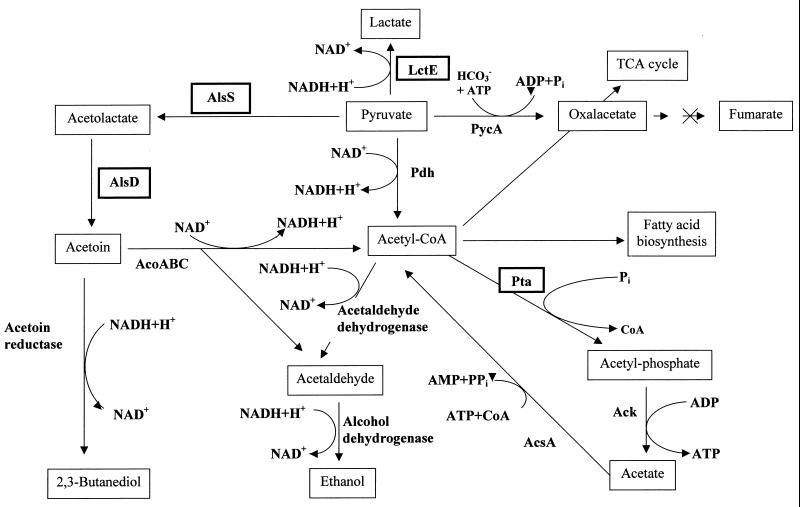
Proposed pathways for anaerobic fermentation and related catabolism in B. subtilis (modified from references 11 and 19). Enzymes with known coding genes are as follows: LctE, lactate dehydrogenase; AlsS, acetolactate synthase; AlsD, acetolactate decarboxylase; Pta, phosphotransacetylase; Ack, acetate kinase; AcoABC, acetoin dehydrogenase; Pdh, pyruvate dehydrogenase; PycA, pyruvate carboxylase; AcsA, acetyl-CoA synthetase. TCA, tricarboxylic acid.
To study the molecular basis for the coordinated induction for anaerobic respiration and fermentation, the completely sequenced B. subtilis genome was analyzed for loci potentially involved in fermentation. Three loci on the B. subtilis chromosome were investigated for their potential involvement in fermentative metabolism and corresponding regulation: lctEP, alsSD, and pta (Fig. 1). The lctE gene, encoding a protein with similarity to known dissimilatory lactate dehydrogenases, was identified during the systematic sequencing of the B. subtilis genome but has not yet been further studied (36). However, 20 years ago, the aerobic regulation of lactate dehydrogenase formation in B. subtilis was investigated using biochemical methods (37). The alsSD operon, which has been shown to encode an acetolactate synthase and an acetolactate decarboxylase, is responsible for acetoin production. Its aerobic catabolite-sensitive and growth phase regulation has been studied (29). Aerobic alsSD transcription is activated in late exponential growth phase. The alsR gene, located upstream of the alsSD operon and transcribed in the opposite direction, encodes a regulator involved in this activation process. As observed for alsSD expression, lactate dehydrogenase synthesis is induced upon the onset of the stationary phase (37). Finally, acetate formation from acetyl coenzyme A (acetyl-CoA) is catalyzed in a two-step reaction by phosphotransacetylase (pta) and acetate kinase (ack). Two-dimensional gel electrophoresis showed that Pta formation is aerobically regulated by various stress conditions (1). The second gene, ack, located in a different position of the genome was found to be subject to catabolite regulation mediated by the catabolite regulator CcpA (6, 27, 32). Here we describe the investigation of the roles played by lctEP, alsSD, and pta in anaerobic metabolism. Their oxygen tension- and nitrate-dependent expression dependent on the regulatory loci resDE and fnr was investigated. An initial regulatory model is proposed.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
All B. subtilis and E. coli strains used throughout this work are listed in Table 1. Luria-Bertani medium was used for standard cultures of B. subtilis and E. coli if not indicated otherwise (16). For the investigation of the expression of the various lacZ fusions, the host strains were grown anaerobically at 37°C on Luria-Bertani medium supplemented with 20 mM K3PO4 (pH 7), 2 mM (NH4)2SO4, 1 mM l-glutamic acid, 1 mM l-tryptophan, 0.8 mM l-phenylalanine, 0.005% (wt/vol) ammonium iron(III) citrate, 1 mM glucose, and, when indicated, 10 mM nitrite or nitrate. The bacteria were incubated in completely filled flasks with rubber stoppers and with shaking at 100 rpm in an incubation shaker to minimize aggregation of the bacteria. Inoculation was performed aerobically at a 1:100 ratio of aerobically grown overnight culture and prewarmed medium. Anaerobic conditions were achieved after a short time through consumption of residual oxygen by the inoculated bacteria. After five doubling times, in the middle of the exponential growth phase, samples for β-galactosidase were taken. The minimal medium for the high-performance liquid chromatography (HPLC) determination of the fermentation products produced by the various investigated strains consisted of 80 mM K2HPO4, 44 mM KH2PO4, 0.8 mM MgSO4 · 7H2O, 1.5 mM thiamine, 40 μM CaCl2 · 2H2O, 68 μM FeCl2 · 4H2O, 5 μM MnCl2 · 4H2O, 12.5 μM ZnCl2, 24 μM CuCl2 · 2H2O, 2.5 μM CoCl2 · 6H2O, 2.5 μM Na2 MoO4 · 2H2O, 50 mM glucose, 50 mM pyruvate, and where indicated, 10 mM nitrate or 10 mM nitrite. Anaerobic growth was performed as described above (4, 8, 20).
TABLE 1.
Strains
| B. subtilis strain | Relevant genotype | Source or reference |
|---|---|---|
| 168 | trpC2 | BGSCa |
| BSIP1104 | trpC2 pta-lacZ cat | 27 |
| BSIP1171 | trpC2 pta::aphA3 | 27 |
| BSIP1173 | trpC2 alsS::alsS-lacZ2 cat | This study |
| BSIP1174 | trpC2 alsS::alsS-lacZ cat pta::aphA3 | This study |
| BSIP1185 | trpC2 lctE-lacZ1 cat | This study |
| BSIP1186 | trpC2 alsR::spc | This study |
| BSIP1187 | trpC2 amyE::alsS-lacZΔfnr cat | This study |
| BSIP1188 | trpC2 lctE-lacZ1 cat alsR::spc | This study |
| BSIP1189 | trpC2 lctE-lacZΔFnr cat | This study |
| BSIP1190 | trpC2 lctE::lctE-lacZ2 cat | This study |
| BSIP1191 | trpC2 lctE::lctP-lacZ cat | This study |
| BSIP1192 | trpC2 alsS-lacZ1 cat | This study |
| BSIP1194 | trpC2 alsS-lacZ1 cat alsR::spc | This study |
| MH5081 | trpC2 pheA1 ΔresDE::tet | 32 |
| MMB100 | trpC2 pta-lacZ cat ΔresDE::tet | This study |
| MMB101 | trpC2 pta-lacZ cat fnr::spc | This study |
| MMB110 | trpC2 lctE-lacZ1 cat ΔnarGH::tet fnr::spc | This study |
| MMB111 | trpC2 lctE-lacZ1 cat ΔresDE::tet fnr::spc | This study |
| MMB112 | trpC2 alsS-lacZ1 cat ΔnarGH::tet fnr::spc | This study |
| MMB113 | trpC2 alsS-lacZ1 cat ΔresDE::tet fnr::spc | This study |
| MMB114 | trpC2 lctE-lacZΔFnr cat fnr::spc | This study |
| MMB115 | trpC2 lctE-lacZΔFnr cat ΔresDE::tet | This study |
| MMB116 | trpC2 lctE-lacZΔFnr cat ΔnarGH::tet | This study |
| THB1 | trpC2 pheA1 ΔnarGH::tet | 9 |
| THB2 | trpC2 pheA1 fnr::spc | 20 |
| THB357 | trpC2 alsS-lacZ1 cat ΔresDE::tet | This study |
| THB361 | trpC2 lctE-lacZ1 cat ΔresDE::tet | This study |
| THB457 | trpC2 alsS-lacZ1 cat fnr::spc | This study |
| THB461 | trpC2 lctE-lacZ1 cat fnr::spc | This study |
| THB557 | trpC2 alsS-lacZ1 cat ΔnarGH::tet | This study |
| THB561 | trpC2 lctE-lacZ1 cat ΔnarGH::tet | This study |
BGSC, Bacillus Genetic Stock Center.
