Abstract
Introduction
Nasopharyngeal carcinoma is the most common cancer originating from the nasopharynx.
Objective
To study the mechanisms of nasopharyngeal carcinoma, we analyzed GSE12452 microarray data.
Methods
GSE12452 was downloaded from the Gene Expression Omnibus database and included 31 nasopharyngeal carcinoma samples and 10 normal nasopharyngeal tissue samples. The differentially expressed genes were screened by ANOVA in the PGS package. Using the BiNGO plugin in Cytoscape and pathway enrichment analysis in the PGS package, functional and pathway enrichment analyses were performed separately to predict potential functions of the differentially expressed genes. Furthermore, Transcription factor-differentially expressed gene pairs were searched, and then the transcription factor-differentially expressed gene regulatory network was visualized using Cytoscape software.
Results
A total of 487 genes were screened as differentially expressed genes between the nasopharyngeal carcinoma samples and the normal nasopharyngeal tissue samples. Enrichment analysis indicated that PTGS2 was involved in the regulation of biological process and small cell lung cancer. ZIC2 and OVOL1 may function in nasopharyngeal carcinoma through targeting significantly up-regulated genes (such as PTGS2, FN1, CXCL9 and CXCL10) in the Transcription factor-differentially expressed gene regulatory network (e.g., ZIC2→PTGS2 and OVOL1→CXCL10).
Conclusion
PTGS2, FN1, CXCL9, CXCL10, ZIC2 and OVOL1 might play roles in nasopharyngeal carcinoma.
Keywords: Nasopharyngeal carcinoma, Differentially expressed genes, Enrichment analysis, Regulatory network
Resumo
Introdução
O carcinoma nasofaríngeo é o câncer mais comum originário da nasofaringe.
Objetivo
Estudar os mecanismos do câncer de nasofaringe; dados do microarray GSE12452 foram analisados.
Método
GSE12452 foi obtido da base de dados Gene Expression Omnibus e inclui 31 amostras de carcinoma nasofaríngeo e 10 amostras de tecido nasofaríngeo normal. Os genes diferencialmente expressos foram analisados por ANOVA no kit PGS. Usando o plugin BiNGO no Cytoscape e análise de enriquecimento da via no kit PGS, análises de enriquecimento funcional e da via foram realizadas separadamente para prever as potenciais funções dos genes diferencialmente expressos. Além disso, os pares Fator de Transcrição - genes diferencialmente expressos foram pesquisados e em seguida a sua rede reguladora foi visualizada usando o programa Cytoscape.
Resultados
Um total de 487 genes foram analisados como genes diferencialmente expressos entre as amostras de carcinoma nasofaríngeo e amostras de tecido nasofaríngeo normal. A análise de enriquecimento indicou que PTGS2 estava envolvido na regulação do processo biológico e câncer pulmonar de pequenas células. ZIC2 e OVOL1 podem funcionar no carcinoma nasofaríngeo almejando-se de maneira significativa os genes suprarregulados (como o PTGS2, FN1, CXCL9 e CXCL10) na rede reguladora de fator de transcrição - genes diferencialmente expressos (p.ex., ZIC2→PTGS2 e OVOL1→CXCL10).
Conclusão
PTGS2, FN1, CXCL9, CXCL10, ZIC2 e OVOL1 podem desempenhar alguns papéis no carcinoma nasofaríngeo.
Palavras-chave: Carcinoma nasofaríngeo, Genes diferencialmente expressos, Análise de enriquecimento, Rede reguladora
Introduction
As the most common cancer originating from the nasopharynx, nasopharyngeal carcinoma (NPC) caused approximately 86,700 new cases and 50,800 deaths globally in 2012.1 NPC is extremely common in Southeast Asia and southern China, with more than 50,000 new cases each year.2, 3 NPC can be induced by multiple factors including heredity, viral factors and environmental influences.4 Most cases of NPC are correlated with Epstein–Barr Virus (EBV) infection, which is a B-lymphotropic herpesvirus possessing growth-transforming properties.5 Therefore, it is of great importance to study the mechanisms of NPC.
Many studies investigating the mechanisms of NPC have been published. There are several genes (such as C-myc, AKT1, p53, MDM2, LMP1 and PTEN) implicated in the pathogenesis of NPC because they are often amplified or altered in patients with this disease.6, 7, 8 Disabled 2 (DAB2) is frequently down-regulated by promoter hypermethylation and may be a potential tumor suppressor in NPC.9 Previous studies show that the potential tumor suppressor gene A disintegrin-like and metalloprotease domain with thrombospondin type 1 motifs 9 (ADAMTS9) is closely related to lymph node metastases, and it can inhibit tumor growth by suppressing angiogenesis in NPC.10, 11 The transcription factor (TF) adaptor-related protein complex 1 (AP-1) activated by the EBV-encoded Nuclear Antigen 1 (EBNA1) can target hypoxia-inducible factor-1α, interleukin 8 and Vascular Endothelial Growth Factor (VEGF), which promotes microtubule formation in NPC cells.12 The TF Forkhead Box M1 (FOXM1) is involved in tumor development, and the adenovirus vector AdFOXM1shRNA, which expresses FOXM1-specific short hairpin RNA, may be used as a therapeutic intervention for the treatment of patients with NPC.13 By down-regulating the expression of Secreted Protein, Acidic and Rich in Cysteine (SPARC), TF sex determining region Y-box 5 (SOX-5) functions in the progression of NPC and may be used as a predictor for poor NPC prognosis.14, 15 Although these studies have been performed to investigate NPC, the mechanisms of NPC still remain unclear.
