Abstract
Avian infectious bronchitis virus (IBV) is a prevalent RNA virus that causes respiratory distress, nephritis, salpingitis, and egg production decline in chickens, resulting in significant economic loss. IBV is composed of complex genotypes and serotypes, which poses a great challenge for disease control. The current study reports 2 IBV outbreaks which were characterized by respiratory symptoms in IBV vaccinated commercial broilers and layers in Guangdong, China, in 2021. Two IBV strains, ZH01 and HH09, were identified via a RT-PCR assay through targeting the N gene and further characterization through full-length spike (S) gene sequence analysis. Phylogenetic analysis of S1 gene revealed that both ZH01 and HH09 belonged to the GI-19 lineage but contained a certain genetic distance from the GI-19 strain. Of note, the ZH01 and HH09 strains share a low homology of 70 and 86%, respectively, with common vaccine strains (H120), resulting in low vaccine protection. Further recombination analysis based on the S1 sequence suggested the newly identified IBV strains emerged through an intragroup recombination events between CK/CH/SCDY2003-2 and I0305/19 from G1-19 lineage. In addition, a number of novel mutations such as T273I, T292A, and S331K were found in the emerging IBV strains. Taken together, this study reports the genetic characteristics of 2 recent IBV outbreaks in southern China and emphasizes the urgent need for enhanced surveillance and development of novel vaccines for the control of IBV.
Key words: infectious bronchitis virus, S1 gene, avian coronavirus, phylogenetic analysis, G1-19 genotype
Introduction
Avian infectious bronchitis virus (IBV) is a common pathogen in poultry. This pathogen causes an acute and highly contagious infectious bronchitis (IB) disease, leading to large economic losses in the poultry breeding industry (Sjaak de Wit et al., 2011). IBV can be transmitted through the respiratory and fecal-oral routes and cause damage to the respiratory, reproductive, and urinary systems. Therefore, the poultry exhibits symptoms such as difficulty breathing, sneezing, rales in the trachea, lethargy, nephritis, fallopian tube disease, reduced egg production, and even death (Houta et al., 2021). Although chicken flocks of all ages are susceptible hosts, the symptoms of infection in chickens at different ages are not always the same (Cavanagh, 2007). Chickens and pheasants were considered as the nature hosts for IBV; however, IBV has also been isolated from other poultry species and wild birds, including peacocks, guinea fowls, turkey, quail, geese, ducks, parrots, and partridges (Cavanagh, 2005).
IBV, a type of coronavirus of the genus γ-coronavirus, family Coronaviridae, is a large enveloped, positive stranded RNA virus with a genome of approximately 27.6 kb in size (Boursnell et al., 1987). About two-thirds of its genome is occupied by two large open reading frames (ORFs), ORF1a and ORF1b, which encode components of virus replicase (Jiang et al., 2017). The remaining one-third of the genome encodes for four structural proteins: the spike (S), membrane (M), envelop (E), and the nucleocapsid (N) protein. It also contains at least 4 accessory genes such as 3a, 3b, 5a, and 5b at the 3’end of the genome (Chen et al., 2017). The precursor S glycoprotein is cleaved into 2 polypeptides (subunits S1 and S2) by proteases. The S1 protein contains important virus-neutralizing epitopes and carries serotype-specific determinants (Wickramasinghe et al., 2011), which are closely related to the antigenicity or pathogenicity of the virus (Domanska-Blicharz et al., 2017). In addition, the S1 gene contains three hypervariable regions (HVR) that were shown to have the highest variability in the entire viral genome (Niesters et al., 1986). Therefore, the mutation and recombination of the S1 gene are crucial for the emergence of new IBV genotypes and serotypes. According to a classification scheme based on the S1 sequence, IBV strains have been classified into 36 lineages in 7 genotypes (Valastro et al., 2016; Jiang et al., 2017; Ma et al., 2019; Hou et al., 2020; Molenaar et al., 2020). Due to this large number of serotypes and genotypes, IBV prevention remains to be a challenge.