DNA methods and genetic techniques.
E. coli was transformed as described by Chung and Miller (3). B. subtilis cells were transformed as described by Kunst and Rapoport (13). RNA extractions were performed as described by Hagen and Young (7). Southern blotting and Northern blotting were performed as described by Sambrook et al. (30). Membranes were further hybridized with nonradioactively digoxigenin-labeled probes (Boehringer, Mannheim, Germany). The lctE- and lctP-specific DNA fragments used as probes in the Northern blotting experiments were amplified by PCR using the following pairs of oligonucleotides: 5′-GCGGAATTCTTTAATCGGAGCGGGT-3′ and 5′-ACCTGCGATCCCTCCGC-3′ for lctE and 5′-TCTCGAATTCCTTTTGGCTTTAACTGT-3′ and 5′-CTCCCGTGACAACCTGC-3′ for lctP. Primer extensions experiments using reverse transcriptase were performed as described by Pikielny and Rosbash (26). The two oligonucleotides used as primers for mapping the lctE promoter were 5′-CGCAAATGCATAACTGCTTCCAAC-3′ and 5′-TGACCACAAGCTCATCTGTGATCCC-3′. The SubtiList database was used to search for sequence patterns in the B. subtilis genome (17).
Construction of fusion and mutant strains.
B. subtilis strains containing transcriptional fusions between the E. coli lacZ gene and the lctE, lctP, pta, and alsS upstream regions were constructed using PCR-amplified chromosomal fragments and the integrative plasmid pJM783 (25) or the pAC5 derivative pDIA5322 (integration at the amy locus) (Table 1) (15). In pDIA5322, the pAC5 EcoRI-SacI DNA fragment encompassing the 3′ part of the lacZ gene was replaced by the equivalent fragment including the spoVG initiation codon from pJM783. The oligonucleotides pairs used and their relative positions with respect to each start codon are described below. During the amplification, EcoRI and BamHI restriction sites were created at opposite ends of the amplified fragment (underlined). After digestion using these two enzymes, the amplified fragment was inserted into pJM783 digested with the same enzymes. The plasmid pDIA5373, containing the lacZ fusion PlctE-lacZ1, was constructed by the integration of the region −206 to +434 (with respect to the start codon of lctE), amplified using the primers 5′-CCGGAATTCTCGGGCTTAAGCGGTTC-3′ and 5′-CGCGGATCCAATCACCCGCTCT-3′, into pJM783. The plasmid pDIA5374 harbors the fusion PlctE-lacZ2, consisting of the region +29 to +434 relative to lctE initiation codon amplified using the primers 5′-GCGGAATTCTTTAATCGGAGCGGGT-3′ and 5′-CGCGGATCCAATCACCCGCTCT-3′, inserted into pJM783. The plasmid pDIA5375, with the fusion PlctE-lacZΔfnr, was constructed using region −383 to −54 of lctE amplified using the primers 5′-GTAATTGAATTCACCGGATCTTGGCCTGGA-3′ and 5′-CCCGGATCCTTTCACATTTATATTGTGCAACACTTCACAAACTTTTGC-3′ and inserted into pJM783 (see Fig. 4). The vector pDIA5376 with PlctP-lacZ contains region +78 to +405 of lctP amplified using the primers 5′-TCTCGAATTCCTTTTGGCTTTAACTGT-3′ and 5′-ATCGGGATCCGAACACCAAAACCGGCC-3′ and integrated into pJM783. Plasmid pDIA5377, containing PalsS-lacZ1 with region −481 to +394 with respect to the start codon of alsS amplified using the primers 5′-AGTTGAATTCCTTGTCCGATTTG-3′ and 5′-CCGTGGATCCTGCCCTGCTGACGCTAT-3′, was constructed using pJM783. Plasmid pDIA5378 contains the fusion PalsS-lacZ2 spanning the region +105 to +394 of alsS, which was amplified using the primers 5′-GCAGGAATTCATGGCCCAAGCAGTC-3′ and 5′-CCGTGGATCCTGCCCTGCTGACGCTAT-3′. Plasmid pDIA5379, harboring PalsS-lacZΔfnr, was constructed using region −481 to −143 of alsS amplified using the primers 5′-AGTTGAATTCCTTGTCCGATTTG-3′ and 5′-GCGAGGATCCGATAAGTTTCACTATACACTC-3′ and integrated into pDIA5322. Pals-lacZΔfnr was inserted at the amyE locus of B. subtilis 168 after a double crossover event to generate strain BSIP1187. PlctE-lacZ1, PlctE-lacZ2, PlctE-lacZΔfnr, PlctP-lacZ, PalsS-lacZ1, and PalsS-lacZ2 were integrated into the corresponding genes of B. subtilis 168, leading in some cases to the inactivation of the gene (PlctE-lacZ2, PlctP-lacZ, and PalsS-lacZ2). The resulting strains were named BSIP1185, BSIP1190, BSIP1189, BSIP1191, BSIP1192, and BSIP1173, respectively. The description of the employed pta-lacZ fusion was given previously (27). B. subtilis strains in which alsR was disrupted by a spectinomycin resistance gene (18) were constructed by homologous recombination using plasmid pDIA5372. pDIA5372 was constructed by the insertion of a spectinomycin cassette into the unique StuI site of the pUC18 derivative pDIA5370, previously obtained by a shotgun cloning experiment (28). In the resulting pDIA5372, the alsR gene was disrupted 270 bp downstream of the translational start codon. Linearized pDIA5372 was used to transform B. subtilis 168 to generate BSIP1186. PlctE-lacZ1 and PalsS-lacZ1 were transferred into BSIP1186 to generate BSIP1188 and BSIP1194, respectively. The strain THB361 was constructed by the transfer of ΔresDE::tet from MH5081 into BSIP1185 via transformation of BSIP1185 using genomic DNA from MH5081 and appropriate antibiotic selection. All newly created strains were checked using PCR and hybridization experiments. THB461 and THB561 resulted from the transfer of fnr::spc (THB2) and ΔnarGH::tet (THB1), respectively, to BSIP1185. BSIP1188 contains alsR::spec from BSIP1186 in BSIP1185. THB357, THB457, THB557, and BSIP1194 were formed by the transfer of ΔresDE::tet, fnr::spc, ΔnarGH::tet, and alsR::spc from MH5081, THB1, THB2, and BSIP1186, into BSIP1192, respectively. B. subtilis MMB100 and MMB101 were created by the transformation of BSIP1104 with genomic DNA prepared from MH5081 and THB2, respectively. BSIP1174 was formed by the transfer of pta::aphA3 to BSIP1173. MMB110, MMB111, MMB112, and MMB113 were created by the transfer of ΔnarGH::tet or ΔresDE::tet into THB461 and THB457. MMB114, MMB115, and MMB116 resulted from the transformation of BSIP1189 with genomic DNA prepared from THB2, MH5081, and THB1, respectively.
