Abstract
FtsH (HflB) is a conserved, highly specific, ATP-dependent protease for which a number of substrates are known. The enzyme participates in the phage λ lysis-lysogeny decision by degrading the lambda CII transcriptional activator and by its response to inhibition by the λ CIII gene product. In order to gain further insight into the mechanism of the enzymatic activity of FtsH (HflB), we identified the peptides generated following proteolysis of the phage λ CII protein. It was found that FtsH (HflB) acts as an endopeptidase degrading CII into small peptides with limited amino acid specificity at the cleavage site. β-Casein, an unstructured substrate, is also degraded by FtsH (HflB), suggesting that protein structure may play a minor role in determining the products of proteolysis. The majority of the peptides produced were 13 to 20 residues long.
Degradation of regulatory proteins by ATP-dependent proteases is an important mechanism for the rapid control of gene activity in all organisms. In bacteria, proteolysis acts on key regulatory transcription factors which regulate the heat shock response, stationary-phase and SOS stress responses, capsular polysaccharide biosynthesis, and the control of the lysis-lysogeny decision of phage λ (12, 14, 27).
FtsH, a membrane-bound Zn2+ metalloprotease, which was originally identified as an hflB mutation (high-frequency lysogenization by phage λ), is highly conserved in all organisms, is the only known essential protease in Escherichia coli, and does not participate in cell division (28, 35). The name FtsH was coined before the discovery of a second mutation in the ftsI gene, in the strain carrying the temperature-sensitive ftsH mutation, that is responsible for cell filamentation (7). Following infection by bacteriophage λ, the λ CII regulatory protein, which activates transcription of the CI repressor from the pE promoter, is rapidly degraded by FtsH (18, 20, 30, 32). The λ CIII peptide extends the half-life of CII, thereby prolonging CI repressor synthesis, which in turn promotes the lysogenic pathway (4, 21).
In higher organisms, FtsH orthologs are found in mitochondria and chloroplasts. It was suggested that in yeast mitochondria FtsH plays a key role in maintaining membrane integrity, possessing an ATP-dependent chaperone-like activity and a protease activity that degrades unassembled membrane components (5, 25). Similar roles have been suggested for FtsH in E. coli (2, 3). Recently, mutations in the gene coding for paraplegin, a human ortholog of FtsH, were demonstrated to be responsible for hereditary spastic paraplegia (9). The mutations lead to mitochondrial defects, suggesting that paraplegin is a nucleus-encoded mitochondrial protein (9, 11).
FtsH belongs to a large class of ATPase-containing proteins belonging to the AAA protein superfamily (ATPases associated with different cellular activities), which is characterized by a highly conserved domain of 230 to 250 amino acid residues that contains an ATP binding consensus and ATPase activity (8, 10). These proteins are found in all organisms and play essential roles in the cell cycle, vesicular transport in the Golgi complex, mitochondrial assembly, proteasomes, and cellular metalloproteases. The crystal structure of the N-ethylmaleimide-sensitive fusion protein (NSF) D2 hexameric AAA motif has been determined recently (24, 37).
FtsH is a highly discriminatory protease; the number of known substrates is rather small and includes the heat shock sigma factor ς32, phage λ CII, CIII, and Xis proteins, SecY, YccA, subunit a of the membrane-embedded F0 part of the H+-ATPase proteins, and SsrA tagged proteins (1, 15, 16, 20, 23, 32, 34). However, little is known about the mechanism by which these substrates are selected. It was proposed that a different process is used by FtsH to select soluble and membrane-bound substrates (19). Nothing is known about the proteolytic products generated by FtsH and the amino acid specificity of FtsH surrounding the cleavage sites.
In this study we monitored the degradation of purified CII in vitro by glutathione S-transferase (GST)-FtsH. We utilized reverse-phase high-performance liquid chromatography (HPLC) and electrospray mass spectrometry (MS) to identify peptide products larger than 3 amino acid residues that were produced by FtsH proteolysis. It was found that CII degradation by FtsH is processive and that only small peptides, 4 to 26 residues long, could be identified. Our results suggest that FtsH can hydrolyze peptide bonds with limited specificity at the cleavage sites.
MATERIALS AND METHODS
Strains.
Strain A9286 is a derivative of BL21(DE3) (33) carrying plasmid pET-CII. Strain A8926 is a derivative of W3110 carrying sfhC zad-220::Tn10 ΔftsH3::kan (28, 29). Strain A9241 is strain A2097 (22) carrying plasmid pLCIIH6.