In 2006, Sengupta et al. analyzed the expression of all latent EBV genes between NPC samples and normal nasopharyngeal epithelium samples, and obtained a panel of differentially expressed genes (DEGs).3 Using the same data that was used by Sengupta et al.,3 we not only screened the DEGs but also performed a comprehensive bioinformatic analysis to identify key genes associated with NPC. The potential functions of the DEGs were predicted by functional and pathway enrichment analyses. In addition, a TF-DEG regulatory network was constructed to investigate the regulatory relationships between TFs and DEGs.
Methods
Microarray data
The GSE12452 expression profile data deposited by Sengupta et al.3 was downloaded from the Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) database, which was based on the platform of the GPL570 [HG-U133_Plus_2] Affymetrix Human Genome U133 Plus 2.0 Array. The specimens included 31 NPC samples and 10 normal nasopharyngeal tissue samples collected from patients from Taiwan with informed consent. The samples were resected, fast frozen and then stored in liquid nitrogen.
Screening for DEGs
For the GSE12452 dataset, the Robust MultiArray Averaging (RMA) method in the Partek® Genomics Suite™ (PGS) (http://www.partek.com/) package16 was used for preprocessing, including background adjustment, quantile normalization and log 2 transformation. After batch correction, the processed expression matrix was obtained. The ANOVA (analysis of variance) method in the PGS package16 was used to compare the DEGs between the NPC samples and the normal nasopharyngeal tissue samples. The p-values obtained were adjusted for multiple testing using the Benjamini and Hochberg method.17 Genes with fold-change (FC) >2 and adjusted p-value < 0.05 were selected as DEGs.
Functional and pathway enrichment analysis
Gene Ontology (GO) terms can be used to describe three types of gene products including biological process in which they implicated, their molecular function and subcellular location.18 The Kyoto Encyclopedia of Genes and Genomes (KEGG) is a database composed of genes and their functional information.19 Using the BiNGO plugin20 in Cytoscape (http://www.cytoscape.org/) and pathway enrichment analysis in the PGS package,16 GO and KEGG pathway enrichment analyses were performed separately for DEGs between the NPC samples and the normal nasopharyngeal tissue samples. The adjusted p-value < 0.05 was used as the cut-off criterion.
TF-DEG regulatory network construction
The Genomatix Software Suite (https://www.genomatix.de/) package21 was used to predict Transcription Factors (TFs). In brief, the Gene2Promoter (http://www.genomatix.de) tool22 was used to extract promoter sequences of corresponding DEGs from the Genomatix database. Then, the TFs were analyzed using the MatInspector tool.21 TFs with a p-value of <0.05 and FC > 2 were selected as differentially expressed TFs, and then their corresponding TF-DEG pairs were screened. Finally, the TF-DEG regulatory network was visualized using Cytoscape software.23
Results
Analysis of DEGs
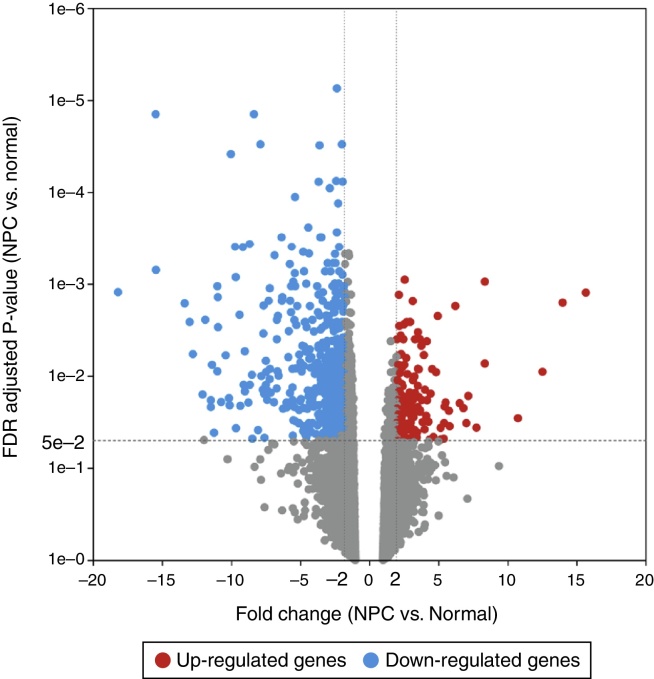
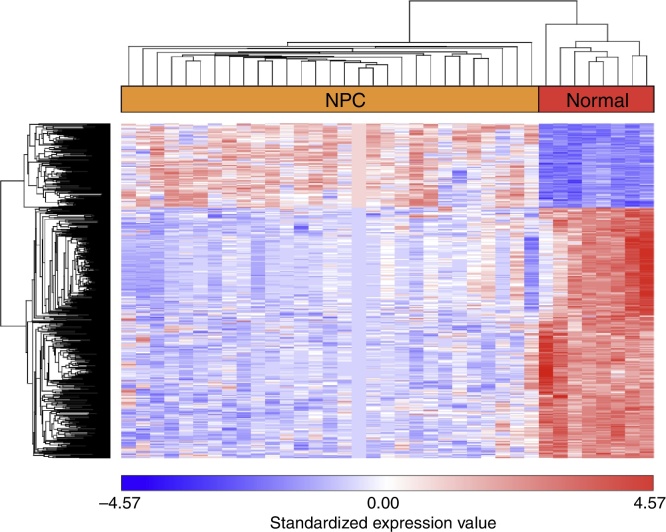