FIG. 4.
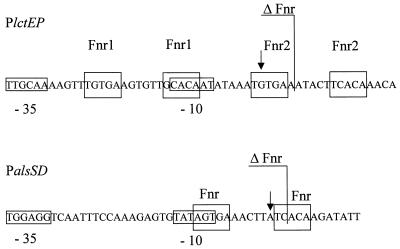
DNA sequences of the B. subtilis lctEP and alsSD promoter regions. The mapped mRNA ends are indicated by arrows. Predicted −10 and −35 regions are boxed. Predicted Fnr binding sites are also boxed. The 3′ ends of the lctE and alsS promoter regions in the Plct-lacZΔFnr and Pals-lacZΔFnr fusions are shown. The alsSR start point was described by Renna et al. (29).
HPLC analysis of fermentation products.
Fermentation products were separated and quantified by HPLC using an LKB-Pharmacia liquid chromatograph (Amersham Pharmacia Biotech, Freiburg, Germany). Metabolites were separated on a Eurocat H 300- by 8-mm, 10-μm-pore-size cation-exchange resin (Knauer, Berlin, Germany). Chromatography was performed at 60°C, with a flow rate of 0.6 ml/min in 0.01N H2SO4. Eluted compounds were registered and quantified by a refractive index detector equipped with a computer-powered integrator. Soluble fermentation products were identified by comparison with retention times and peak areas of corresponding standards.
β-Galactosidase assays.
β-Galactosidase activity was assayed as previously described (16, 20).
RESULTS AND DISCUSSION
Analysis of B. subtilis fermentation products using HPLC.
Previously, Nakano and coworkers demonstrated by nuclear magnetic resonance the formation of acetate, ethanol, lactate, acetoin, and, under certain conditions, small amounts of 2,3-butanediol from the carbon source glucose in combination with pyruvate (19). To monitor more precisely the flow of carbon from glucose and pyruvate into the various fermentation products, a semiquantitative HPLC analysis was established. A cation-exchange column was calibrated with glucose (retention time, 12.1 min), pyruvate (12.8 min), succinate (16.7 min), lactate (18.3 min), acetate (22.2 min), 2,3-butanediol (25.8 min), and ethanol (28.1 min). B. subtilis was grown anaerobically in minimal medium containing 50 mM glucose and 50 mM pyruvate as carbon sources. Ammonia served as a nitrogen source. Where indicated, 10 mM nitrate or nitrite was added. Since fermentation products are excreted by B. subtilis, metabolites present in the growth medium before and after anaerobic growth were quantified. Fermentative growth in the presence of 50 mM glucose and 50 mM pyruvate led to the detection of lactate (38.4 mM), 2,3-butanediol (9.5 mM), and acetate (13.3 mM). Surprisingly, the addition of the electron acceptors nitrate and nitrite did not drastically change the overall formation of fermentation products (Table 2). In both cases lactate formation was found to be slightly reduced, while the formation of acetate and 2,3-butanediol was slightly increased. These observations indicate the presence of the various fermentation processes during anaerobic respiration in B. subtilis. In all cases only a minor part of the initial added glucose was consumed, while the pyruvate was no longer detectable at the end of the growth (data not shown). At a higher pyruvate concentration (80 mM), excretion of acetoin was observed (data not shown). Ethanol was not detected in the growth media under any of the employed conditions using the HPLC method.
TABLE 2.
Fermentation product formation in wild-type B. subtilis and various fermentation gene mutantsa
| B. subtilis strain (relevant genotype) | Electron acceptor (10 mM) | Lactate (mM) | Acetate (mM) | 2,3-Butanediol (mM) | Total product concn (mM) | Concn of cells (g [wet wt]/liter) |
|---|---|---|---|---|---|---|
| 168 (wild type) | Nitrate | 23.3 | 16.4 | 16.7 | 56.4 | 8.0 |
| BSIP1190 (lct) | Nitrate | 1.1 | 14.3 | 13.4 | 28.8 | 2.0 |
| BSIP1171 (pta) | Nitrate | 9.0 | 3.7 | 4.2 | 16.9 | 4.3 |
| BSIP1173 (als) | Nitrate | 38.3 | 18.1 | NDb | 56.4 | 7.8 |
| BSIP1174 (pta als) | Nitrate | 2.7 | 1.7 | ND | 4.4 | 2.4 |
| 168 (wild type) | Nitrite | 25.9 | 16.0 | 11.1 | 53.0 | 7.3 |
| BSIP1190 (lct) | Nitrite | 0.9 | 14.4 | 13.1 | 28.4 | 2.7 |
| BSIP1171 (pta) | Nitrite | 12.3 | 3.0 | 1.1 | 16.4 | 3.0 |
| BSIP1173 (als) | Nitrite | 38.0 | 15.5 | ND | 53.5 | 7.5 |
| BSIP1174 (pta als) | Nitrite | 2.0 | 1.0 | ND | 3.0 | 2.1 |
| 168 (wild type) | 38.4 | 13.3 | 9.5 | 61.2 | 7.3 | |
| BSIP1190 (lct) | 3.0 | 7.0 | 7.5 | 16.5 | 1.8 | |
| BSIP1171 (pta) | 28.8 | 2.5 | 18.2 | 49.5 | 3.2 | |
| BSIP1173 (als) | 39.4 | 12.5 | ND | 51.9 | 7.4 | |
| BSIP1174 (pta als) | 15.0 | 1.1 | ND | 16.1 | 2.0 |
The indicated B. subtilis strains were incubated anaerobically for 10 h at 37°C in defined minimal media using 50 mM glucose and 50 mM pyruvate as carbon sources. Fermentation products were isolated from the growth media and quantified using HPLC as described in Materials and Methods.
ND, not detectable.
Functional identification of genes involved in anaerobic lactate, 2,3-butanediol, and acetate formation.
Predicted metabolic pathways for the synthesis of lactate, acetate, and 2,3-butanediol are shown in Fig. 1 (11, 19). To identify the genetic loci involved in anaerobic fermentation product formation, we investigated mutants with mutations in potential fermentation genes previously identified by the sequencing project or other approaches.
