Abstract
Helicobacter pylori is the most prevalent pathogen causing chronic gastritis, gastroduodenal ulcers, and gastric tumors and is asymptomatically present in 50% of the world's population. This research is focused on investigating the effect of Lactobacillus paracasei ZFM 54 (CCTCC NO:2016667) on attenuating H. pylori-induced gastritis. H. pylori ZJC03 isolated from a patient with gastritis harbored the virulence genes of vacA and cagA and was highly resistant to metronidazole (MIC > 256 μg/mL). In vitro analysis revealed that the potential anti-H. pylori characteristics of L. paracasei ZFM54 in terms of 65.57 ± 1.87% survival rate in simulated gastric juices at a pH of 2.0, 69.00 ± 2.73% auto-aggregation, 30.28 ± 2.24% co-aggregation, 70.27 ± 2.23% urease inhibition, and 57.89 ± 1.27% radical scavenging. In H. pylori infectious mice, L. paracasei ZFM54 pre- and post-treatment reduced the levels of malondialdehyde in liver tissues to 0.71 ± 0.04 nmol/mgprot (p < 0.05) and 0.70 ± 0.06 nmol/mgprot (p < 0.05), respectively. Glutathione levels were increased to 1.78 ± 0.02 μmol/gprot (p < 0.05) and 1.76 ± 0.52 μmol/gprot (p < 0.05), respectively. L. paracasei ZFM54 significantly inhibited H. pylori-mediated inflammation observed in gastric mucosal repair and downregulated the mRNA expression of pro-inflammatory cytokines IFN-γ, IL-1β, and IL-6 (p < 0.01). Importantly, L. paracasei ZFM54 increased Firmicutes and Actinobacteriota and decreased the relative abundance of bacterial taxa belonging to Campilobacterota and Proteobacteria. With the preventive and therapeutic administration of L. paracasei ZFM54, significant reductions in the average relative abundance of genera Helicobacter, Muribaculum, Staphylococcus, Lachnospiraceae_NK4A136_group, Prevotellaceae_UCG-001, Alloprevotella, and Oscillibacter were observed compared to infected mice. These findings suggest that L. paracasei ZFM 54 has the potential to protect against H. pylori infection by ameliorating inflammation and restoring the gastric microbiota.
Keywords: Lactobacillus paracasei ZFM54, Helicobacter pylori, gastritis, gastric microbiota, inflammation cytokines
Introduction
Helicobacter pylori is the only bacterium reported to cause cancer and has been grouped as a class I carcinogen by the International Agency for Research on Cancer (IARC). H. pylori infection is the main risk factor for gastric duodenal ulcer, tumor, and lymphoma. Approximately, half of the population on the earth is infected with H. pylori (1). A high prevalence of H. pylori infection was reported in developing countries with approximately 80% prevalence in China and some states of South America and Eastern Europe (2). The major virulence genes of H. pylori are vacuolating cytotoxin A (vac A) and cytotoxin-associated gene A (cagA), which are closely related to pathogenesis (3). VacA exists in almost all H. pylori, including the signaling region (s1 and s2 alleles) and the middle region (m1 and m2 alleles), while cagA, regarded as a virulence marker, is positive in 60–70% of H. pylori strains (4). Treatment of H. pylori infection includes two antibiotics and a proton pump inhibitor (PPI), but the emergence of antibiotic resistance in H. pylori presents a great challenge (5). Side effects of antibiotics (nausea, vomiting, and diarrhea) often lead to patients' non-compliance. Recurrence of infection after treatment is also a threat, especially in high prevalence areas (6). The declining rate of successful treatment due to antibiotic resistance, side effects, and patients' non-compliance prompts researchers to seek alternative therapy to control the risk of H. pylori infection and associated gastroduodenal ulcers and tumors (7).
Lactobacillus species are the most commonly studied genera to have potential probiotic characteristics and have been extensively studied to control H. pylori infection (8, 9). In Deng et al.'s study, H. pylori could only infect specific pathogen-free mice, while pre-gavage with L. paracasei FJ861111.1 inhibited the colonization of H. pylori in gastric tissue (10). L. paracasei HP7 associated with extracts of Perilla frutescens and Glycyrrhiza glabra inhibited H. pylori with virulence genes such as alpA, cagA, flaA, and ureA and significantly reduced the infection rate of H. pylori in mice (11). L. paracasei strain 06TCa19 was shown to suppress H. pylori-mediated inflammatory chemokines of IL-8 and regulated the activation of normal T-cell expression and secretion in MKN45 cells (12).
The mechanism of action of Lactobacillus has been investigated, but the exact underlying mechanism is largely unknown. Lactobacillus has been demonstrated to inhibit the urease enzyme activity of H. pylori in in vitro, in vivo, and clinical trials (9). Inhibition of urease would not allow H. pylori to colonize the gastric mucosa and further protect from H. pylori-mediated inflammation. Secretion of antimicrobial substances (H2O2, lactic acid, bacteriocin, etc.), competition for adhesion receptors, co-aggregation ability, strengthening epithelial barrier, and immunomodulation are the main actions of Lactobacillus against pathogens (13, 14). H. pylori also induced oxidative damage to the host using its multiple antigens such as lipopolysaccharides and the type IV secretory system (15). As a consequence, neutrophils produce free radicals to combat H. pylori and cause the accumulation of different reactive oxygen species (ROS), which subsequently damage the host's cellular membranes producing malondialdehyde (MDA), which is a hallmark of an H. pylori infection (16). Furthermore, the production of anti-oxidant metabolites such as glutathione (GSH) and butyrate helps Lactobacillus block free radicals with glutathione peroxidase (GSH-Px) (17). Not all Lactobacilli possess probiotic potential, and even the differences occur within the same species; therefore, the characteristics of newly identified strains should be studied in detail.
The present study study is aimed at investigating the potential of L. paracasei ZFM54 (CCTCC NO:M 2016667) to ameliorate gastric inflammation, oxidative stress, and microbiota dysbiosis in the mice model by administering L. paracasei ZFM54 before (prevention) and after (therapy) H. pylori challenge. The probiotic potential of L. paracasei ZFM54 was first examined in in vitro experiments to get insight into the strain's efficacy to withstand the harsh gastric environment and inhibition against H. pylori.
Materials and methods
Bacterial culture
L. paracasei ZFM 54 isolated from healthy infant's feces was stored at the China Center for Type Culture Collection (Wuhan, China) and sub-cultured in DeMan–Rogosa–Sharpe (MRS) broth at 37°C for 24 h. Cell-free supernatant (CFS) was obtained by centrifuging at 8,000 ×g at 4°C for 15 min.
H. pylori ZJC03 (CCTCC NO: M 20211218) was provided by the Zhejiang University School of Medicine and was previously isolated from patients with gastritis. H. pylori was inoculated on Columbia Blood Agar (CBA) broth supplemented with sheep's blood (7%, v/v) (Hopebio, Qingdao, China) and antibiotics (10 mg/mL vancomycin, 5 mg/mL amphotericin, 2.5 units/mL polymyxin B, and 5 mg/mL trimethoprim) (Sangon Biotech, Shanghai, China). The plates were kept in an anaerobic box containing a micro-aerobic pack (Anaeropack-MicroAero, Mitsubishi, Japan) and incubated at 37°C for 48–72 h. For liquid culture, the bacteria were scraped off and sub-cultured in Brain Heart Infusion (BHI) broth in microaerophilic condition at 37°C for 48 h.
Simulated gastric juice tolerance
The simulated gastric juice tolerance assay of L. paracasei ZFM54 was detected according to Charteris et al. (18) with some modifications. Bacterial cells were centrifuged (4°C, 8,000 ×g) for 10 min and suspended in an equal volume of PBS (pH 7) after 18–20 h of culture. The pH of 1 g/L pepsin (Sigma Aldrich, USA) was set to 2, 3, and 4, respectively. A mixture of 100 μL cell suspension, 300 μL NaCl solution (0.5% w/v), and 1 mL pepsin with different pH was cultured at 37°C. Aliquots were removed at 0 h, 2 h, and 4 h, diluted, and enumerated for viability by plate count. The survival rate (%) was calculated as below:
| (1) |
Auto-aggregation assay
Auto-aggregation of L. paracasei ZFM54 was assayed as reported by Ahire et al. (19) with slight modifications. The initial optical density of the cell suspension at 600 nm was set to 0.5 and incubated at 37°C for 4 h. The OD600 was recorded every 1 h. The auto-aggregation rate (%) was measured by the following formula:
| (2) |
where A0 is the absorbance at 0 h, and At is the absorbance at time t = 1, 2, 3, and 4 h.