After GSE12452 was downloaded, the expression profile data were preprocessed and then the DEGs were identified by ANOVA using the PGS package. The volcano plot for DEGs separately is shown in Fig. 1. A total of 487 genes were selected as DEGs between the NPC samples and the normal nasopharyngeal tissue samples, including 122 up-regulated and 365 down-regulated genes. The number of down-regulated genes was greater than the number of up-regulated genes. In the heat map of the hierarchical cluster analysis for the DEGs, the NPC samples and normal nasopharyngeal tissue samples were clearly divided into two groups (Fig. 2).
Figure 1.
The volcano plot for differentially expressed genes (DEGs) (FC > 2 and adjusted p-value < 0.05). The horizontal axis represents the fold change, and the vertical axis represents the adjusted p-value. The red and blue circles indicate up- and down-regulated genes, respectively.
Figure 2.
The heat map of the hierarchical cluster analysis for differentially expressed genes (DEGs).
Functional and pathway enrichment analysis
The top 10 GO functions for the up-regulated genes included collagen fibril organization (p = 1.44E−06), extracellular matrix organization (p = 6.73E−08) and regulation of biological process (p = 3.35E−06, which involved prostaglandin-endoperoxide synthase 2, PTGS2) (Table 1A). The enriched KEGG pathways for the up-regulated genes are listed in Table 1C, and these included amoebiasis (p = 1.25E−07), ECM-receptor interaction (p = 9.01E−10) and small cell lung cancer (p = 3.10E−06, which involved PTGS2).
Table 1.
The enriched GO functions and KEGG pathways for the up-regulated and the down-regulated genes. (A) The top 10 GO functions enriched for the up-regulated genes. (B) The top 10 GO functions enriched for the down-regulated genes. (C) The top 10 KEGG pathways enriched for the up-regulated genes. (D) The top 10 KEGG pathways enriched for the down-regulated genes.
| Term | Description | Gene number | Gene symbol | p-Value |
|---|---|---|---|---|
| A | ||||
| 30198 | Extracellular matrix organization | 9 | COL18A1, COL4A2…… | 6.73E−08 |
| 30199 | Collagen fibril organization | 5 | COL3A1, COL1A2…… | 1.44E−06 |
| 43062 | Extracellular structure organization | 9 | COL3A1, COL1A2…… | 2.85E−06 |
| 50789 | Regulation of biological process | 72 | PTGS2, EZH2…… | 3.35E−06 |
| 32964 | Collagen biosynthetic process | 3 | COL1A1, COL5A1, COL5A1 | 3.91E−06 |
| 43588 | Skin development | 5 | COL5A2, COL5A1…… | 4.77E−06 |
| 65007 | Biological regulation | 74 | SLC9A7, PTGS2…… | 6.95E−06 |
| 6950 | Response to stress | 30 | C3AR1, PTGS2…… | 8.65E−06 |
| 9611 | Response to wounding | 15 | BMP2, TNFSF4…… | 1.03E−05 |
| 6954 | Inflammatory response | 11 | CXCL9, PLA2G7…… | 2.15E−05 |
| B | ||||
| 10880 | Regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum | 4 | CALM3, FKBP1B…… | 2.32E−07 |
| 30855 | Epithelial cell differentiation | 13 | EREG, ELF3…… | 1.30E−06 |
| 60316 | Positive regulation of ryanodine-sensitive calcium-release channel activity | 3 | CALM2, CALM1, CALM1 | 3.21E−06 |
| 60314 | Regulation of ryanodine-sensitive calcium-release channel activity | 4 | FKBP1B, CALM2…… | 9.20E−06 |