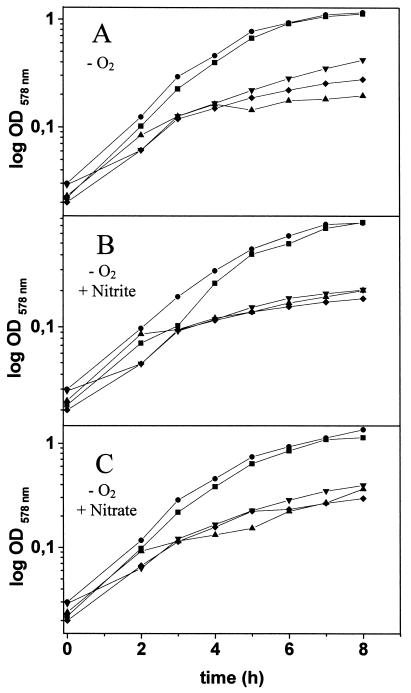
First, the compounds excreted by those mutants when grown under anaerobic fermentative and respiratory conditions were compared using HPLC. Lactate is usually produced by reduction of pyruvate in a single step (Fig. 1). This reaction is catalyzed by lactate dehydrogenase, with the simultaneous oxidation of one molecule of NADH per molecule of pyruvate reduced. The lctE gene, potentially encoding lactate dehydrogenase from B. subtilis, was identified through systematic sequencing (36). In a lctE mutant (strain BSIP1190), almost no lactate accumulation was observed under any tested anaerobic condition in minimal (Table 2) or rich (data not shown) medium. The lack of lactate dehydrogenase led to a severe defect in anaerobic growth in rich and minimal media, although the lct mutant still accumulates significant amounts of acetate and 2,3-butanediol (Fig. 2; Table 2). In agreement with the observed lactate accumulation pattern for the wild-type strain, the observed reduction in anaerobic growth of the lctE mutant was independent of the presence of alternative electron acceptors, such as nitrate and nitrite (Fig. 2). These observations indicate that the lactate dehydrogenase encoded by lctE is generally important for anaerobic growth. Its function in the reoxidation of reducing equivalents is not limited to fermentation but is also important for respiratory growth with nitrate and nitrite.
FIG. 2.
Growth of wild-type B. subtilis (■); mutants with mutations in the alsS (●), lctE (▴), and pta (▾) genes; and a pta alsS double mutant (⧫) under fermentative conditions (A) and nitrite (B) and nitrate (C) respiratory conditions. Growth was monitored by determination of the optical density at 578 nm (OD578 nm) at the indicated time points. Values reported are the averages from at least five independent experiments performed in triplicate.
Aerobic acetoin synthesis from pyruvate in B. subtilis was studied before (29). It involves two steps catalyzed by acetolactate synthase and acetolactate decarboxylase, which are encoded by alsS and alsD, respectively (Fig. 1). The two genes are organized in an operon (29). An additional step catalyzed by acetoin reductase converts acetoin to 2,3-butanediol. The corresponding B. subtilis gene is presently unknown. The insertional inactivation of the als operon (strain BSIP1173) totally abolished 2,3-butanediol production under all tested conditions (Table 2). However, the mutation had only a small effect on the growth behavior of B. subtilis under the tested anaerobic conditions (Fig. 2).
The energetically most efficient fermentative pathway is acetate production. After conversion of pyruvate to acetyl-CoA, the phosphotransacetylase (Pta) and acetate kinase (Ack) form acetate in a two-step reaction (Fig. 1). Usually, one ATP molecule per molecule of acetate is produced. Significantly reduced amounts of acetate were produced in a pta background (BSIP1171) under all tested conditions (Table 2). A significant reduction of growth was observed under all tested anaerobic conditions independent of the presence of alternative electron acceptors. Similar to the conclusions drawn from the growth behavior of the lctE mutant, pta plays a general role in the anaerobic energy metabolism. Recent investigation of the aerobic function of pta revealed its contribution to the growth in the presence of oxygen (27, 32). The pta gene is subject to a complex catabolite regulation involving ccpA, hprK, ptsH1, and crh (27).
We have also investigated the effect of a pta als double mutation (BSIP1174). In agreement with the results of the investigation of the strains with single mutations, the double mutant was unable to produce acetate or 2,3-butanediol and its growth was severely reduced compared to that of the wild type strains (Table 2; Fig. 2).
Organization of the lctEP operon.
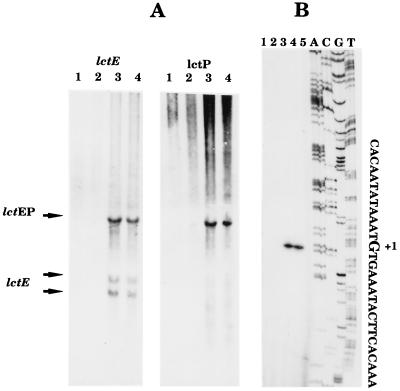
Initial inspection of the DNA sequence of lctE, encoding lactate dehydrogenase, and lctP, encoding a putative lactate permease, suggested the presence of a single transcriptional unit. Northern blot experiments confirmed this expectation (Fig. 3A). Total cellular RNA was prepared from wild-type B. subtilis grown under aerobic conditions (Fig. 3A, lanes 1) and under anaerobic conditions in the presence (Fig. 3A, lanes 2) and absence of nitrate (Fig. 3A, lanes 3). The addition of fumarate reflects fermentative conditions (Fig. 3A, lanes 4). Probes specific to lctE and lctP were employed for the detection of specific mRNA. The lctE probe detected three bands of approximately 2,700, 1,050, and 650 nt. The lctP probe revealed only the single band of higher molecular weight of approximately 2,700 nt, which was also seen with the lctE probe. First, the observed pattern indicated that the two genes are indeed organized in an operon and that no internal promoter upstream of lctP was active under the conditions tested. The large transcript of approximately 2,700 nt has exactly enough coding capacity for LctE and LctP. The observed transcriptional polarity may contribute to a lower synthesis of the potential lactate permease compared to lactate dehydrogenase. Second, comparison of band intensities for the RNAs prepared from B. subtilis grown under various growth conditions showed that the lctEP operon is significantly induced on the transcriptional level under anaerobic conditions in the absence of nitrate (Fig. 3A, lanes 3 and 4). The presence of nitrate significantly reduced anaerobic lctEP expression. However, the utilization of increased amounts of total cellular RNA for the Northern blot and primer extension experiments revealed the presence of lctEP mRNA even in the presence of nitrate in the growth medium (data not shown). The molecular basis of the observed regulatory phenomena was further investigated as outlined below.
FIG. 3.
mRNA analysis of the lctEP operon. (A) Northern blot analysis. The lctE- and lctP-specific probes were generated and the blotting was performed as outlined in Materials and Methods. (B) Primer extension mapping. The location of the 5′ end of the lctEP mRNA was deduced from the lengths of the cDNA bands. The length was obtained by comparison with the sequencing reaction products (in the order A, C, G, and T) performed with the primer used for the extension reaction. For both panels, the RNA used in each experiment was extracted from B. subtilis grown under the following conditions: lane 1, exponential phase in complex medium in the presence of oxygen; lane 2, without oxygen and with 10 mM nitrate; 3, without oxygen or further additions; 4, without oxygen and with 5 mM fumarate; 5, control without RNA.
Promoter structure of the lctEP operon.
The 5′ end of the lctEP mRNA was investigated using the primer extension technique. As shown in Fig. 3B, the mRNA appeared to start at a guanosine residue 62 nt upstream of the translational start codon (Fig. 2B and 3). Potential −10 (CACAAT) and −35 (TTGCAA) regions for a ςA-dependent promoter, separated by 17 bp, were deduced (Fig. 4). No other mRNA 5′ ends were detected by these mapping experiments, indicating the absence of other active promoters under the conditions tested. In agreement with the Northern experiment results and reporter gene fusion experiments (see below), the strongest primer extension signal was observed using RNA prepared from fermentatively grown B. subtilis.