Virulence genes
Genomic DNA of H. pylori ZJC03 was extracted by genome extraction kits (Takara, Beijing, China) according to the manufacturer's instructions. Virulence genes of cagA, vacA s1/s2, and vacA m1/m2 were amplified, and the primers were synthesized (Sangon, Shanghai, China) and are listed in Table 1. PCR cycling procedure is as follows: initial denaturation step at 95°C for 10 min; 35 cycles of denaturation at 95°C for 50 s, annealing at 55°C for 30 s, extension at 72°C for 30 s; final extension at 72°C for 5 min; and incubation at 4°C. The amplified products were verified by gel electrophoresis (1% agarose). The size of DNA fragment was estimated using a 500-bp DNA Marker.
Table 1.
Primer sequences for virulence genes.
| Primer | DNA sequence (5′-3′) | DNA size (bp) | |
|---|---|---|---|
| cagA | F | TTGACCAACAACCACAAACCGAAG | 183 |
| A | CTTCCCTTAATTGCGAGATTCC | ||
| vacA s1/s2 | F | ATGGAAATACAACAAACACAC | 259/286 |
| A | CTGCTTGAATGCGCCAAAC | ||
| vacA m1/m2 | F | CAATCTGTCCAATCAAGCGAG | 567/642 |
| A | GCGTCAAAATAATTCCAAGG | ||
F, forward; R, reverse.
Antibiotic sensitivity assay
Susceptibility of H. pylori ZJC03 to amoxicillin, metronidazole, tetracycline, and clarithromycin was evaluated by the epsilometric method (E-test) (20). The E-test strip containing antibiotics was attached to the surface of the agar medium containing H. pylori (108 CFU/mL) and cultured in a microaerophilic tank (85 N2, 10 CO2, and 5% O2) at 37°C for 48 h−72 h. The minimal inhibitory concentration (MIC) was determined by the point of the antibacterial ring and the strip.
Co-aggregation assay
Co-aggregation ability was assayed according to Kos et al. (21). The cells were washedhree times with PBS (pH 7.0), and the initial optical density was set to 0.50 at 600 nm. Same volumes of L. paracasei ZFM54 (109 CFU/mL) and H. pylori ZJC03 (108 CFU/mL) cell suspension were mixed, vortexed for 10 s, and incubated at 37°C for 4 h. OD600 was measured every 1 h. The co-aggregation rate (%) was measured by the following formula:
| (3) |
where Ax is the absorbance of L. paracasei ZFM54 at 0 h, Ay is the absorbance of H. pylori at 0 h, and At is the absorbance of the mixture at 1, 2, 3, and 4 h.
Anti-H. pylori activity in vitro
As previously described by Hirano et al., antibacterial activity was determined by the Oxford cup assay with slight modifications (22). H. pylori ZJC03 cell cultured in a semi-liquid BHI medium containing 7% sheep's blood was poured onto agar plates in sterilized Oxford cups. After solidification, the Oxford cups were pulled out to form wells, and 100 μL of L. paracasei ZFM 54 cell (109 CFU/mL) in phosphate-buffered solution (PBS) and CFS was added into different wells.
Urease inhibition assay
Urease activity was used as the activity index of H. pylori ZJC03 by the phenol red method (23) with modification. In a 96-well microtiter plate, the bacterial cultures were added: (i) 40 μL H. pylori cells (108 CFU/mL) and 10 μL L. paracasei 54 cells (109 CFU/mL), (ii) 40 μL H. pylori cells (108 CFU/mL) and 10 μL L. paracasei ZFM54 CFS, and (iii) 40 μL H. pylori cells (108 CFU/mL) and 10 μL PBS. The 96-well microtiter plate was incubated at 37°C for 48 h under microaerophilic condition (85 N2, 10 CO2, and 5% O2). Urease indicator (0.9% NaC1, 20 mmo1/L Urea, 14 μg/mL phenol red, pH 6.8) was added to each well, and the absorbance was measured at 540 nm. The inhibition rate was calculated by the following formula:
| (4) |
Free radical scavenging activity
The anti-oxidant activity was assessed by the 2,2-diphenyl-1-picrylhydrazyl (DPPH) free radical scavenging activity according to Saravanakumar et al. (24) with some modifications. About 1 mL L. paracasei ZFM54 cell culture (109 CFU/mL) and 1 mL CFS were cultured in combination with 1 ml DPPH ethanol solution (0.2 mM) respectively. Then, 1 mL DPPH mixed with 1 mL saline was used as the control. All tubes were vortexed and placed in the dark at room temperature for 30 min. The absorbance was measured at 517 nm. The DPPH scavenging activity (%) was calculated by the following formula:
| (5) |
where A1 is the absorbance of the control and A2 is the absorbance of the sample.
Animal model
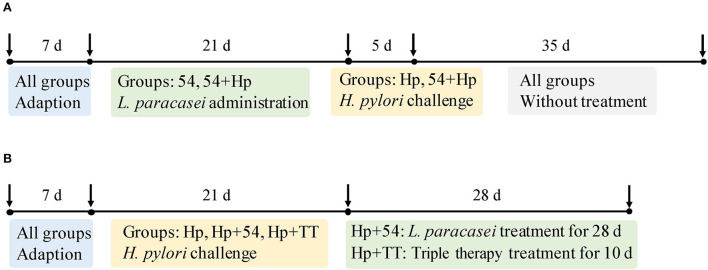
The animal experiment was conducted in accordance with the ethical guidelines for animal use and with permission from Shanghai Public Health Clinical Center Animal Ethics Committee. Specific pathogen-free C57BL/6 mice (6-week-old, 18–20 g) were obtained from Sino-British SIPPR/BK Lab Animal Ltd. (Shanghai, China), and the experiment was carried out in the Laboratory Animal Department of Shanghai Public Health Clinical Center. Mice were adapted to the controlled condition (24 ± 2°C room temperature and 50 ± 5% relative humidity) for a week prior to the experiment. All mice were grouped into two experimental approaches including preventive administration and therapeutic administration, as seen in Figure 1. Six mice in each subgroup were randomly scarificed for sampling and testing.
Figure 1.
Grouping of C57BL/6 mice for prevention and therapeutic models. (A) The preventive groups of control (normal mice without prevention and infection in the preventive group), Hp (mice infected with H. pylori without prevention in the preventive group), 54 (only treated with L. paracasei ZFM54), and 54+Hp (pretreatment with L. paracasei ZFM54 before H. pylori challenge). (B) The therapeutic groups of control' (normal mice without prevention and infection in the therapeutic group), Hp' (mice infected with H. pylori without treatment in the therapeutic group), Hp+54 (treatment with L. paracasei ZFM54 after H. pylori challenge), and Hp+TT (treatment with triple therapy after H. pylori challenge).
Preventive trials
Mice in the control group (normal mice) were free to eat and drink without infection. On a group of 54 mice, with only L. paracasei ZFM54 (109 CFU/mL) was administrated for 21 days without H. pylori challenge. After 21-day intragastrical administration of L. paracasei ZFM54, mice in the groups of Hp (negative control) and 54+Hp (prevention with L. paracasei ZFM54) were challenged with H. pylori ZJC03 (108 CFU/mL, 400 μL) three times (every other day). After infection, all mice were fed with a normal diet for 35 days and finally sacrificed by cervical dislocation.
Therapeutic trials
Mice in the control' group (normal mice) were free to eat and drink without H. pylori ZJC03 infection. Mice in the groups of Hp' (negative control), Hp+54 (therapy with L. paracasei ZFM54), and Hp+TT (therapy with triple therapy) were challenged intragastrically with H. pylori (108 CFU/mL, 400 μL) three times a week for 21 days. Then, mice in the groups of Hp+54 and Hp+TT were fed intragastrically with 400 μL L. paracasei ZFM54 (109 CFU/mL) for 28 days and triple therapy for 10 days, respectively. Triple therapy (TT) consists of PPI (5.2 mg/kg), amoxicillin (260 mg/kg), and clarithromycin (65 mg/kg).
MDA and GSH assays
Liver tissues of mice were homogenized and centrifuged at 4000 ×g for 10 min. The MDA level was determined using thiobarbituric acid (TBA) assay kit according to the manufacturer's instructions. One molecule of MDA reacts with two molecules of TBA to form thiobarbituric acid reactive substance (TBARS) at high temperature and acidic conditions and was measured at 532 nm. The following formula was used to calculate MDA content in liver tissues:
| (6) |
where Cs is the standard concentration (10 nmol/mL), and Cp is the protein concentration of samples before being measured (mgprot/mL).
