Abstract
The avian infectious bronchitis virus (IBV) is a highly mutable coronavirus that causes an acute and highly contagious disease responsible for economic losses to the poultry industry worldwide. Preventing and controlling bronchitis disease is difficulted by the numerous IBV circulating types with limited antigenic cross-protection that hamper the prevention and control by heterologous vaccines. The coding region of the variable spike S1 receptor-attachment domain is used to classify IBV in 7 genotypes (GI–GVII) comprising 35 viral lineages (1–35). Knowledge of the circulating IBV types causing outbreaks in a specific geographic region is beneficial to select better the appropriate vaccine(s) and contribute to disease control. In the study, 17 avian infectious bronchitis virus strains were obtained from chickens showing signs of illness in Mexico from 2007 to 2021. We detected 4 lineages within genotype I, three already known (GI-3, GI-9, GI-13) and one newly described (GI-30). In addition, we identified 2 divergent monophyletic groups that are tentatively described as lineages of new genotypes (GVIII-1 and GIX-1). Our findings revealed that Mexico's high genetic IBV diversity results from the co-circulation of divergent lineages belonging to different genotypes. Mexican IBV lineages differ significantly from Massachusetts and Connecticut vaccine strains, indicating that the currently used vaccines may need to be updated.
Key words: IBV, Mexico, genetic lineage, respiratory illness, poultry
INTRODUCTION
The avian gammacoronavirus Infectious Bronchitis Virus (IBV) is the causative agent of an acute and highly contagious disease that affects chickens and poses a major economic burden on the poultry industry (Cavanagh, 2007). The virus can affect the upper respiratory and reproductive tracts, and some strains cause nephritis.
IBV has a single-stranded positive-sense RNA of nearly 27 kb genome surrounded by a lipid envelope. The membrane spike glycoprotein (∼1,145 amino acids), which forms club-shaped projections on the viral surface, is the major inducer of neutralizing and serotype-specific antibodies and is responsible for virus binding and entry to host cells. The precursor S protein is post-translationally cleaved by proteases into 2 non-covalently bound polypeptides (S1 and S2). The amino-terminal S1 (∼535 amino acids) subunit forms the tip of the spike, and the carboxyl-terminal S2 (∼627 amino acids) subunit anchors the S1 into the viral membrane.
The virus has extraordinary genetic variability, promoting pathogen complexity and challenging prevention and control. Variant strains and new genotypes of IBV are continuously emerging worldwide.
IBV has many serotypes, challenging vaccine efficacy due to poor cross-protection. An effective control strategy involves identifying the virus type causing outbreaks, followed by vaccination with a closely-related vaccine strain. However, only a few different kinds of IBV vaccines are available, in contrast with the numerous types and variants of the virus worldwide. Considering the IBV antigenic variability, the characterization of strains based on genotype or serotype is beneficial to elucidating disease epidemiology. The genotype classification based on the S1 coding region becomes the primary method for categorizing IBV strains because of the rapid evolution of the S1 gene and the importance of S1 in virus-host interactions, induction of poor cross-protection of commercial vaccines, and the emergence of IBV variants.
The most recent comprehensive classification uses the entire S1 region and identified 6 main genotypes (GI–GVI), 32 viral lineages (1–32), and a few interlineage recombinants in globally circulating strains (Valastro et al., 2016). This classification system originally included 27 lineages for genotype 1 and one for the remaining 5. However, the number of viral types has increased over the years; 2 new lineages for genotype 1 (GI-28 and GI-29) and one additional genotype (GVII-1) have been described (Jiang et al., 2017; Ma et al., 2019; Zhang et al., 2020).
IBV is one of Mexico's most important respiratory diseases in poultry. Despite wide vaccination with the Massachusetts (Mass) and Connecticut (Conn) vaccine strains, it is common to find IBV-caused disease in vaccinated chickens. The only 2 Mexican complete S1 sequences available in the Databases were included in Valastro et al.’s (2016) classification. The 98-07484 (AF288467) strain was denoted as a unique variant (UV), and the BL-56 strain (AF352831) was classified within the GI-3 lineage. To date, no other reports describe the IBV genotypes and lineages circulating in Mexico; knowing IBV viral types would be crucial to preventing the disease and selecting the most appropriate vaccine.
The present study identified the IBV lineages circulating in commercial poultry farms in Mexico to contribute to the knowledge of the molecular epidemiology of this relevant poultry pathogen.