Analysis of the DNA sequence upstream of the lctEP operon revealed a palindromic sequence spanning positions 89 to 44 bp upstream of the translational start codon and overlapping the determined transcriptional start site. Each symmetrical arm contains a highly conserved potential binding site for the redox regulator Fnr (Fnr box 1, 5′-TGTGA-AGTGTT-GCACA-3′; Fnr box 2, 5′-TGTGA-AATACT-TCACA-3′) (4). The deduced promoter region is partly within the inverted repeat sequence (Fig. 4). The predicted −10 sequence is relatively poorly conserved and coincides with the second half of the first potential Fnr site. An initial characterization of this promoter region is described.
Redox- and nitrate-regulated expression of the lctEP operon.
In order to further substantiate the transcriptional regulation highlighted by the mRNA analysis, we have studied the expression of the lctEP and alsSD operons by using transcriptional fusions with the E. coli lacZ gene as a reporter gene. The lacZ fusions were constructed as described in Materials and Methods. In the strain carrying PlctE-lacZ1 or PalsS-lacZ1, the wild-type copy of the corresponding gene was maintained. Integration of PlctE-lacZ2, PlctP-lacZ, and PalsS-lacZ2 resulted in the disruption of lctE, lctP, and alsS, respectively. Expression of these fusions was analyzed during aerobiosis and after a shift to anaerobiosis in the presence or absence of nitrate or nitrite.
The β-galactosidase activity of the PlctE-lacZ1 fusion was rapidly induced (more than 70-fold) after a shift to anaerobic conditions (Table 3). Anaerobic induction of the PlctE-lacZ1 fusion was suppressed fivefold if nitrate was present in the medium. The presence of nitrite had no inhibitory effect on anaerobic PlctE-lacZ1 expression. The PlctP-lacZ fusion induction profile was similar to that of Plct-lacZ1. Inactivation of lctE did not significantly change the observed anaerobic induction of lctE. However, the nitrate repression of lctE transcription was reduced. Changes in lctE-lacZ expression due to structural differences between the employed fusions cannot be excluded.
TABLE 3.
Regulation of lctEP expression by oxygen tension and nitrate, the regulatory loci resDE, fnr, and alsR, and the nitrate reductase genes narGHJIa
| Strain (relevant genotype) | Fusion | β-Galactosidase activity (Miller units)b
|
|||
|---|---|---|---|---|---|
| Aerobic | Anaerobic
|
||||
| Fermentative | With nitrate | With nitrite | |||
| BSIP1185 (wild type) | PlctE-lacZ1 | <10 | 730 | 140 | 630 |
| THB361 (ΔresDE) | PlctE-lacZ1 | 15 | 180 | 130 | 170 |
| THB461 (fnr) | PlctE-lacZ1 | <10 | 330 | 320 | 320 |
| THB561 (ΔnarGH) | PlctE-lacZ1 | <10 | 630 | 620 | 610 |
| BSIP1188 (alsR) | PlctE-lacZ1 | <10 | 60 | 30 | 60 |
| BSIP1190 (lctE) | PlctE-lacZ2 | <10 | 780 | 510 | 600 |
| MMB110 (fnr ΔnarGH) | PlctE-lacZ1 | <10 | 480 | 470 | 320 |
| MMB110 (fnr ΔresDE) | PlctE-lacZ1 | 20 | 220 | 240 | 170 |
| BSIP1191 (lctP) | PlctP-lacZ | <10 | 380 | 40 | 390 |
| BSIP1189 (wild type) | PlctE-lacZΔfnr | <10 | 1,200 | 520 | 1,430 |
| MMB114 (fnr) | PlctE-lacZΔfnr | <10 | 1,315 | 1,230 | 1,020 |
| MMB115 (resDE) | PlctE-lacZΔfnr | 30 | 450 | 320 | 460 |
| MMB116 (ΔnarGH) | PlctE-lacZΔfnr | <10 | 1,220 | 1,210 | 1,480 |
Construction of the various B. subtilis strains carrying the indicated lacZ fusions is described in Materials and Methods. The indicated strains were grown either aerobically or anaerobically with 50 mM glucose as a carbon source and ammonia as a nitrogen source, with the indicated additions (10 mM nitrate or nitrite), to the mid-exponential growth phase as outlined in detail before (8, 20).
Oxygen tension- and nitrate-regulated expression of the alsSD operon.
Similar to the case for lctE expression, an approximately 60-fold anaerobic induction of β-galactosidase activity was observed for the PalsS-lacZ1 fusion in the B. subtilis wild-type strain grown under fermentative conditions and in the presence of nitrite (Table 4). Again, nitrate reduced alsS expression threefold. Mutation of alsS did not influence anaerobic PalsS-lacZ2 expression (Table 4). To investigate the participation of various known redox regulators and of AlsR in redox- and nitrate-dependent alsSD and lctEP expression, lacZ fusions were tested in B. subtilis strains carrying mutations in the fnr, resDE, and alsR loci.
TABLE 4.
Regulation of als expression by oxygen tension and nitrate, the regulatory loci resDE, fnr, and alsR, and the nitrate reductase genes narGHJIa
| Strain (relevant genotype) | Reporter gene fusion | β-Galactosidase activity (Miller units)b
|
|||
|---|---|---|---|---|---|
| Aerobic | Anaerobic
|
||||
| Fermentative | With nitrate | With nitrite | |||
| BSIP1192 (wild type) | PalsS-lacZ1 | <10 | 610 | 190 | 600 |
| THB357 (ΔresDE) | PalsS-lacZ1 | <10 | 740 | 880 | 940 |
| THB457 (fnr) | PalsS-lacZ1 | <10 | 390 | 380 | 330 |
| THB557 (ΔnarGH) | PalsS-lacZ1 | <10 | 530 | 630 | 790 |
| MMB112 (fnr ΔnarGH) | PalsS-lacZ1 | <10 | 270 | 280 | 250 |
| MMB113 (fnr ΔresDE) | PalsS-lacZ1 | <10 | 410 | 390 | 440 |
| BSIP1194 (alsR) | PalsS-lacZ1 | <10 | 40 | 20 | 30 |
| BSIP1173 (alsS) | PalsS-lacZ2 | <10 | 610 | 200 | 610 |
| BSIP1187 (wild type) | PalsS-lacZΔfnr | <10 | <10 | <10 | <10 |
Construction of the various B. subtilis strains carrying the indicated lacZ fusions is described in Materials and Methods. The indicated strains were grown either aerobically or anaerobically with 50 mM glucose as a carbon source and ammonia as a nitrogen source, with the indicated additions (10 mM nitrate or nitrite), to the mid-exponential growth phase as outlined in detail before (8, 20).
Participation of fnr, resDE, and alsR in lctEP and alsSD expression.
Mutation of fnr reduced anaerobic PlctE-lacZ1 and PalsS-lacZ1 expression by half (Tables 3 and 4). Expression of lctEP and alsSD is subject to arfM (ywiD) regulation (10; M. Marino, H. Cruz Ramos, T. Hoffman, P. Glaser, and D. Jahn, unpublished observations). Transcription of arfM (ywiD) was found to be completely fnr dependent. Due to the similar degrees of arfM and fnr regulation, it was concluded that there is an indirect participation of fnr in lctEP and alsSD expression via arfM induction (10; M. Marino et al., unpublished observations). Moreover, the location and initial analysis of the potential Fnr binding sites (see below) made their participation in a direct Fnr-mediated anaerobic induction of both operons very unlikely.