Tissue GSH level was determined using the GSH assay kit by the manufacturer's instructions. The reaction of 5, 5-Dithiobis(2-nitrobenzoic acid) (DTNB) with GSH takes place to produce a yellow chromogen, and the absorbance is measured at 405 nm. The calculation formula is as follows:
| (7) |
where Cs is the standard concentration (20 mM), Cp is the protein concentration of samples before being measured (mgprot/mL), and the dilution factor of samples is 2 times.
Hematoxylin and eosin (H&E) staining
Histopathological alterations of gastric mucosa in preventive and therapeutic groups were evaluated by H&E staining. Gastric tissues of mice were fixed in 4% paraformaldehyde for 48 h at room temperature, were dehydrated, waxed, embedded, and sectioned according to Ahire et al. (19). H&E images were observed under a NIKON Eclipse Ci microscope assisted with a NIKON digital sight DS-FI2 (NIKON Corporation, Tokyo, Japan).
Real-time quantitative polymerase chain reaction (RT-qPCR)
The mRNA expression of IL-1β, IFN-γ, IL-6, and IL-10 in the stomach tissues in the preventive and therapeutic groups was quantitatively analyzed by RT-qPCR (25). Total RNA from gastric tissue was extracted by TRIzol reagent (Servicebio, Wuhan, China) and reversed-transcribed into single-strand cDNA by RevertAid First Strand cDNA Synthesis Kit according to the manufacturer's instruction (Thermo Scientific, Vilnius, Lithuania). qPCR was conducted using FastStart Universal SYBR Green Master (Rox) (Servicebio, Wuhan, China) with the primers of inflammatory cytokines (Table 2). The cycling program consisted of initial denaturation at 95°C for 10 min followed by 40 cycles at 95°C for 15s, 60°C for 15 s, 60°C for 60 s, and 95°C for 15 s. The gene expression was calculated using the 2−ΔΔCt method, with glyceraldehyde-3-phosphate dehydrogenase (GADPH) as the reference gene (21).
Table 2.
Primers used for inflammatory cytokines.
| Gene | Primers | DNA sequences (5'-3') | Segment length (bp) | RefSeq ID |
|---|---|---|---|---|
| GAPDH | S | CCTCGTCCCGTAGACAAAATG | 133 | NM_008084.2 |
| A | TGAGGTCAATGAAGGGGTCGT | |||
| IFN-γ | S | CTCAAGTGGCATAGATGTGGAAG | 251 | NM_008337.4 |
| A | TGACCTCAAACTTGGCAATACTC | |||
| IL-1β | S | TCAAATCTCGCAGCAGCACATC | 206 | NM_008361 |
| A | CGTCACACACCAGCAGGTTATC | |||
| IL-6 | S | CCCCAATTTCCAATGCTCTCC | 141 | NM_031168.2 |
| A | CGCACTAGGTTTGCCGAGTA | |||
| IL-10 | S | TTTAAGGGTTACTTGGGTTGCC | 106 | NM_010548.2 |
| A | AATGCTCCTTGATTTCTGGGC |
16S rRNA sequencing
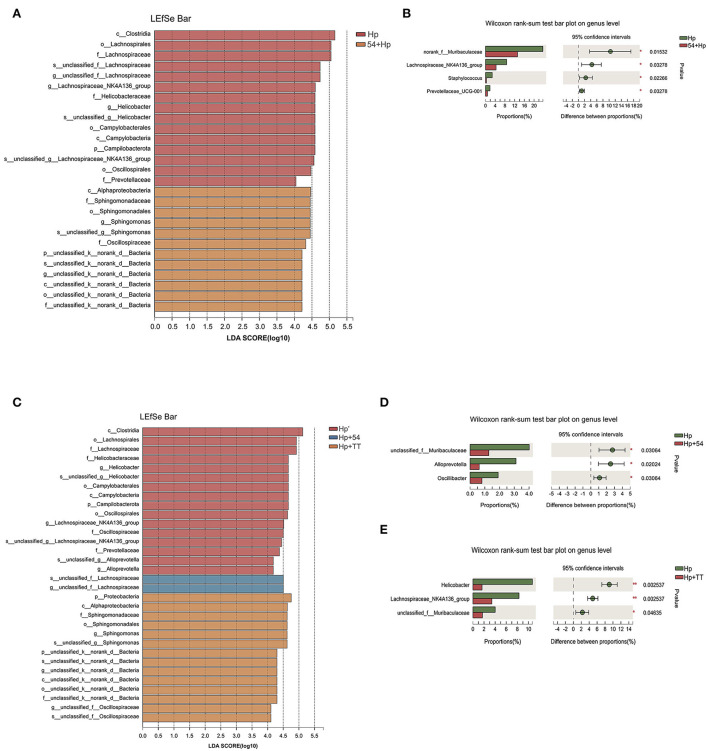
The V3-V4 region of the 16S rRNA gene of gastric microbiota was sequenced using the QIIME pipeline (26). According to SILVA 138/16s database, paired-end reads were inferred by DADA2, linked to tags, and clustered to Amplicon Sequence Variant (ASV) with 100% similarity. Alpha diversity-related community richness (Chao index and Ace index) and community diversity (Shannon index and Simpson index) were analyzed. Beta diversity analysis was performed by principal coordinate analysis (PCoA) based on unweighted-uniFrac distances. The difference between the classes and the features such as organism, clades, and ASV was analyzed by the linear discriminant analysis effect size (LEfSe) method (LDA > 4.0). The relative abundance differences of the genus Helicobacter between the Hp group and the preventive or therapeutic groups were further analyzed.
Statistical analysis
All results are expressed as mean value ± standard deviation (SD). Graphs of in vitro indexes, MDA, GSH, and inflammatory cytokines were drawn using GraphPad Prism 9.0, and the differences between the two groups were compared by Tukey's test or Student's t-test. The differences in gastric microbiota were analyzed using the Wilcoxon rank-sum test. A p-value < 0.05 was considered statistically significant.
Results
pH tolerance, auto-aggregation, and co-aggregation properties of L. paracasei ZFM54
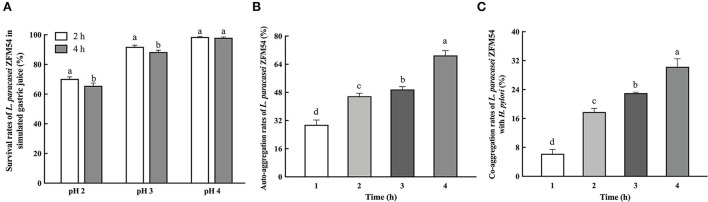
Probiotic strains need to resist low pH to survive and colonize in the host's stomach. The survival rate of L. paracasei ZFM54 was 65.57 ± 1.87%, 88.37 ± 1.19%, and 97.99 ± 0.50% at pH 2, 3, and 4 for 4 h, respectively (Figure 2A). These findings indicated that L. paracasei ZFM54 can normally grow and meet the requirements for probiotic growth in vivo. The auto-aggregation ability of L. paracasei ZFM54 was checked, which is the ability to interact non-specifically with own cells. The auto-aggregation rate was 29.60 ± 2.73% after 1 h, which gradually increased to 45.80 ± 1.75% after 2 h and 69.00 ± 2.73% after 4 h (Figure 2B). L. paracasei ZFM54 was also able to co-aggregate with H. pylori ZJC03 in vitro at 6.23 ± 1.23% after 1 h, 17.78 ± 1.02% after 2 h, and 30.28 ± 2.24% after 4 h, respectively (Figure 2C).
Figure 2.
Tolerance to simulated gastric juice (A), auto-aggregation rate (B), and co-aggregation rate (C) of L. paracasei ZFM54. The results are expressed as mean ± standard deviation (n = 3), and columns with different lowercase letters were significantly different (p < 0.05) using Tukey's test.
Virulence genes and antibiotic susceptibility of H. pylori ZJC03
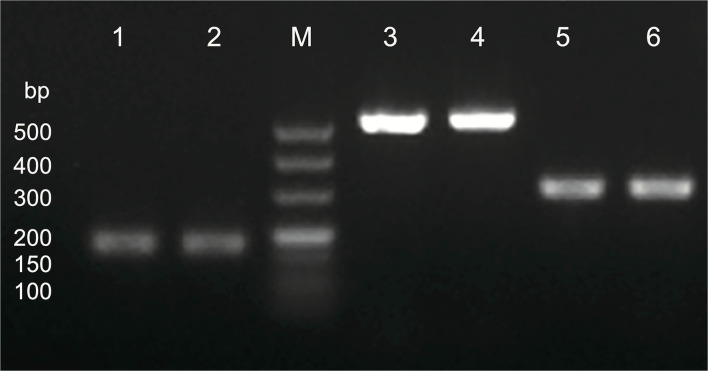
As seen in Figure 3, virulence genes of cagA, vacA s1/s2, and vacA m1/m2 were all expressed in H. pylori ZJC03. E-test results showed that H. pylori ZJC03 was sensitive to clarithromycin, amoxicillin, and tetracycline with the MIC values of 0.80 μg/mL, 0.016 μg/mL, and 0.023 μg/mL, respectively. However, H. pylori ZJC03 was not sensitive to metronidazole, with an MIC value exceeding 256 μg/mL.
