Abstract
FnrL, the homolog of the global anaerobic regulator Fnr, is required for the induction of the photosynthetic apparatus in Rhodobacter sphaeroides 2.4.1. Thus, the precise role of FnrL in photosynthesis (PS) gene expression and its interaction(s) with other regulators of PS gene expression are of considerable importance to our understanding of the regulatory circuitry governing spectral complex formation. Using a CcoP and FnrL double mutant strain, we obtained results which suggested that FnrL is not involved in the transduction of the inhibitory signal, by which PS gene expression is “silenced,” emanating from the cbb3 oxidase encoded by the ccoNOQP operon under aerobic conditions. The dominant effect of the ccoP mutation in the FnrL mutant strain with respect to spectral complex formation under aerobic conditions and restoration of a PS-positive phenotype suggested that inactivation of the cbb3 oxidase to some extent bypasses the requirement for FnrL in the formation of spectral complexes. Additional analyses revealed that anaerobic induction of the bchE, hemN, and hemZ genes, which are involved in the tetrapyrrole biosynthetic pathways, requires FnrL. Thus, FnrL appears to be involved at multiple loci involved in the regulation of PS gene expression. Additionally, bchE was also shown to be regulated by the PrrBA two-component system, in conjunction with hemN and hemZ. These and other results to be discussed permit us to more accurately describe the role of FnrL as well as the interactions between the FnrL, PrrBA, and other regulatory circuits in the regulation of PS gene expression.
Rhodobacter sphaeroides 2.4.1 is a purple nonsulfur photosynthetic bacterium which displays a versatile metabolic lifestyle. It is able to grow by aerobic and anaerobic respiration and photosynthetically in the light under anaerobic conditions. The major determinant for the synthesis of the photosynthetic apparatus is oxygen. When oxygen tensions fall below a certain threshold value (∼2.5%), the photosynthetic apparatus is synthesized. In addition, the levels and the ratios of the light-harvesting complexes are determined by the incident light intensity.
Transcriptional regulation of the photosynthesis (PS) genes in R. sphaeroides involves the coordinate action of at least four major regulatory systems (the PrrBA two-component activation system, the AppA-PpsR antirepressor-repressor system, FnrL, and TspO) which are responsible for redox sensing (25). Previous studies in our laboratory have shown that mutations in the ccoNOQP operon encoding the cbb3 oxidase result in the induction of PS gene expression under aerobic growth conditions (18, 19, 28). This observation was interpreted to suggest that a pathway involving the cbb3 oxidase in response to O2 serves as an oxygen sensor which generates an inhibitory signal for PS gene expression. This signal is mediated by the PrrBA two-component regulatory system, which is a major switch controlling PS gene expression (17).
However, we could not rule out the possibility that FnrL, at least in part, is somehow involved in transmission of the inhibitory signal emanating from the cbb3 oxidase, since expression of the puc operon and hemA gene, which are in part regulated by FnrL, is partially derepressed in the Cco mutants under aerobic conditions (18, 27–29).
The construction of a CcoP and FnrL double mutant enabled us to answer this question. In this study, we demonstrate that FnrL does not mediate the derepression signal derived from the inactivation of the cbb3 oxidase under aerobic conditions. Furthermore, we do present evidence that the bchE, hemN, and hemZ genes, which have previously been identified as potential targets on the basis of sequence analyses of their upstream regions for regulation by FnrL, are in fact regulated by both FnrL and the PrrBA two-component system. These observations, as well as earlier results, will be discussed in conjunction with the role of FnrL and its relation to other regulatory circuits in controlling PS gene expression.
MATERIALS AND METHODS
Strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. R. sphaeroides 2.4.1 strains were grown at 30°C on Sistrom's medium A (SIS) (3) containing succinate as the carbon source and supplemented, as required, with the following antibiotics: tetracycline, 1 μg/ml; kanamycin, 25 μg/ml; trimethoprim, 50 μg/ml; and streptomycin and spectinomycin, 50 μg/ml each. Chemoheterotrophic cultures were grown either aerobically, sparged with 30% O2–69% N2–1% CO2, or semiaerobically, sparged with 2% O2–97% N2–1% CO2. Photosynthetic cultures were grown at a low incident light intensity of 3 W/m2 in completely filled screw-cap glass tubes. The tubes containing photosynthetic cultures were rotated by using a rotary drum to keep cells suspended and mixed. Photosynthetic growth of R. sphaeroides was monitored with a Klett-Summerson colorimeter with a no. 66 filter. Anaerobic growth with dimethyl sulfoxide (DMSO) as a terminal electron acceptor was performed in SIS medium supplemented with 0.1% (wt/vol) yeast extract in the presence of DMSO (0.5% vol/vol) in screw-cap tubes in the dark. Escherichia coli strains were grown at 37°C on Luria-Bertani medium supplemented, when required, with the following antibiotics: ampicillin, 50 μg/ml; kanamycin, 50 μg/ml; tetracycline, 20 μg/ml; and streptomycin and spectinomycin, 50 μg/ml each.
TABLE 1.