| 60315 | Negative regulation of ryanodine-sensitive calcium-release channel activity | 3 | CALM3, CALM2, CALM1 | 1.27E−05 |
| 7017 | Microtubule-based process | 14 | TPPP3, DNAH9…… | 1.92E−05 |
| 8544 | Epidermis development | 12 | CST6, EREG…… | 2.15E−05 |
| 1539 | Ciliary or flagellar motility | 4 | TEKT2, DNAH2…… | 3.02E−05 |
| 7018 | Microtubule-based movement | 9 | DNAH2, DNAH7…… | 3.72E−05 |
| 30216 | Keratinocyte differentiation | 7 | PPL, CNFN…… | 3.90E−05 |
| C | ||||
| ECM-receptor interaction | 11 | COL1A1, COL1A2…… | 9.01E−10 | |
| Protein digestion and absorption | 10 | COL1A2, COL3A1…… | 9.05E−09 | |
| Amoebiasis | 10 | CD14, COL1A2…… | 1.25E−07 | |
| Small cell lung cancer | 8 | PTGS2, FN1…… | 3.10E−06 | |
| Staphylococcus aureus infection | 5 | C3AR1, FCGR2A…… | 2.66E−04 | |
| Osteoclast differentiation | 6 | SOCS1, TYK2…… | 2.23E−03 | |
| Toll-like receptor signaling pathway | 5 | CXCL9, NFKBIA…… | 4.54E−03 | |
| DNA replication | 3 | PCNA, MCM2…… | 6.36E−03 | |
| Leishmaniasis | 4 | FCGR2A, PTGS2…… | 6.94E−03 | |
| Cell cycle | 5 | CCNE2, SMC1A…… | 9.53E−03 | |
| D | ||||
| Drug metabolism-cytochrome P450 | 9 | ADH1 C, ADH7…… | 3.02E−07 | |
| Metabolism of xenobiotics by cytochrome P450 | 8 | ADH7, ALDH1A3…… | 8.48E−06 | |
| Tyrosine metabolism | 5 | ALDH3A1, ALDH3B2…… | 1.69E−04 | |
| Glycolysis/gluconeogenesis | 6 | ADH7, ALDH1A3…… | 2.51E−04 | |
| Histidine metabolism | 4 | ALDH3A1, ALDH3A2…… | 5.74E−04 | |
| beta-Alanine metabolism | 4 | ALDH3A2, ALDH3B2…… | 6.59E−04 | |
| Phenylalanine metabolism | 3 | ALDH1A3, ALDH3A1, ALDH3B2 | 1.88E−03 | |
| Tight junction | 7 | CGN, CLDN7…… | 2.40E−03 | |
| Phototransduction | 3 | CALM1, CALM2, CALM3 | 7.52E−03 | |
| Retinol metabolism | 4 | DHRS9, SDR16C5…… | 1.03E−02 | |
Similarly, functional and pathway enrichment analyses were performed for the down-regulated genes. As shown in Table 1B, the functions including the regulation of release of sequestered calcium ion into the cytosol by sarcoplasmic reticulum (p = 2.32E−07), epithelial cell differentiation (p = 1.30E−06) and positive regulation of ryanodine-sensitive calcium-release channel activity (p = 3.21E-06) were enriched. The enriched pathways included drug metabolism-cytochrome P450 (p = 3.02E−07), metabolism of xenobiotics by cytochrome P450 (p = 8.48E−06) and tyrosine metabolism (p = 1.69E−04) (Table 1D).
TF-DEG regulatory network analysis
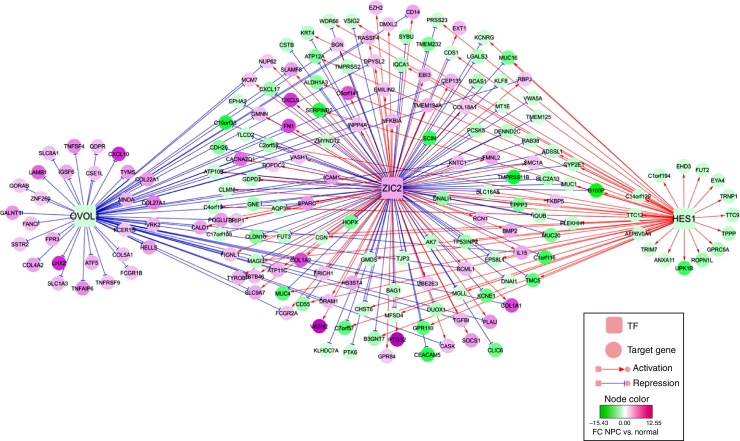
Using the Genomatix Software Suite package, TFs were predicted for the 122 up-regulated and 365 down-regulated genes. There were up-regulated TFs targeting up-regulated or down-regulated genes, as well as down-regulated TFs targeting up-regulated or down-regulated genes. However, only TFs that targeted both up-regulated and down-regulated genes were selected. The TF-DEG regulatory network of TFs (Zic family member 2, ZIC2; ovo-like 1, OVOL1; and hairy and enhancer of split 1, HES1) and their targeted DEGs are shown in Fig. 3. In particular, significantly up-regulated PTGS2, fibronectin (FN1) and chemokine (C-X-C motif) ligand 9 (CXCL9) were targeted and activated by ZIC2. Nevertheless, significantly up-regulated chemokine (C–X–C motif) ligand 10 (CXCL10) was targeted and suppressed by OVOL1.