Figure 3.
Virulence genes of H. pylori ZJC03. Lane M, maker; lane 1 and 2, cagA; lane 3 and 4, vacA s1/s2; lane 5 and 6, vacA m1/m2.
Anti-H. pylori activity of L. paracasei ZFM54
The cells of L. paracasei 54 showed stronger anti-H. pylori ability compared to CFS. The inhibition zone of the cells was 11.25 ± 0.78 mm, and the zone of the CFS was 9.02 ± 0.20 mm (Table 3). L. paracasei ZFM 54 cells inhibited urease activity of H. pylori ZJC03 with a rate of 70.27 ± 2.23%, while CFS showed an increased inhibition rate of 89.51 ± 0.72%. L. paracasei ZFM54 exhibited strong radical scavenging ability in DPPH scavenging assay. L. paracasei ZFM 54 cells showed remarkable oxidative activity calculated to be 57.89 ± 1.27%, and CFS was 51.36 ± 0.78%.
Table 3.
Anti-H. pylori activity, urease inhibition, and DPPH scavenging of L. paracasei 54.
|
Anti-H. pylori activity (mm) |
Urease inhibition (%) |
DPPH scavenging (%) |
|---|---|---|---|
| Cells of L. paracasei 54 | 11.25 ± 0.78 | 70.27 ± 2.23 | 57.89 ± 1.27 |
| CFS of L. paracasei 54 | 9.02 ± 0.20 | 89.51 ± 0.72 | 51.36 ± 0.78 |
MDA and GSH levels in liver
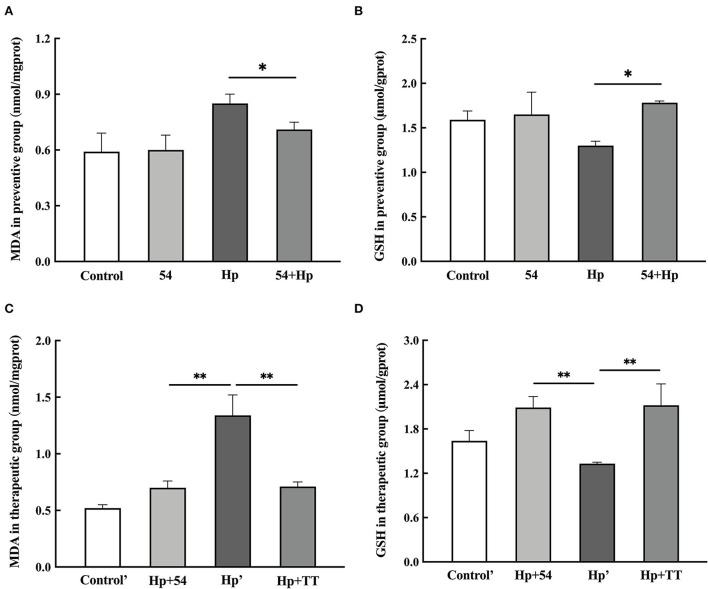
The MDA level in liver tissue of H. pylori-infected mice in the preventive group (0.85 ± 0.05 nmol/mgprot) was higher than that of the control group (p < 0.05) (Figure 4A). Pretreatment with L. paracasei ZFM54 significantly reduced MDA to 0.71 ± 0.04 nmol/mgprot (p < 0.05), thus protecting the mice mucosal layer from oxidative damage of ROS. Treatment with L. paracasei ZFM54 also significantly reduced the MDA level to 0.70 ± 0.06 nmol/mgprot (p < 0.01), when MDA contents were also significantly reduced in mice treated with triple therapy (TT) (0.71 ± 0.04 nmol/mgprot, p < 0.01) (Figure 4B).
Figure 4.
Levels of MDA and GSH during prevention and therapy. (A) MDA levels in preventive groups. (B) GSH levels in preventive groups. (C) MDA levels in therapeutic groups. (D) GSH levels in therapeutic groups. Data are shown as the mean ± SD (n = 6). Significant differences between two groups based on Student's t-test, *p < 0.05, **p < 0.01.
GSH content in liver tissues of H. pylori-infected mice significantly decreased (1.30 ± 0.05 μmol/gprot) compared to control (1.59 ± 0.10 μmol/gprot, p < 0.05) (Figure 4C), while significant increased level of GSH was observed in mice pretreated with L. paracasei ZFM54 (1.78 ± 0.02 μmol/gprot, p < 0.05), treated with L. paracasei ZFM54 (2.09 ± 0.15 μmol/gprot, p < 0.01), and triple therapy (TT) (2.12 ± 0.29 μmol/gprot, p < 0.01) (Figure 4D).
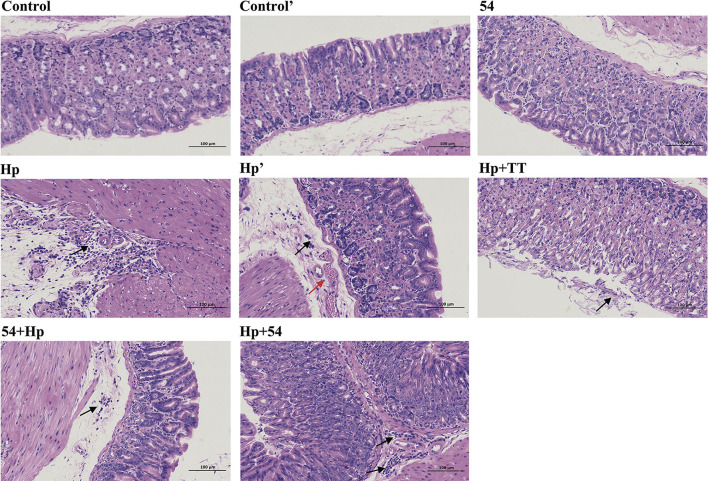
Histopathological changes of gastric tissues
Mice in the control and control' groups exhibited normal appearance of gastric mucosal morphologically, including the normal glands and structure of gastric epithelial cells, and little inflammatory infiltration in gastric epithelium (Figure 5). In contrast, the gastric mucosa of H. pylori-infected mice (Hp and Hp') showed irregular arrangement of mucosal glands, enlarged and hyperchromatic nuclei, and severe infiltration of inflammatory cells in the lamina propria. L. paracasei ZFM54 did not induce mucosal inflammation and other pathological changes in stomach. Inflammatory infiltration was significantly inhibited in mice pretreated with L. paracasei ZFM54 before H. pylori challenge (54+Hp). As expected, no obvious signs of tissue lesion and inflammation were also observed in the therapeutic groups of L. paracasei ZFM54 (Hp+54) and triple therapy (Hp+TT). These observations suggest that L. paracasei ZFM54 can inhibit or even reverse the injury process and protect gastric mucosa.
Figure 5.
Hematoxylin–eosin staining of the gastric tissues. Control, normal mice without prevention and infection in the preventive group; Control', normal mice without infection and therapy in the therapeutic group; Hp, mice infected with H. pylori without prevention in the preventive group; Hp', mice infected with H. pylori without therapy in the therapeutic group; 54+Hp, pretreatment with L. paracasei ZFM54 before H. pylori challenge; Hp+54, treatment with L. paracasei ZFM54 after H. pylori challenge; Hp+TT, treatment with triple therapy after H. pylori challenge. Black arrow, infiltration of inflammatory cells in lamina propria; red arrow, vascular congestion in the submucosa.