Bacterial strains and plasmids used in this work
| Strain or plasmid | Relevant phenotype or genotype | Source or reference |
|---|---|---|
| Strains | ||
| R. sphaeroides | ||
| 2.4.1 | Wild type | 23 |
| FNRL | 2.4.1 derivative, ΔfnrL::ΩKmr; formerly JZ1678 | 27 |
| CCOP1 | 2.4.1 derivative, ccoP::ΩTpr | 18 |
| CCOP1FNRL | FNRL derivative, ΔfnrL::ΩKmrccoP::ΩTpr | This study |
| PRRBCA2 | 2.4.1 derivative, ΔprrCBA::ΩTpr | This study |
| E. coli | ||
| DH5α | (φ80dlacZΔM15) ΔlacU169 recA1 endA1 hsdR17 supE44 thi-1 gyrA96 relA1 | 8 |
| S17-1 | Pro− Res− Mod+recA; integrated plasmid RP4-Tc::Mu-Km::Tn7 | 21 |
| Plasmids | ||
| pCF1010 | Spr Str Tcr; lacZYA′ IncQ | 10 |
| pCFL | pCF1010 in which ∼1.5-kb HindIII fragment containing Tc resistance gene is deleted; Spr Str | M. Gomelsky |
| pSUP203 | pBR325 derivative; Mob+ Apr Cmr Tcr | 21 |
| pUI1637 | pUI1188 containing the ΩKm | 6 |
| pUI1680 | pUI1637 with a 1,017-bp BglII-NruI deletion replaced with a ∼1.7-kb EcoRI-DraI fragment from pSUP5TpMCS containing a gene encoding Tpr | This study |
| pJE2077 | pSUP203 derivative containing a 5,232-bp fragment with the prr region, with a 2,069-bp Tth111I-BspEI deletion replaced with an ΩTp | This study |
| pJE3153 | pBSII SK+::0.75-kb SmaI fragment with the hemZ control region from pUI1970 (26) | This study |
| pUI2801 | ccoP::ΩTp in pSUP203 | 18 |
| pCF200Km | Spr Str Kmr; IncQ, puc::lacZYA′ | 9 |
| pUI1063 | Spr Str Tcr; IncQ, hemA::lacZYA′ | 27 |
| pJE3170 | Spr Str Tcr; IncQ, hemZ::lacZYA′ | This study |
| pUI2762 | Spr Str; IncQ, hemN::lacZYA′ | 24 |
| pBCHE | Spr Str; IncQ, bchE::lacZYA′ | This study |
DNA manipulations and conjugation techniques.
Genomic DNA from R. sphaeroides was isolated by the method of Ausubel et al. (1). Standard protocols or the manufacturer's instructions were followed for recombinant DNA manipulations (20).
Plasmids were mobilized by either biparental or triparental matings from E. coli strains into R. sphaeroides strains as described elsewhere (5).
Construction of mutants and lacZ transcriptional fusions.
For the construction of the CCOP1FNRL double mutant, the ccoP gene was mutated in the FNRL background using plasmid pUI2801, which had been used for the construction of the original CCOP1 mutant (18).
PrrBCA2 was constructed by crossing pJE2077 into R. sphaeroides 2.4.1 and selecting for the double-crossover recombinant. Plasmid pJE2077 is a suicide vector in R. sphaeroides, and it contains the prr region, with a 2,069-bp deletion extending from a Tth111I site within prrA to a BspEI site within prrB. In addition, the ΩTp cassette from pUI1680 was cloned within these boundaries as a selectable marker. The structure was verified by Southern hybridization.
In order to construct the bchE::lacZ transcriptional fusion, the bchE promoter region was amplified with primers 5′-CTGTCGCTGCAGTCGCTTTATGTG-3′ and 5′-CGGCCTTCTAGAGCGCGCCACACA-3′ to generate a 318-bp product with PstI and XbaI restriction sites at both ends. The PCR product was restricted with PstI and XbaI and cloned into the promoterless lacZ vector pCFL digested with the same enzymes, yielding plasmid pBCHE. The sequence of the insert in pBCHE was confirmed by DNA sequencing.
To construct the hemZ::lacZ transcriptional fusion, the 0.75-kb EcoRV-XbaI fragment containing the promoter region of hemZ was cloned into pCF1010 restricted with StuI and XbaI from pJE3153 to give plasmid pJE3170.
Quantitative analysis of spectral complexes.
The harvested cells were resuspended in ICM buffer (10 mM KH2PO4/K2HPO4, 1 mM EDTA [pH 7.2]) and disrupted by passage through a French pressure cell (ca. 0.9-cm-diameter piston) at 90 MPa. Cell-free crude extracts were obtained following centrifugation in a benchtop microcentrifuge at 13,000 rpm for 15 min at 4°C to remove unbroken cells and cell debris. Spectra were recorded with a UV 1601PC spectrophotometer (Shimadzu Corp., Columbia, Md.). The B800-850 and B875 complex levels were determined by the method of Meinhardt et al. (12) from the spectral data collected.
Crt and Bchl analyses.
For extraction of photopigments, 0.5 ml of crude cell extract was mixed with 1.5 ml of acetone-methanol (7:2, vol/vol), followed by vigorous vortexing for 1 min. After centrifugation in a benchtop microcentrifuge at 13,000 rpm for 15 min, the supernatant was used for quantitation of carotenoid (Crt) and bacteriochlorophyll (Bchl) as described previously (3).
β-Galactosidase assays and protein determinations.
β-Galactosidase assays were performed on crude cell extracts as described elsewhere (19). Protein concentration was determined by the bicinchoninic acid protein assay (Pierce, Rockford, Ill.) using bovine serum albumin as the standard protein.
RESULTS AND DISCUSSION
Formation of spectral complexes in the CCOP1FNRL double mutant under aerobic conditions.
In order to address the question of whether the signal emanating from the cbb3 oxidase involves the participation of the FnrL protein, a double mutant carrying mutations in both fnrL and ccoP was constructed (strain CCOP1FNRL). When grown on SIS agar plates aerobically, the double mutant formed colonies with red pigmentation, the extent of which was intermediate between the appearance of the wild type and the high levels of pigmentation of the CCOP1 mutant (18). To confirm that the increased pigmentation of the CCOP1FNRL mutant under aerobic conditions was due to the presence of photosynthetic spectral complexes, crude extracts from cultures of this mutant strain grown aerobically were prepared, and the levels of the light-harvesting complexes (B800-850 and B875) as well as the photopigments Bchl and Crt were determined spectrophotometrically (Table 2). For comparison, crude extracts were also prepared from the wild-type, CCOP1, and FNRL strains grown under the same conditions. As expected, the wild-type and FNRL mutant strains produced virtually no light-harvesting complexes under aerobic conditions. In contrast, the CCOP1FNRL mutant strain exhibited 50 and 64% of the levels of the B800-850 and B875 complexes, respectively, relative to the levels which were detected in the CCOP1 mutant. In agreement with these results, the cellular levels of the photopigments Bchl and Crt also increased in both CCOP1 and CCOP1FNRL strains compared with those levels in the wild-type and FNRL strains. These data demonstrated that the mutation in ccoP led to the oxygen-insensitive formation of the photosynthetic complexes even in the FnrL-minus background. It should be recalled that an FnrL mutant strain of R. sphaeroides cannot produce spectral complexes even under inducing conditions such as 2% oxygen (26).
