Abstract
Growth of Corynebacterium glutamicum on mixtures of the carbon sources glucose and acetate is shown to be distinct from growth on either substrate alone. The organism showed nondiauxic growth on media containing acetate-glucose mixtures and simultaneously metabolized these substrates. Compared to those for growth on acetate or glucose alone, the consumption rates of the individual substrates were reduced during acetate-glucose cometabolism, resulting in similar total carbon consumption rates for the three conditions. By 13C-labeling experiments with subsequent nuclear magnetic resonance analyses in combination with metabolite balancing, the in vivo activities for pathways or single enzymes in the central metabolism of C. glutamicum were quantified for growth on acetate, on glucose, and on both carbon sources. The activity of the citric acid cycle was high on acetate, intermediate on acetate plus glucose, and low on glucose, corresponding to in vivo activities of citrate synthase of 413, 219, and 111 nmol · (mg of protein)−1 · min−1, respectively. The citric acid cycle was replenished by carboxylation of phosphoenolpyruvate (PEP) and/or pyruvate (30 nmol · [mg of protein]−1 · min−1) during growth on glucose. Although levels of PEP carboxylase and pyruvate carboxylase during growth on acetate were similar to those for growth on glucose, anaplerosis occurred solely by the glyoxylate cycle (99 nmol · [mg of protein]−1 · min−1). Surprisingly, the anaplerotic function was fulfilled completely by the glyoxylate cycle (50 nmol · [mg of protein]−1 · min−1) on glucose plus acetate also. Consistent with the predictions deduced from the metabolic flux analyses, a glyoxylate cycle-deficient mutant of C. glutamicum, constructed by targeted deletion of the isocitrate lyase and malate synthase genes, exhibited impaired growth on acetate-glucose mixtures.
In their natural environments microorganisms often encounter situations when not a single carbon source but mixtures of carbon and energy sources are present. Under such conditions, bacteria often utilize one carbon source preferentially, with the further carbon source(s) being consumed only, when the preferred one is exhausted. As already shown by Monod (29), the preferred carbon source in general supports the best growth rate and/or growth yield, and the successive utilization of the substrates is often represented by a biphasic growth behavior (29). The classical example of this phenomenon is the diauxic growth of Escherichia coli on glucose plus lactose, and the study of the underlying principles initiated the era of research on gene regulation. On the other hand, by analysis of growth and carbon source consumption, it was shown that, e.g., Leuconostoc oenos cometabolizes glucose with citrate or fructose (38). Also, E. coli cometabolizes hexoses under carbon limitation conditions (reviewed in reference 20). Several other bacteria use two carbon sources in parallel (reviewed in reference 16); among these is Corynebacterium glutamicum, a gram-positive bacterium known for its ability to excrete amino acids. C. glutamicum grows aerobically on a variety of carbohydrates and organic acids as carbon sources (23). The organism cometabolizes glucose and fructose, glucose and lactate, and glucose and pyruvate (6, 7), whereas it shows diauxic growth on glucose-glutamate mixtures (21). The carbon sources glucose and acetate have been shown to serve as substrates for amino acid production by C. glutamicum (17). There is considerable knowledge about the enzymes and genes involved in acetate and glucose metabolism as well as their regulation (35, 36, 52, 53), whereas neither growth on acetate-glucose mixtures nor metabolite fluxes during growth on acetate have been studied in detail.
The utilization of acetate involves its uptake and subsequent activation to acetyl coenzyme A (acetyl-CoA) and, when acetate is the sole carbon source, also requires the operation of the glyoxylate cycle as an anaplerotic pathway (5). The key glyoxylate cycle enzymes isocitrate lyase and malate synthase have been purified from C. glutamicum, and biochemical characterization showed that both enzymes are subject to allosteric regulation by several intermediates of the central metabolism (35, 36). In addition, the genes encoding isocitrate lyase (aceA) and malate synthase (aceB), as well as the operon coding for the acetate-activating enzymes acetate kinase and phosphotransacetylase (the pta-ack operon), have been isolated and characterized, and, by gene-directed mutagenesis and analysis of the mutants, all four enzymes have been shown to be essential for the growth of C. glutamicum on acetate as the sole carbon and energy source (35, 36, 37). Further studies revealed that all four enzymes are coordinatedly and specifically up-regulated by the presence of acetate in the growth medium, and in all four cases, cat fusion and Northern blot experiments revealed that this regulation is due to transcriptional control of the respective genes (37, 53).
The tight regulation of the enzymes involved in acetate metabolism of C. glutamicum is expected to cause significant changes of the carbon flux within the central metabolism of this organism when acetate instead of glucose is the sole carbon source. Although the up-regulation of the enzymes during growth on acetate or on acetate-glucose mixtures (53) suggests that the respective pathways could also be active under this condition, this cannot be regarded as proof for in vivo activities and certainly cannot be used to predict the in vivo fluxes quantitatively. Not only have 13C-labeling experiments been used to identify metabolic pathways (see, e.g., references 24, 25, and 43), but by quantitative analyses of positional isotopic labeling information Walsh and Koshland (50) pioneered the determination of relative metabolic fluxes (reviewed in reference 19). The combination of metabolite balancing and 13C-labeling experiments allowed the quantification of absolute metabolite fluxes within the cellular central metabolism (see, e.g., references 26 and 41). In order to study the cometabolism of acetate plus glucose by C. glutamicum in a quantitative manner, we performed 13C-labeling experiments in combination with nuclear magnetic resonance (NMR) analyses and metabolite balancing to determine the in vivo activities of pathways or single enzymes in the central metabolism, with a particular focus on the tricarboxylic acid (TCA) cycle and anaplerosis. From the flux analysis results, a hypothesis regarding the physiological significance of the glyoxylate cycle for C. glutamicum was deduced and tested experimentally.
MATERIALS AND METHODS
Materials.
Sodium [1-13C]acetate (99% atom enrichment) was purchased from Cambridge Isotope Laboratories, Andover, Mass., d-[5-13C]glucose (99% atom enrichment) from Isotec, Miamisburg, Ohio, and sodium 3-trimethylsilyl-[2,2′,3,3′-D4]propionate (99% atom enrichment) from Aldrich Chemicals, Milwaukee, Wis.
Microorganisms and cultivation conditions.
The wild-type (WT) strain of C. glutamicum ATCC 13032 and the isocitrate lyase- and malate synthase-negative double mutant C. glutamicum WTΔAB, described in this work, were used. All C. glutamicum strains were precultured on Luria-Bertani complex medium (39), with kanamycin (50 μg/ml) added when appropriate. Exponentially growing cells were harvested by centrifugation (5,000 × g, 5 min, 4°C), washed twice in 50 mM NaCl–50 mM Tris-HCl (pH 6.3), and used to inoculate CgC minimal medium (11). The carbon and energy sources were either unlabeled sodium acetate and/or glucose at the concentrations indicated in Results or, for the 13C-labeling experiments, sodium [1-13C]acetate (99% atom enrichment) (10 g/liter), [5-13C]glucose (99% atom enrichment) (20 g/liter), or sodium [1-13C]acetate (99% atom enrichment) plus unlabeled glucose (10 g/liter each). All cultivations were done as 60-ml cultures in 500-ml baffled Erlenmeyer flasks at 30°C and with agitation at 140 rpm. After about 4 generations, exponentially growing cells were harvested and washed as described above. The cell pellet was then used for the extraction of amino acids.
Extraction, purification, and quantification of amino acids.
Amino acids were extracted by acid hydrolysis of cell pellets and purified by cation-exchange chromatography as described previously (31). They were quantified by reversed-phase liquid chromatography with precolumn ortho-phthaldialdehyde derivatization (42).
NMR spectroscopy.
High-resolution 1H NMR spectra of amino acids were obtained on an AMX-400 WB spectrometer (Bruker, Karlsruhe, Germany) operating at 400.13 MHz and equipped with a multichannel interface and a 5-mm inverse probehead. 13C enrichments were determined using the parameters and the methods described previously (44, 52).