Figure 3.
The regulatory network of Transcription Factors (TFs, such as ZIC2, OVOL and HES1) and their targeted differentially expressed genes (DEGs).
Discussion
In this study, a total of 487 genes were selected as DEGs between NPC samples and normal nasopharyngeal tissue samples, including 122 up-regulated and 365 down-regulated genes. There were many functions enriched for the DEGs, such as extracellular matrix organization and extracellular structure organization. PTGS2, FN1, CXCL9 and CXCL10 were significantly up-regulated genes involved in the TF-DEG regulatory network, and they may function in NPC through ZIC2 or OVOL1 (e.g., ZIC2→PTGS2 and OVOL1→CXCL10).
Enrichment analysis indicated that PTGS2 was involved in the regulation of biological process and small cell lung cancer. As a downstream gene involved in the NF-κB pathway, PTGS2 is also referred to as cyclooxygenase-2 (COX-2). It is known that COX2 can regulate cancer stem-like cells of NPC cells and promote their characteristics. Furthermore, parthenolide may function in targeted chemotherapy via the NF-κB/COX2 pathway.24 Based on PCR and restriction fragment length polymorphism analysis, the COX2 −765 G>C functional promoter polymorphism may be related to risk and neoplastic progression of NPC.25 The results of immunohistochemistry and semiquantitative assessment show that COX-2 expression increases as the nasopharyngeal epithelium develops from normal to dysplastic and then into NPC, indicating that COX-2 promotes the development of NPC.26, 27 This suggests that PTGS2 might be associated with NPC.
It is known that FN1 contributes to tumorigenesis by promoting the growth and migration of tumor cells, as well as by enhancing resistance to therapy.28, 29 Up-regulated FN1 correlates with the epithelial to mesenchymal transition (EMT) and contributes to tumor cell metastasis; therefore, it may be used as a potential diagnostic marker and therapeutic target of NPC.30 Based on a multiplex suspension array system, a previous study found that CXCL9 expression is significantly up-regulated in patients with NPC and oral cavity squamous cell carcinoma.31, 32 Quantitative real-time PCR and immunohistochemistry suggested that up-regulated CXCL9 is related to the aggressiveness of NPC, and an enzyme-linked immunosorbent assay indicated that its serum level may be a valuable prognostic indicator.33 The ZIC genes, which consist of five Cys2His2 zinc-finger domains, encode zinc-finger Transcription Factors.34 Among the family members, ZIC2 mediates the tissue-specific expression of the G-protein coupled receptor dopamine receptor D1.35 ZIC2 expression is up-regulated in several malignant tumors, including synovial sarcoma, pediatric medulloblastoma and endometrial cancers.36, 37, 38 Therefore, the expression levels of FN1, CXCL9 and ZIC2 might be correlated with NPC. In the TF-DEG regulatory network, we found that ZIC2 targeted and activated FN1, CXCL9 and PTGS2, indicating that ZIC2 might also function in NPC by regulating FN1, CXCL9 and PTGS2.
It is reported that the TFs OVOL1 and OVOL2 play critical roles in inducing the transition from mesenchymal to epithelial (MET) in several types of human cancers.39 As a member of the CXC chemokine family, CXCL10 binds to its receptor CXCR3 to exert biological functions in infectious diseases, immune dysfunction, chronic inflammation, tumor development and metastasis.40 The ELR-negative CXC chemokine CXCL10 can attenuate angiogenesis and suppress tumors.41 Overexpression of CXCL10 and CXCR3 play essential roles in advanced cancers, such as basal cell carcinoma,42 B-cell lymphoma,43 multiple myeloma44 and ovarian carcinoma.45 Inflammatory cytokines including CXCL10, Macrophage-Inhibitory-Protein 1 (MIP1) and interleukin 1 alpha are overexpressed in malignant NPC cells, which may promote the leukocyte infiltrate.46 These results suggest that OVOL1 and CXCL10 might be correlated with NPC. In the TF-DEG regulatory network, we also found that OVOL1 targeted and repressed CXCL10, suggesting that OVOL1 might also play a role in NPC by mediating CXCL10.
Conclusion
In summary, we used GSE12452 expression profile data downloaded from GEO to study the mechanisms of NPC. A total of 487 genes were selected as DEGs between the NPC samples and the normal nasopharyngeal tissue samples. Several genes (PTGS2, FN1, CXCL9, CXCL10, ZIC2 and OVOL1) might play roles in NPC and might also be used for targeted therapy of NPC in clinical practice. However, further experimental validation is still required to unravel their mechanisms of action in NPC.