TABLE 2.
Levels of spectral complexes and photopigments in R. sphaeroides strains grown under aerobic conditionsa
| Strain | Spectral complexes (nmol/mg of protein)
|
Photopigments (μg/mg of protein)
|
||
|---|---|---|---|---|
| B800-850 | B875 | Bchl | Crt | |
| 2.4.1 | 0.1 ± 0.0 | <0.05 | <0.05 | 0.4 ± 0.1 |
| CCOP1 | 0.4 ± 0.1 | 4.5 ± 0.2 | 1.3 ± 0.2 | 1.0 ± 0.1 |
| FNRL | 0.1 ± 0.0 | <0.05 | <0.05 | 0.4 ± 0.1 |
| CCOP1FNRL | 0.2 ± 0.0 | 2.9 ± 0.2 | 0.9 ± 0.1 | 0.8 ± 0.2 |
Strains were grown aerobically by sparging with 30% O2–69% N2–1% CO2 to an OD600 of 0.4 to 0.5. All values are the averages of two independent determinations.
The reduced levels of spectral complexes in the CCOP1FNRL mutant grown aerobically compared with those in the CCOP1 mutant grown under the same conditions suggested that FnrL of R. sphaeroides is “active” under aerobic conditions. This had been predicted previously after the observation that R. sphaeroides FnrL contains the two residues which, when changed in E. coli Fnr, enable the latter protein to be active under aerobic conditions of growth, which is not the normal physiological situation (27). This finding is in agreement with a similar conclusion which was reached under different experimental conditions by Zeilstra-Ryalls and Kaplan (27, 29). Mouncey and Kaplan (14) reported that FnrL is a positive regulator of genes encoding the cbb3 oxidase. The fact that the FNRL mutant is incapable of spectral complex formation under aerobic conditions indicates that sufficient cbb3 oxidase is produced in order to be able to repress spectral complex formation in this mutant under the same conditions.
FnrL is not involved in the cbb3-mediated signal transduction pathway under aerobic conditions.
Previous studies revealed that mutations of the cco operon in R. sphaeroides lead to the aerobic derepression of not only those PS genes regulated by the PrrBA two-component system, such as puc and puf, but also hemA, which encodes the 5-aminolevulinic acid (ALA) synthase and which is, at least in part, under the control of FnrL (6, 18, 28). An interpretation of these results raised the possibility that FnrL is an additional regulator which communicates with the cbb3 oxidase. To address this question, we compared the promoter activities of genes which are both regulated by FnrL and derepressed by mutations in the cco operon under aerobic conditions. These experiments were performed with the wild-type, CCOP1, and CCOP1FNRL mutant strains grown under aerobic conditions. We speculated that if FnrL mediates the derepression signal resulting from inactivation of the cbb3 oxidase under aerobic conditions, then the derepression of PS gene expression observed in the CCOP1 mutant strain should be abolished in the CCOP1FNRL mutant. As shown in Table 3, the puc operon and hemA gene were strongly derepressed under aerobic conditions in the CCOP1 mutant by factors of 2.8- and 7.2-fold, respectively, compared with the wild type. The disruption of fnrL in the background of the CCOP1 mutant resulted in a further 1.4- and 2.3-fold increase in puc and hemA promoter activities, respectively, compared with the CCOP1 mutant. This result suggested that FnrL does not mediate the derepression signal resulting from inactivation of the cbb3 oxidase under aerobic conditions, which is in good agreement with the oxygen-insensitive formation of the spectral complexes in the CCOP1FNRL mutant under aerobic conditions. The higher promoter activity of the hemA gene in the CCOP1FNRL mutant grown under aerobic conditions than in the CCOP1 mutant can be explained by the fact that the transcription start point of hemA under aerobic conditions lies within the predicted FnrL binding motif (16), and therefore suggests that FnrL might be a repressor of hemA under aerobic conditions. In the case of increased puc operon expression, the FnrL binding sequence overlaps the integration host factor binding sequence, and removal of FnrL could permit enhanced activation of puc operon expression by integration host factor (11). These results raise an additional question. If hemA and puc operon expression is increased in the CCOP1FNRL mutant background, why do we not see at least the level of spectral complexes in the double mutant as observed for the CCOP1 mutant? One possible answer is that there might be some PS genes which are regulated by FnrL and which are not derepressed by the inactivation of the cbb3 oxidase under aerobic conditions. Thus, these gene products might be limiting factors for spectral complex formation.
TABLE 3.
Promoter activities of reporter genes in R. sphaeroides strains grown under aerobic conditionsa
| Strain | β-Galactosidase activity (nmol/min/mg of protein)
|
||
|---|---|---|---|
| puc::lacZ | hemA::lacZ | bchE::lacZ | |
| 2.4.1 | 49.4 ± 1.9 | 6.1 ± 0.5 | 4.3 ± 1.7 |
| CCOP1 | 136.3 ± 4.2 | 44.0 ± 3.3 | 73.8 ± 8.1 |
| CCOP1FNRL | 186.6 ± 1.0 | 99.8 ± 1.4 | 68.8 ± 5.0 |
Strains were grown aerobically by sparging with 30% O2–69% N2–1% CO2 to an OD600 of 0.4 to 0.5. Plasmids pCF200Km, pUI1063, and pBCHE, carrying transcriptional fusions puc::lacZ, hemA::lacZ, and bchE::lacZ, respectively, were used to determine the promoter activities of the corresponding gene. All values are the averages of two independent determinations.