Plasmids.
Plasmid pET-CII, which carries the cII gene fused to a His6 tag at the region corresponding to the N terminus, was derived from pET-15b (Novagen). In this plasmid a thrombin cleavage site is engineered between the His6 and CII sequences. The PCR product of the cII gene was inserted between the NdeI and BamHI restriction sites. The oligonucleotides used for the PCR were 5′-CGAATTCAACCACACCTA-3′ and 5′-CGGGATCCTCAGAACTCCATCTGG-3′. Plasmid pLCII-H6 was constructed by inserting a PCR product carrying the cII gene fused to the His6 sequence at the region corresponding to the C terminus between the NdeI and HindIII restriction sites of plasmid pTG44 (a λ pL expression plasmid from our collection). The oligonucleotides used for the PCR were 5′-GGGCATCAAATTAAACCAC-3′ and 5′-AACCAAGCTTAGTGGTGATGGTGATGGTG-3′, and pHG326 (from our collection) was used as the template. Plasmid pGST-FtsH was constructed by fusion of the FtsH gene with the first 9 bp deleted to pGEX-2T by insertion of a BglII-EcoRI PCR fragment of FtsH into the BamHI-EcoRI sites of the vector. This plasmid was introduced into a strain in which FtsH had been deleted (A8926) to generate strain A9390. The sequences of all inserts were confirmed by DNA sequencing.
Protein purification.
His6-CII, expressed in strain A9286 following 3 h of isopropyl-β-d-thiogalactopyranoside (IPTG) induction at 37°C, was bound to Ni-nitrilotriacetic acid (NTA) beads (Qiagen RA 97008). This protein was eluted by thrombin cleavage to generate CII. To obtain CII protein carrying additional residues at the amino terminus, His6-CII protein was eluted with 500 mM imidazole in 20 mM Tris (pH 8.0)–150 mM NaCl. To obtain CII protein carrying additional residues at the carboxy terminus, CII-His6, expressed in strain A9241 following a 30-min heat induction at 42°C, was bound to Ni-NTA beads and obtained by elution with 500 mM imidazole in 20 mM Tris (pH 6.8)–300 mM NaCl.
GST-FtsH, expressed in strain A9390 following induction by IPTG, was purified (about 90%) by binding to glutathione (GSH)-agarose beads followed by elution with GSH. A detailed description of the expression and properties of the GST-FtsH protein fusion will be presented elsewhere.
Proteolysis experiments.
Degradation reactions were performed as previously described (32). For the addition of “Complete” inhibitor mix (Boehringer Mannheim), a tablet was dissolved in 1 ml, and 0.5 μl was added to the reaction mixture. For the identification of peptides following proteolysis of CII, the proteolysis reactions were carried out at 42°C with 50 pmol of CII and 10 pmol of GST-FtsH.
Liquid chromatography and MS.
The peptides produced by FtsH proteolysis were resolved by reverse-phase HPLC on a 1- by 150-mm Vydac C18 column with a linear gradient of 4 to 65% (1%/min) acetonitrile in 0.025% trifluoroacetic acid (TFA), at a flow rate of 40 ml/min. The MS analysis was done in the positive-ion mode using repetitively a full MS scan followed by an MS-MS experiment (collision-induced fragmentation) on the most abundant ion of the MS scan. The MS and MS-MS data from the run were compared to the simulated proteolysis and fragmentation of the substrates by using Sequest software (J. Eng and J. Yates, University of Washington).
RESULTS
Purification and characterization of CII protein.