MATERIALS AND METHODS
Nine samples were collected from different outbreaks in commercial broilers and layers with respiratory signs during 2019–2021 in Jalisco, Mexico (Table 1). Eight samples were previous virus isolates stored at the Universidad Nacional Autónoma de México (UNAM) collected in 2007 from Central Mexico. The clinical samples used in this study were collected according to the guidelines for Animal Ethics Committees.
Table 1.
S1 IBV genetic variants in Mexico. Name, Accession number, classification (genotype_lineage), bird type, collection year, and geographic origin are indicated from Mexican strains.
| Strain | Accession | Classification | Type | Year | Origin |
|---|---|---|---|---|---|
| Mex-56-10 | ON470378 | GI-3 | Hen | 2007 | Central Mexico |
| Mex-56-6 | ON470379 | GI-3 | Hen | 2007 | Central Mexico |
| Mex-56-8 | ON470380 | GI-3 | Hen | 2007 | Central Mexico |
| Mex-20 | ON470381 | GI-9 | Broiler | 2019 | Jalisco |
| Mex-Ark1 | ON470382 | GI-9 | Hen | 2018 | Jalisco |
| Mex-327 | ON470383 | GI-9 | Hen | 2019 | Jalisco |
| Mex-15 | ON470384 | GI-13 | Hen | 2020 | Jalisco |
| Mex-1 | ON470385 | GI-30 | Hen | 2019 | Jalisco |
| Mex-07-1 | ON470386 | GI-30 | Hen | 2007 | Central Mexico |
| Mex-07-2 | ON470387 | GI-30 | Hen | 2007 | Central Mexico |
| Mex-07-3 | ON470388 | GI-30 | Hen | 2007 | Central Mexico |
| Mex-3-1 | ON470389 | GI-30 | NA | 2019 | Jalisco |
| Mex-430 | ON470390 | GI-30 | Hen | 2019 | Jalisco |
| Mex-12 | ON470391 | GVIII-1 | Hen | 2020 | Jalisco |
| Mex-3009 | ON470392 | GVIII-1 | NA | 2021 | Jalisco |
| Mex-14P | ON470393 | GIX-1 | Hen | 2020 | Jalisco |
| Mex-56-7 | ON470394 | GIX-1 | NA | 2007 | Central Mexico |
NA: not available, Central Mexico: states of Queretaro, Morelos, Hidalgo, or Puebla.
The entire S1 coding region was obtained by RT-PCR in 2 overlapping fragments using previously described primers and procedures (Marandino et al., 2015). Primers (S5 and S4) for the second fragment were updated to improve the amplification of most divergent lineages: S5 (TTTATAAACGGCACTGCACA) for GI-30, S5 (TTTATAAATGGMASWGTGCA), and S4 (CATAAGACACATAAGRGCAA) for GVIII and GIX-1.
A full-length S1 dataset was built using the newly obtained Mexican sequences (Table 1), prototype IBVs (n = 199) from Valastro et al. (2016), and new identified lineages and genotypes: 5 strains for GI-28 (Chen et al., 2017), 2017), 3 for GI-29 (Jiang et al., 2017), and 2 for GVII-1 (Ma et al., 2019).
DNA alignments and maximum-likelihood phylogenetic trees with 1,000-replicates bootstrap support were inferred using MAFFT and FastTree plugins in Geneious (https://www.geneious.com/). Tree visualization was performed with the ggTree package in RStudio (https://www.rstudio.com/). Evolutionary divergence (p-distance) within and between lineages was obtained with MEGA (https://www.megasoftware.net/)
The S1 coding region of Mexican IBV strains was analyzed using the Recombination Detection Program (http://web.cbio.uct.ac.za/∼darren/rdp.html). Recombinant events were considered positive when supported by at least 2 methods with a P-value adjusted to 0.05 and confirmed by significant phylogenetic incongruence among trees estimated on either side of the putative recombination breakpoints (Valastro et al., 2016).
RESULTS AND DISCUSSION
Seventeen complete S1 nucleotide sequences were obtained from chickens with typical respiratory clinical signs (Table 1). The S1 sequence had 9 to 11 nucleotides more than the GI-1 Baudette reference strain (GenBank: M95169.1). In addition, the nucleotide identity varied from 58 to 99.9% among the Mexican strains and was 80.0% or less with the reference sequence. This diversity results from single nucleotide polymorphisms (SNPs), insertion, and deletions (indels) occurring along the entire S1. No recombination was observed in the Mexican strains, and all of them were included in the phylogenetic analysis.