Regulation of the bchE gene.
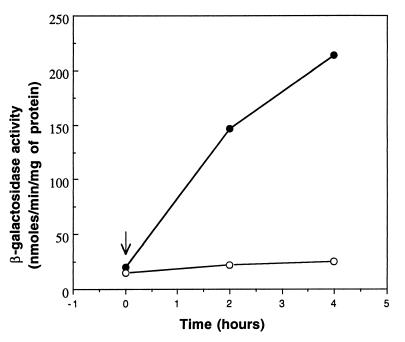
A putative FnrL-binding motif is located 53 nucleotides upstream of the translational start codon of the bchE gene, whose product catalyzes the conversion of Mg-protoporphyrin monomethyl ester to divinyl-protochlorophyllide in the Bchl biosynthetic pathway. The presence of an FnrL-binding motif upstream of the bchE gene prompted us to examine whether this gene is regulated by FnrL. To address this question, we assayed the promoter activity of bchE in the FnrL-minus background. Since the FNRL mutant is unable to grow under anaerobic conditions, we performed an oxygen shift experiment in which cultures grown with 30% oxygen were shifted to 2% oxygen (Fig. 1). At the time intervals indicated, samples were harvested, and their β-galactosidase activities were determined. In the wild-type strain, the levels of bchE::lacZ expression increased steadily and appreciably after the cultures were shifted to 2% oxygen. In contrast, the promoter activity of bchE in the FNRL mutant showed a marginal increase up to 4 h after the shift to 2% oxygen. This indicated a clear requirement for FnrL for the induction of bchE expression in response to lowering of the oxygen tension.
FIG. 1.
Induction kinetics of bchE expression from the bchE::lacZ transcriptional fusion plasmid pBCHE in the wild-type strain (●) and FNRL mutant strain (○) following a shift from 30 to 2% oxygen (indicated by the vertical arrow). Strains were grown under 30% O2 to an optical density at 600 nm (OD600) of 0.15 to 0.2, and the partial pressure of O2 was reduced to 2%. Cultures were sampled at the times indicated, and crude extracts were assayed for β-galactosidase activity.
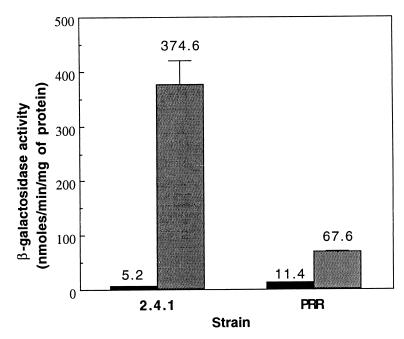
A determination of bchE promoter activity using the bchE::lacZ transcriptional fusion revealed that expression of bchE was substantially increased under aerobic conditions (30% O2) in the CCOP1 and CCOP1FNRL mutants compared with that in the wild type (Table 3). The derepression of bchE in the CCOP1 mutant under aerobic conditions also indicated the involvement of the PrrBA two-component system in the regulation of bchE. To examine this possibility, we measured the promoter activities of bchE in the PrrCBA-minus background (PRRBCA2 mutant) as well as in the wild-type strain. Under 30% oxygen conditions, both wild-type and mutant strains exhibited basal levels of promoter activity (Fig. 2). In the wild type grown under dark-DMSO conditions, the expression of bchE was induced by a factor of 72 compared to that found in the same strain grown aerobically. In contrast, the promoter activity increased only sixfold in the PRRBCA2 mutant strain grown under dark-DMSO conditions. Taken together, these results clearly suggest that bchE is regulated by both FnrL and the PrrBA two-component system. They are both needed for the full induction of bchE under oxygen-limiting conditions, although either of them alone can induce the gene only partially. The approximately sixfold induction of bchE under dark-DMSO conditions in the PrrBCA-minus background suggests that FnrL might produce an approximately sixfold increase in bchE gene expression under the same conditions.
FIG. 2.
β-Galactosidase activities in R. sphaeroides strains containing the bchE::lacZ transcriptional fusion plasmid pBCHE. The wild-type (2.4.1) and prrCBA-minus (PRR) strains were grown aerobically (30% oxygen, black bars) to an OD600 of 0.3 to 0.4 or anaerobically in the dark with 0.5% DMSO (gray bars). All values are the averages of two independent determinations. Vertical bars represent the standard deviations from the mean.
Regulation of the hemZ and hemN genes.
Sequence analyses have also revealed the presence of FnrL binding sequences 35 and 34 bp upstream of the presumed translational start codons of the hemN and hemZ genes, respectively, encoding isoenzyme forms of the coproporphyrinogen III oxidase involved in the tetrapyrrole biosynthetic pathway. Previous work by Yeliseev and Kaplan (24) revealed that both genes are derepressed under semiaerobic and anaerobic conditions and that the hemN gene appears to encode a form of the enzyme that may be active aerobically. To examine the role of FnrL in the regulation of hemZ and hemN, the corresponding transcriptional lacZ fusions were introduced into the wild-type strain 2.4.1 as well as the FNRL mutant strain. The strains were grown either under 30% oxygen conditions or under 2% oxygen conditions, in which many of the genes known to be under the control of FnrL appear to be fully induced.