Quantitation of metabolite fluxes.
For the quantitation of carbon fluxes in the central metabolism of C. glutamicum, NMR spectroscopic and metabolite balancing data were combined in a nonlinear least-squares fitting procedure as described previously (26, 27, 28, 45). C. glutamicum was cultured on labeled carbon sources, and the 13C-labeling patterns of amino acids purified from exponentially growing cells were determined. The 13C-labeling patterns of the precursors pyruvate, oxaloacetate, 2-oxoglutarate, and 3-phosphoglycerate were deduced from the labeling patterns of alanine and valine, threonine and aspartate, glutamate and arginine, and serine and glycine, respectively. For flux calculations, a representation of the central metabolism of C. glutamicum essentially according to that described by Marx et al. (26) was used. The (sets of) enzyme reactions and metabolite pools used to construct the model are listed in Table 1. The precursor requirements for C. glutamicum biomass synthesis were taken from Marx et al. (26) except that, since in our experiments leucine was not supplemented, the precursor requirements were calculated based on the determined leucine content of 439 μmol · (g [dry weight])−1. Flux calculations were based on the mathematical approach and the computational tools described by Wiechert and de Graaf (54, 55). The metabolic steady-state condition in the carbon flux analyses presented is based on the exponential growth of batch cultures. Precultivation was optimized such that the growth on 13C-labeled carbon sources was exponential without an initial lag phase. To account for the unlabeled carbon content in the harvested cells derived from the inoculum (i.e., 5%), the 13C label of the positionally labeled carbon source was corrected (i.e., 94.9% instead of 99.9% [5-13C]glucose) as has been described previously (26).
TABLE 1.
Description of fluxes and metabolites constituting the flux model of C. glutamicum
| Flux | Conversion of metabolitesa | Enzyme reaction(s)b |
|---|---|---|
| v1 | Glucose-6-P↔fructose-6-P | Glucose-6-phosphate isomerase (EC 5.3.1.9) |
| v2 | Fructose-6-P↔2 glyceraldehyde-3-P's | Phosphofructokinase (EC 2.7.1.11), fructose-1,6-bisphosphate aldolase (EC 4.1.2.13), triose phosphate isomerase (EC 5.3.1.1) |
| v3 | Glyceraldehyde-3-P↔PEP/pyruvate | Glyceraldehyde-3-phosphate dehydrogenase (EC 1.2.1.12), 3-phosphoglycerate kinase (EC 2.7.2.3), phosphoglycerate mutase (EC 2.7.5.3), enolase (EC 4.2.1.11) |
| v4 | Glucose-6-P↔pentose-5-P + CO2 | Glucose-6-phosphate dehydrogenase (EC 1.1.1.49), lactonase (EC 3.1.1.31), 6-phosphogluconate dehydrogenase (EC 1.1.1.44) |
| v5 | 2 Pentose-5-P's↔sedoheptulose-7-P + glyceraldehyde-3-P | Transketolase (EC 2.2.1.1) |
| v6 | Sedoheptulose-7-P + glyceraldehyde-3-P↔fructose-6-P + erythrose-4-P | Transaldolase (EC 2.2.1.2) |
| v7 | Erythrose-4-P + pentose-5-P↔fructose-6-P + glyceraldehyde-3-P | Transketolase (EC 2.2.1.1) |
| v8 | PEP/pyruvate↔AcCoA + CO2 | Pyruvate dehydrogenase complex (EC 1.2.4.1; EC 2.3.1.12; EC 1.8.1.4) |
| v9 | AcCoA + malate/oxaloacetate↔isocitrate | Citrate synthase (EC 4.1.3.7), aconitase (EC 4.2.1.3) |
| v10 | Isocitrate↔oxoglutarate + CO2 | Isocitrate dehydrogenase (EC 1.1.1.42) |
| v11 | Oxoglutarate↔succinate + CO2 | 2-Oxoglutarate dehydrogenase (EC 1.2.4.2; EC 2.3.1.61; EC 1.8.1.4), succinyl CoA synthetase (EC 6.2.1.5) |
| v12 | Succinate↔malate/oxaloacetate | Succinate dehydrogenase (EC 1.3.5.1), fumarase (EC 4.2.1.2) |
| v13 | Isocitrate + AcCoA↔succinate + malate/oxaloacetate | Isocitrate lyase (EC 4.1.3.1), malate synthase (EC 4.1.3.2), succinate dehydrogenase (EC 1.3.5.1), fumarase (EC 4.2.1.2) |
| v14 | PEP/pyruvate + CO2↔malate/oxaloacetate | PEP carboxylase (EC 4.1.1.31), PEP carboxykinase (EC 4.1.1.32), pyruvate carboxylase (EC 6.4.1.1), oxaloacetate decarboxylase (EC 4.1.1.3), malic enzyme (EC 1.1.1.40) |
Pentose-5-phosphate combines ribose-5-phosphate, ribulose-5-phosphate, and xylulose-5-phosphate. P, phosphate; AcCoA, acetyl-CoA.
Treating PEP and pyruvate, malate and oxaloacetate, and ribose-5-phosphate, ribulose-5-phosphate, and xylulose-5-phosphate as single pools precludes the assignment of fluxes for enzyme reactions interconverting these metabolites (pyruvate kinase [EC 2.7.1.40] and the PEP:glucose-phosphotransferase system [EC 2.1.1.69; EC 2.7.3.9] for PEP/pyruvate, malate dehydrogenase [EC 1.1.1.37] for malate/oxaloacetate, and ribose-5-phosphate isomerase [EC 5.3.1.6] and ribulose-5-phosphate epimerase [EC 5.1.3.1] for pentose-5-phosphates).
Construction of the isocitrate lyase- and malate synthase-deficient C. glutamicum WTΔAB.
To generate a glyoxylate cycle-deficient mutant strain of C. glutamicum WT, the chromosomal region comprising the genes aceA and aceB, coding for isocitrate lyase and malate synthase, respectively, was deleted using standard methods (40). For the construction of the gene replacement vector pK19mobsacB-3′AB3′, the 3′-terminal parts of genes aceA and aceB were amplified using the PCR Core Kit from Boehringer Mannheim according to the manufacturer's instructions. To amplify the 3′-terminal region of aceB and aceA, primers VW1 and VW2 and primers VW3 and VW4, respectively, were used. The sequences of the primers are as follows (underlined nucleotides are derived from the aceA-aceB locus, and boldfaced nucleotides correspond to either a BamHI, an EcoRI, or an XbaI restriction site): VW1, 5′-CGGGATCCGCCATGATGTTG-3′ (nucleotides [nt] 2567 to 2584 in EMBL accession no. 78491); VW2, 5′-CGGAATTCGCATCATCACCATTG-3′ (nt 3021 to 3005 in EMBL accession no. 78491); VW3, 5′-CGGGATCCGCAGGCTACTTCGAC-3′ (nt 1719 to 1734 in EMBL accession no. 75504); VW4, 5′-GCTCTAGAAGTTGGGTTCTGAGAAG-3′ (nt 2170 to 2151 in EMBL accession no. 75504). The amplification products were subcloned into vector pGEM-T (Promega), resulting in the vectors pGEM-T-aceA3′ and pGEM-T-aceB3′, respectively. In the next step, the 1.6-kb BamHI-ScaI fragment of pGEM-T-aceA3′ was ligated with the 2.3-kb BamHI-ScaI fragment of pGEM-T-aceB3′, resulting in vector pGEM-T-3′AB3′. Vector pGEM-T-3′AB3′ contains the 3′-terminal regions of genes aceA and aceB adjacent to each other and in opposite directions. The 0.9-kb XbaI-EcoRI fragment of pGEM-T-3′AB3′ was ligated into the XbaI- and EcoRI-restricted gene replacement vector pK19mobsacB (40), which is not replicable in C. glutamicum. Applying the procedure described by Peters-Wendisch et al. (30), the resulting vector pK19mobsacB-3′AB3′ was used to delete the chromosomal aceA-aceB locus. The deletion of the chromosomal aceA-aceB locus was verified by Southern blot hybridization (data not shown). The aceA-aceB mutant was designated C. glutamicum WTΔAB. When it was grown on minimal medium containing either glucose or glucose plus acetate, neither isocitrate lyase nor malate synthase activity could be detected in crude extracts (data not shown), thus proving that the respective genes in C. glutamicum WTΔAB had in fact been knocked out.