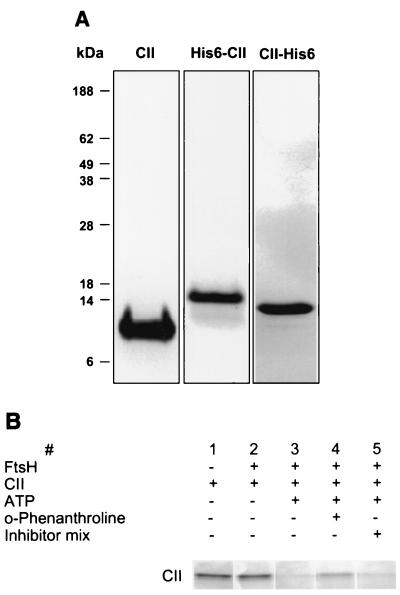
For rapid and simple purification of CII, the cII gene was cloned into pET15b (Novagen) expressing a His6-CII protein fusion in which 6 histidines were added to the N terminus (see Materials and Methods). Soluble CII protein was purified by binding to agarose-Ni beads and released from the beads by thrombin cleavage (Fig. 1A). This highly purified CII protein was rapidly degraded by FtsH (data not shown). Degradation of CII is absolutely dependent on the presence of ATP and is partially inhibited by the addition of o-phenanthroline (Fig. 1B). As expected, no inhibition was observed in the presence of a protease inhibitor cocktail that is known to inhibit a large variety of serine, aspartate, and cysteine proteases. We estimated, based on circular dichroism (CD) measurements, that CII in this preparation is highly structured and is made of 65% α helix, which is similar to the predicted α-helical content of about 70% (31).
FIG. 1.
Purification of CII protein. (A) Wild-type CII (10 μg), His6-CII (5 μg), and CII-His6 (5 μg) were resolved by SDS-PAGE and stained with Coomassie brilliant blue. Wild-type CII carries 3 additional residues, GSH, at the N terminus (Mw, 11,337); His6-CII carries 20 additional residues, MGSSHHHHHHSSGLVPRGSH, at the N terminus (Mw, 13,219) and CII-His6 carries 12 additional residues, EFIEGRHHHHHH. at the C terminus (Mw, 12,610). (B) Analysis of FtsH protease activity, its dependence on ATP, and the effect of inhibitors. Reactions were carried under standard conditions for 60 min. Lane 1, CII alone; lane 2, reaction in the absence of ATP; lane 3, complete reaction; lane 4, reaction in the presence of 10 mM o-phenanthroline; lane 5, reaction in the presence of the inhibitor mix Complete (Boehringer Mannheim).
Peptides recovered following proteolysis of CII.
The degradation of CII by FtsH in vitro leads to the disappearance of the full-length protein. No intermediates or partially cleaved proteins were ever observed by gel electrophoresis (see also references 20 and 32). These results suggest that, once initiated, proteolysis by FtsH is rapid and complete and that substrate recognition may be the rate-limiting step. Attempts to obtain degradation intermediates by increasing the ratio of CII to FtsH or by diluting FtsH, a condition known to reduce enzyme activity (T. Ogura, personal communication), proved unsuccessful. No degradation of CII was obtained in the absence of ATP.
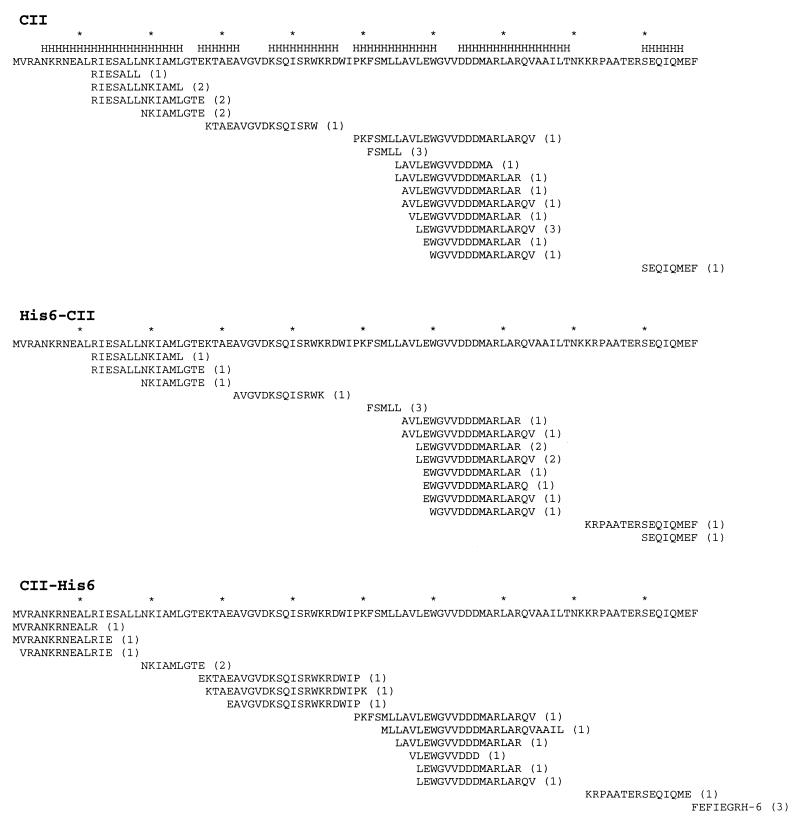
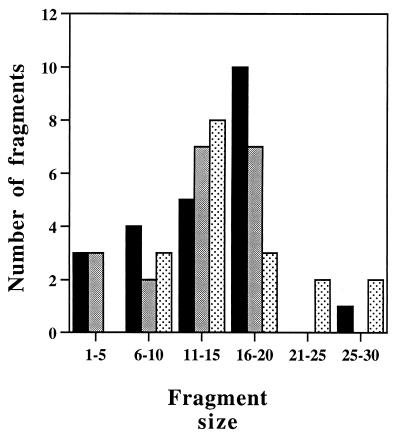
To identify CII degradation products, the reaction products were separated by reverse-phase HPLC and analyzed by electrospray MS analysis (liquid chromatography and MS [LC-MS]). Peptides larger than 3 residues were identified by their mass/charge ratio followed by collision-induced fragmentation (MS-MS). Peptides consisting of more than 4 residues that were found following complete digestion of CII in three independent experiments are shown in Fig. 2. These peptides account for most of CII. The peptides cluster in groups with minor differences at the N or C terminus. Only seven peptides were shorter than 10 residues, and about 65% (15 of 23) were 13 to 19 residues long (Fig. 3). We do not know why the N-terminal peptide was not obtained. Interestingly, for many of the peptides (10 of 23), the N-terminal residue is located in the highly hydrophobic region LLAVLEW. However, no obvious cleavage site consensus could be found.