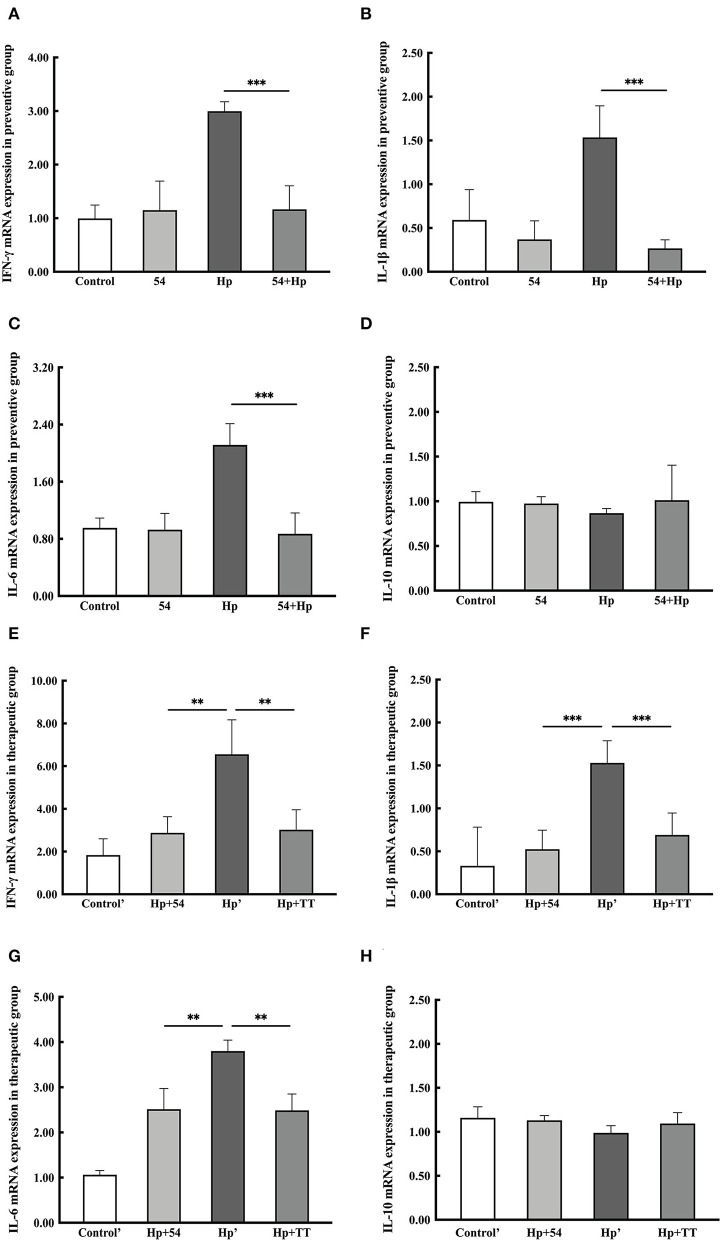
mRNA expression level of inflammatory cytokines
The levels of pro-inflammatory cytokines (IFN-γ, IL-1β, and IL-6) and anti-inflammatory cytokine (IL-10) were significantly altered after H. pylori infection, as displayed in Figure 4. In groups of Hp and Hp', the upregulation levels ranged between 3.00 ± 0.18 and 6.55 ± 1.86 log fold change of the INF-γ gene after H. pylori challenge, respectively (Figures 6A,E). IL-1β gene expression was significantly stimulated to 1.54 ± 0.36 and 1.53 ± 0.26 log folds, respectively (Figures 6B,F). Another pro-inflammatory cytokine, IL-6, was also increased to 2.11 ± 0.30 and 3.80 ± 0.24 log folds, respectively (Figures 6C,G). All pro-inflammatory cytokines genes were notably downregulated in L. paracasei ZFM54 prevention (54+Hp) and therapy (Hp+54) groups compared to probiotically unprotected mice (p < 0.01). No changes in the anti-inflammatory cytokine IL-10 were observed in either group (Figures 6D,H).
Figure 6.
Expression levels of mRNA for pro-inflammatory or anti-inflammatory cytokines in preventive and therapeutic groups. (A–D) IFN-γ, IL-1β, IL-6, and IL-10 mRNA expression levels in the preventive group. (E–H) IFN-γ, IL-1β, IL-6, and IL-10 mRNA expression levels in the therapeutic group. Data are shown as the mean ± SD (n = 6). Significant differences between two groups based on Student's t-test, **p < 0.01, ***p < 0.001.
Difference analysis of gastric microbiota
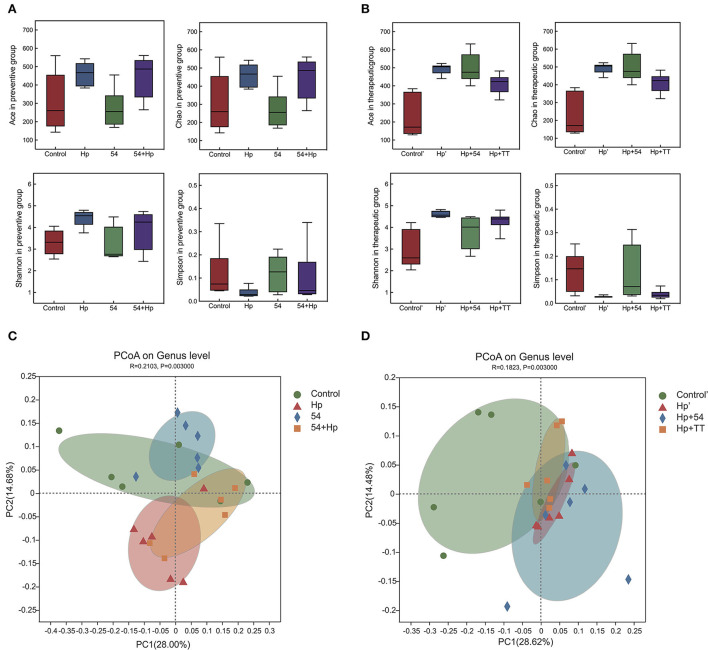
To compare the gastric microbial composition in various groups of mice, we randomly selected the gastric contents for 16S high-throughput sequencing and taxonomic classification analysis. H. pylori infection increased the abundance and diversity of gastric microbiota by analysis of the Ace, Chao, Shannon, and Simpson indices of alpha diversity (Figures 7A,B). PCoA analysis showed apparent clustering of gastric microbiota in different groups based on genus-level unweighted-uniFrac distances (Figures 7C,D). Preventive and therapeutic administration of L. paracasei ZFM54 improved gastric microbiota structure to near normal levels.
Figure 7.
Bacterial alpha and beta diversity in the preventive and therapeutic groups. (A) Alpha diversity (Ace, Chao, Shannon, and Simpson) in the preventive group. (B) alpha diversity (Ace, Chao, Shannon, and Simpson) in the therapeutic group. (C) PCoA on genus level in the preventive group. (D) PCoA on the genus level in the therapeutic group.
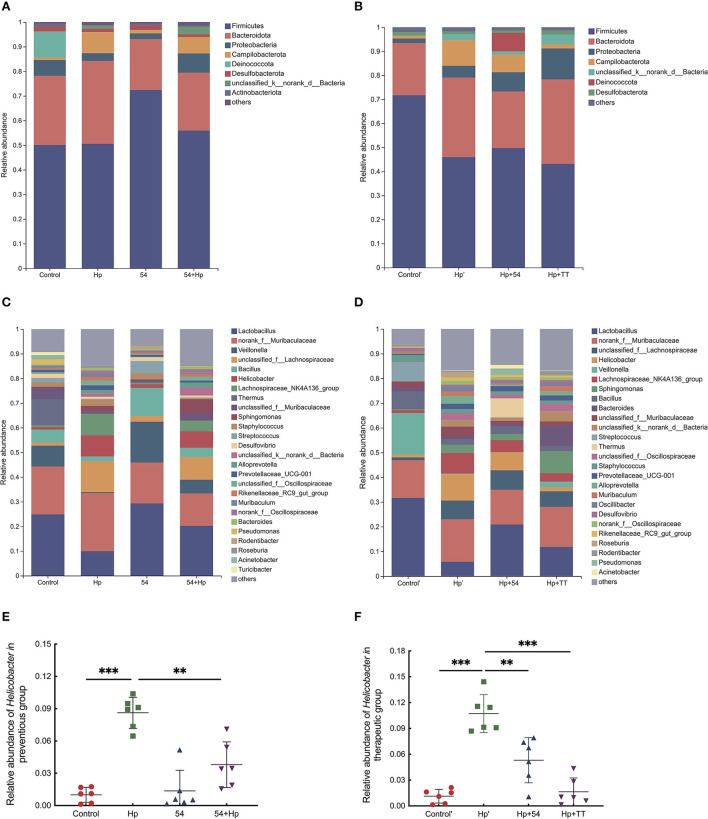
At the phylum level, it was found that the gavage of L. paracasei ZFM54 significantly increased the relative abundances of Firmicutes and Actinobacteriota and significantly decreased the abundance of Bacteroidota and Campilobacterota (Figures 8A,B). Instead, triple therapy treatment promoted the growth of Bacteroidota and Proteobacteria (Figure 8B). At the genus level, the relative abundances of bacterial groups such as Lactobacillus, Veillonella, and Bacillus in the H. pylori group decreased significantly, while Helicobacter, Lachnospiraceae_NK4A136_group, Sphingomonas, Staphylococcus, Alloprevotella, and Prevotellaceae_UCG-001 increased significantly (Figures 8C,D). Compared with infected mice, L. paracasei ZFM54 prevention and treatment significantly decreased the relative abundance of Helicobacter, Lachnospiraceae_NK4A136_group, Staphylococcus, and Prevotellaceae_UCG-001 and increased Lactobacillus, Veillonella, and Thermus. Although triple therapy treatment significantly inhibited the growth of gastric Helicobacter, it resulted in a significant increase in relative abundances of Sphingomonas, Bacteroides, Streptococcus, Oscillibacter, and Desulfovibrio (Figure 8D). The relative abundance of Helicobacter of 54, 54+Hp, Hp+54, and Hp+TT groups was lower than that of the H. pylori group (Hp and Hp', p < 0.01) (Figures 8E,F).