RESULTS
Characterization of the growth of C. glutamicum on acetate, glucose, and acetate-glucose mixtures.
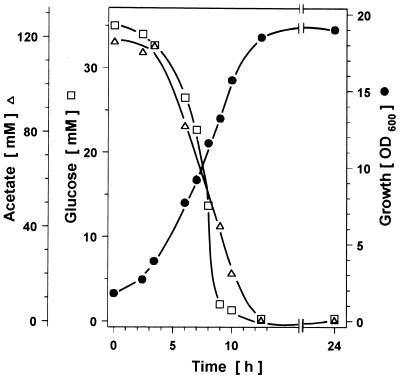
C. glutamicum WT was cultured on CgC minimal medium containing various concentrations of acetate (20 to 180 mM) plus glucose (5 to 110 mM). The growth of C. glutamicum on all acetate-glucose mixtures was monophasic; an example is shown in Fig. 1. The concentrations of both substrates in the culture broth decreased concomitantly, indicating that they were metabolized simultaneously. Thus, it was likely that the parameters characterizing the growth of C. glutamicum on mixtures of glucose and acetate are distinct from those for cells growing on either carbon source alone. In comparative growth experiments with C. glutamicum WT on CgC minimal media containing acetate and/or glucose, the growth rates, the biomass yields, and the substrate uptake rates were determined (Table 2). On glucose as a sole carbon source, but not on acetate or on acetate-glucose mixtures, the dependence of the growth rate on the substrate concentration followed Monod kinetics, with a maximal growth rate of 0.38 h−1 and a half-maximal growth rate at 4.5 mM glucose. In the presence of acetate in the medium, the growth rate decreased with increasing acetate concentrations (from about 0.32 h−1 at 60 mM acetate to about 0.24 h−1 at 180 mM acetate), probably reflecting the detrimental effect of acetate as an uncoupler of the transmembrane pH gradient (2). Comparison of the growth rates on acetate, acetate plus glucose, and glucose showed that the growth rate on acetate was lower than that of cells growing on glucose or glucose-acetate mixtures (Table 2). The biomass yield (calculated as grams of carbon [C] [dry weight] per grams of C in the substrate) for growth of C. glutamicum on any acetate-glucose mixture was the same as that for growth on glucose alone, whereas that for growth on acetate was significantly lower (Table 2). During growth on acetate plus glucose, the acetate consumption rate was always decreased compared to that for growth on acetate alone, as was the glucose consumption rate compared to that for growth on glucose as the sole carbon source (Table 2). Interestingly, the consumption rate of total carbon was similar under the three conditions (about 900 to 1,100 nmol of C · [mg of protein]−1 · min−1). Thus, simultaneous utilization of acetate and glucose by C. glutamicum is clearly distinct from growth on either substrate alone, and discernible carbon fluxes in the central metabolism should reflect these differences.
FIG. 1.
Growth of C. glutamicum WT on mineral medium with 35 mM glucose and 120 mM acetate.
TABLE 2.
Growth characteristics of C. glutamicum grown on minimal medium with sodium [1-13C]acetate, sodium [1-13C]acetate plus unlabeled glucose, or [5-13C]glucose as the carbon and energy source
| Growth parametera (unit) | Result for growth on minimal medium with:
|
||
|---|---|---|---|
| 120 mM acetate | 120 mM acetate + 55 mM glucose | 110 mM glucose | |
| Growth rate (h−1) | 0.28 | 0.36 | 0.32 |
| Biomass yield (g of C [dry wt] · [g of C in substrate]−1) | 0.29 | 0.41 | 0.41 |
| Acetate consumption rate (nmol · [mg of protein]−1 · min−1) | 540 | 270 | |
| Glucose consumption rate (nmol · [mg of protein]−1 · min−1) | 72 | 148 | |
| Carbon consumption rate (nmol of C · [mg of protein]−1 · min−1) | 1,080 | 972 | 888 |
Errors were <5% for determinations of growth rates, <5% for determinations of biomass yields, and <10% for determinations of carbon consumption rates. These data coincided with values obtained from at least three independent cultures with unlabeled substrates.
Quantitative determination of metabolite fluxes in C. glutamicum during growth on acetate, glucose, and a mixture of acetate and glucose.
In order to quantify metabolic fluxes in the central metabolism of C. glutamicum, we performed 13C-labeling experiments combined with metabolite balancing as described in Materials and Methods. C. glutamicum WT was cultured on minimal medium with either sodium [1-13C]acetate, [5-13C]glucose, or a mixture of sodium [1-13C]acetate and unlabeled glucose as the sole carbon source. The growth characteristics of the three cultures are summarized in Table 2. No significant by-product formation was detected in NMR analyses of culture supernatants. Exponentially growing cells of the cultures were harvested and hydrolyzed, and amino acids were purified from the lysate by liquid chromatography. The 13C-labeling patterns of alanine and valine, aspartic acid and threonine, glutamic acid and arginine, and serine and glycine were determined by NMR, and the corresponding labeling patterns of pyruvate, oxaloacetate, 2-oxoglutarate, and 3-phosphoglycerate were deduced (Table 3). Using these data, those for biomass accumulation, and those for acetate, glucose, and acetate-plus-glucose consumption, the in vivo carbon metabolite fluxes within the central metabolism of C. glutamicum were then quantitatively determined as summarized in Fig. 2, 3, and 4, respectively.
TABLE 3.
13C enrichments in carbon atoms of central metabolites of C. glutamicum WT cultured on CgC mineral medium containing [1-13C]acetate, [5-13C]glucose, or [1-13C]acetate plus unlabeled glucose
| Central metabolite | Carbon atom |
13C enrichment (%)a
|
|||||
|---|---|---|---|---|---|---|---|
| Glucose
|
Acetate
|
Acetate + glucose
|
|||||
| Exp. | Calc. | Exp. | Calc. | Exp. | Calc. | ||
| Pyruvate | C-1 | 3.6 ± 0.2 | 5.7 | 46.8 ± 0.8 | 48.8 | 22.7 ± 1.0 | 23.0 |
| C-2 | 49.3 ± 0.8 | 49.8 | 1.1 ± 0.4 | 1.1 | 1.1 ± 0.4 | 1.1 | |
| C-3 | 3.1 ± 0.2 | 4.2 | 1.1 ± 0.3 | 1.1 | 1.1 ± 0.4 | 1.1 | |
| Oxaloacetate | C-1 | 16.8 ± 1.0 | 16.8 | 49.3 ± 0.3 | 48.8 | 45.5 ± 1.5 | 45.4 |
| C-2 | 35.7 ± 0.8 | 37.6 | 1.1 ± 0.4 | 1.1 | 1.1 ± 0.4 | 1.1 | |
| C-3 | 11.1 ± 0.2 | 11.2 | 1.1 ± 0.4 | 1.1 | 1.1 ± 0.4 | 1.1 | |
| C-4 | 11.8 ± 0.5 | 19.0 | 49.1 ± 0.3 | 50.2 | 46.9 ± 0.6 | 47.0 | |
| 2-Oxoglutarate | C-1 | 11.8 ± 0.6 | 19.0 | 48.1 ± 0.5 | 50.2 | 47.3 ± 0.3 | 47.0 |
| C-2 | 9.9 ± 0.6 | 11.5 | 1.1 ± 0.4 | 1.1 | 1.0 ± 0.4 | 1.1 | |
| C-3 | 34.9 ± 3.0 | 37.5 | 1.1 ± 0.4 | 1.1 | 1.0 ± 0.4 | 1.1 | |
| C-4 | 3.0 ± 0.3 | 4.4 | 1.1 ± 0.4 | 1.1 | 1.0 ± 0.4 | 1.1 | |
| C-5 | 50.8 ± 0.7 | 49.4 | 85.2 ± 4.6 | 79.0 | 82.0 ± 0.8 | 82.0 | |
| 3-Phosphoglycerate | C-1 | 4.1 | 50.8 ± 0.5 | 48.8 | 18.5 ± 2.0 | 19.4 | |
| C-2 | 52.4 ± 0.5 | 51.5 | 1.1 ± 0.4 | 1.1 | 1.1 ± 0.4 | 1.1 | |
| C-3 | 3.2 ± 0.2 | 3.1 | 1.1 ± 0.4 | 1.1 | 1.1 ± 0.4 | 1.1 | |
Determined by NMR (Exp.) and calculated (Calc.) by the flux analysis program. Experimental values given are means of two to four determinations. 13C enrichments were deduced from the labeling patterns of alanine and valine for pyruvate, aspartic acid and threonine for oxaloacetate, glutamic acid and arginine for 2-oxoglutarate, and glycine and serine for 3-phosphoglycerate.