Conflicts of interest
The authors declare no conflicts of interest.
Footnotes
Please cite this article as: Jiang X, Feng L, Dai B, Li L, Lu W. Identification of key genes involved in nasopharyngeal carcinoma. Braz J Otorhinolaryngol. 2017;83:670–6.
Peer Review under the responsibility of Associação Brasileira de Otorrinolaringologia e Cirurgia Cérvico-Facial.
References
- 1.Torre L.A., Bray F., Siegel R.L., Ferlay J., Lortet-Tieulent J., Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Chang E.T., Adami H-O. The enigmatic epidemiology of nasopharyngeal carcinoma. Cancer Epidemiol Biomark Prev. 2006;15:1765–1777. doi: 10.1158/1055-9965.EPI-06-0353. [DOI] [PubMed] [Google Scholar]
- 3.Sengupta S., Den Boon J.A., Chen I.-H., Newton M.A., Dahl D.B., Chen M., et al. Genome-wide expression profiling reveals EBV-associated inhibition of MHC class I expression in nasopharyngeal carcinoma. Cancer Res. 2006;66:7999–8006. doi: 10.1158/0008-5472.CAN-05-4399. [DOI] [PubMed] [Google Scholar]
- 4.Tsao S.W., Yip Y.L., Tsang C.M., Pang P.S., Lau V.M.Y., Zhang G., et al. Etiological factors of nasopharyngeal carcinoma. Oral Oncol. 2014;50:330–338. doi: 10.1016/j.oraloncology.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 5.Tay J.K., Chan S.H., Lim C.M., Siow C.H., Goh H.L., Loh K.S. The role of Epstein–Barr virus DNA load and serology as screening tools for nasopharyngeal carcinoma. Otolaryngol Head Neck Surg. 2016 doi: 10.1177/0194599816641038. [DOI] [PubMed] [Google Scholar]
- 6.Dawson C.W., Port R.J., Young L.S. Seminars in cancer biology. Elsevier; 2012. The role of the EBV-encoded latent membrane proteins LMP1 and LMP2 in the pathogenesis of nasopharyngeal carcinoma (NPC) [DOI] [PubMed] [Google Scholar]
- 7.Jiang Q., Zhang Y., Zhao M., Li Q., Chen R., Long X., et al. miR-16 induction after CDK4 knockdown is mediated by c-Myc suppression and inhibits cell growth as well as sensitizes nasopharyngeal carcinoma cells to chemotherapy. Tumor Biol. 2016;37:2425–2433. doi: 10.1007/s13277-015-3966-1. [DOI] [PubMed] [Google Scholar]
- 8.Zhang X., Chen X., Zhai Y., Cui Y., Cao P., Zhang H., et al. Combined effects of genetic variants of the PTEN, AKT1, MDM 2 and p53 genes on the risk of nasopharyngeal carcinoma. PLoS ONE. 2014;9:e92135. doi: 10.1371/journal.pone.0092135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tong J.H., Ng D.C., Chau S.L., So K.K., Leung P.P., Lee T.L., et al. Putative tumour-suppressor gene DAB2 is frequently down regulated by promoter hypermethylation in nasopharyngeal carcinoma. BMC Cancer. 2010;10:253. doi: 10.1186/1471-2407-10-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lung H.L., Lo P.H.Y., Xie D., Apte S.S., Cheung A.K.L., Cheng Y., et al. Characterization of a novel epigenetically-silenced, growth-suppressive gene, ADAMTS9, and its association with lymph node metastases in nasopharyngeal carcinoma. Int J Cancer. 2008;123:401–408. doi: 10.1002/ijc.23528. [DOI] [PubMed] [Google Scholar]
- 11.Lo P.H.Y., Lung H.L., Cheung A.K.L., Apte S.S., Chan K.W., Kwong F.M., et al. Extracellular protease ADAMTS9 suppresses esophageal and nasopharyngeal carcinoma tumor formation by inhibiting angiogenesis. Cancer Res. 2010;70:5567–5576. doi: 10.1158/0008-5472.CAN-09-4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O’Neil J.D., Owen T.J., Wood V.H., Date K.L., Valentine R., Chukwuma M.B., et al. Epstein–Barr virus-encoded EBNA1 modulates the AP-1 transcription factor pathway in nasopharyngeal carcinoma cells and enhances angiogenesis in vitro. J Gen Virol. 2008;89:2833–2842. doi: 10.1099/vir.0.2008/003392-0. [DOI] [PubMed] [Google Scholar]
- 13.Chen H., Yang C., Yu L., Xie L., Hu J., Zeng L., et al. Adenovirus-mediated RNA interference targeting FOXM1 transcription factor suppresses cell proliferation and tumor growth of nasopharyngeal carcinoma. J Gene Med. 2012;14:231–240. doi: 10.1002/jgm.2614. [DOI] [PubMed] [Google Scholar]
- 14.Huang D.Y., Lin Y.T., Jan P.S., Hwang Y.C., Liang S.T., Peng Y., et al. Transcription factor SOX-5 enhances nasopharyngeal carcinoma progression by down-regulating SPARC gene expression. J Pathol. 2008;214:445–455. doi: 10.1002/path.2299. [DOI] [PubMed] [Google Scholar]