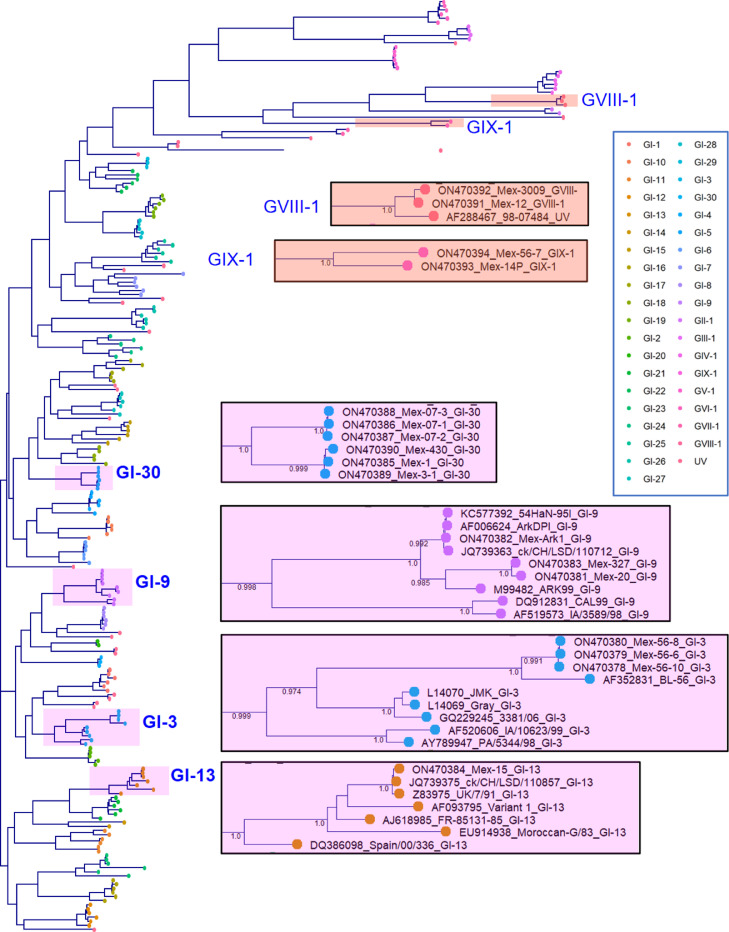
The genetic relationships were analyzed using the nucleotide sequences of 226 S1 strains, including IBV references from the existing genotypes and lineages. The phylogenetic tree clustered the strains according to their genotype classification and lineages in well-supported clades (support values >0.99; Figure 1). Genotype I includes the greater number of strains divided into several lineages. The remaining genotypes containing one lineage are clustered; they have longer branch lengths, reflecting more genetic changes and suggesting low relatedness.
Figure 1.
Maximum-likelihood tree method based on 226 S1 sequences from Mexican and reference IBV strains of the existing genotypes and lineages. Details of the Mexican lineages are zoomed in on the right.
The Mexican IBV strains were clustered into 6 IBV lineages divided into 3 putative genotypes. Seven Mexican strains fall into 3 lineages of genotype 1 (GI-3, GI-9, and GI-13) by phylogenetic clustering within the corresponding reference strains and genetic distances (Table 1, Figure 1).
The three strains of the GI-3 are associated in a subclade with the Mexican BL-56 (AF352831) strain previously assigned to GI-3 by Valastro et al. (2016). In a previous study, the Mexican strains 5697/99, 7483/98, and 5700/99 appeared closely related to BL-56 (Gelb et al., 2001). Only partial S1 sequences (not deposited in the Genbank database) were obtained from these strains avoiding proper assignation to GI-3. The BL-56 strain was collected in central Mexico more than 15 yr ago. The first GI-3 strains were described in the United States in the 1960s and the late 1990s before being identified in Taiwan in 2006. They were originally designated as JMK or the Gray serotype based on the reference type strains (Accessions: L14070 and L14069).