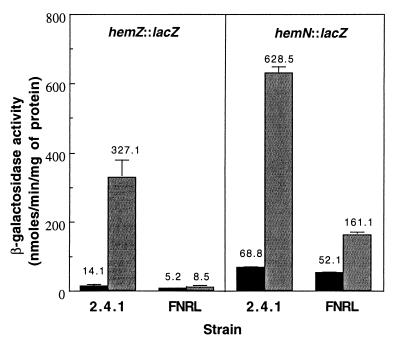
The expression of hemZ is strictly regulated by FnrL (Fig. 3). Under 30% oxygen conditions, the promoter activity of hemZ is at basal levels in both the wild-type and FNRL mutant strains. A 23-fold increase in expression of hemZ was observed in the wild type grown under 2% oxygen conditions, whereas the gene was not induced in the FNRL mutant grown under the same conditions, which is indicative of the essential role of FnrL in the induction of hemZ. These results are in agreement with those of Yeliseev and Kaplan (24), suggesting that hemZ encodes an anaerobic coproporphyrinogen III oxidase. Furthermore, the PrrBA two-component system is in part responsible for the induction of this gene under anaerobic conditions (J. M. Eraso and S. Kaplan, unpublished data). Although the hemZ gene is positively regulated by the PrrBA two-component system, derepression of the gene under 30% oxygen conditions by the inactivation of the cbb3 oxidase was not observed in the FnrL-minus background, which confirms the absolute requirement for FnrL for the expression of hemZ (Table 4).
FIG. 3.
β-Galactosidase activities in R. sphaeroides strains containing either the hemZ::lacZ or hemN::lacZ transcriptional fusion plasmid. The wild-type (2.4.1) and fnrL-minus (FNRL) strains were grown aerobically (30% oxygen, black bars) to an OD600 of 0.3 to 0.4 or semiaerobically (2% oxygen, gray bars) to an OD600 of 0.4 to 0.5. All values provided are the averages of two independent determinations. Vertical bars represent the standard deviations from the mean.
TABLE 4.
Promoter activities of hemZ and hemN in R. sphaeroides FNRL and CCOP1FNRL mutants grown under aerobic conditionsa
| Strain | β-Galactosidase activity (nmol/min/mg of protein)
|
|
|---|---|---|
| hemZ::lacZ | hemN::lacZ | |
| FNRL | 5.2 ± 2.0 | 52.1 ± 1.8 |
| CCOP1FNRL | 5.2 ± 1.0 | 195.2 ± 1.4 |
Strains were grown aerobically by sparging with 30% O2–69% N2–1% CO2 to an OD600 of 0.4 to 0.5. Plasmids pJE3170 and pUI2762 carrying transcriptional fusions hemZ::lacZ and hemN::lacZ, respectively, were used to determine promoter activities of the corresponding gene. All values are the averages of two independent determinations.
As shown in Fig. 3, these same experiments were performed with the hemN::lacZ fusion. When the promoter activity of hemN was compared in strains grown under 2% oxygen conditions with that found in the strains grown under 30% oxygen, a ninefold induction of hemN expression was observed in the wild-type strain grown under 2% oxygen conditions. Only a threefold induction was found in the FNRL mutant grown under the same conditions. The promoter activity found in the FNRL mutant grown under 2% oxygen conditions amounted to ∼25% of that present in the wild-type strain grown under the same conditions. Furthermore, a significant level of hemN promoter activity was detected in both strains even under 30% oxygen conditions compared with that of hemZ. This finding is also in agreement with that of Yeliseev and Kaplan, suggesting that hemN is both an aerobically and anaerobically active enzyme (24). Taken together, these results indicated that hemN is also positively regulated by FnrL under oxygen-limiting conditions and that the gene is less stringently regulated by FnrL than is hemZ. The threefold induction of hemN under 2% oxygen conditions in the FnrL-minus background suggests that another regulatory system(s) is involved in the regulation of the gene. In fact, the anaerobic induction of hemN in part requires the PrrBA two-component system (Eraso and Kaplan, unpublished data). Furthermore, the hemN gene is derepressed under 30% O2 conditions in the CCOP1FNRL mutant, again indicating that FnrL is not essential for the expression of the gene (Table 4).
We can probably rule out the regulation of hemN by the PpsR repressor, since there is no PpsR binding sequence upstream of the hemN gene.
Regulation of the tetrapyrrole biosynthetic pathway.
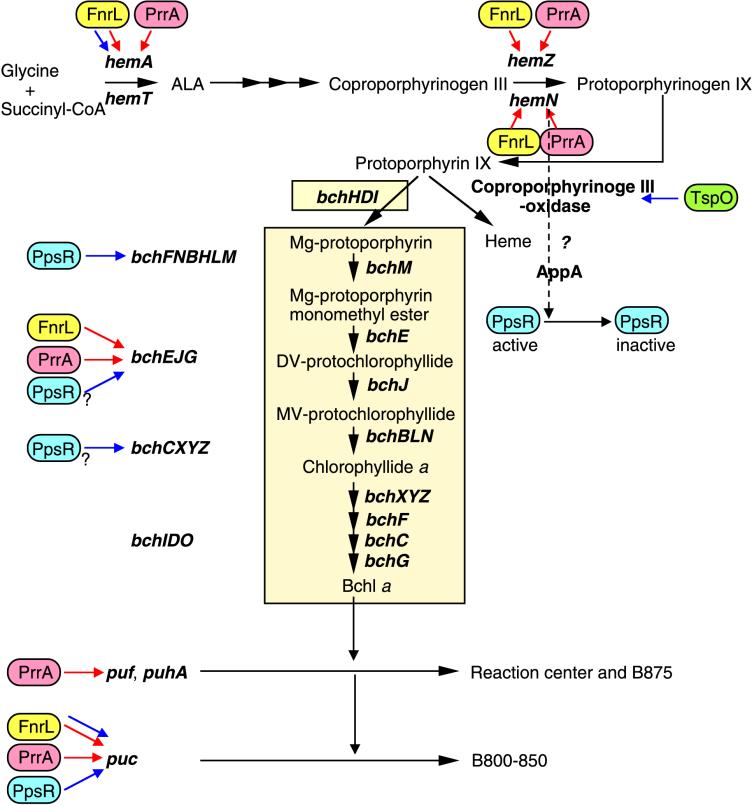
As outlined in Fig. 4, both the Bchl and heme biosynthetic pathways have the same intermediates from ALA through protoporphyrin IX, where they branch (22). Hemes are absolutely required for both aerobic and anaerobic growth of R. sphaeroides. In contrast, the synthesis of Bchl should be inhibited in cells grown under high-oxygen conditions, since free Bchl in the presence of light and O2 produces toxic free radicals and the expression of the puf and puc operons is minimal under these conditions. In fact, the levels of detectable Bchl found in R. sphaeroides grown under high-oxygen conditions is minimal. Therefore, the “opening” of the pathway from protoporphyrin IX to Bchl a must be stringently controlled and exquisitely responsive to the levels of O2 as well as light present in the environment. The results presented here and those published earlier (25) indicate that R. sphaeroides has developed overlapping regulatory circuits to enable such controls (Fig. 4). R. sphaeroides has two isoenzymes for the synthesis of ALA (HemA and HemT) (15, 16) and two coproporphyrinogen III oxidases (HemN and HemZ) (4, 27), which have been identified as points of FnrL regulation in the tetrapyrrole biosynthetic pathway prior to protoporphyrin IX. The hemT product was shown to be fully functional under both aerobic and anaerobic conditions in the absence of HemA (in the HemA-minus mutant), and hemT seems not to be regulated by FnrL due to the lack of an observed FnrL-binding motif in the control region (15, 16).