The present study reports 2 cases of IBV outbreaks in southern China that caused serious respiratory symptoms in chickens. The genotype, recombination events and evolutionary characteristics of the novel emerging IBV strains were investigated, which will be beneficial to the control of IBV in the poultry industry.
MATERIALS AND METHODS
Case History
In January 2021, a sudden respiratory disease emerged from a farm containing laying hens and broiler chickens which received H120 vaccine in Zhuhai City, Guangdong Province. Respiratory disease was detected in laying hens that are 200 days old and in broilers that are 20 days old, wherein the laying hens showed a decreased laying rate. Broilers were revealed to have typical clinical symptoms such as weakness, nasal discharge, dyspnea, and bronchial blockage, leading to some deaths. At the same time, it was observed that there was no obvious effect after treatment with antibacterial drugs.
In May 2021, an outbreak of broiler respiratory disease occurred in 6 poultry farms in Shaoguan City, Guangdong Province. The main symptoms shown were difficulty breathing, paralysis, and loss of appetite. In addition, one 40-day-old chick was unable to stand and appeared depressed. All broilers have been vaccinated with the H120 and LaSota vaccine. Since May 2021, an average of 1/1,000 broilers aged 20 to 50 days have died every day.
RNA Extraction, IBV Detection, and Virus Isolation
Multiple tissue samples, including the lungs, trachea, and kidneys, were collected and homogenized with 1 mL of phosphate-buffered saline containing 1,000 U/mL penicillin and 100 µg/mL streptomycin. The homogenate was then frozen and thawed 3 times, followed by clarification via centrifugation at 1,500 × g for 10 min at 4°C. The total RNA was extracted from 200 µL of the supernatant using Trizol reagent (Takaka, Dalian, China) following the manufacturer's instructions. Then, the RNA was transcribed to cDNA using HiScript III 1st Strand cDNA Synthesis Kit (Vazyme, Nanjing, China) according to the manufacturer's instructions. Specific primer pairs (forward primer: 5′-GTGAT (G/C)AGTGTGT(T/C)GATGGTGT-3′; reverse primer: 5′-CCTGCTTCTT (T/G)G(G/A)CTTTCTCC-3′) were then designed based on the conserved regions of the N gene sequence of IBV as taken from GenBank. RT-PCR with an expected 150 bp product was then carried out using a 2 × Taq Plus Master Mix II (Dye Plus) (Vazyme, Nanjing, China) under the following conditions: 95°C for 5 min; 34 cycles of 95°C for 15 s, 55°C for 20 s, 72°C for 30 s; and 72°Cfor 10 min. The supernatants of the IBV-positive samples were then filtered using a 0.45-µm filter (Millipore, Bedford, MA) and propagated in 10-day-old specific pathogen free (SPF) chicken eggs at 37°C for 72 h.
Virus Sequencing
Specific primers for amplifying the S1 gene (forward primer, IBV-F: 5'-GAAAAGACCGACTTAGT-3′; reverse primer, IBV-R: 5'-ATTAAGTAAAGGTGCCAC-3′) were designed based on the conserved regions of the S gene of IBV sequences deposited in GenBank. RT-PCR was then performed using a 2 × Taq Plus Master Mix II (Dye Plus) kit in a 1 µL reaction volume containing 10 pM of each primer under the following conditions: 95°C for 5 min; 34 cycles of 95°C for 15 s, 55°C for 20 s, 72°C for 2 min; and 72°C for 10 min. The PCR product of S1 with an expected size of 1,800 bp was then purified using a GeneJET gel extraction kit (Thermo Fisher Scientific, Waltham, MA) according to the manufacturer's instructions. The purified PCR product was ligated with the pMD18-T vector (Takara, Dalian, China) and then transfected into Top10 competent cells (AngYuBio, Shanghai, China). The positive clones were then cultured and treated with the Gene JET plasmid miniprep kit (Thermo Fisher Scientific, Waltham, MA) for plasmid extraction. The extracted plasmids were then sequenced via Sanger sequencing (Sangon Biotech; Shanghai, China).