Figure 8.
Bacterial community distribution among different groups at the phylum and genus levels. (A) Distribution of abundant phyla in the preventive group. (B) Distribution of abundant phyla in the preventive group. (C) Distribution of abundant genera in the preventive group. (D) distribution of abundant genera in the therapeutic group. (E) The relative abundance of Helicobacter in the preventive group. (F) The relative abundance of Helicobacter in the therapeutic group. Phyla with a mean abundance <1.0% and genera with a mean abundance <1.5% among all samples were consolidated. **p < 0.01, ***p < 0.001.
Gastric microbiota in the normal, Hp infection, and intervention groups were compared using LEfSe. Abundant bacterial taxa observed in the Hp group were Lachnospiraceae, Helicobacteraceae, and Prevotellaceae, most of which belong to gram-negative human opportunistic pathogens (Figure 9A). Bacterial taxa were observed in abundance in pretreatment with L. paracasei ZFM54 including Sphingomonadaceae and Oscillospiraceae. According to the analysis of the Wilcoxon rank-sum test, significant reductions in the average relative abundance of Muribaculum, Staphylococcus, Lachnospiraceae_NK4A136_group, and Prevotellaceae_UCG-001 were observed in the preventive group of L. paracasei ZFM54 on genus level compared to the Hp group (Figure 9B). In therapeutic groups (Figure 9C), 16 bacterial taxa were identified to be in abundance in the H. pylori group including Helicobacteraceae, Lachnospiraceae, Oscillospiraceae, and Prevotellaceae. Lachnospiraceae were observed in significant abundance in treatment with L. paracasei ZFM54 (Hp+54), while Sphingomonadaceae were abundant in triple therapy group (Hp+TT). In addition, significant reductions in average relative abundance of Muribaculaceae, Alloprevotella, and Oscillibacter were observed by L. paracasei ZFM54 treatment (Hp+54) (Figure 9D), while Helicobacter, Lachnospiraceae_NK4A136_group, and Muribaculaceae were significantly reduced by triple therapy treatment (Hp+TT) on the genus level using the Wilcoxon rank-sum test (Figure 9E).
Figure 9.
Difference in bacterial taxa among preventive and therapeutic groups. (A) Bacterial taxa significantly differentiated among the preventive group identified by the linear discriminant analysis (LDA). (B) Bacterial communities with significant relative abundance differences at the genus level between Hp and 54+Hp groups by the Wilcoxon rank-sum test (n = 6). (C) Bacterial taxa significantly differentiated among the therapeutic group by LDA. (D) Bacterial communities with significant relative abundance differences at the genus level between Hp and Hp+54 and Hp and Hp+TT groups by the Wilcoxon rank-sum test (n = 6). Only taxa meeting an LDA significance threshold of > 4 were shown, *p < 0.05, **p < 0.01.
Discussion
Usually, host innate and adaptive immune responses are stimulated shortly after H. pylori colonization of gastric epithelial cells (27). Various clinical complications of H. pylori infection can be attributed to changes in bacterial genotypic and phenotypic characteristics. Infection with vacA- and cagA-positive H. pylori leads to severe gastric inflammation and a high risk for cancer development (28). H. pylori ZJC03 in this study was previously isolated from a patient with gastritis, which has virulence genes of vacA and cagA and is highly resistant to metronidazole. Lactobacillus strains are among the most prominent members of probiotics, mostly normal residents of different regions of human body. Our previous study examined the prebiotic properties of L. paracasei ZFM54 by genome sequence analysis and related prebiotic properties, including antagonism against H. pylori ZJC03, antibiotic susceptibility, and no adverse effects during acute oral toxicity in mice (29–31), suggesting that L. paracasei ZFM54 can be a potential candidate for use in the health or diet field.
For an effective strain to be called a probiotic, it must be able to survive in a hostile gastric environment (32). To reach host epithelial tissues of stomach, the acidic pH (1–2) in stomach is the foremost challenge to overcome. The ingested food buffers the acidic environment to pH 3, so it is considered as the optimum pH for the survival of a potential probiotic strain (33). L. paracasei ZFM 54 exhibited good tolerance as it showed considerable growth at low pH (2.0). Sornsenee et al. found that the survival rates of eight L. paracasei strains isolated from fermented palm sap in Thailand were 40–60% at pH 2 for 4 h (34). L. paracasei L1 was highly resistant to low pH (viability of 98.73% at pH 2 for 3 h) and was fed as probiotics to improve the intestinal microflora of chicken (35). The auto-aggregation ability of a probiotic strain is another important factor correlated with the bacterium's ability to form biofilm, which is an important property of probiotic to prevent pathogenic microorganisms from invading the host (1). The auto-aggregation rate of L. paracasei ZFM 54 rose to 69.00 ± 2.73% after 4 h. The 2- and 24-h auto-aggregation values of L. paracasei L1 were 20.4 ± 2.3% and 47.2 ± 0.8%, respectively (35). Co-aggregation is considered a key strategy for Lactobacillus strains to prevent pathogenic bacteria from adhering to biological surfaces (36). Barache et al. analyzed that L. paracasei FB1 showed stronger co-aggregation with S. aureus (28.34% ± 2.56%) after 2 h of incubation, followed by E. coli (23.39 ± 2.12%) and L. monocytogenes (20.10 ± 2.04%) (37). L. paracasei ZFM 54 displayed a co-aggregation rate of over 30% and exhibited their capability to agglutinate H. pylori in vitro.
Inhibition of urease enzyme is an important ability of Lactobacillus to decrease H. pylori density in the host and inhibit dysbiosis. H. pylori uses urease enzyme to survive in the acidic environment of the stomach by utilizing urease enzyme activity to convert urea into ammonia, thus raising the pH and creating a favorable environment for its growth and colonization (38). Infection with H. pylori stimulates phagocytic cells at inflammation sites, producing ROS to combat H. pylori (17). The increased level of ROS is detrimental for host tissues, causes lipid peroxidation, and produces MDA. MDA is a biomarker of H. pylori infection (17). GSH, a thiol-based anti-oxidant, is reduced in H. pylori infection, which is naturally present in all cells and contributes in defending against oxidative injury. As observed in this study, L. paracasei ZFM 54 significantly reduced H. pylori-mediated MDA level in mice liver tissues, while significantly increasing the GSH level in preventive and therapeutic studies. L. paracasei ZFM 54 appears to have potential anti-oxidation ability in vivo as well, validating the anti-oxidation potential shown in in vitro experiment.
H. pylori infection stimulates adaptive immune response mainly through the activation of T-helper cells. Th1 cells further produce cytokines IL-2, IL-22, and IFN-γ with subsequent stimulation of neutrophil infiltration and macrophage activation (39). H. pylori modulate pro-inflammatory and regulatory immune responses, which stimulate the activation of IL-1β, a strong pro-inflammatory cytokine which promotes gastric tumor cancer through genetic polymorphism (40). Consistent results in this study were observed through H. pylori-mediated enhanced level of pro-inflammatory cytokines (IFN-γ, IL-1β, and IL-6) and severe infiltration of inflammatory cells in gastric mucosa. Prevention and therapy with L. paracasei ZFM 54 significantly downregulated the pro-inflammatory cytokines in gastric mucosa. Previously, similar results were obtained in pretreatment with L. plantarum ZDY 2013 against H. pylori in mice (41). L. plantarum ZDY 2013 showed a significant increase in IL-10 against H. pylori in AGS cell lines (42). No increase in the level of anti-inflammatory cytokine IL-10 was observed in this research, suggesting the different mechanisms to be involved in reducing inflammation. Other mechanisms responsible for the protective effect may be contributed by competitive exclusion of H. pylori or inhibition by secreting antimicrobial substances or blocking urease activity, which further hinder H. pylori colonization.