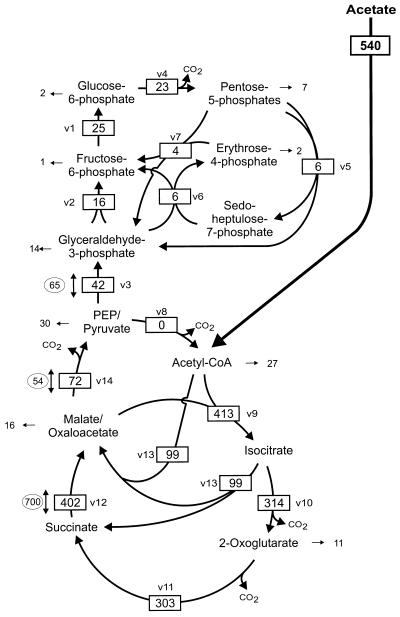
FIG. 2.
Metabolite fluxes in the central metabolism of C. glutamicum during growth on acetate. Fluxes are given in milliunits per milligram of protein. Fluxes for central metabolic pathways/reactions are boxed, fluxes towards biomass formation are given as pure numbers, and fluxes for exchange reactions are given in ovals. Abbreviations are as in Table 1.
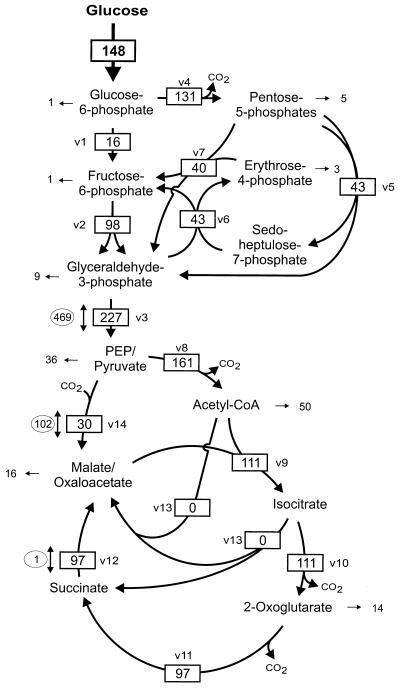
FIG. 3.
Metabolite fluxes in the central metabolism of C. glutamicum during growth on glucose. Fluxes are given in milliunits per milligram of protein. Fluxes for central metabolic pathways/reactions are boxed, fluxes towards biomass formation are given as pure numbers, and fluxes for exchange reactions are given in ovals. Abbreviations are as in Table 1.
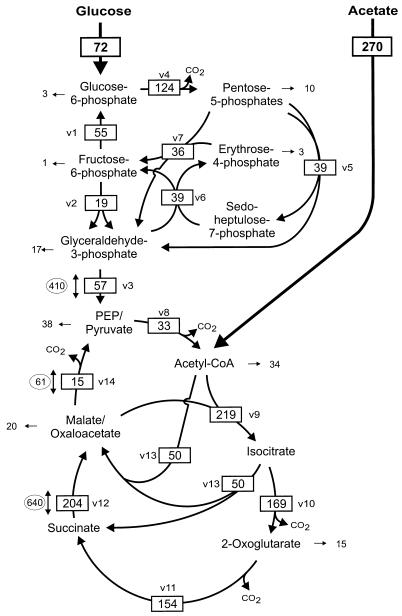
FIG. 4.
Metabolite fluxes in the central metabolism of C. glutamicum during growth on acetate plus glucose. Fluxes are given in milliunits per milligram of protein. Fluxes for central metabolic pathways/reactions are boxed, fluxes towards biomass formation are given as pure numbers, and fluxes for exchange reactions are given in ovals. Abbreviations are as in Table 1.
In C. glutamicum cells growing on acetate as a sole carbon source, the acetate was activated to acetyl-CoA with a specific activity of 540 mU · (mg of protein)−1 (Fig. 2). About 5% of the acetyl-CoA was used directly in anabolic reactions, e.g., for fatty acid synthesis, whereas 18% was converted by malate synthase in the glyoxylate cycle and 76% was converted by citrate synthase and aconitase to isocitrate. Isocitrate was metabolized to about 24% via isocitrate lyase in the glyoxylate cycle, whereas the rest was further oxidized in the reactions of the citric acid cycle. The malate formed by the glyoxylate cycle (99 mU · [mg of protein]−1) served to replenish the citric acid cycle. The majority (72 mU · [mg of protein]−1, i.e., 73%) of biosynthetic precursors withdrawn from the citric acid cycle was used for the generation of cell material derived from phosphoenolpyruvate and/or pyruvate (PEP/pyruvate)- and glyceraldehyde-3-phosphate. In summary, metabolization of acetate by C. glutamicum is characterized by a high in vivo activity of the citric acid cycle, a high in vivo activity of the glyoxylate cycle as the anaplerotic reaction, a high rate of PEP/pyruvate formation from oxaloacetate/malate, and gluconeogenesis with a 42-mU · (mg of protein)−1 conversion of PEP to glyceraldehyde-3-phosphate.
In C. glutamicum cells growing on glucose as a sole carbon source, the glucose was consumed at a rate of 148 mU · (mg of protein)−1, about 25% of the glucose 6-phosphate was shuttled into biomass and CO2 (directly, via fructose-6-phosphate, glyceraldehyde-3-phosphate, and pentose phosphate cycle intermediates), and the glycolytic flux from glyceraldehyde-3-phosphate to PEP/pyruvate was 227 mU · (mg of protein)−1 (Fig. 3). The formation of acetyl-CoA by pyruvate dehydrogenase (161 mU · [mg of protein]−1) amounted to about 70% of the PEP/pyruvate formation. Citrate synthase fueled acetyl-CoA into the citric acid cycle with an activity of 111 mU · (mg of protein)−1, whereas about 30% of the acetyl-CoA was used as precursors, mainly for fatty acid synthesis. The glyoxylate cycle was inactive during the growth of C. glutamicum on glucose. The anaplerotic reactions PEP carboxylase and pyruvate carboxylase replenished the citric acid cycle with a net conversion of PEP/pyruvate to oxaloacetate/malate of 30 mU · (mg of protein)−1. The combination of the [5-13C]glucose labeling experiment with metabolite balancing also allowed the determination of exchange rates between glyceraldehyde-3-phosphate and PEP/pyruvate and between PEP/pyruvate and oxaloacetate/malate. The exchange rate in the interconversion of PEP/pyruvate and oxaloacetate/malate was about twofold higher than the net flux of the carboxylating reaction, i.e., the net conversion of PEP/pyruvate to oxalacetate-malate (30 mU · [mg of protein]−1) is the sum of the PEP/pyruvate carboxylation of 132 mU · (mg of protein)−1 and the simultaneously occurring malate-oxalacetate decarboxylation of 102 mU · (mg of protein)−1. Similarly, an interconversion of glyceraldehyde-3-phosphate and PEP/pyruvate of about four times the net conversion was found (i.e., the exchange rate was 469 mU · [mg of protein]−1 versus a net glyceraldehyde-3-phosphate formation of 227 mU · [mg of protein]−1). To summarize, the central metabolism of C. glutamicum cells growing on glucose is characterized by a low activity of the citric acid cycle, the absence of glyoxylate cycle activity, and anaplerosis solely by carboxylation of PEP or pyruvate.