FIG. 2.
Map of the peptides identified following CII proteolysis. The results of degradation of CII, His6-CII, and CII-His6 are shown. The peptides obtained from three independent experiments for each substrate are shown below the CII amino acid sequence. The number of experiments in which each peptide was observed is given in parentheses. Asterisks above the CII sequence mark intervals of 10 residues, and the letter H denotes regions predicted to form α helices (PHD program).
FIG. 3.
Size distribution of CII peptides. The histogram shows a plot of the data presented in Fig. 2. Filled bars, CII; shaded bars, His6-CII; stippled bars, CII-His6.
We attempted to test whether degradation by FtsH may proceed from the N or C terminus of CII by the addition of amino acid residues at either end. Two CII proteins, His6-CII and CII-His6, were purified (Fig. 1) and analyzed. The addition of His6 at the N terminus was found to reduce the rate of degradation from about 1 to 2 min (half-life [t1/2]) under our standard conditions to 20 to 40 min. However, this increase in stability had a minor effect on the nature of the peptides recovered following proteolysis (Fig. 2 and 3). The addition of His6 to the C-terminal end had an even stronger stabilization effect (t1/2 = 60 to 90 min). The composition of the peptides recovered following proteolysis of this protein was also affected; about half of the peptides identified, including peptides from the N terminus, were not found following the degradation of wild-type CII or His6-CII (Fig. 2 and 3). In addition, a number of peptides that were present in the degradation products of wild-type CII or His6-CII were absent following degradation of CII-His6. The addition of 20 amino acid residues to the N terminus had only a minor effect on the collection of peptides recovered. In contrast, about two-thirds of the peptides recovered following the addition of 12 residues to the C terminus were new and were not found in reactions with CII. Our results, especially with CII-His6 as a substrate, suggest the presence of five major clusters for cleavage by FtsH. Although the rate of degradation of the tagged CII proteins was greatly reduced, no intermediate, partially degraded proteins were observed. It appears that the addition of amino acid tails at either end of CII drastically reduces the affinity to FtsH.
FtsH degrades β-casein.
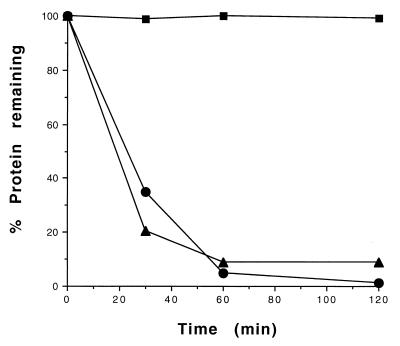
In search of additional substrates, we found that purified FtsH is capable of degrading β-casein. This protein is known to be present as a noncompact, largely unstructured protein that is highly modified at the N-terminal region by methylation and phosphorylation (17). The proteolysis of β-casein is ATP dependent and is inhibited by the Zn2+ chelator o-phenanthroline (data not shown). The rate of proteolysis of β-casein is similar to that of CII (Fig. 4). In contrast, we found that FtsH is unable to degrade bovine serum albumin (BSA) in vitro.