The microbiota in normal healthy stomach has been reported to be mainly composed of Firmicutes, Bacteroidetes, Actinobacteria, and Proteobacteria, and the most abundant genera have been reported to include Streptococcus, Prevotella, Fusobacteria, Veillonella, and Rothia (43, 44). Previous reports provided evidence of reduced bacterial diversity and an increase in the bacterial taxa belonging to the phyla Campilobacterota and Proteobacteria in subjects with H. pylori-positive infection (43). Preventive and therapeutic experiments with L. paracasei ZFM 54 significantly suppressed the relative abundance of H. pylori, while increasing the abundance of bacterial taxa belonging to the phyla Firmicutes and Actinobacteriota. Interestingly, treatment with triple therapy reduced high levels of inflammation cytokines and MDA, but the LEfSe analysis showed increased abundance of Rodentibacter and Escherichia-Shigella Mucispirillum, members of which are opportunistic human pathogens in the stomach. It is also suggestive of disruption of gastric microbiota due to antibiotics. Suppression of acid by PPI triggers dysbiosis in gastric microbiota composition (45). An increased abundance of non-H. pylori bacteria such as Escherichia-Shigella, Nitrospirae, and Burkholderia were found in patients with gastric cancer (46).
Conclusion
The strain L. paracasei ZFM 54 demonstrated remarkable probiotic characteristics owing to its ability to survive in the simulated gastric acid, auto-aggregation, and hydrophobic activities. The potential inhibition against H. pylori and urease enzyme, anti-oxidizing, and co-aggregation activities further validate the efficacy of the strain to be effective against H. pylori. During preventive and therapeutic experiments in the H. pylori-induced gastritis mouse model, L. paracasei ZFM 54 can effectively reduce inflammation, MDA level, and inhibition of microbiota dysbiosis. The use of L. paracasei ZFM 54 against H. pylori challenge would be more advantageous over antibiotics, as it more favorably improves gastric microbiota caused by H. pylori infection. The potential to prevent and treat H. pylori inflammation is highly indicative of its use not only as a prophylaxis but also as a therapeutic agent to eradicate H. pylori.
Data availability statement
The original contributions presented in the study are included in the article, further inquiries can be directed to the corresponding author.
Ethics statement
The animal study was reviewed and approved by mice in vivo Experiments was conducted under approval of by Shanghai Public Health Clinical Center Laboratory Animal Welfare and Ethics Committee with protocol No. 2019-A003-01.
Author contributions
PL and QG: conceptualization and supervision. QZ, NQ, and BX: methodology, investigation, and data curation. QZ and NQ: writing—original draft preparation. QZ and ZX: writing—review and editing. QG: funding acquisition. All authors have read and agreed to the published version of the manuscript.
Funding
The work was supported by the Chinese Academy of Engineering Academy-Locality Cooperation Project (No. 2019-ZJ-JS-02) and the Natural Science Foundation of Zhejiang Province of China (No. LZ21C200001).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- 1.Dunne C, Dolan B, Clyne M. Factors that mediate colonization of the human stomach by Helicobacter pylori. World J Gastroenterol. (2014) 20:5610–24. 10.3748/wjg.v20.i19.5610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roberts SE, Morrison-Rees S, Samuel DG, Thorne K, Akbari A, Williams JG, et al. Review article: the prevalence of Helicobacter pylori and the incidence of gastric cancer across Europe. Aliment Pharmacol Ther. (2016) 43:334–45. 10.1111/apt.13474 [DOI] [PubMed] [Google Scholar]
- 3.Valdez-Salazar HA, Ares MA, Fernández FJ, Ibarra JA, Torres J, Bustamante VH, et al. Long-chain fatty acids alter transcription of Helicobacter pylori virulence and regulatory genes. PeerJ. (2021) 9:e12270. 10.7717/peerj.12270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tegtmeyer N, Harrer A, Schmitt V, Singer BB, Backert S. Expression of CEACAM1 or CEACAM5 in AZ-521 cells restores the type IV secretion deficiency for translocation of CagA by Helicobacter pylori. Cell Microbiol. (2019) 21:e12965. 10.1111/cmi.12965 [DOI] [PubMed] [Google Scholar]
- 5.Toracchio S, Cellini L, Di Campli E, Cappello G, Malatesta MG, Ferri A, et al. Role of antimicrobial susceptibility testing on efficacy of triple therapy in Helicobacter pylori eradication. Aliment Pharmacol Ther. (2000) 14:1639–43. 10.1046/j.1365-200000870.x [DOI] [PubMed] [Google Scholar]
- 6.Choi IJ, Kook MC, Kim YI, Cho SJ, Lee JY, Kim CG, et al. Helicobacter pylori therapy for the prevention of metachronous gastric cancer. N Engl J Med. (2018) 378:1085–95. 10.1056/NEJMoa1708423 [DOI] [PubMed] [Google Scholar]
- 7.Chen L, Xu W, Lee A, He J, Huang B, Zheng W, et al. The impact of Helicobacter pylori infection, eradication therapy and probiotic supplementation on gut microenvironment homeostasis: an open-label, randomized clinical trial. EBio Medicine. (2018) 35:87–96. 10.1016/j.ebiom.2018.08.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deng K, Chen T, Wu Q, Xin H, Wei Q, Hu P, et al. In vitro and in vivo examination of anticolonization of pathogens by Lactobacillus paracasei FJ861111.1. J Dairy Sci. (2015) 98:6759–66. 10.3168/jds.2015-9761 [DOI] [PubMed] [Google Scholar]
- 9.Qureshi N, Li P, Gu Q. Probiotic therapy in Helicobacter pylori infection: a potential strategy against a serious pathogen? Appl Microbiol Biotechnol. (2019) 103:1573–88. 10.1007/s00253-018-09580-3 [DOI] [PubMed] [Google Scholar]
- 10.Kabir AM, Aiba Y, Takagi A, Kamiya S, Miwa T, Koga Y, et al. Prevention of Helicobacter pylori infection by lactobacilli in a gnotobiotic murine model. Gut. (1997) 41:49–55. 10.1136/gut.41.1.49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee HA, Kim JY, Kim J, Nam B, Kim O. Anti-Helicobacter pylori activity of acomplex mixture of Lactobacillus paracasei HP7 including the extract of Perilla frutescens var. acuta and Glycyrrhiza glabra Lab. Anim Res. (2020) 36:40. 10.1186/s42826-020-00073-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takeda S, Igoshi K, Tsend-Ayush C, Oyunsuren T, Sakata R, Koga Y, et al. Lactobacillus paracasei strain 06TCa19 suppresses inflammatory chemokine induced by Helicobacter pylori in human gastric epithelial cells. Hum Cell. (2017) 30:258–66. 10.1007/s13577-017-0172-z [DOI] [PubMed] [Google Scholar]
- 13.Chen YH, Tsai WH, Wu HY, Chen CY, Yeh WL, Chen YH, et al. Probiotic Lactobacillus spp. act against Helicobacter pylori-induced Inflammation. J Clin Med. (2019) 8:90. 10.3390./jcm8010090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reuben RC, Roy PC, Sarkar SL, Rubayet Ul Alam ASM, Jahid IK. Characterization and evaluation of lactic acid bacteria from indigenous raw milk for potential probiotic properties. J Dairy Sci. (2020) 103:1223–37. 10.3168/jds.2019-17092 [DOI] [PubMed] [Google Scholar]
- 15.Gobert AP, Verriere T, de Sablet T, Peek RM, Jr, Chaturvedi R, Wilson KT. Haem oxygenase-1 inhibits phosphorylation of the Helicobacter pylori oncoprotein CagA in gastric epithelial cells. Cell Microbiol. (2013) 15:145–56. 10.1111/cmi.12039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang YK, Chiang WC, Kuo FC, Wu MC, Shih HY, Wang SSW, et al. Levels of malondialdehyde in the gastric juice: its association with Helicobacter pylori infection and stomach diseases. Helicobacter. (2018) 23:e12460. 10.1111/hel.12460 [DOI] [PubMed] [Google Scholar]
- 17.Khanzode SS, Khanzode SD, Dakhale GN. Serum and plasma concentration of oxidant and antioxidants in patients of Helicobacter pylori gastritis and its correlation with gastric cancer. Cancer Lett. (2003) 195:27–31. 10.1016/S0304-3835(03)00147-2 [DOI] [PubMed] [Google Scholar]