In C. glutamicum cells growing on a mixture of acetate and glucose, the consumption of both carbon sources (72 mU · [mg of protein]−1 for glucose and 270 mU · [mg of protein]−1 for acetate) was reduced approximately twofold compared to growth on either carbon source alone (Fig. 4). Acetyl-CoA was formed predominantly by activation of acetate (270 mU · [mg of protein]−1) and only with 33 mU · (mg of protein)−1 by the pyruvate dehydrogenase complex. About 73% of the acetyl-CoA was funneled into the citric acid cycle, and 11% was used directly in biosynthetic reactions. Surprisingly, the glyoxylate cycle was active under this growth condition, with isocitrate lyase and malate synthase activities of 50 mU · (mg of protein)−1. Isocitrate was metabolized to 23% by isocitrate lyase, the rest being oxidized by isocitrate dehydrogenase and the further reactions of the citric acid cycle. The glyoxylate cycle provided malate not only to replenish the citric acid cycle for intermediates withdrawn for 2-oxoglutarate- and oxaloacetate-derived biosyntheses, but to an extent of 30% to generate PEP or pyruvate by decarboxylation. PEP carboxylase and pyruvate carboxylase did not serve an anaplerotic function under this growth condition. During growth on glucose plus acetate, the metabolization of glucose served primarily to generate the precursors for biosyntheses derived from intermediates of glycolysis and the pentose phosphate pathway. Taken together, the central metabolism of C. glutamicum cells growing on acetate plus glucose is characterized by a relatively low conversion rate of glucose to PEP/pyruvate, an intermediate activity of the citric acid cycle, and a glyoxylate cycle completely fulfilling the anaplerotic function.
Construction and analysis of a glyoxylate cycle-deficient C. glutamicum strain.
From the finding that in C. glutamicum cells growing on the mixture of acetate and glucose, the glyoxylate cycle completely fulfills the anaplerotic function, it might be deduced that this cycle is essential for the growth of C. glutamicum on minimal medium containing both substrates. In order to test this hypothesis, we constructed and analyzed a defined glyoxylate-deficient C. glutamicum mutant. By gene-directed mutagenesis, the aceA and aceB genes, encoding isocitrate lyase and malate synthase, respectively, were deleted from the chromosome of C. glutamicum WT (see Materials and Methods). The deletion of the chromosomal aceA-aceB locus was verified by Southern blot hybridization (data not shown), and the aceA-aceB mutant was designated C. glutamicum WTΔAB. In crude extracts of C. glutamicum WTΔAB grown in the presence of acetate, neither isocitrate lyase nor malate synthase activity was detectable (<0.01 U · [mg of protein]−1), whereas crude extracts of the parental strain, C. glutamicum WT, showed activities of both isocitrate lyase (1.05 U · [mg of protein]−1) and malate synthase (0.56 U · [mg of protein]−1). Comparative growth experiments on minimal media containing acetate, glucose, and acetate plus glucose as carbon sources were performed with C. glutamicum WT and the glyoxylate cycle-deficient strain C. glutamicum WTΔAB. Both strains grew comparably well on glucose, with growth rates of 0.33 and 0.31 h−1, respectively. In contrast to the parental WT strain, the glyoxylate cycle-deficient strain WTΔAB was not able to grow on acetate as a sole carbon source, corroborating our previous finding that the activities of isocitrate lyase and malate synthase are essential for growth on this substrate as a sole carbon source (36, 37). On minimal medium containing acetate plus glucose, C. glutamicum WTΔAB was able to grow; however, the growth rate was 0.27 h−1 compared to 0.34 h−1 (mean values from three independent cultivations by two determinations per experiment with relative errors of less than 5%) for the parental WT strain and thus was significantly lower. These results show that the glyoxylate cycle is essential for optimal growth of C. glutamicum when both acetate and glucose are present in the growth medium; however, they also show that the anaplerotic function of the glyoxylate cycle can be partly taken over by PEP carboxylase and/or pyruvate carboxylase.
DISCUSSION
The carbon and energy source acetate can serve as a substrate in fermentative amino acid production with C. glutamicum (17). The quantitative determination of metabolic fluxes during the growth of C. glutamicum WT on acetate as a sole carbon source reported here complements flux analyses of different C. glutamicum strains metabolizing either glucose or fructose (see, e.g., references 8, 26, 27, 46). During growth on acetate, the acetate consumption rate was in the range of 540 nmol · (mg of protein)−1 · min−1, which is much higher than expected from the previous kinetic characterization of the acetate carrier in glucose-grown cells (9). However, in acetate-grown cells, it might well be that the acetate carrier gene is up-regulated, as are the acetate utilization genes in C. glutamicum (53). The low biomass yield on acetate (Table 2) indicated an increased energy metabolism, as most of the substrate carbon is oxidized to carbon dioxide. Consistently, the TCA cycle activity was high in vivo (isocitrate dehydrogenase activity in vivo, 314 mU · [mg of protein]−1 [Fig. 2]), which represents an increase of approximately threefold compared to growth on glucose (isocitrate dehydrogenase activity in vivo, 111 mU · [mg of protein]−1 [Fig. 3]). The increased generation of energy equivalents by the TCA cycle satisfies the demand for gluconeogenesis and, moreover, might compensate for energy lost due to uncoupling of the membrane potential by acetate. Uncoupling by weak acids and its deleterious effect on growth have been demonstrated for E. coli and Clostridium thermoaceticum (1, 2) and may explain our finding that growth rates of C. glutamicum on acetate decrease with increasing acetate concentrations in the medium.
Acetate-glucose mixtures were coutilized by C. glutamicum and supported monophasic growth. During growth on glucose-acetate mixtures, the consumption rates for the individual carbon sources were reduced in such a way that the total carbon consumption was about the same as that during growth on either carbon source alone. This result indicates that either the uptake of acetate and of glucose or the in vivo carbon fluxes from glucose to acetyl-CoA and from acetate to acetyl-CoA are directly or indirectly regulated by the carbon source in the growth medium. In contrast to C. glutamicum, Azotobacter vinelandii preferentially uses acetate when grown on glucose-acetate mixtures, which is due to acetate-dependent inhibition of glucose uptake and glycolysis (15, 48). E. coli, too, does not coutilize glucose and acetate but preferentially uses glucose (4) due to carbon catabolite repression of the acetate-activating acetyl-CoA synthetase and probably also of the glyoxylate cycle enzymes isocitrate lyase and malate synthase in the presence of glucose (reviewed in reference 5). The metabolism of E. coli during growth on glucose-acetate mixtures is very similar in the first phase to growth on glucose as a sole carbon source and in the second phase to growth on acetate as a sole carbon source (50, 51). To determine whether, for C. glutamicum, metabolism of acetate-glucose mixtures is simply an overlay of glucose and acetate metabolism or whether metabolism of these substrate mixtures is characterized by a unique metabolic pattern, we performed metabolic flux analyses.