FIG. 4.
Proteolysis of β-casein by FtsH. In vitro kinetic degradation of β-casein (40 pmol), CII (68 pmol), and BSA (30 pmol) by GST-FtsH (14 pmol) was carried out at 42°C, and samples were taken at 30, 60, and 120 min. The proteins were resolved by SDS-PAGE (4 to 20% polyacrylamide), visualized by Coomassie staining, and quantitated as described in Materials and Methods. Squares, BSA; triangles, β-casein; circles, CII.
Peptides recovered following proteolysis of β-casein.
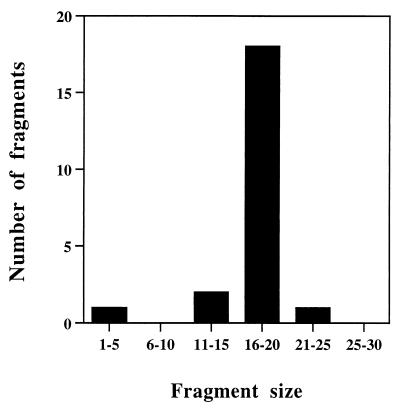
As was found with CII, no degradation intermediates of β-casein could be detected by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The modifications present in the N-terminal region of β-casein make the identification of the peptide by electrospray MS difficult. We therefore focused on identifying the peptides generated from the nonmodified C terminus following cleavage by FtsH. The general profile of the peptides obtained is similar to that found with CII, with peptide clustering and with no obvious cleavage site consensus (Fig. 5). The size distribution of the peptides obtained is similar to that obtained with CII; most peptides were found to be 16 to 20 residues long (Fig. 6). About half of the peptides resulted from cleavage in the highly hydrophobic region AFLLY.
FIG. 5.
Map of the peptides identified following β-casein proteolysis. The amino acid sequence of the C-terminal domain of β-casein from residue 136 to residue 224 is shown at the top. The peptides obtained from three independent experiments are shown below the β-casein amino acid sequence. The number of experiments in which each peptide was observed is given in parentheses. Asterisks above the β-casein sequence mark intervals of 10 residues.
FIG. 6.
Size distribution of β-casein peptides. The histogram shows a plot of the data presented in Fig. 5.
DISCUSSION
Electron microscopy revealed that purified FtsH forms ring-shaped structures with a diameter of 6 to 7 nm, suggesting that the active site is hidden in the central cavity (32). This organization of ATP-dependent proteases was also shown for ClpA/ClpP, HslU/HslV, and the 26S proteasome (see references 6 and 26 for an overview). The active site may be accessible via a narrow channel that prevents accidental protein degradation. The role of the ATPase activity in the degradation of proteins is not known. Its activity is probably utilized in providing the energy needed for conformational changes of the protease and for substrate unfolding to allow the passage of polypeptides into the catalytic chamber (6, 13).
The experiments described in this report were directed to the detailed analysis of FtsH digestion products. Our inability to obtain degradation intermediates may suggest that the rate-limiting step in proteolysis is the formation of substrate-enzyme complexes. Accordingly, rapid and complete proteolysis proceeds once degradation is initiated. The generation of peptides common to the CII and β-casein substrates substantiates the hypothesis that FtsH is an endopeptidase. For some unknown reason, the addition of tails containing His6 greatly stabilized CII against degradation by FtsH. Unfortunately, no information on the proteolysis of other substrates by FtsH is available. It is also possible that the peptide distribution found following proteolysis is determined by the rate of the peptides' escape from the enzyme.
The identification of peptides following proteolysis by FtsH leads to a number of findings, set forth below.
(i) Proteolysis does not appear to be random. A number of restricted sites for degradation were observed.
(ii) Many of the peptides identified appear as clusters where the cleavage sites are separated by 1, 2, or 3 residues. This phenomenon is found at both the N and C termini of CII derivatives and was observed in all reactions, suggesting either a degree of relaxation of the cleavage sites or that the enzyme “chews” on the ends of the peptides by an auxiliary secondary activity. Similar clusters were found in the analysis of the degradation products of 20-residue-long peptides (our unpublished results).