- 15.Wang H.Y., Li Y.Y., Shao Q., Hou J.H., Wang F., Cai M.B., et al. Secreted protein acidic and rich in cysteine (SPARC) is associated with nasopharyngeal carcinoma metastasis and poor prognosis. J Transl Med. 2012;10:10–17. doi: 10.1186/1479-5876-10-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maiti S. 2012. Comprehensive assessment of CNV calling algorithms: a family based study involving monozygotic twins discordant for schizophrenia. [Google Scholar]
- 17.Haynes W. Encyclopedia of systems biology. Springer; 2013. Benjamini–Hochberg method; p. 78. [Google Scholar]
- 18.Consortium G.O. Gene ontology consortium: going forward. Nucleic Acids Res. 2015;43:D1049–D1056. doi: 10.1093/nar/gku1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kanehisa M., Sato Y., Kawashima M., Furumichi M., Tanabe M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2015 doi: 10.1093/nar/gkv1070. gkv1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maere S., Heymans K., Kuiper M. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics. 2005;21:3448–3449. doi: 10.1093/bioinformatics/bti551. [DOI] [PubMed] [Google Scholar]
- 21.Cartharius K., Frech K., Grote K., Klocke B., Haltmeier M., Klingenhoff A., et al. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics. 2005;21:2933–2942. doi: 10.1093/bioinformatics/bti473. [DOI] [PubMed] [Google Scholar]
- 22.Scherf M., Klingenhoff A., Werner T. Highly specific localization of promoter regions in large genomic sequences by PromoterInspector: a novel context analysis approach. J Mol Biol. 2000;297:599–606. doi: 10.1006/jmbi.2000.3589. [DOI] [PubMed] [Google Scholar]
- 23.Smoot M.E., Ono K., Ruscheinski J., Wang P.-L., Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27:431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liao K., Xia B., Zhuang Q.-Y., Hou M.-J., Zhang Y.-J., Luo B., et al. Parthenolide inhibits cancer stem-like side population of nasopharyngeal carcinoma cells via suppression of the NF-κB/COX-2 pathway. Theranostics. 2015;5:302. doi: 10.7150/thno.8387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ben Nasr H., Chahed K., Bouaouina N., Chouchane L. PTGS2 (COX-2)− 765 G>C functional promoter polymorphism and its association with risk and lymph node metastasis in nasopharyngeal carcinoma. Mol Biol Rep. 2009;36:193–200. doi: 10.1007/s11033-007-9166-3. [DOI] [PubMed] [Google Scholar]
- 26.Tan K.B., Putti T.C. Cyclooxygenase 2 expression in nasopharyngeal carcinoma: immunohistochemical findings and potential implications. J Clin Pathol. 2005;58:535–538. doi: 10.1136/jcp.2004.021923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shi D., Xiao X., Tian Y., Qin L., Xie F., Sun R., et al. Activating enhancer-binding protein-2α induces cyclooxygenase-2 expression and promotes nasopharyngeal carcinoma growth. Oncotarget. 2015;6:5005. doi: 10.18632/oncotarget.3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jia D., Entersz I., Butler C., Foty R.A. Fibronectin matrix-mediated cohesion suppresses invasion of prostate cancer cells. BMC Cancer. 2012;12:1. doi: 10.1186/1471-2407-12-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bae Y.K., Kim A., Kim M.K., Choi J.E., Kang S.H., Lee S.J. Fibronectin expression in carcinoma cells correlates with tumor aggressiveness and poor clinical outcome in patients with invasive breast cancer. Hum Pathol. 2013;44:2028–2037. doi: 10.1016/j.humpath.2013.03.006. [DOI] [PubMed] [Google Scholar]
- 30.Ma L.J., Lee S.W., Lin L.C., Chen T.J., Chang I.W., Hsu H.P., et al. Fibronectin overexpression is associated with latent membrane protein 1 expression and has independent prognostic value for nasopharyngeal carcinoma. Tumor Biol. 2014;35:1703–1712. doi: 10.1007/s13277-013-1235-8. [DOI] [PubMed] [Google Scholar]
- 31.Chang K.P., Chang Y.T., Liao C.T., Yen T.C., Chen I.H., Chang Y.L., et al. Prognostic cytokine markers in peripheral blood for oral cavity squamous cell carcinoma identified by multiplexed immunobead-based profiling. Clin Chim Acta. 2011;412:980–987. doi: 10.1016/j.cca.2011.02.002. [DOI] [PubMed] [Google Scholar]