FIG. 4.
Bchl and heme biosynthetic pathways. The relevant genes, the regulation of which is either established or predicted on the basis of sequence analyses, are depicted. The question mark to the right of a regulator symbol indicates that regulation by the corresponding regulator is inferred from sequence analyses and is therefore not proven experimentally. The Bchl biosynthetic branch is shown in a yellow box. To the left of the yellow box, the regulation of four inferred operons which are involved in Bchl biosynthesis is represented. The red and blue arrows represent induction under oxygen-limiting conditions (semiaerobic or anaerobic conditions) and repression under high-oxygen conditions, respectively. The outer membrane TspO protein affects the expression of PpsR target genes by acting on HemN posttranscriptionally (24). At the bottom, the regulation of the puc and puf operons as well as the puhA gene, which encode the structural polypeptides of the reaction center and light-harvesting complexes, is presented. Abbreviations: MV, monovinyl; DV, divinyl; CoA, coenzyme A.
As shown above, the hemN gene is transcribed at quite significant levels under aerobic conditions, and although clearly under FnrL control, it is less stringently regulated by FnrL than is hemZ. This observation helps to explain the “less stringent” regulation of heme biosynthesis than of Bchl. In addition, hemA, hemZ, and hemN are also regulated by the PrrBA system, and from the data provided here, we can provide some quantitative measures as to the contributions of each of these systems. Thus, for the first time we begin to describe an overlapping regulatory network in the control of PS gene expression in R. sphaeroides.
With regard to the regulation of the Bchl biosynthetic branch, at least one additional regulatory point was reported in R. sphaeroides. The repressor PpsR represses the bchF gene, whose product catalyzes the conversion of chlorophyllide a to 2-hydroxyethyl bacteriochlorophyllide a under aerobic conditions (oxidized conditions) (7). In addition, sequence analyses of the PS gene cluster of R. sphaeroides, whose sequence was recently determined, revealed that there are PpsR binding sequences 16 bp upstream of the bchE gene as well as 72 and 98 bp upstream of the bchC genes (2). The genes involved in the Bchl biosynthetic branch appear to be clustered into four transcriptional units (bchFNBHLM, bchEJG, bchCXYZ, and bchLDO), which also suggests that regulation of the bchC, bchE, bchL, and bchF genes governs the expression of the corresponding downstream genes. Considering the data presented above, we conclude that the stringent control of the biosynthesis of Bchl appears to be achieved throughout the Bchl biosynthetic branch by the coordinate action of the regulators FnrL, AppA-PpsR, and the PrrBA two-component activation system without interrupting the flow of tetrapyrrole intermediates to heme, depending on the availability of oxygen, light intensity, and/or the cellular redox state (Fig. 4).
Inactivation of the cbb3 oxidase bypasses the requirement for FnrL in the formation of spectral complexes.
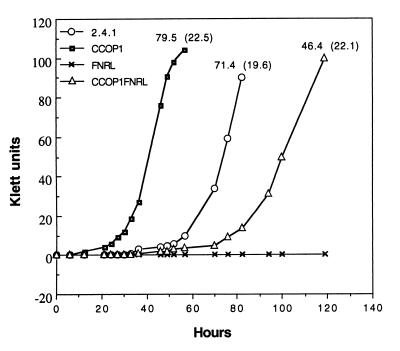
Spectral complex formation under inducing conditions, such as 2% oxygen, is severely affected in the FNRL mutant of R. sphaeroides, indicating that FnrL is an essential regulator in the formation of spectral complexes (26). However, the oxygen-insensitive formation of spectral complexes in the CCOP1FNRL double mutant also suggested that the requirement for FnrL in spectral complex formation can be at least partially alleviated by the inactivation of the cbb3 oxidase. This finding raised the question of whether the CCOP1FNRL mutant is able to grow photosynthetically. Zeilstra-Ryalls and Kaplan (27) have reported that the FNRL mutant can grow neither photosynthetically nor anaerobically with DMSO as a terminal electron acceptor for anaerobic respiration. Surprisingly, we found that the CCOP1FNRL double mutant regains the ability to grow, albeit slowly, under photosynthetic conditions. In liquid medium in completely filled screw-cap glass tubes, a continuous increase in cell density was observed for the CCOP1FNRL mutant after approximately the same lag phase as for the wild type when the medium was inoculated with aerobically grown cells, although its growth rate was slower than that of the wild-type and CCOP1 mutant strains (Fig. 5). As expected, the FNRL mutant cannot grow photosynthetically.