(iii) The sites of cleavage do not reveal a consensus sequence. Hydrophobic residues are found to be the preferred residues at the P1 site (the residue N-terminal to the cleavage site). This is especially evident in the LLAVL cluster in CII and the AFLLY cluster in β-casein. It is possible that the presence of hydrophobic residues at the peptide improves the stability of these peptides vis-à-vis secondary proteolytic events.
(iv) Cleavage sites in CII were found within regions predicted to form α helices and also at the ends of the predicted helical stretches, making it difficult to determine the importance of the secondary and tertiary structures in determining the preferential sites of cleavage. It is possible that CII, like β-casein, is highly unstructured during enzymatic cleavage.
(v) We have previously found that chemical cross-linking of CII produces a collection of monomers, dimers, trimers, and tetramers. Following incubation with FtsH, the monomers and dimers were degraded, the trimeric form was partially degraded, and the tetrameric form was fully stable (31).
It is not known what determines the size distribution of the peptides produced by FtsH. For both CII and β-casein, the most prevalent size is 16 to 20 residues, suggesting that peptide size is determined mainly by the enzyme. Proteolysis may be viewed as a process that occurs when the substrate is threaded through the enzyme cavity. Alternatively, it is possible that peptide size is determined by a “molecular ruler” which reflects the distance between catalytic sites within the enzyme complex. Such a model has been previously suggested for the proteasome (36). The oligomeric state of FtsH has not been established. However, based on the hexameric form of the highly related AAA domain (the NSF D2 hexameric AAA motif [24, 37]), it is possible that the protease active site of FtsH exists as a hexamer. Proteolysis may be viewed as a concerted reaction at six catalytic sites taking place after the substrate enters the catalytic cavity. Both models account for the peptide size distribution and for the low degree of neighboring-peptide overlaps observed in this work. Additional experimental data are required to distinguish between these models for FtsH activity.
ACKNOWLEDGMENTS
We thank Yossi Shlomai and Marika Lindahl for stimulating discussions and suggestions, and we thank Teru Ogura, Bernd Bukau, and Koriaki Ito for bacterial strains, plasmids, and unpublished information. We thank Marty Gonzales for CII protein and antibodies, Hilla Giladi and Ariella Oppenheim for critical reading of the manuscript, and Susan Gottesman for stimulating discussions.
This work was supported by a grant from the Israel Science Foundation and was performed, in part, in the Irene and Davide Sala Laboratory for Molecular Genetics.
REFERENCES
- 1.Akiyama Y, Kihara A, Ito K. Subunit a of proton ATPase F0 sector is a substrate of the FtsH protease in Escherichia coli. FEBS Lett. 1996;399:26–28. doi: 10.1016/s0014-5793(96)01283-5. [DOI] [PubMed] [Google Scholar]
- 2.Akiyama Y, Shirai Y, Ito K. Involvement of FtsH in protein assembly into and through the membrane. II. Dominant mutations affecting FtsH functions. J Biol Chem. 1994;269:5225–5229. [PubMed] [Google Scholar]
- 3.Akiyama Y, Yoshihisa T, Ito K. FtsH, a membrane-bound ATPase, forms a complex in the cytoplasmic membrane of Escherichia coli. J Biol Chem. 1995;270:23485–23490. doi: 10.1074/jbc.270.40.23485. [DOI] [PubMed] [Google Scholar]
- 4.Altuvia S, Oppenheim A B. Translational regulatory signals within the coding region of the bacteriophage λ cIII gene. J Bacteriol. 1986;167:415–419. doi: 10.1128/jb.167.1.415-419.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arlt H, Tauer R, Feldmann H, Neupert W, Langer T. The YTA10-12 complex, an AAA protease with chaperone-like activity in the inner membrane of mitochondria. Cell. 1996;85:875–885. doi: 10.1016/s0092-8674(00)81271-4. [DOI] [PubMed] [Google Scholar]