The second consequence of fnr mutation was the complete loss of nitrate repression on both operons (Tables 3 and 4). Interestingly, mutation of resDE also led to the loss of nitrate repression. These findings prompted us to investigate the participation of narGHJI, encoding respiratory nitrate reductase, in nitrate repression, since narGHJI expression is strictly dependent on fnr and resDE. As shown in Tables 3 and 4, narGH deletion also completely abolished nitrate repression of both operons. These results suggest that the observed nitrate regulation depends on the enzymatic nitrate reductase activity. Consequently, only an indirect participation of fnr and resDE via narGHJI induction in nitrate repression of anaerobic lctEP and alsSD transcription is suggested.
A clear difference was observed for the consequences of a resDE mutation on the overall anaerobic lctEP and alsSD expression. While PlctE-lacZ1 expression was reduced three- to fourfold in a resDE mutant, PalsS-lacZ1 expression remained mainly unchanged. This observation indicated an additional direct or indirect participation of resDE in anaerobic lctEP induction.
In agreement with the results obtained for the investigation of lctEP expression in single fnr and resDE mutants, analysis of a PlctE-lacZ1 fusion in an fnr resDE double mutant revealed significantly reduced anaerobic induction with the complete loss of nitrate repression (Table 3). An fnr resDE double mutant led to the reduction of alsSD expression slightly below the level determined for the fnr mutant alone and to the complete loss of nitrate repression. This is again in agreement with the analysis of single regulatory mutants, where the redox regulation of alsSD did not significantly respond to resDE mutation (Table 4). In agreement with the observed effects of single fnr and narGH mutation on lctEP and alsSD expression, an fnr narGH double mutation reduced anaerobic lctEP and alsSD induction and completely abolished nitrate repression (Table 3).
The regulatory gene alsR, located upstream of the alsSD operon, was found to be essential for both PlctE-lacZ and PalsS-lacZ expression under all tested anaerobic conditions. Previous investigations suggested the intracellular pH as well as the acetate concentration in the growth medium as possible signals for AlsR-dependent regulation (29).
Initial analysis of the potential Fnr binding site in the lctEP and alsSD promoters.
The analysis of the lct promoter region revealed a complex structure. In order to assess the role of the apparent Fnr binding sites in regulation, a truncated fusion was constructed (PlctE-lacZΔfnr [Fig. 4]). In this fusion only the first five bases after the transcription start point were kept, while the downstream half of the second putative Fnr site was removed as shown in Fig. 4. The expression of this PlctE-lacZΔfnr fusion was monitored under aerobic and various anaerobic growth conditions (Table 3). The loss of the second half site led to a higher anaerobic induction of lctE expression and a significant decrease of nitrate repression. These results indicate that this region might be involved in a mechanism of repression the lct transcription. In agreement with the data obtained for the analysis of the wild-type lctE promoter, resDE mutation significantly reduced anaerobic lctEP induction via the mutated promoter. However, the values obtained for the resDE mutant still documented the same degree of derepression observed for the wild-type strain (Table 3). Moreover, similar to the wild-type lctEP promoter, the mutated promoter revealed significantly reduced nitrate repression in a resDE mutant. In complete agreement with the case for the wild-type lctEP promoter, the mutated promoter responded to a narGH mutation with complete loss of nitrate repression. The degree of anaerobic depression of the mutated promoter in the narGH mutant was identical to that of the wild-type strain. Surprisingly, mutation of fnr did not decrease transcription from the mutated lctEP promoter, while the native promoter partially lost its anaerobic induction capacity. This observation indicated the importance of the mutated promoter sequence for direct or indirect influence of fnr on lctEP expression.
Interestingly, a similar Fnr-like binding site is found in the vicinity of the als operon transcription starting point (Fig. 4). In order to assess if this sequence is involved in als regulation, the Pals-lacZΔfnr fusion was constructed. In this fusion only the first three bases downstream of the transcription starting point were kept (Fig. 4). In the resulting strain, BSIP1187, the transcription of the Pals-lacZΔfnr fusion was abolished during both aerobiosis and anaerobiosis (Table 4). These results could be interpreted in two ways: either the binding of an essential activator has been abolished or the integrity of the overall promoter structure has been disturbed in this construct. In both cases the effect of an fnr gene mutation on lctEP and alsSD expression contrasts with the results of the deleted-promoter study. In the case of lctEP, an fnr mutation decreased lctEP transcription, while the mutation of the putative Fnr box derepressed lctEP transcription. The location of the putative Fnr box with respect to the transcriptional start site of alsSD suggested that a protein binding to this site would act as a repressor. However, an fnr mutation again led to decreased transcription, and deletion of the putative Fnr box totally abolished transcription.
These results indicate that the Fnr box-like elements of the lctEP and alsSD promoters do not serve known Fnr-dependent regulatory functions.
Expression of pta under anaerobic conditions.
The third fermentative locus, pta, was investigated for its potential oxygen- or nitrate-dependent control of transcription (Table 5). In agreement with its general aerobic and anaerobic function, no obvious regulation by oxygen tension, nitrate, or nitrite was observed. Mutation of resDE and fnr did not change pta transcription. These data in combination with the observed pta growth phenotypes and a recent analysis of anaerobic pta function (27) demonstrated the overall importance of acetate formation for aerobic and anaerobic growth.
TABLE 5.
Investigation of dependence of pta expression on changing oxygen tension and nitrate as well as nitritea
| Strain (relevant genotype) | Reporter gene fusion | β-Galactosidase activity (Miller units)b
|
|||
|---|---|---|---|---|---|
| Aerobic | Anaerobic
|
||||
| Fermentative | With nitrate | With nitrite | |||
| BSIP1104 (wild type) | Ppta-lacZ | 210 | 185 | 200 | 200 |
| MMB100 (ΔresDE) | Ppta-lacZ | 180 | 175 | 170 | 170 |
| MMB101 (fnr) | Ppta-lacZ | 215 | 185 | 170 | 170 |
Construction of the various B. subtilis strains carrying the indicated lacZ fusions is described in Materials and Methods. The indicated strains were grown either aerobically or anaerobically with 50 mM glucose as a carbon source and ammonia as a nitrogen source, with the indicated additions (10 mM with nitrate or nitrite), to the mid-exponential growth phase as outlined in detail before (8, 20).
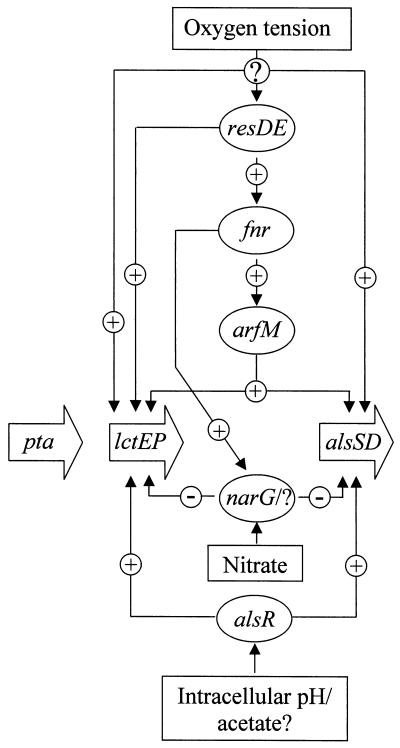
A model for the regulation of anaerobic fermentation genes in B. subtilis.
The lctEP and alsSD operons are an integral part of the anaerobic modulon of B. subtilis. Under anaerobic conditions, both are induced via a regulatory cascade from an unknown sensor via ResDE, Fnr, and ArfM (Fig. 5). However, ArfM activation is responsible for only a part of the observed degree of anaerobic lctEP and alsSD induction. Additional, yet-unknown, redox regulatory components are required for full anaerobic gene expression. A similar regulatory cascade was recently determined for B. subtilis hemN and hemZ transcription (10).