Phylogenetic Analysis
The IBV sequences obtained in this study were then assembled using DNAStar Lesergene SeqmanPro software (Version 11.0). The assembled full-length sequences of the S1 gene were then aligned with the sequences obtained from Genbank using the BLAST tool in the NCBI database. A total of 80 representative IBV strains from different lineages and genotypes were extracted from the Genbank to investigate the evolution of the IBV strains identified in this study. A multiple sequence alignment was then performed using MAFFT (Multiple Alignment using Fast Fourier Transform) embedded in the UGENE software (Version 36.0). The distance matrix of IBV strains identified in this study and the reference strains was generated by the UGENE software (Version 36.0). The maximum likelihood phylogenetic tree of the S1 gene of IBV was constructed as previously described (Wen et al., 2021).
Recombination Analysis
The S1 gene sequence of the IBV strain identified in this study was then compared with the reference strain to study the possible recombination events obtained from recombination breakpoints. The recombination events were then predicted using RDP 4.0 as previously described (Wen et al., 2021). A total of nine methods, including RDP, GENECONV, BootScan, MaxChi, Chimaera, SiScan, PhyLPro, LARD, and 3Seq were then applied, and the predicted recombination events were further verified using SimPlot software (Version 3.5.1).
Amino Acid Polymorphism Analysis
The amino acid sequence of the S1 glycoprotein of the newly identified IBV in this study was aligned with the IBV strains from 7 genotypes by MAFFT and was viewed through the UGENE software. Sequence motif logos and the amino acid frequency at each position of the S1 glycoprotein were determined using Seq2Logo 2.0 program (http://www.cbs.dtu.dk/biotools/Seq2Logo) with the default settings (Thomsen and Nielsen, 2012).
RESULTS
Clinical Features and IBV Detection
Outbreaks of respiratory disease with clinical signs such as rough feathers, sneezing, expiratory dyspnea, nasal discharge, lethargy, or even death were found in chickens that received the H120 vaccine at different ages from 7 different farms in Guangdong China. It was observed that there was no noticeable improvement after antibiotic treatment. All samples were positive for IBV by targeting the conserved region of the N gene. Meanwhile, all samples were detected negative for avian influenza virus and Newcastle disease virus using the aforementioned method (Wen et al., 2020; Yang et al., 2021).
Sequencing of the S1 Gene
The S1 gene has an important antigenic site of the IBV virus that may be involved in serotyping. Therefore, we amplified the S1 gene and conducted sequencing for analysis. Among the 2 identified strains, the nucleic acid sequences of ZH01 and HH09 were both shown to be 1,620 nt in length. The corresponding amino acid sequence derived by MEGA 5.10 is determined to be 540 aa in length. A BLAST search from GenBank showed that S1 gene of the ZH01 and HH08 viruses shared the highest homology with HeN-2/China/2019 (MN055628.1) and ck/CH/LSD/150311 (KX219795.1), with an identity of 98.8% and 97.9%, respectively. The cleavage site of the S protein was located at the end of the S1 protein, which is an important part of its sequence segmentation. In addition, the split points showed a certain degree of preference in different genotypes that may be of important research significance. Through MEGA 5.10 software translation, the cleavage sites of ZH01 and HH09 were determined as HRRRR, which is consistent with most QX IBV cleavage sites (Table 1).
Table 1.