- 6.Baumeister W, Walz J, Zuhl F, Seemuller E. The proteasome: paradigm of a self-compartmentalizing protease. Cell. 1998;92:367–380. doi: 10.1016/s0092-8674(00)80929-0. [DOI] [PubMed] [Google Scholar]
- 7.Begg K J, Tomoyasu T, Donachie W D, Khattar M, Niki H, Yamanaka K, Hiraga S, Ogura T. Escherichia coli mutant Y16 is a double mutant carrying thermosensitive ftsH and ftsI mutations. J Bacteriol. 1992;174:2416–2417. doi: 10.1128/jb.174.7.2416-2417.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beyer A. Sequence analysis of the AAA protein family. Protein Sci. 1997;6:2043–2058. doi: 10.1002/pro.5560061001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Casari G, De Fusco M, Ciarmatori S, Zeviani M, Mora M, Fernandez P, De Michele G, Filla A, Cocozza S, Marconi R, Durr A, Fontaine B, Ballabio A. Spastic paraplegia and OXPHOS impairment caused by mutations in paraplegin, a nuclear-encoded mitochondrial metalloprotease. Cell. 1998;93:973–983. doi: 10.1016/s0092-8674(00)81203-9. [DOI] [PubMed] [Google Scholar]
- 10.Confalonieri F, Duguet M. A 200-amino-acid ATPase module in search of a basic function. Bioessays. 1995;17:639–650. doi: 10.1002/bies.950170710. [DOI] [PubMed] [Google Scholar]
- 11.DiMauro S, Schon E A. Nuclear power and mitochondrial disease. Nat Genet. 1998;19:214–215. doi: 10.1038/883. [DOI] [PubMed] [Google Scholar]
- 12.Gottesman S. Proteases and their targets in Escherichia coli. Annu Rev Genet. 1996;30:465–506. doi: 10.1146/annurev.genet.30.1.465. [DOI] [PubMed] [Google Scholar]
- 13.Gottesman S, Maurizi M R, Wickner S. Regulatory subunits of energy-dependent proteases. Cell. 1997;91:435–438. doi: 10.1016/s0092-8674(00)80428-6. [DOI] [PubMed] [Google Scholar]
- 14.Gottesman S, Wickner S, Maurizi M R. Protein quality control: triage by chaperones and proteases. Genes Dev. 1997;11:815–823. doi: 10.1101/gad.11.7.815. [DOI] [PubMed] [Google Scholar]
- 15.Herman C, Thevenet D, Bouloc P, Walker G C, D'Ari R. Degradation of carboxy-terminal-tagged cytoplasmic proteins by the Escherichia coli protease HflB (FtsH) Genes Dev. 1998;12:1348–1355. doi: 10.1101/gad.12.9.1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Herman C, Thevenet D, D'Ari R, Bouloc P. Degradation of sigma 32, the heat shock regulator in Escherichia coli, is governed by HflB. Proc Natl Acad Sci USA. 1995;92:3516–3520. doi: 10.1073/pnas.92.8.3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holt C, Sawyer L. Primary and predicted secondary structures of the caseins in relation to their biological functions. Protein Eng. 1988;2:251–259. doi: 10.1093/protein/2.4.251. [DOI] [PubMed] [Google Scholar]
- 18.Hoyt M A, Knight D M, Das A, Miller H I, Echols H. Control of phage λ development by stability and synthesis of cII protein: role of the viral cIII and host hflA, himA, and himD genes. Cell. 1982;31:565–573. doi: 10.1016/0092-8674(82)90312-9. [DOI] [PubMed] [Google Scholar]
- 19.Kihara A, Akiyama Y, Ito K. Different pathways for protein degradation by the FtsH/HflKC membrane-embedded protease complex: an implication from the interference by a mutant form of a new substrate protein, YccA. J Mol Biol. 1998;279:175–188. doi: 10.1006/jmbi.1998.1781. [DOI] [PubMed] [Google Scholar]
- 20.Kihara A, Akiyama Y, Ito K. Host regulation of lysogenic decision in bacteriophage lambda: transmembrane modulation of FtsH (HflB), the cII degrading protease, by HflKC (HflA) Proc Natl Acad Sci USA. 1997;94:5544–5549. doi: 10.1073/pnas.94.11.5544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kornitzer D, Altuvia S, Oppenheim A B. The activity of the CIII regulator of lambdoid bacteriophages resides within a 24-amino-acid protein domain. Proc Natl Acad Sci USA. 1991;88:5217–5221. doi: 10.1073/pnas.88.12.5217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kornitzer D, Teff D, Altuvia S, Oppenheim A B. Genetic analysis of bacteriophage lambda cIII gene: mRNA structural requirements for translation initiation. J Bacteriol. 