Catabolism of glucose to PEP/pyruvate during growth on acetate-glucose mixtures differed considerably from that during growth on glucose alone. During growth on glucose, the glycolytic formation of PEP and pyruvate from glyceraldehyde-3-phosphate (227 mU · [mg of protein]−1) was about 1.5-fold higher than the glucose uptake rate, which is in good agreement with previous studies on glucose-grown strains of C. glutamicum (26, 27, 28, 34, 49). Comparison of growth on glucose to growth on acetate plus glucose shows that the glucose uptake was reduced twofold, but the glycolytic formation of PEP/pyruvate from glyceraldehyde-3-phosphate was reduced fourfold. This reflects the fact that on acetate-glucose mixtures, glucose predominantly serves to generate precursors for biosyntheses starting with glycolytic intermediates, and the proportion of carbon oxidized to acetyl-CoA is greatly diminished compared to that during growth on glucose as a sole carbon source. Acetyl-CoA was derived almost exclusively from acetate when C. glutamicum was grown on acetate plus glucose (Fig. 4). The flux via the pentose phosphate shunt has been shown to be low when fructose is the sole carbon source (8) but high during growth on glucose (see also reference 26). Whereas it was low with acetate as the sole carbon source, flux via the pentose phosphate shunt is also high during growth on acetate-glucose mixtures.
The TCA cycle enzymes showed increasing in vivo activities in comparisons of growth on glucose, on glucose plus acetate, and on acetate (e.g., citrate synthase activity, 111, 219, and 413 mU · [mg of protein]−1, respectively). In C. glutamicum, citrate synthase levels vary little with the carbon source (specific activities in crude extracts, about 0.8 U · [mg of protein]−1) and the enzyme is inhibited by high concentrations of ATP and cis-aconitate (12). As its Km value for acetyl-CoA of 51 μM (12) is higher than the intracellular concentration of acetyl-CoA during growth on glucose but lower than that during growth on acetate (24 and 145 μM, respectively [53]), the in vivo activities of citrate synthase can be explained at least in part by the availability of the substrate acetyl-CoA.
C. glutamicum possesses two anaplerotic enzymes, PEP carboxylase and pyruvate carboxylase (31, 32, 33). During growth on glucose, PEP carboxylase and/or pyruvate carboxylase replenishes the TCA cycle with precursors for biosyntheses requiring TCA cycle intermediates. Besides the net conversion of PEP and pyruvate to oxaloacetate and malate of 30 mU · (mg of protein)−1, an exchange rate of 102 mU · (mg of protein)−1 could be determined, corroborating the findings of Marx et al. (27), who showed that during growth of C. glutamicum strain LE4 on glucose, the PEP/pyruvate-oxaloacetate/malate interconversion amounts to about twice the net flux towards PEP/pyruvate (27). It remains to be shown whether these exchange reactions constitute a futile cycle or whether a reaction cycle of ATP-dependent pyruvate carboxylase, NAD-dependent malate dehydrogenase, and NADPH-dependent malic enzyme replaces transhydrogenase as has been proposed (8, 18). The exchange reactions interconverting PEP/pyruvate and malate/oxaolacetate were also present when C. glutamicum was grown on either acetate or glucose plus acetate (54 and 61 mU · [mg of protein]−1, respectively). However, during growth on acetate or glucose plus acetate, there was no net formation of oxaloacetate/malate from PEP/pyruvate, but opposite, gluconeogenetic fluxes of 72 and 15 mU · (mg of protein)−1 were determined (Fig. 3 and 4). The fact that pyruvate carboxylase and PEP carboxylase did not serve an anaplerotic role during the growth of C. glutamicum on glucose plus acetate is surprising and could not be predicted from the measurements of enzyme levels, as both pyruvate carboxylase and PEP carboxylase show similar enzyme levels during growth on acetate compared to growth on glucose. Our finding is consistent with the acetyl-CoA inhibition of pyruvate carboxylase from C. glutamicum (Ki, 110 μM [32]) and an intracellular acetyl-CoA concentration of about 150 μM during growth on glucose plus acetate (53). PEP carboxylase of C. glutamicum is inhibited by aspartate (10), and it is possible that during growth on acetate or on glucose plus acetate, the intracellular aspartate pool is high due to sufficient formation of malate by the glyoxylate cycle and thus PEP carboxylase would also be inhibited. In order to completely comprehend the complex PEP/pyruvate branch point in corynebacterial metabolism, an experimental approach allowing the quantitative determination of which enzymes, and to which extent, are involved in the (inter)conversion of PEP/pyruvate and oxaloacetate/malate, will have to be pursued. Such an approach will include genetic analyses as well as 13C isotopomer analyses (3, 47; A. A. de Graaf, V. F. Wendisch, and H. Sahm, Abstr. XVIIth Int. Conf. Magn. Reson. Biol. Syst., abstr. TP25, 1996) that still need optimization before their application to complex metabolic systems.
The glyoxylate cycle constitutes an anaplerotic sequence alternative to carboxylation of PEP and/or pyruvate (18). In C. glutamicum the glyoxylate cycle is inactive during growth on glucose, and the metabolization of isocitrate in the TCA cycle mainly serves energy generation (Fig. 3). During growth on acetate, isocitrate is a metabolic branch point; 24% of isocitrate is fueled into the glyoxylate cycle, and 76% is converted further in the TCA cycle (Fig. 2). Metabolic fluxes at this branch point have also been determined in acetate-grown E. coli, and a similar relative conversion of isocitrate was found (28% via the glyoxylate cycle and 72% via isocitrate dehydrogenase [24, 50]). However, for growth on glucose plus acetate, the metabolic fluxes at the isocitrate branch point in both organisms clearly differ. In E. coli, the glyoxylate cycle is inactive in vivo under this condition (51), but surprisingly, the glyoxylate cycle is active in C. glutamicum (isocitrate conversion by the glyoxylate cycle and isocitrate dehydrogenase shows a relation of 23/77 [Fig. 4]). This difference between E. coli and C. glutamicum metabolism cannot be accounted for by the substrate affinities of the respective enzymes, as those are comparable (13, 35, 50), but is most probably due to differing regulatory control. In E. coli, the genes coding for isocitrate lyase (aceA) and malate synthase (aceB) are part of the aceBAK operon, which is repressed in the presence of glucose but derepressed during growth on acetate or fatty acids (reviewed in reference 5). aceK codes for the bifunctional isocitrate dehydrogenase kinase/phosphatase, which controls the phosphorylation status of isocitrate dehydrogenase (14, 22). Thus, during growth on acetate, there are high levels of isocitrate lyase and malate synthase, while isocitrate dehydrogenase is predominantly in its inactive, phosphorylated form. During the growth of E. coli on glucose-acetate mixtures, lower isocitrate lyase and malate synthase levels are present and isocitrate dehydrogenase is mostly in its active, unphosphorylated form. In C. glutamicum, however, there is no evidence for a similar modificatory control of isocitrate dehydrogenase, and transcription of the isocitrate lyase and malate synthase genes, which are not part of an operon, is increased in the presence of acetate regardless of the presence or absence of glucose (53). The relative metabolization of isocitrate by isocitrate dehydrogenase and by isocitrate lyase (76 to 24%) is consistent with their Km values of 12 and 280 μM, respectively, and allosteric regulation by glyoxylate and oxaloacetate (13, 35).
The quantitative determination of metabolic fluxes presented here led to the hypothesis that a glyoxylate cycle-deficient strain of C. glutamicum should be impaired in growth on glucose-acetate mixtures. To test this hypothesis, strain WTΔAB, which is devoid of isocitrate lyase and malate synthase activities, was constructed by targeted deletion of the aceA-aceB gene cluster on the C. glutamicum chromosome. In this strain accumulation of glyoxylate and side reactions of malate synthase such as might occur in the malate synthase-deficient strain ALB1 and the isocitrate lyase-deficient strain ASK1, respectively (35, 36), are avoided. Indeed, the glyoxylate cycle-deficient strain WTΔAB could cometabolize glucose and acetate but showed longer doubling times during growth on glucose-acetate mixtures than the WT. Thus, during growth on glucose plus acetate, PEP carboxylase and pyruvate carboxylase can only partly fulfill the anaplerotic function in C. glutamicum. Clearly, the glyoxylate cycle is required for optimal growth of C. glutamicum on glucose-acetate mixtures, verifying the physiological effect of this pathway alteration as predicted from our metabolic flux analyses.