IBV reference strains used in this study.
| Strain | Origin | Year | Genotype/serotype | Cleavage site | Accession No. |
|---|---|---|---|---|---|
| ZH01 | China | 2021 | QX | HRRRR | OL332822 |
| HH09 | China | 2021 | QX | HRRRR | OL332823 |
| LX4 | China | 1999 | QX | HRRRR | AY189157 |
| CK/CH/LJL/08III | China | 2008 | QX | HRRRR | GQ258318 |
| ck/CH/LJL/05I | China | - | QX | HRRRR | KX252778 |
| IBS138/2015 | Malaysia | 2015 | QX | HRRRR | KU949745 |
| ck/CH/LHB/111190 | China | 2011 | QX | HRRRR | JQ739276 |
| I0305/19 | China | 2019 | QX | HRRRR | MN794188 |
| CK/CH/SCDY2003-2 | China | 2020 | QX | HRRKR | MW042848 |
| 4/91 | UK | vaccine | 4/91 | RRSRR | KF377577 |
| CK/CH/HBXN08-1 | China | - | 4/91 | RRSRR | MT766955 |
| ck/CH/LBJ/140402 | China | 2014 | 4/91 | RRSRR | KP118882 |
| LDT3-A | China | vaccine | LDT3 | RRFRR | KR608272 |
| GX-NN120091 | China | 2012 | LDT3 | HRRKR | KJ999806 |
| H52 | Netherlands | vaccine | Mass | RRFRR | EU817497 |
| H120 | Netherlands | vaccine | Mass | RRFRR | KF188436 |
| M41 | USA | vaccine | Mass | RRFRR | AY561711 |
| Ark99 | USA | 1989 | Arkansas | HRSRR | M99482 |
Abbreviations: HRRRR, His-Arg-Arg-Arg-Arg; HRRKR, His-Arg-Arg-Lys-Arg; HRHKR, His-Arg-His-Lys-Arg; RRSRR, Arg-Arg-Ser-Arg-Arg; RRFRR, Arg-Arg-Phe-Arg-Arg.
Phylogenetic Analysis of the S1 Gene
UGENE software was used to analyze the genetic characteristics of the S1 gene of the newly identified IBV strains. The nucleotide and amino acid similarity of the S1 gene sequences between ZH01 and HH09 were 96.6 and 96.1%, respectively (Supplementary Figure 1). The genetic distance matrix showed that the S1 gene of ZH01 and HH09 share high homology with the GX-19 representative strain of QX-type (LX4 AY338732) in the nucleotides (95.7 and 94.8%, respectively) and amino acid (94.4 and 93.9%, respectively) sequences. However, the ZH01 and HH09 shared low nucleotide homology with the common vaccine strains H120 (77.0 and 77.3%, respectively), 4/91 (78.2 and 77.8%, respectively), ldt3-a (85.2% and 86.0%, respectively), and M41 (76.8 and 77.1%, respectively). Similarly, low homologies of the amino acids ranging from 74.8 to 85% were observed between the newly identified IBV strains and the commercial vaccine strains. The maximum likelihood phylogenetic tree based on the S1 gene showed that both ZH01 and HH09 are clustered within the GI-19 lineage (Figure 1). However, there was a certain genetic distance between the IBV strain and GI-19 strain QX, indicating that the IBV strain in southern China tended to evolve during transmission. This statement is supported by the long genetic distance from the vaccine strains H120, H52, and 4/91.
Figure 1.
Phylogenetic tree of S1 genes of emerging IBV strains identified in this study and 80 reference strains. The maximum likelihood methods with 1,000 bootstrap replicates were applied for the phylogenetic analysis. The newly identified IBV strains are marked with red. The commercial vaccine strains are marked with blue. Abbreviation: IBV, infectious bronchitis virus.