Three Mexican strains belong to GI-9, a lineage containing vaccine and virulent field strains known as Arkansas (Ark) and Ark DPI-like types. The Ark virus is one of the most commonly reported types to cause widespread disease in the United States. When it first emerged in Arkansas in 1973, it was described as genetically distinct from all the known IBV serotypes recognized and was referred to as Ark99. During the 1980s, an attenuated vaccine derived from an Ark-type virus isolated in the Delmarva Peninsula (Ark DPI strain) was extensively used in the United States. The similarity of Mexican strains (Mex-20 and Mex-327) to M99482 (ARK99), collected in the United States in 1973, suggests it most likely represents the vaccine strain's re-isolations; the Mex-Ark1 is more divergent and might be an Ark field type.
A single Mexican strain was classified as GI-13. This lineage occurs in many parts of the world and has been previously denoted as 793/B, 4/91, and CR88 (Cook et al., 1996). The 793/B-type (4/91) and the Massachusetts-based vaccines are 2 major antigenic types that have been commonly used in several countries for a long time. GI-13 has never been genetically reported in the Mexican poultry industry, but there is serological evidence for its circulation during the 1990s (Cook et al., 1996). However, the strain here detected is very similar (99%) to the 4/91 vaccine strain (KF377577.1), supporting that it corresponded to a vaccine-derived strain, although we do not have evidence that vaccination was implemented with this variant.
Six strains did not fall within established lineages within genotype 1. They were then classified as a new lineage (GI-30) by clustering in a well-supported clade. The GI-30 seems to be an indigenous lineage in Mexico that has persisted for at least 20 yr in different geographic regions in central and western Mexico (Table 1). GI-30 has low intragroup divergence (3% nucleotides and 6% amino acids) and differs by 17% nucleotides and 25% amino acids from the closest GI-27 lineage. These values lie within the range observed for other IBV lineages of genotype 1 (Valastro et al., 2016).
Four Mexican strains clustered apart from the GI viruses and likely belong to the first lineages of new IBV genotypes, lineage 1 of genotype VIII (GVIII-1) and lineage 1 of genotype IX (GIX-1).
The new GVIII includes 2 Mexican samples clustered with the unique 98-0748 Mexican variant (AF288467 or UNAM-97) previously described by Valastro et al. (2016). The GVIII-1 has a low intralineage distance (2% nucleotides and 3% amino acids). The designation as a new genotype is supported by their independent detection in the field, clustering in the phylogenetic tree, the high distance (28% nucleotides and 45% amino acids) to the closest GIV-1, and 30 unique amino acid changes. IBV genotypes differ by more than 29% in nucleotides and amino acids (Valastro et al., 2016). Strains similar to the unique 98-0748 Mexican variant were described by partial S1 sequence and PCR-RFLP (Escorcia et al., 2000; Gelb et al., 2001). These original viruses were collected from broilers between 1997 and 1999 in the central Mexican states of Queretaro, Guanajuato, and San Luis Potosi. Although no sequences are available to confirm the classification, these strains might correspond to putative ancient GVIII-1.
The GIX-1 includes 2 Mexican samples that appear separated in the phylogenetic tree. Samples belonged to different geographic areas and were collected 13 yr apart. This new genotype has an intralineage divergence of 6% nucleotides and 10% amino acids and differs extensively from the closest GVII-1 (35% nucleotides and nearly 50% amino acids). The GIX-1 has 24 unique residues and 2 characteristic insertions of 5 and 3 amino acids.
All the lineages identified in Mexico (GI-3, GI-9, GI-13, GI-30, GVIII-1, and GIX-1) have a wide geographic distribution and were collected across time and geographic location (Table 1) supporting that they are persistent and not atypical or sporadic strains.
Our findings revealed that Mexico's high genetic diversity results from the co-circulation of divergent lineages belonging to different genotypes. Determining if these lineages are also different serotypes is of paramount importance. Although there are some exceptions, there was a strong correlation between the genotype (sequence identity), serotype, and immunotype/protectotype (the level of cross-protection). IBV strains within the same serotype usually share more than 95% amino acid similarity in S1 and belong to the same immunotype. In contrast, IBV serotypes differ by 20 to 25% at the amino acid level in S1 but may vary by up to 50% (Cavanagh, 2007). However, in some cases, as little as 2% variation can lead to a new IBV serotype due to changes in the epitopes eliciting cross-neutralizing antibodies. In other cases, highly similar viruses show only limited cross-protection, while a high level of cross-protection may exist for strains with a much lower homology. The high amino acid divergence observed in the lineages here described (>25% in all cases) strongly suggests that they are also different serotypes, particularly GVIII-1 and GIX-1, with divergence levels higher than 45%. Serological evidence suggests that strains similar to GVIII-1 are antigenically unique (Escorcia et al., 2000). Virus cross-neutralization results are needed to establish if these Mexican IBV lineages and genotypes circulating in the field have specific antigenic profiles.