ACKNOWLEDGMENTS
We thank Petra Peters-Wendisch and Lothar Eggeling for critical reading of the manuscript and Achim Marx for discussions.
This work was supported by EU grant BIO 4-CT96-0145. V.F.W. was a fellow of the Deutsche Forschungsgemeinschaft (DFG)-Graduiertenkolleg 'Molekulare Physiologie: Stoff- und Energieumwandlung' at the Heinrich-Heine-Universität Düsseldorf, Düsseldorf, Germany.
Footnotes
Dedicated to Rudolf K. Thauer on the occasion of his 60th birthday.
REFERENCES
- 1.Axe D D, Bailey J E. Transport of lactate and acetate through the energized cytoplasmic membrane of Escherichia coli. Biotechnol Bioeng. 1995;47:8–19. doi: 10.1002/bit.260470103. [DOI] [PubMed] [Google Scholar]
- 2.Baronofsky J J, Schreurs W J A, Kashket E R. Uncoupling by acetic acid limits growth of and acetogenesis by Clostridium thermoaceticum. Appl Environ Microbiol. 1984;48:1134–1139. doi: 10.1128/aem.48.6.1134-1139.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beale J M, Foster J L. Carbohydrate fluxes into alginate biosynthesis in Azotobacter vinelandii NCIB 8789: NMR investigations of the triose pools. Biochemistry. 1996;35:4492–4501. doi: 10.1021/bi951922v. [DOI] [PubMed] [Google Scholar]
- 4.Brown T D K, Jones-Mortimer M C, Kornberg H L. The enzymatic interconversion of acetate and acetyl-coenzyme A in Escherichia coli. J Gen Microbiol. 1977;102:327–336. doi: 10.1099/00221287-102-2-327. [DOI] [PubMed] [Google Scholar]
- 5.Clark D P, Cronan J E. Two-carbon compounds and fatty acids as carbon sources. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: American Society for Microbiology; 1996. pp. 343–357. [Google Scholar]
- 6.Cocaign M, Monnet C, Lindley N D. Batch kinetics of Corynebacterium glutamicum during growth on various substrates: use of substrate mixtures to localize metabolic bottlenecks. Appl Microbiol Biotechnol. 1993;40:526–530. [Google Scholar]
- 7.Dominguez H, Cocaign-Bousquet M, Lindley N D. Simultaneous consumption of glucose and fructose from sugar mixtures during batch growth of Corynebacterium glutamicum. Appl Microbiol Biotechnol. 1993;47:600–603. [Google Scholar]
- 8.Dominguez H, Rollin C, Guyonvarch A, Guerquin-Kern J L, Cocaign-Bousquet M, Lindley N D. Carbon flux distribution in the central metabolic pathways of Corynebacterium glutamicum during growth on fructose. Eur J Biochem. 1998;254:96–102. doi: 10.1046/j.1432-1327.1998.2540096.x. [DOI] [PubMed] [Google Scholar]
- 9.Ebbighausen H, Weil B, Krämer R. Carrier-mediated acetate uptake in Corynebacterium glutamicum. Arch Microbiol. 1991;155:505–510. [Google Scholar]
- 10.Eikmanns B J, Follettie M T, Griot M U, Sinskey A J. The phosphoenolpyruvate carboxylase gene of Corynebacterium glutamicum: molecular cloning, nucleotide sequence, and expression. Mol Gen Genet. 1989;218:330–339. doi: 10.1007/BF00331286. [DOI] [PubMed] [Google Scholar]
- 11.Eikmanns B J, Metzger M, Reinscheid D J, Sahm H. Amplification of three threonine biosynthesis genes in Corynebacterium glutamicum and its influence on carbon flux in different strains. Appl Microbiol Biotechnol. 1991;102:93–98. doi: 10.1007/BF00167910. [DOI] [PubMed] [Google Scholar]
- 12.Eikmanns B J, Thum-Schmitz N, Eggeling L, Lüdtke K, Sahm H. Nucleotide sequence, expression, and transcriptional analysis of the Corynebacterium glutamicum gltA gene encoding citrate synthase. Microbiology. 1994;140:1817–1828. doi: 10.1099/13500872-140-8-1817. [DOI] [PubMed] [Google Scholar]
- 13.Eikmanns B J, Rittmann D, Sahm H. Cloning, sequence analysis, expression, and inactivation of the Corynebacterium glutamicum icd gene encoding isocitrate dehydrogenase and biochemical characterization of the enzyme. J Bacteriol. 1995;177:774–782. doi: 10.1128/jb.177.3.774-782.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garnak M, Reeves H C. Phosphorylation of isocitrate dehydrogenase of Escherichia coli. Science. 1979;203:1111–1112. doi: 10.1126/science.34215. [DOI] [PubMed] [Google Scholar]
- 15.George S E, Costenbader C J, Melton T. Diauxic growth in Azotobacter vinelandii. J Bacteriol. 1985;164:866–871. doi: 10.1128/jb.164.2.866-871.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harder W, Dijkhuizen L. Strategies of mixed substrate utilization in microorganisms. Phil Trans R Soc Lond. 1982;297:459–480. doi: 10.1098/rstb.1982.0055. [DOI] [PubMed] [Google Scholar]
- 17.Kinoshita S, Tanaka K. Glutamic acid. In: Yamada K, editor. The microbial production of amino acids. New York, N.Y: John Wiley; 1972. pp. 263–324. [Google Scholar]
- 18.Kornberg H L. The role and control of the glyoxylate cycle in Escherichia coli. Biochem J. 1966;99:1–11. doi: 10.1042/bj0990001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koshland D E., Jr The era of pathway quantification. Science. 1998;280:852–853. doi: 10.1126/science.280.5365.852. [DOI] [PubMed] [Google Scholar]
- 20.Kovarova-Kovar K, Egli T. Growth kinetics of suspended microbial cells: from single-substrate-controlled growth to mixed-substrate kinetics. Microbiol Mol Biol Rev. 1998;62:646–666. doi: 10.1128/mmbr.62.3.646-666.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krämer R, Lambert C, Hoischen C, Ebbighausen H. Uptake of glutamate in Corynebacterium glutamicum. 1. Kinetic properties and regulation by internal pH and potassium. Eur J Biochem. 1990;194:929–935. doi: 10.1111/j.1432-1033.1990.tb19488.x. [DOI] [PubMed] [Google Scholar]
- 22.LaPorte D C, Koshland D E., Jr A protein with kinase and phosphatase activities involved in regulation of the tricarboxylic acid cycle. Nature. 1982;300:458–460. doi: 10.1038/300458a0. [DOI] [PubMed] [Google Scholar]
- 23.Liebl W. The genus Corynebacterium—nonmedical. In: Balows A, Trüper H G, Dworkin M, Harder W, Schleifer K H, editors. The procaryotes. Vol. 2. New York, N.Y: Springer; 1991. pp. 1157–1171. [Google Scholar]
- 24.London R E. 13C labeling in studies of metabolic regulation. Prog NMR Spectrosc. 1988;20:337–383. [Google Scholar]
- 25.Lundberg P, Harmsen E, Ho C, Vogel H J. Nuclear magnetic resonance studies of cellular metabolism. Anal Biochem. 1990;191:193–222. doi: 10.1016/0003-2697(90)90210-z. [DOI] [PubMed] [Google Scholar]
- 26.Marx A, de Graaf A A, Wiechert W, Eggeling L, Sahm H. Determination of the fluxes in the central metabolism of Corynebacterium glutamicum by nuclear magnetic resonance spectroscopy combined with metabolite balancing. Biotechnol Bioeng. 1996;49:111–129. doi: 10.1002/(SICI)1097-0290(19960120)49:2<111::AID-BIT1>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 27.Marx A, Striegel K, de Graaf A A, Sahm H, Eggeling L. Response of the central metabolism of Corynebacterium glutamicum to different flux burdens. Biotechnol Bioeng. 1997;56:168–180. doi: 10.1002/(SICI)1097-0290(19971020)56:2<168::AID-BIT6>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 28.Marx A, Eikmanns B J, Sahm H, de Graaf A A. Response of the central metabolism in Corynebacterium glutamicum to the use of an NADH-dependent glutamate dehydrogenase. Metab Eng. 1999;1:35–48. doi: 10.1006/mben.1998.0106. [DOI] [PubMed] [Google Scholar]