Amino Acid Analysis
The S1 gene of IBV has important viral neutralization epitopes and 3 hypervariable regions (HVRs; Figure 2): HVR I (38 aa-67 aa), HVR II (91 aa-141 aa), and HVR III (274 aa-387 aa) (Valastro et al., 2016; Zhang et al., 2020). To further study the amino acid changes of the IBV S1 gene, analysis of the amino acid changes of the S1 gene were conducted in the two identified strains and 80 reference strains used for nucleotide analysis (Table 2). The results show that most of the amino acid mutations of ZH01 and HH09 occur in the HVR region, while the specific amino acid mutations of ZH01 (T278L) and HH09 (T115A, and F280L) occur in conserved region of the GI-19 genotype. However, some of the amino acid changes may occur in the receptor-binding domain, affecting its antigen-binding ability. Unique mutations such as T273I, T292A, and S331K were found in ZH01 strain.
Figure 2.
Sequence weighting of HVR I (A), HVR II (B), and HVR III (C) are displayed by Seq2Logo 2.0 program (http://www.cbs.dtu.dk/biotools/Seq2Logo). The symbols at each position represent the amino acids.
Table 2.
Amino acid polymorphism of the S1 protein from the newly identified IBV strains and the reference strains.
| Position | IBV Strains |
Location | |||
|---|---|---|---|---|---|
| ZH01 | HH09 | GI-19 genotype | Other genotypes | ||
| 23 | La | P | SPDFY | D/S | RBD |
| 39 | N | T | NTD | C/T/Y/N/G/I/M/L | RBD/HVRI |
| 90 | P | S | S/L | S/L/C/E/T/A/H/F/D | RBD |
| 115 | T | Ab | T | T | RBD/HVRII |
| 122 | T | T | S/A/P/R/I | H/S/Y/C/A/V/R/G | RBD/HVRII |
| 161 | R | R | N/T/S/K | K/V/T/N | RBD |
| 278 | Ia | T | T | H/R/T/N | HVRIII |
| 280 | F | Lb | F | F/Y/V/H/I | HVRIII |
| 283 | T | Sb | F/H/T | H/Y/V/R/F/A | HVRIII |
| 298 | Aa | T | T | N/T/S | HVRIII |
| 338 | K | S | S/I/T/R | A/H/S/C/R/N | HVRIII |
| 360 | T | A | T/A/G/S | T/A/G/S | HVRIII |
| 376 | K | R | R/K | M/R/I/S/K | HVRIII |
| 390 | T | V | V/R/I | H/S/L/V/R/I | / |
| 397 | S | Ac | S/R | L/A/R/T/N | / |
| 403 | T | N | N/D | N/D/V/C/L/K | / |
Abbreviations: RBD-respected the receptor-binding domain.
Amino acid mutation only occurred in ZH01.
Amino acid mutation only occurred in HH09.
The amino acid mutation often occurred in other genotypes.
Recombination Analysis
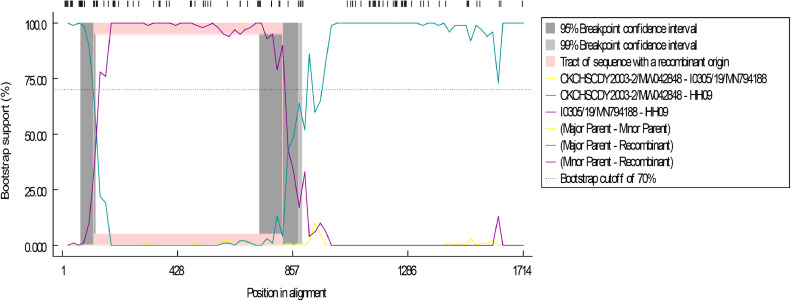
The S1 gene of IBV has important hypervariable regions where recombination events may occur. In this study, RDP5 software was used to perform S1 sequence recombination analysis on the identified ZH01 and HH09 strains (Table 3). The results show that ZH01 was produced through the recombination of ck/CH/LHB/111190 (major parent) and IBS138/2015 (minor parents in between 674 bp and 852 bp.) In addition, the strain is highly homologous to both ck/CH/LHB/111190 (98.2%) and IBS138/2015 (97.9%). The strain HH09 emerged through recombination event between CK/CH/SCDY2003-2 (major parent) and I0305/19 (minor parent) located in 120 bp to 764 bp (Figure 3). It shows high similarity to I0305/19 (95.5%) and CK/CH/SCDY2003-2 (97%).