- 32.Chang K.P., Wu C.C., Chen H.C., Chen S.J., Peng P.H., Tsang N.M., et al. Identification of candidate nasopharyngeal carcinoma serum biomarkers by cancer cell secretome and tissue transcriptome analysis: potential usage of cystatin A for predicting nodal stage and poor prognosis. Proteomics. 2010;10:2644–2660. doi: 10.1002/pmic.200900620. [DOI] [PubMed] [Google Scholar]
- 33.Hsin L.J., Kao H.K., Chen I.H., Tsang N.M., Hsu C.L., Liu S.C., et al. Serum CXCL9 levels are associated with tumor progression and treatment outcome in patients with nasopharyngeal carcinoma. PLoS ONE. 2013;8:e80052. doi: 10.1371/journal.pone.0080052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grinberg I., Millen K. The ZIC gene family in development and disease. Clin Genet. 2005;67:290–296. doi: 10.1111/j.1399-0004.2005.00418.x. [DOI] [PubMed] [Google Scholar]
- 35.Yang Y., Hwang C.K., Junn E., Lee G., Mouradian M.M. ZIC2 and Sp3 repress Sp1-induced activation of the human D1A dopamine receptor gene. J Biol Chem. 2000;275:38863–38869. doi: 10.1074/jbc.M007906200. [DOI] [PubMed] [Google Scholar]
- 36.Bidus M.A., Risinger J.I., Chandramouli G.V., Dainty L.A., Litzi T.J., Berchuck A., et al. Prediction of lymph node metastasis in patients with endometrioid endometrial cancer using expression microarray. Clin Cancer Res. 2006;12:83–88. doi: 10.1158/1078-0432.CCR-05-0835. [DOI] [PubMed] [Google Scholar]
- 37.Pfister S., Schlaeger C., Mendrzyk F., Wittmann A., Benner A., Kulozik A., et al. Array-based profiling of reference-independent methylation status (aPRIMES) identifies frequent promoter methylation and consecutive downregulation of ZIC2 in pediatric medulloblastoma. Nucleic Acids Res. 2007;35:e51. doi: 10.1093/nar/gkm094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fernebro J., Francis P., Edén P., Borg Å., Panagopoulos I., Mertens F., et al. Gene expression profiles relate to SS18/SSX fusion type in synovial sarcoma. Int J Cancer. 2006;118:1165–1172. doi: 10.1002/ijc.21475. [DOI] [PubMed] [Google Scholar]
- 39.Roca H., Hernandez J., Weidner S., McEachin R.C., Fuller D., Sud S., et al. Transcription factors OVOL1 and OVOL2 induce the mesenchymal to epithelial transition in human cancer. PLoS ONE. 2013;8:e76773. doi: 10.1371/journal.pone.0076773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu M., Guo S., Stiles J.K. The emerging role of CXCL10 in cancer. Oncol Lett. 2011;2:583–589. doi: 10.3892/ol.2011.300. [review] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Persano L., Crescenzi M., Indraccolo S. Anti-angiogenic gene therapy of cancer: current status and future prospects. Mol Aspects Med. 2007;28:87–114. doi: 10.1016/j.mam.2006.12.005. [DOI] [PubMed] [Google Scholar]
- 42.Lo B.K.K., Yu M., Zloty D., Cowan B., Shapiro J., McElwee K.J. CXCR3/ligands are significantly involved in the tumorigenesis of basal cell carcinomas. Am J Pathol. 2010;176:2435–2446. doi: 10.2353/ajpath.2010.081059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ansell S.M., Maurer M.J., Ziesmer S.C., Slager S.L., Habermann T.M., Link B.K., et al. Elevated pretreatment serum levels of interferon-inducible protein-10 (CXCL10) predict disease relapse and prognosis in diffuse large B-cell lymphoma patients. Am J Hematol. 2012;87:865–869. doi: 10.1002/ajh.23259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pellegrino A., Antonaci F., Russo F., Merchionne F., Ribatti D., Vacca A., et al. CXCR3-binding chemokines in multiple myeloma. Cancer Lett. 2004;207:221–227. doi: 10.1016/j.canlet.2003.10.036. [DOI] [PubMed] [Google Scholar]
- 45.Furuya M., Suyama T., Usui H., Kasuya Y., Nishiyama M., Tanaka N., et al. Up-regulation of CXC chemokines and their receptors: implications for proinflammatory microenvironments of ovarian carcinomas and endometriosis. Hum Pathol. 2007;38:1676–1687. doi: 10.1016/j.humpath.2007.03.023. [DOI] [PubMed] [Google Scholar]
- 46.Teichmann M., Meyer B., Beck A., Niedobitek G. Expression of the interferon-inducible chemokine IP-10 (CXCL10), a chemokine with proposed anti-neoplastic functions, in Hodgkin lymphoma and nasopharyngeal carcinoma. J Pathol. 2005;206:68–75. doi: 10.1002/path.1745. [DOI] [PubMed] [Google Scholar]