The commercial attenuated vaccines against IBV (including Mass and Conn strains of GI-1) have been widely used in Mexico. However, infectious bronchitis still frequently occurs in commercial chicken farms due to poor cross-protection. Considering that Mass and Conn strains have a low amino acid identity (<75%) with the IBV lineages circulating in the Mexican poultry industry, it is possible that the currently used vaccine strains need to be updated. In this sense, long-term molecular epidemiological surveillance acquires increased relevance for preventing and controlling infectious bronchitis disease.
ACKNOWLEDGMENTS
This work was supported by the Fondo Conjunto de Cooperación México-Uruguay (2019). Ana Marandino, Gonzalo Tomás, Yanina Panzera and Ruben Pérez belong to the Programa de Desarrollo de las Ciencias Básicas (PEDECIBA, Uruguay).
DISCLOSURES
We do not have any commercial or associative interest representing a conflict of interest with the submitted work.
REFERENCES
- Cavanagh D. Coronavirus avian infectious bronchitis virus. Vet. Res. 2007;38:281–297. doi: 10.1051/vetres:2006055. [DOI] [PubMed] [Google Scholar]
- Chen Y., Jiang L., Zhao W., Liu L., Zhao Y., Shao Y., Li H., Han Z., Liu S. Identification and molecular characterization of a novel serotype infectious bronchitis virus (GI-28) in China. Vet. Microbiol. 2017;198:108–115. doi: 10.1016/j.vetmic.2016.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook J.K., Orbell S.J., Woods M.A., Huggins M.B. A survey of the presence of a new infectious bronchitis virus designated 4/91 (793B) Vet. Rec. 1996;138:178–180. doi: 10.1136/vr.138.8.178. [DOI] [PubMed] [Google Scholar]
- Escorcia M., Jackwood M.W., Lucio B., Petrone V.M., López C., Fehervari T., Téllez G. Characterization of Mexican strains of avian infectious bronchitis isolated during 1997. Avian Dis. 2000;44:944–947. [PubMed] [Google Scholar]
- Gelb J., Ladman B.S., Tamayo M., Gonzalez M., Sivanandan V. Novel infectious bronchitis virus S1 genotypes in Mexico 1998-1999. Avian Dis. 2001;45:1060–1063. [PubMed] [Google Scholar]
- Jiang L., Zhao W., Han Z., Chen Y., Zhao Y., Sun J., Li H., Shao Y., Liu L., Liu S. Genome characterization, antigenicity and pathogenicity of a novel infectious bronchitis virus type isolated from south China. Infect. Genet. Evol. 2017;54:437–446. doi: 10.1016/j.meegid.2017.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma T., Xu L., Ren M., Shen J., Han Z., Sun J., Zhao Y., Liu S. Novel genotype of infectious bronchitis virus isolated in China. Vet. Microbiol. 2019;230:178–186. doi: 10.1016/j.vetmic.2019.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marandino A., Pereda A., Tomás G., Hernández M., Iraola G., Craig M.I., Hernaández D., Banda A., Villegas P., Panzera Y., Pérez R., Hernández D., Banda A., Villegas P., Panzera Y., Pérez R., Hernaández D., Banda A., Villegas P., Panzera Y., Pérez R. Phylodynamic analysis of avian infectious bronchitis virus in South America. J. Gen. Virol. 2015;96:1340–1346. doi: 10.1099/vir.0.000077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valastro V., Holmes E.C., Britton P., Fusaro A., Jackwood M.W., Cattoli G., Monne I. S1 gene-based phylogeny of infectious bronchitis virus: an attempt to harmonize virus classification. Infect. Genet. Evol. 2016;39:349–364. doi: 10.1016/j.meegid.2016.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Deng T., Lu J., Zhao P., Chen L., Qian M., Guo Y., Qiao H., Xu Y., Wang Y., Li X., Zhang G., Wang Z., Bian C. Molecular characterization of variant infectious bronchitis virus in China, 2019: implications for control programmes. Transbound. Emerg. Dis. 2020;67:1349–1355. doi: 10.1111/tbed.13477. [DOI] [PMC free article] [PubMed] [Google Scholar]