FIG. 5.
Growth characteristics of the wild-type and mutant strains under photosynthetic conditions. Photosynthetic cultivations were performed at low light intensity (3 W/m2) in completely filled screw-cap glass tubes to minimize the repression effect of PpsR. Cultures were inoculated with aerobically grown cells. The levels of spectral complexes in the strains grown under the same conditions to an OD600 of 0.3 to 0.4 are given above the growth curves for B800-850 and (B875). OD600 = (Klett unit + 3.06)/102.04.
In order to examine whether the fnrL mutation affects spectral complex formation under photosynthetic conditions, the levels of the spectral complexes were determined with the wild-type and mutant strains grown photosynthetically (3 W/m2). As shown in Fig. 5, the levels of B800-850 complex were substantially decreased in the CCOP1FNRL mutant compared with those found in the CCOP1 mutant and the wild type. The CCOP1FNRL mutant exhibited only 58% of the B800-850 levels detected in the CCOP1 mutant under low light conditions, while the levels of B875 remained relatively fixed in all three strains. The reduced levels of the spectral complexes in the CCOP1FNRL mutant can account for the slow growth of this mutant under photosynthetic conditions.
The CCOP1FNRL mutant is unable to grow anaerobically with DMSO in the dark, as is the case for the FNRL mutant. When immunoblot analysis was performed with an antibody against DorA, the catalytic subunit of DMSO reductase, from Rhodobacter capsulatus, only a trace amount of DorA was detected in both FNRL and CCOP1FNRL mutant strains grown semiaerobically in the presence of 0.5% (vol/vol) DMSO, while a large induction of DMSO reductase expression was observed in the wild-type and CCOP1 mutant strains grown under the same conditions (data not shown). This result confirmed earlier studies in this laboratory (13, 26) that the fnrL mutation impairs the expression of DMSO reductase, and unlike PS gene expression, the inactivation of the cbb3 oxidase cannot bypass the requirement for FnrL in the induction of the genes encoding DMSO reductase, as is the case for hemZ.
A plausible explanation for the partial bypass of the FnrL requirement in spectral complex formation and photosynthetic growth by the inactivation of the cbb3 oxidase can now be formulated. It is based on the observation that many genes involved in Bchl biosynthesis or in the formation of the spectral complexes are coregulated by both FnrL and the PrrBA two-component system, and the complete absence of the cbb3-generated inhibitory signal provides for the full activation of the PrrBA system (17) and thereby partially compensates for the lack of FnrL. This rationale is supported by the data shown here and can be applied to gene regulation of the hemA, hemN, and bchE genes as well as the puc operon.
Recently Yeliseev and Kaplan (24) reported that the overexpression of hemN in R. sphaeroides leads to the aerobic derepression of genes which are regulated by the AppA/PpsR antirepressor/repressor system and the outer membrane protein TspO negatively controls the activity of coproporphyrinogen III oxidase posttranscriptionally. Since hemN is positively regulated by both FnrL and PrrBA, these regulators can indirectly affect the expression of PpsR target genes. These observations imply that the increased expression of hemN as the result of the combined effect of the removal of the cbb3-generated inhibitory signal (in the CCOP1 and CCOP1FNRL mutants) can bring about the partial alleviation of PpsR repression. Thus, the data obtained here and elsewhere (29) suggest that FnrL control of PS gene expression is additive and cooperative, acting together with other regulatory circuits in R. sphaeroides. This network of interacting regulatory circuits may permit R. sphaeroides to more finely “tune” PS gene expression to a wider array of environmental condition than R. capsulatus, which does not require FnrL for PS gene expression (26). Therefore, for the first time we are able to bring together, at least preliminarily, all of the regulatory pathways (Prr, FnrL, AppA/PpsR, and TspO) operating in R. sphaeroides to control PS gene expression into a coherent whole.
ACKNOWLEDGMENTS
This work was supported by grants GM15590 and GM55481 to S.K.
We thank M. Choudhary for providing sequence information on the PS gene cluster of R. sphaeroides 2.4.1.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: Greene Publishing Associates and John Wiley & Sons; 1988. [Google Scholar]
- 2.Choudhary M, Kaplan S. DNA sequence analysis of the photosynthetic region of Rhodobacter sphaeroides 2.4.1 (T) Nucleic Acids Res. 2000;28:862–867. doi: 10.1093/nar/28.4.862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cohen-Bazire G, Sistrom W R, Stanier R Y. Kinetic studies of pigment synthesis by non-sulfur purple bacteria. J Cell Comp Physiol. 1956;49:25–68. doi: 10.1002/jcp.1030490104. [DOI] [PubMed] [Google Scholar]
- 4.Coomber S A, Jones R M, Jordan P M, Hunter C N. A putative anaerobic coproporphyrinogen III oxidase in Rhodobacter sphaeroides. I. Molecular cloning, transposon mutagenesis and sequence analysis of the gene. Mol Microbiol. 1992;6:3159–3169. doi: 10.1111/j.1365-2958.1992.tb01772.x. [DOI] [PubMed] [Google Scholar]