- 29.Monod J. The growth of bacterial cultures. Annu Rev Microbiol. 1949;3:371–394. [Google Scholar]
- 30.Peters-Wendisch P G, Eikmanns B J, Thierbach G, Bachmann B, Sahm H. Phosphoenolpyruvate carboxylase in Corynebacterium glutamicum is dispensable for growth and lysine production. FEMS Microbiol Lett. 1993;112:269–274. [Google Scholar]
- 31.Peters-Wendisch P G, Wendisch V F, de Graaf A A, Eikmanns B J, Sahm H. C3-carboxylation as an anaplerotic reaction in phosphoenolpyruvate carboxylase-deficient Corynebacterium glutamicum. Arch Microbiol. 1996;165:387–396. doi: 10.1007/s002030050342. [DOI] [PubMed] [Google Scholar]
- 32.Peters-Wendisch P G, Wendisch V F, Paul S, Eikmanns B J, Sahm H. Pyruvate carboxylase as an anaplerotic enzyme in Corynebacterium glutamicum. Microbiology. 1997;143:1095–1103. doi: 10.1099/00221287-143-4-1095. [DOI] [PubMed] [Google Scholar]
- 33.Peters-Wendisch P G, Kreutzer C, Kalinowski J, Patek M, Sahm H, Eikmanns B J. Pyruvate carboxylase from Corynebacterium glutamicum: characterization, expression and inactivation of the pyc gene. Microbiology. 1998;144:915–927. doi: 10.1099/00221287-144-4-915. [DOI] [PubMed] [Google Scholar]
- 34.Pons A, Dussap C G, Pequinot C, Gros J B. Metabolic flux distribution in Corynebacterium melassecola ATCC 17965 for various carbon sources. Biotechnol Bioeng. 1996;51:177–189. doi: 10.1002/(SICI)1097-0290(19960720)51:2<177::AID-BIT7>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 35.Reinscheid D J, Eikmanns B J, Sahm H. Characterization of the isocitrate lyase gene from Corynebacterium glutamicum and biochemical analysis of the enzyme. J Bacteriol. 1994;176:3474–3483. doi: 10.1128/jb.176.12.3474-3483.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reinscheid D J, Eikmanns B J, Sahm H. Malate synthase from Corynebacterium glutamicum: sequence analysis of the gene and biochemical characterization of the enzyme. Microbiology. 1994;140:3099–3108. doi: 10.1099/13500872-140-11-3099. [DOI] [PubMed] [Google Scholar]
- 37.Reinscheid D J, Schnicke S, Rittmann D, Zahnow U, Sahm H, Eikmanns B J. Cloning, sequence analysis, expression and inactivation of the Corynebacterium glutamicum pta-ack operon encoding phosphotransacetylase and acetate kinase. Microbiology. 1999;145:503–513. doi: 10.1099/13500872-145-2-503. [DOI] [PubMed] [Google Scholar]
- 38.Salou P, Loubiere P, Pareilleux A. Growth and energetics of Leuconostoc oenos during cometabolism of glucose with citrate or fructose. Appl Environ Microbiol. 1994;60:1459–1466. doi: 10.1128/aem.60.5.1459-1466.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 40.Schäfer A, Tauch A, Jäger W, Kalinowski J, Thierbach G, Pühler A. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene. 1994;145:69–73. doi: 10.1016/0378-1119(94)90324-7. [DOI] [PubMed] [Google Scholar]
- 41.Scharfstein S T, Tucker S N, Mancuso A, Blanch H W, Clark D S. Quantitative in vivo nuclear magnetic resonance studies of hybridoma metabolism. Biotechnol Bioeng. 1994;43:1059–1074. doi: 10.1002/bit.260431109. [DOI] [PubMed] [Google Scholar]
- 42.Schrumpf B, Schwarzer A, Kalinowski J, Pühler A, Eggeling L, Sahm H. A functional split pathway for lysine synthesis in Corynebacterium glutamicum. J Bacteriol. 1991;173:4510–4516. doi: 10.1128/jb.173.14.4510-4516.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sequin U, Scott A I. Carbon-13 as a label in biosynthetic studies. Science. 1974;186:101–107. doi: 10.1126/science.186.4159.101. [DOI] [PubMed] [Google Scholar]
- 44.Sonntag K, Eggeling L, de Graaf A A, Sahm H. Flux partitioning in the split pathway of lysine synthesis in Corynebacterium. Eur J Biochem. 1995;213:1325–1331. doi: 10.1111/j.1432-1033.1993.tb17884.x. [DOI] [PubMed] [Google Scholar]
- 45.Sonntag K, Schwinde J, de Graaf A A, Marx A, Eikmanns B J, Wiechert W, Sahm H. 13C NMR studies of the fluxes in the central metabolism of Corynebacterium glutamicum during growth and overproduction of amino acids in batch cultures. Appl Microbiol Biotechnol. 1995;44:489–495. [Google Scholar]
- 46.Stephanopoulos G, Vallino J J. Network rigidity and metabolic engineering in metabolite overproduction. Science. 1991;252:1675–1681. doi: 10.1126/science.1904627. [DOI] [PubMed] [Google Scholar]
- 47.Szyperski T. Biosynthetically directed fractional 13C-labeling of proteinogenic amino acids. Eur J Biochem. 1996;232:433–448. doi: 10.1111/j.1432-1033.1995.tb20829.x. [DOI] [PubMed] [Google Scholar]
- 48.Tauchert K, Jahn A, Oelze J. Control of diauxic growth of Azotobacter vinelandii on acetate and glucose. J Bacteriol. 1990;172:6447–6451. doi: 10.1128/jb.172.11.6447-6451.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vallino J J, Stephanopoulos G. Metabolic flux distributions in Corynebacterium glutamicum during growth and lysine overproduction. Biotechnol Bioeng. 1993;41:633–646. doi: 10.1002/bit.260410606. [DOI] [PubMed] [Google Scholar]
- 50.Walsh K, Koshland D E., Jr Determination of flux through the branch point of two metabolic cycles. J Biol Chem. 1984;259:9646–9654. [PubMed] [Google Scholar]
- 51.Walsh K, Koshland D E., Jr Branch point control by the phosphorylation state of isocitrate dehydrogenase. J Biol Chem. 1985;260:8430–8437. [PubMed] [Google Scholar]
- 52.Wendisch V F, de Graaf A A, Sahm H. Accurate determination of 13C enrichments in nonprotonated carbon atoms of isotopically enriched amino acids by 1H nuclear magnetic resonance. Anal Biochem. 1997;245:196–202. doi: 10.1006/abio.1996.9977. [DOI] [PubMed] [Google Scholar]
- 53.Wendisch V F, Spies M, Reinscheid D J, Schnicke S, Sahm H, Eikmanns B J. Regulation of acetate metabolism in Corynebacterium glutamicum: transcriptional control of the isocitrate lyase and malate synthase genes. Arch Microbiol. 1997;168:262–269. doi: 10.1007/s002030050497. [DOI] [PubMed] [Google Scholar]
- 54.Wiechert W, de Graaf A A. Bidirectional reaction steps in metabolic networks. Part I. Modeling and simulation of carbon labelling experiments. Biotechnol Bioeng. 1997;55:101–117. doi: 10.1002/(SICI)1097-0290(19970705)55:1<101::AID-BIT12>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 55.Wiechert W, Siefke C, Marx A, de Graaf A A. Bidirectional reaction steps in metabolic networks. Part II. Flux estimation and statistical analysis. Biotechnol Bioeng. 1997;55:118–135. doi: 10.1002/(SICI)1097-0290(19970705)55:1<118::AID-BIT13>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]