- 18.Charteris WP, Kelly PM, Morelli L, Collins JK. Development and application of an in vitro methodology to determine the transit tolerance of potentially probiotic Lactobacillus and Bifidobacterium species in the upper human gastrointestinal tract. J Appl Microbiol. (1998) 84:759–68. 10.1046/j.1365-2672.1998.00407.x [DOI] [PubMed] [Google Scholar]
- 19.Ahire JJ, Jakkamsetty C, Kashikar MS, Lakshmi SG, Madempudi RS. In vitro evaluation of probiotic properties of Lactobacillus plantarum UBLP40 isolated from traditional indigenous fermented food. Probiotics Antimicro. (2021) 13:1413–24. 10.1007/s12602-021-09775-7 [DOI] [PubMed] [Google Scholar]
- 20.Aguilar-Zamora E, Weimer BC, Torres RC, Gómez-Delgado A, Ortiz-Olvera N, Aparicio-Ozores G, et al. Molecular epidemiology and antimicrobial Resistance of Clostridioides difficile in hospitalized patients from Mexico. Front Microbiol. (2021) 12:787451. 10.3389/fmicb.2021.787451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kos B, Susković J, Vuković S, Simpraga M, Frece J, Matosić S, et al. Adhesion and aggregation ability of probiotic strain Lactobacillus acidophilus M92. J Appl Microbiol. (2003) 94:981–87. 10.1046/j.1365-2672.2003.01915.x [DOI] [PubMed] [Google Scholar]
- 22.Hirano J, Yoshida T, Sugiyama T, Koide N, Mori I, Yokochi T, et al. The effect of Lactobacillus rhamnosus on enterohemorrhagic Escherichia coli infection of human intestinal cells in vitro. Microbiol Immunol. (2003) 47:405–9. 10.1111/j.1348-0421.2003.tb03377.x [DOI] [PubMed] [Google Scholar]
- 23.Korona-Glowniak I, Glowniak-Lipa A, Ludwiczuk A, Baj T, Malm A. The in vitro activity of essential oils against Helicobacter pylori growth and urease activity. Molecules. (2020) 25:586. 10.3390/molecules25030586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saravanakumar K, Chellia R, Hu X, Kathiresan K, Oh DH, Wang MH, et al. Eradication of Helicobacter pylori through the inhibition of urease and peptide deformylase: Computational and biological studies. Microb Pathog. (2019) 128:236–44. 10.1016/j.micpath.2019.01.001 [DOI] [PubMed] [Google Scholar]
- 25.Zhou Q, Xue B, Gu R, Li P, Gu Q. Lactobacillus plantarum ZJ316 attenuates Helicobacter pylori-induced gastritis in C57BL/6 mice. J Agric Food Chem. (2021) 69:6510–23. 10.1021/acs.jafc.1c01070 [DOI] [PubMed] [Google Scholar]
- 26.Bao W, He Y, Yu J, Liu M, Yang X, Ta N, et al. Regulatory effect of Lactiplantibacillus plantarum 2-33 on intestinal microbiota of mice with antibiotic-associated diarrhea. Front Nutr. (2022) 9:921875. 10.3389/fnut.2022.921875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cadamuro AC, Rossi AF, Maniezzo NM, Silva AE. Helicobacter pylori infection: host immune response, implications on gene expression and microRNAs. World J Gastroenterol. (2014) 20:1424–37. 10.3748/wjg.v20.i6.1424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang L, Zhou J, Xin Y, Geng C, Tian Z, Yu X, et al. Bacterial overgrowth and diversification of microbiota in gastric cancer. Eur J Gastroenterol Hepatol. (2016) 28:261–6. 10.1097/MEG.0000000000000542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ye P, Wang J, Liu M, Li P, Gu Q. Purification and characterization of a novel bacteriocin from Lactobacillus paracasei ZFM54. LWT-Food Sci Technol. (2021) 143:111125. 10.1016/j.lwt.2021.11112534453561 [DOI] [Google Scholar]
- 30.Zhu Y, Zhou Q, Li P, Gu Q. Purification, characterization, and mode of action of Paracin 54, a novel bacteriocin against Staphylococci. Appl Microbiol Biotechnol. (2021) 105:6735–48. 10.1007/s00253-021-11505-6 [DOI] [PubMed] [Google Scholar]
- 31.Qureshi N, Gu Q, Whole LiP. Genome sequence analysis and in vitro probiotic characteristics of a Lactobacillus strain Lactobacillus paracasei ZFM54. J Appl Microbiol. (2020) 129:422–33. 10.1111/jam.14627 [DOI] [PubMed] [Google Scholar]
- 32.Ren DY, Li C, Qin YQ, Yin RL, Du SW, Ye F, et al. Lactobacilli reduce chemokine IL-8 production in response to TNF-α and Salmonella challenge of Caco-2 cells. Biomed Res Int. (2013) 2013:925219. 10.1155/2013/925219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu WJ, Chen YF, Kwok LY, Li MH, Sun T, Sun CL, et al. Preliminary selection for potential probiotic Bifidobacterium isolated from subjects of different Chinese ethnic groups and evaluation of their fermentation and storage characteristics in bovine milk. J Dairy Sci. (2013) 96:6807–17. 10.3168/jds.2013-6582 [DOI] [PubMed] [Google Scholar]
- 34.Sornsenee P, Singkhamanan K, Sangkhathat S, Saengsuwan P, Romyasamit C. Probiotic properties of Lactobacillus species isolated from fermented palm sap in Thailand. Probiotics Antimicro. (2021) 13:957–69. 10.1007/s12602-021-09754-y [DOI] [PubMed] [Google Scholar]
- 35.Xu Y, Tian Y, Cao Y, Li J, Guo H, Su Y, et al. Probiotic properties of Lactobacillus paracasei subsp. paracasei L1 and its growth performance-promotion in chicken by improving the intestinal microflora. Front Physiol. (2019) 10:937. 10.3389/fphys.2019.00937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Merino L, Trejo FM, De Antoni G, Golowczyc MA. Lactobacillus strains inhibit biofilm formation of Salmonella sp. isolates from poultry. Food Res Int. (2019) 123:258–65. 10.1016/j.foodres.2019.04.067 [DOI] [PubMed] [Google Scholar]
- 37.Barache N, Belguesmia Y, Ladjouzi R, Bendali F, Drider D. Clusters of Lactobacillus strains from vegetal origins are associated with beneficial functions: experimental data and statistical interpretations. Foods. (2020) 9:985. 10.3390./foods9080985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eaton KA, Brooks CL, Morgan DR, Krakowka S. Essential role of urease in pathogenesis of gastritis induced by Helicobacter pylori in gnotobiotic piglets. Infect Immun. (1991) 59:2470–5. 10.1128/iai.59.7.2470-2475.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bamford KB, Fan X, Crowe SE, Leary JF, Gourley WK, Luthra GK, et al. Lymphocytes in the human gastric mucosa during Helicobacter pylori have a T helper cell 1 phenotype. Gastroenterology. (1998) 114:482–92. 10.1016/S0016-5085(98)70531-1 [DOI] [PubMed] [Google Scholar]
- 40.Filaly HE, Outlioua A, Medyouf H, Guessous F, Akarid K. Targeting IL-1β in patients with advanced Helicobacter pylori infection: a potential therapy for gastric cancer. Future Microbiol. (2022) 17:633–41. 10.2217/fmb-2021-0242 [DOI] [PubMed] [Google Scholar]
- 41.Pan M, Wan C, Xie Q, Huang R, Tao X, Shah NP, et al. Changes in gastric microbiota induced by Helicobacter pylori infection and preventive effects of Lactobacillus plantarum ZDY 2013 against such infection. J Dairy Sci. (2016) 99:970–81. 10.3168/jds.2015-10510 [DOI] [PubMed] [Google Scholar]
- 42.Zhao K, Xie Q, Xu D, Guo Y, Tao X, Wei H, et al. Antagonistics of Lactobacillus plantarum ZDY2013 against Helicobacter pylori SS1 and its infection in vitro in human gastric epithelial AGS cells. J Biosci Bioeng. (2018) 126:458–63. 10.1016/j.jbiosc.2018.04.003 [DOI] [PubMed] [Google Scholar]
- 43.Sheh A, Fox JG. The role of the gastrointestinal microbiome in Helicobacter pylori pathogenesis. Gut Microbes. (2013) 4:505–31. 10.4161/gmic.26205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brawner KM, Kumar R, Serrano CA, Ptacek T, Lefkowitz E, Morrow CD, et al. Helicobacter pylori infection is associated with an altered gastric microbiota in children. Mucosal Immunol. (2017) 10:1169–77. 10.1038/mi.2016.131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parsons BN, Ijaz UZ, D'Amore R, Burkitt MD, Eccles R, Lenzi L, et al. Comparison of the human gastric microbiota in hypochlorhydric states arising as a result of Helicobacter pylori-induced atrophic gastritis, autoimmune atrophic gastritis and proton pump inhibitor use. PLoS Pathog. (2017) 13:e1006653. 10.1371/journal.ppat.1006653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bakhti SZ, Latifi-Navid S. Interplay and cooperation of Helicobacter pylori and gut microbiota in gastric carcinogenesis. BMC Microbiol. (2021) 21:258. 10.1186/s12866-021-02315-x [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The original contributions presented in the study are included in the article, further inquiries can be directed to the corresponding author.