- 5.Davis J, Donohue T J, Kaplan S. Construction, characterization, and complementation of a Puf− mutant of Rhodobacter sphaeroides. J Bacteriol. 1988;170:320–329. doi: 10.1128/jb.170.1.320-329.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eraso J M, Kaplan S. prrA, a putative response regulator involved in oxygen regulation of photosynthesis gene expression in Rhodobacter sphaeroides. J Bacteriol. 1994;176:32–43. doi: 10.1128/jb.176.1.32-43.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gomelsky M, Kaplan S. Genetic evidence that PpsR from Rhodobacter sphaeroides 2.4.1 functions as a repressor of puc and bchF expression. J Bacteriol. 1995;177:1634–1637. doi: 10.1128/jb.177.6.1634-1637.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jessee J. New subcloning efficiency competent cells: >1 × 106 transformants/μg. Focus. 1986;8:9. [Google Scholar]
- 9.Lee J K, Kaplan S. cis-Acting regulatory elements involved in oxygen and light control of puc operon transcription in Rhodobacter sphaeroides. J Bacteriol. 1992;174:1146–1157. doi: 10.1128/jb.174.4.1146-1157.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee J K, Kaplan S. Transcriptional regulation of puc operon expression in Rhodobacter sphaeroides. Analysis of the cis-acting downstream regulatory sequence. J Biol Chem. 1995;270:20453–20458. [PubMed] [Google Scholar]
- 11.Lee J K, Wang S, Eraso J M, Gardner J, Kaplan S. Transcriptional regulation of puc operon expression in Rhodobacter sphaeroides. Involvement of an integration host factor-binding sequence. J Biol Chem. 1993;268:24491–24497. [PubMed] [Google Scholar]
- 12.Meinhardt S W, Kiley P J, Kaplan S, Crofts A R, Harayama S. Characterization of light-harvesting mutants of Rhodopseudomonas sphaeroides. I. Measurement of the efficiency of energy transfer from light-harvesting complexes to the reaction center. Arch Biochem Biophys. 1985;236:130–139. doi: 10.1016/0003-9861(85)90612-5. [DOI] [PubMed] [Google Scholar]
- 13.Mouncey N J, Kaplan S. Cascade regulation of dimethyl sulfoxide reductase (dor) gene expression in the facultative phototroph Rhodobacter sphaeroides 2.4.1T. J Bacteriol. 1998;180:2924–2930. doi: 10.1128/jb.180.11.2924-2930.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mouncey N J, Kaplan S. Oxygen regulation of the ccoN gene encoding a component of the cbb3 oxidase in Rhodobacter sphaeroides 2.4.1T: involvement of the FnrL protein. J Bacteriol. 1998;180:2228–2231. doi: 10.1128/jb.180.8.2228-2231.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Neidle E L, Kaplan S. 5-Aminolevulinic acid availability and control of spectral complex formation in hemA and hemT mutants of Rhodobacter sphaeroides. J Bacteriol. 1993;175:2304–2313. doi: 10.1128/jb.175.8.2304-2313.1993. . (Erratum, 175:7123.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Neidle E L, Kaplan S. Expression of the Rhodobacter sphaeroides hemA and hemT genes, encoding two 5-aminolevulinic acid synthase isozymes. J Bacteriol. 1993;175:2292–2303. doi: 10.1128/jb.175.8.2292-2303.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O'Gara J P, Eraso J M, Kaplan S. A redox-responsive pathway for aerobic regulation of photosynthesis gene expression in Rhodobacter sphaeroides 2.4.1. J Bacteriol. 1998;180:4044–4050. doi: 10.1128/jb.180.16.4044-4050.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O'Gara J P, Kaplan S. Evidence for the role of redox carriers in photosynthesis gene expression and carotenoid biosynthesis in Rhodobacter sphaeroides 2.4.1. J Bacteriol. 1997;179:1951–1961. doi: 10.1128/jb.179.6.1951-1961.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oh J-I, Kaplan S. The cbb3 terminal oxidase of Rhodobacter sphaeroides 2.4.1: structural and functional implications for the regulation of spectral complex formation. Biochemistry. 1999;38:2688–2696. doi: 10.1021/bi9825100. [DOI] [PubMed] [Google Scholar]
- 20.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 21.Simon R, Priefer U, Puhler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 22.Suzuki J Y, Bollivar D W, Bauer C E. Genetic analysis of chlorophyll biosynthesis. Annu Rev Genet. 1997;31:61–89. doi: 10.1146/annurev.genet.31.1.61. [DOI] [PubMed] [Google Scholar]
- 23.van Neil C B. The culture, general physiology, morphology, and classification of the non-sulfur purple and brown bacteria. Bacteriol Rev. 1944;8:1–118. doi: 10.1128/br.8.1.1-118.1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yeliseev A A, Kaplan S. A novel mechanism for the regulation of photosynthesis gene expression by the TspO outer membrane protein of Rhodobacter sphaeroides 2.4.1. J Biol Chem. 1999;274:21234–21243. doi: 10.1074/jbc.274.30.21234. [DOI] [PubMed] [Google Scholar]
- 25.Zeilstra-Ryalls J, Gomelsky M, Eraso J M, Yeliseev A, O'Gara J, Kaplan S. Control of photosystem formation in Rhodobacter sphaeroides. J Bacteriol. 1998;180:2801–2809. doi: 10.1128/jb.180.11.2801-2809.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zeilstra-Ryalls J H, Gabbert K, Mouncey N J, Kaplan S, Kranz R G. Analysis of the fnrL gene and its function in Rhodobacter capsulatus. J Bacteriol. 1997;179:7264–7273. doi: 10.1128/jb.179.23.7264-7273.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zeilstra-Ryalls J H, Kaplan S. Aerobic and anaerobic regulation in Rhodobacter sphaeroides 2.4.1: the role of the fnrL gene. J Bacteriol. 1995;177:6422–6431. doi: 10.1128/jb.177.22.6422-6431.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zeilstra-Ryalls J H, Kaplan S. Control of hemA expression in Rhodobacter sphaeroides 2.4.1: regulation through alterations in the cellular redox state. J Bacteriol. 1996;178:985–993. doi: 10.1128/jb.178.4.985-993.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zeilstra-Ryalls J H, Kaplan S. Role of the fnrL gene in photosystem gene expression and photosynthetic growth of Rhodobacter sphaeroides 2.4.1. J Bacteriol. 1998;180:1496–1503. doi: 10.1128/jb.180.6.1496-1503.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]