1989;171:2563–2572. doi: 10.1128/jb.171.5.2563-2572.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leffers G G, Gottesman S. Lambda Xis degradation in vivo by Lon and FtsH. J Bacteriol. 1998;180:1573–1577. doi: 10.1128/jb.180.6.1573-1577.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lenzen C U, Steinmann D, Whiteheart S W, Weis W I. Crystal structure of the hexamerization domain of N-ethylmaleimide-sensitive fusion protein. Cell. 1998;94:525–536. doi: 10.1016/s0092-8674(00)81593-7. [DOI] [PubMed] [Google Scholar]
- 25.Leonhard K, Stiegler A, Neupert W, Langer T. Chaperone-like activity of the AAA domain of the yeast Yme1 AAA protease. Nature. 1999;398:348–351. doi: 10.1038/18704. [DOI] [PubMed] [Google Scholar]
- 26.Lupas A, Flanagan J M, Tamura T, Baumeister W. Self-compartmentalizing proteases. Trends Biochem Sci. 1997;22:399–404. doi: 10.1016/s0968-0004(97)01117-1. [DOI] [PubMed] [Google Scholar]
- 27.Miller C G. Protein degradation and proteolytic modification. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella. Vol. 1. Washington, D.C.: ASM Press; 1996. pp. 938–954. [Google Scholar]
- 28.Ogura T, Inoue K, Tatsuta T, Suzaki T, Karata K, Young K, Su L H, Fierke C A, Jackman J E, Raetz C R, Coleman J, Tomoyasu T, Matsuzawa H. Balanced biosynthesis of major membrane components through regulated degradation of the committed enzyme of lipid A biosynthesis by the AAA protease FtsH (HflB) in Escherichia coli. Mol Microbiol. 1999;31:833–844. doi: 10.1046/j.1365-2958.1999.01221.x. [DOI] [PubMed] [Google Scholar]
- 29.Qu J N, Makino S I, Adachi H, Koyama Y, Akiyama Y, Ito K, Tomoyasu T, Ogura T, Matsuzawa H. The tolZ gene of Escherichia coli is identified as the ftsH gene. J Bacteriol. 1996;178:3457–3461. doi: 10.1128/jb.178.12.3457-3461.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rattray A, Altuvia S, Mahajna J, Oppenheim A B, Gottesman M. Control of bacteriophage lambda cII activity by bacteriophage and host functions. J Bacteriol. 1984;159:238–242. doi: 10.1128/jb.159.1.238-242.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shotland Y. FtsH—an ATP-dependent protease. PhD thesis. Jerusalem, Israel: Hebrew University; 2000. [Google Scholar]
- 32.Shotland Y, Koby S, Teff D, Mansur N, Oren D A, Tatematsu K, Tomoyasu T, Kessel M, Bukau B, Ogura T, Oppenheim A B. Proteolysis of the phage lambda CII regulatory protein by FtsH (HflB) of Escherichia coli. Mol Microbiol. 1997;24:1303–1310. doi: 10.1046/j.1365-2958.1997.4231796.x. [DOI] [PubMed] [Google Scholar]
- 33.Studier F W, Rosenberg A H, Dunn J J, Dubendorff J W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 34.Tomoyasu T, Gamer J, Bukau B, Kanemori M, Mori H, Rutman A J, Oppenheim A B, Yura T, Yamanaka K, Niki H, et al. Escherichia coli FtsH is a membrane-bound, ATP-dependent protease which degrades the heat-shock transcription factor sigma 32. EMBO J. 1995;14:2551–2560. doi: 10.1002/j.1460-2075.1995.tb07253.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tomoyasu T, Yuki T, Morimura S, Mori H, Yamanaka K, Niki H, Hiraga S, Ogura T. The Escherichia coli FtsH protein is a prokaryotic member of a protein family of putative ATPases involved in membrane functions, cell cycle control, and gene expression. J Bacteriol. 1993;175:1344–1351. doi: 10.1128/jb.175.5.1344-1351.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wenzel T, Eckerskorn C, Lottspeich F, Baumeister W. Existence of a molecular ruler in proteasomes suggested by analysis of degradation products. FEBS Lett. 1994;349:205–209. doi: 10.1016/0014-5793(94)00665-2. [DOI] [PubMed] [Google Scholar]
- 37.Yu R C, Hanson P I, Jahn R, Brünger A T. Structure of the ATP-dependent oligomerization domain of N-ethylmaleimide sensitive factor complexed with ATP. Nat Struct Biol. 1998;5:803–811. doi: 10.1038/1843. [DOI] [PubMed] [Google Scholar]