Table 3.
RDP software detects ZH01 and HH09 recombination events.
| Breakpoint positions |
Major parenta |
Minor parentb |
Detection methods |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Strain | Begin | END | Strain | Genotype | Similarity | Strain | Genotype | Similarity | P value* | R | G | B | M | C | S | T |
| ZH01 | 674 | 856 | ck/CH/LHB/111190 | QX | 97.5 % | IBS138/2015 | QX | 94.6 % | 2.620 × 10−06 | + | + | + | + | + | - | + |
| HH09 | 120 | 764 | CKCHSCDY2003-2 | QX | 97 % | I0305/19 | QX | 95.4 % | 1.986 × 10−11 | + | + | + | + | + | - | + |
R: RDP G: GENECONV B: Bootscan M: Maxchi C: Chimaera S: SiScan T: Tree Topo.
P value* of RDP method.
Major parent = Sequence closely related to the transferred fragment in the recombinant.
Minor parent = Sequence most closely related to the sequence surrounding the transferred fragment.
Figure 3.
Recombination event analysis of S1 gene of HH09 using the recombination detection software (RDP v5). The S1 gene of hh09 strain comes from the intragroup recombination events of I0305 / 19 (minor parent) and CK / CH / SCDY2003-2 (major parent). The X aixs presents the position in alignment with a 20-nt sliding window. The The Y-axis represents the bootstrap support (%) between the recombinant and its putative parents.
DISCUSSION
IBV is one of the leading causes of economic loss in the poultry industry as it affects the production performance of broilers and laying hens. Since IBV was isolated in the United States in 1930, different serotypes and genotypes of the virus have been identified worldwide and have been found to have poor cross-protection (Jackwood, 2012). The frequent emergence of new IBV variants and the absence of a cross-protection often leads to the vaccination failure. During our surveillance program of avian pathogens in southern China, 7 commercial chicken farms in Guangdong province reported respiratory disease outbreaks and the pathogen was identified to be IB variant. IBV variant strains which shared low amino acid homology ranged from 74.8 to 85% of vaccine strains (H120, 4/91, ldt3-a, and M41) were detected from both vaccinated broilers and laying hens in all chicken farms. The two identified IBV strains in this study, ZH01 and HH09, were genetically similar, which implied the same IBV serotypes/lineages were involved in the 7 chicken farms.
The S gene is involved in antigen neutralization, hemagglutination, and cell tropism determination of IBV (Lin and Chen, 2017; Brown Jordan et al., 2020). In addition, most phylogenetic analyses of IBV are also based on the S1 gene, which has 3 HVRs that provide a platform for genetic evolution (Valastro et al., 2016). In this study, genetic evolution and homology analysis based on the S1 gene showed that the newly identified strains ZH01 and HH09 belong to the GI-19 lineage in QX genotype, which still remains as the most dominant genotype of IBV circulating in southern China (Lian et al., 2021). Novel mutations such as T273I, T292A, and S331K were found in the emerging IBV strains and further studies based on reverse genetic techniques are needed to study their role in the biology of IBV. It is worth noting that a strain of IBV with a nucleotide homology of 99.3% to the vaccine strain 4/91 was identified in case 2, which is more likely derived from the live vaccine strain.
IBV undergo frequent recombination events between field strains and vaccine strains, leading to the emergence of novel variants. The GI-19 genotype is the most widely distributed IBV and previous studies have shown evidence of recombination events between GI-19 vaccine strain and field strains, which indicates that recombination serves as a major evolutionary mechanism for IBV (Abozeid et al., 2017; Huang et al., 2020; Bali et al., 2021). The emerging IBV strain HH09 in this study was found to be generated through a recombination events between CK/CH/SCDY2003-2 and I0305/19. Of note, I0305/19 has been reported to be a highly pathogenic strain and has broader tissue tropism, including both the respiratory tissues and digestive tract tissues (Hou et al., 2020).