FIG. 5.
Regulatory model for the anaerobic expression of the lctEP, alsSD, and pta loci.
Nitrate only partially represses lctEP and alsSD expression. The nitrate regulatory system involved is still unknown. Intact nitrate reductase and lactate dehydrogenase are required to allow nitrate regulation. One could speculate that the redox status of the cell (NAD-to-NADH ratio) or nitrate-dependent membrane-localized electron flow is a signal for this unknown system. Alternatively, an already-known regulatory system responding to the outlined changes in intracellular parameters could indirectly mediate nitrate regulation in B. subtilis.
Lactate and 2,3-butanediol formation was clearly detectable in the presence of nitrate and nitrite, indicating the importance of NADH reoxidation even for anaerobic respiratory growth. The obvious absence of a proton translocating NADH dehydrogenase of the Nuo type from the B. subtilis genome and the observed strictly aerobic expression of the non-proton-pumping NADH dehydrogenase of the Ndh type explain the observed general anaerobic lctEP and alsSD expression (13, 17; Marino et al., unpublished observations).
Both operons are additionally subject to AlsR regulation. The postulated signal(s) for AlsR activity is the intracellular pH and/or acetate or derived metabolites (29). The observed rapid anaerobic induction of alsSD and lctEP could result from the accumulation of acidic compounds like pyruvate and acetate. Further experiments should shed light on the complex regulatory systems involved in the control of anaerobic metabolism of B. subtilis.
Finally, acetate formation is a general part of the aerobic and anaerobic B. subtilis metabolism (27). In agreement with this observation, pta expression is not subject to redox regulation.
ACKNOWLEDGMENTS
This work was supported by grants from the Deutsche Forschungsgemeinschaft, the Max-Planck-Gesellschaft, the Sonderforschungsbereich 388, Fonds der Chemischen Industrie, and the Graduiertenkolleg “Biochemie der Enzyme.” E.-P.-S. is a fellow of the European Union Biotech Programme (contract ERBB102 CT930272). This research was also supported by grants from the Ministère de l'Education Nationale de la Recherche et de la Technologie, Centre National de la Recherche Scientifique (URA1129), Institut National de la Recherche Agronomique, Institut Pasteur, Université Paris 7, and European Union Biotech Programme (contracts ERBB102 CT930272 and ERBB104 CT960655).
We are indebted to M. Nakano (Louisiana State University) and F. M. Hulett (University of Illinois at Chicago) for the gift of B. subtilis mutants. We thank R. K. Thauer (Max-Planck-Institute, Marburg, Germany) for continuous support.
REFERENCES
- 1.Antelmann H, Bernhardt J, Schmid R, Mach H, Volker U, Hecker M. First steps from a two-dimensional protein index towards a response-regulation map for Bacillus subtilis. Electrophoresis. 1997;18:1451–1463. doi: 10.1002/elps.1150180820. [DOI] [PubMed] [Google Scholar]
- 2.Böck A, Sawers G. Fermentation. In: Neidhardt F C, et al., editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 262–282. [Google Scholar]
- 3.Chung C T, Miller R H. A rapid and convenient method for the preparation and storage of competent bacterial cells. Nucleic Acids Res. 1988;16:3580. doi: 10.1093/nar/16.8.3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cruz Ramos H, Boursier L, Moszer I, Kunst F, Danchin A, Glaser P. Anaerobic transcription activation in Bacillus subtilis: identification of distinct FNR-dependent and -independent regulatory mechanisms. EMBO J. 1995;14:5984–5994. doi: 10.1002/j.1460-2075.1995.tb00287.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Glaser P, Danchin A, Kunst F, Zuber P, Nakano M M. Identification and isolation of a gene required for nitrate assimilation and anaerobic growth of Bacillus subtilis. J Bacteriol. 1995;177:1112–1115. doi: 10.1128/jb.177.4.1112-1115.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grundy F J, Waters D A, Takova T Y, Henkin T M. Identification of genes involved in utilization of acetate and acetoin in Bacillus subtilis. Mol Microbiol. 1993;10:259–271. doi: 10.1111/j.1365-2958.1993.tb01952.x. [DOI] [PubMed] [Google Scholar]
- 7.Hagen F S, Young E T. Effect of RNase III on the size of bacteriophage T7 lysozyme mRNA. J Virol. 1978;26:783–792. doi: 10.1128/jvi.26.3.783-792.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoffmann T, Frankenberg N, Marino M, Jahn D. Ammonification in Bacillus subtilis utilizing dissimilatory nitrite reductase is dependent on resDE. J Bacteriol. 1998;180:186–189. doi: 10.1128/jb.180.1.186-189.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoffmann T, Troup B, Szabo A, Hungerer C, Jahn D. The anaerobic life of Bacillus subtilis. Cloning and characterization of the genes encoding the respiratory nitrate reductase system. FEMS Microbiol Lett. 1995;131:219–225. doi: 10.1111/j.1574-6968.1995.tb07780.x. [DOI] [PubMed] [Google Scholar]
- 10.Homuth G, Rompf A, Schumann W, Jahn D. Characterization of Bacillus subtilis hemZ. J Bacteriol. 1999;181:5922–5929. doi: 10.1128/jb.181.19.5922-5929.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang M, Oppermann-Sanio F B, Steinbüchel A. Biochemical and molecular characterization of the Bacillus subtilis acetoin catabolic pathway. J Bacteriol. 1999;181:3837–3841. doi: 10.1128/jb.181.12.3837-3841.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kunst F, Ogasawara N, Moszer I, Albertini A M, Alloni G, Azevedo V, Bertero M G, Bessieres P, Bolotin A, Borchert S, Borriss R, Boursier L, Brans A, Braun M, Briginell S C, Bron S, Brouillet S, Bruschi C V, Caldwell B, Capuano V, Carter N M, Choi S K, Codani J J, Connerton I F, Cummings N J, Danie R A, Denizot F, Devine K M, Dusterhoft A, Ehrlich S D, Emmerson P T, Entian K D, Errington J, Fabret C, Ferrari E, Foulger D, Fritz C, Fujita M, Fujita Y, Fuma S, Galizzi A, Galleron N, Ghim S Y, Glaser P, Goffeau A, Golightly E J, Grandi G, Guiseppi G, Guy B J, Haga K, Haiech J, Harwood C R, Hasahara Y, Henaut A, Hilbert H, Holsappel S, Hoson S, Hullo M F, Itaya M, Jones L, Joris B, Karamata D, Klaerr-Blanchard M, Klein C, Kobayashi Y, Koetter P, Koningstein G, Krogh S, Kumano M, Kurita K, Lapidus A, Lardinois S, Lauer J, Lazarevic V, Lee S M, Levine A, Liu H, Masuda S, Mauel C, Medigue C, Medina N, Medallo R P, Mizuno M, Moestl D, Nakai S, Noback M, Noone D, O'Reilly M, Ogawa K, Ogiwara A, Oudega B, Park S H, Parro V, Pohl T M, Portetelle D, Porwollik S, Prescott A M, Presecan E, Pujic P, Purnelle B, Rapoport G, Rey M, Reynolds S, Rieger M, Rivolta C, Rocha E, Roche B, Rose M, Sadaie Y, Sato T, Scanlan E, Schleich S, Schroeter R, Scoffone F, Sekiguchi J, Sekowska A, Seror S J, Serror P, Shin B S, Soldo B, Sorokin A, Tacconi E, Takagi T, Takahashi H, Takemaru K, Takeuchi M, Tamakoshi A, Tanaka T, Terpstra P, Tognini A, Tosato V, Uchiyama S, Vandenbol M, Vannier F, Vassarotti A, Viari A, Wambutt R, Wedler E, Wedler H, Weitzenegger T, Winters P, Wipat A, Yamamoto H, Yamane K, Yasumoto K, Yata K, Yoshida K, Yoshikawa H, Yoshikawa H F, Zumstein E, Danchin A. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature. 1998;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 13.Kunst F, Rapoport G. Salt stress is an environmental signal affecting degradative enzyme synthesis in Bacillus subtilis. J Bacteriol. 1995;177:2403–2407. doi: 10.1128/jb.177.9.2403-2407.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mach H, Hecker M, Mach F. Physiological studies on cAMP synthesis in Bacillus subtilis. FEMS Microbiol Lett. 1988;52:189–192. [Google Scholar]
- 15.Martin-Verstraete I, Debarbouille M, Klier A, Rapoport G. Mutagenesis of the Bacillus subtilis “−12, −24” promoter of the levanase operon and evidence for the existence of an upstream activating sequence. J Mol Biol. 1992;226:85–89. doi: 10.1016/0022-2836(92)90126-5. [DOI] [PubMed] [Google Scholar]
- 16.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 17.Moszer I, Glaser P, Danchin A. SubtiList: a relational database for the Bacillus subtilis genome. Microbiology. 1995;141:261–268. doi: 10.1099/13500872-141-2-261. [DOI] [PubMed] [Google Scholar]
- 18.Murphy E. Nucleotide sequence of a spectinomycin adenyltransferase AAD(9) determinant from Staphylococcus aureus and its relationship to AAD(3")(9) Mol Gen Genet. 1985;200:33–39. doi: 10.1007/BF00383309. [DOI] [PubMed] [Google Scholar]
- 19.Nakano M M, Dailly Y P, Zuber P, Clark D P. Characterization of anaerobic fermentative growth in Bacillus subtilis: identification of fermentation end products and genes required for the growth. J Bacteriol. 1997;179:6749–6755. doi: 10.1128/jb.179.21.6749-6755.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakano M M, Hoffmann T, Zhu Y, Jahn D. Nitrogen and oxygen regulation of Bacillus subtilis nasDEF encoding NADH-dependent nitrite reductase by TnrA and ResDE. J Bacteriol. 1998;180:5344–5350. doi: 10.1128/jb.180.20.5344-5350.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakano M M, Hulett F M. Adaptation of Bacillus subtilis to oxygen limitation. FEMS Microbiol Lett. 1997;157:1–7. doi: 10.1111/j.1574-6968.1997.tb12744.x. [DOI] [PubMed] [Google Scholar]
- 22.Nakano M M, Zuber P. Anaerobic growth of a “strict aerobe” (Bacillus subtilis) Annu Rev Microbiol. 1998;52:165–190. doi: 10.1146/annurev.micro.52.1.165. [DOI] [PubMed] [Google Scholar]
- 23.Nakano M M, Zuber P, Glaser P, Danchin A, Hulett M. Two-component regulatory proteins ResD-ResE are required for transcriptional activation of fnr upon oxygen limitation in Bacillus subtilis. J Bacteriol. 1996;178:3796–3802. doi: 10.1128/jb.178.13.3796-3802.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ogawa K, Akagawa E, Yamane K, Sun Z W, LaCelle M, Zuber P, Nakano M M. The nasB operon and nasA gene are required for nitrate/nitrite assimilation in Bacillus subtilis. J Bacteriol. 1995;177:1409–1413. doi: 10.1128/jb.177.5.1409-1413.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perego M. Integrational vectors for genetic manipulation in Bacillus subtilis. In: Sonenshein A L, Hoch J A, Losick R, editors. Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. Washington, D.C.: American Society for Microbiology; 1993. pp. 615–624. [Google Scholar]
- 26.Pikielny C W, Rosbash M. mRNA splicing efficiency in yeast and the contribution of nonconserved sequences. Cell. 1985;41:119–126. doi: 10.1016/0092-8674(85)90066-2. [DOI] [PubMed] [Google Scholar]
- 27.Presecan-Siedel E, Galinier A, Longin R, Deutscher J, Danchin A, Glaser P, Martin-Verstraete I. The catabolite regulation of the pta gene as part of the carbon flow pathways in Bacillus subtilis. J Bacteriol. 1999;181:6889–6897. doi: 10.1128/jb.181.22.6889-6897.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Presecan E, Moszer I, Boursier L, Cruz Ramos H C, de la Fuente V, Hullo M F, Lelong C, Schleich S, Sedowska A, Song B H, Villani G, Kunst F, Danchin A, Glaser P. The Bacillus subtilis genome from gerBC (311 degrees) to licR (334 degrees) Microbiology. 1997;143:3318–3328. doi: 10.1099/00221287-143-10-3313. [DOI] [PubMed] [Google Scholar]
- 29.Renna M C, Najimudin N, Winik L R, Zahler S A. Regulation of the Bacillus subtilis alsS, alsD, and alsR genes involved in post-exponential-phase production of acetoin. J Bacteriol. 1993;175:3863–3875. doi: 10.1128/jb.175.12.3863-3875.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 31.Schirawski J, Unden G. Anaerobic respiration of Bacillus macerans with fumarate, TMAO, nitrate and nitrite and regulation of the pathways by oxygen and nitrate. Arch Microbiol. 1995;163:148–154. [Google Scholar]
- 32.Shin B S, Choi S K, Park S H. Regulation of the Bacillus subtilis phosphotransacetylase gene. J Biochem. 1999;126:333–339. doi: 10.1093/oxfordjournals.jbchem.a022454. [DOI] [PubMed] [Google Scholar]
- 33.Sun G, Sharkova E, Chesnut R, Birkey S, Duggan M F, Sorokin A, Pujic P, Ehrlich S D, Hulett F M. Regulators of aerobic and anaerobic respiration in Bacillus subtilis. J Bacteriol. 1996;178:1374–1385. doi: 10.1128/jb.178.5.1374-1385.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Unden G, Becker S, Bongaerts J, Schirawski J, Six S. Oxygen regulated gene expression in facultatively anaerobic bacteria. Antonie Leeuvenhoek. 1994;66:3–22. doi: 10.1007/BF00871629. [DOI] [PubMed] [Google Scholar]
- 35.Unden G, Becker S, Bongaerts J, Holighaus G, Schirawski J, Six S. O2-sensing and O2-dependent gene regulation in facultatively anaerobic bacteria. Arch Microbiol. 1995;164:81–90. [PubMed] [Google Scholar]
- 36.Yamane K, Kumano M, Kurita K. The 25 degrees-36 degrees region of the Bacillus subtilis chromosome: determination of the sequence of a 146 kb segment and identification of 113 genes. Microbiology. 1996;142:3047–3056. doi: 10.1099/13500872-142-11-3047. [DOI] [PubMed] [Google Scholar]
- 37.Yashphe J, Hoch J A, Kaplan N O. Regulation of lactate dehydrogenase synthesis in Bacillus subtilis. Biochim Biophys Acta. 1978;544:1–7. doi: 10.1016/0304-4165(78)90203-9. [DOI] [PubMed] [Google Scholar]