Overall, we reported the genetic characterization of emerging IBV strains which caused outbreaks in vaccinated broiler and layer chicken flocks in southern China, 2021. The emerging IBV strains were classified into GI-19 genotype strains via phylogenetic analysis but had a certain genetic distance from the classical strain. The current observations suggest that MAS series vaccines such as H120 and H52 have a low protection against epidemic strains, making it an urgent need to accelerate the development of novel vaccines. In addition, the findings highlighted the importance of enhanced surveillance and further studies on the pathogenicity of those emerging IBV variants.
Acknowledgments
We thank all researchers who shared related IBV genome sequences in GenBank. This study was supported by Education Bureau of Guangdong province (2020KZDZX1209); Natural Science Foundation of Guangdong Province, China (Grant No. 2022A1515012462); Postgraduate exploration fundation of Foshan University (Grant No. 2021ZYTS34); The funders of this study had no role in the design, execution, interpretation, or writing of the study.
Availability of data and materials: The viral sequences obtained in this study are deposited in GenBank under the accession numbers: OL332822- OL332823. The data of this study were available from the corresponding author upon reasonable request.
DISCLOSURES
The authors declare that they have no competing interests. All authors have read and approved the final manuscript.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.psj.2022.102082.
Appendix. Supplementary materials
Supplementary Figure 1. The nucleotide (A) and amino acid (B) homology of the S1 gene between the emerging IBV strain identified in this study and the reference strain. The multiple sequence alignment and genetic distance matrix were performed using the UGENE software (Version 36.0).
References
- Abozeid H.H., Paldurai A., Khattar S.K., Afifi M.A., El-Kady M.F., El-Deeb A.H., Samal S.K. Complete genome sequences of two avian infectious bronchitis viruses isolated in Egypt: evidence for genetic drift and genetic recombination in the circulating viruses. Infect Genet. Evol. 2017;53:7–14. doi: 10.1016/j.meegid.2017.05.006. [DOI] [PubMed] [Google Scholar]
- Bali K., Balint A., Farsang A., Marton S., Nagy B., Kaszab E., Belak S., Palya V., Banyai K. Recombination events shape the genomic evolution of infectious bronchitis virus in Europe. Viruses. 2021;13 doi: 10.3390/v13040535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boursnell M.E., Brown T.D., Foulds I.J., Green P.F., Tomley F.M., Binns M.M. Completion of the sequence of the genome of the coronavirus avian infectious bronchitis virus. J. Gen. Virol. 1987;68:57–77. doi: 10.1099/0022-1317-68-1-57. [DOI] [PubMed] [Google Scholar]
- Brown Jordan A., Fusaro A., Blake L., Milani A., Zamperin G., Brown G., Carrington C.V.F., Monne I., Oura C.A.L. Characterization of novel, pathogenic field strains of infectious bronchitis virus (IBV) in poultry in Trinidad and Tobago. Transbound Emerg. Dis. 2020;67:2775–2788. doi: 10.1111/tbed.13637. [DOI] [PubMed] [Google Scholar]
- Cavanagh D. Coronaviruses in poultry and other birds. Avian Pathol. 2005;34:439–448. doi: 10.1080/03079450500367682. [DOI] [PubMed] [Google Scholar]
- Cavanagh D. Coronavirus avian infectious bronchitis virus. Vet. Res. 2007;38:281–297. doi: 10.1051/vetres:2006055. [DOI] [PubMed] [Google Scholar]
- Chen Y., Jiang L., Zhao W., Liu L., Zhao Y., Shao Y., Li H., Han Z., Liu S. Identification and molecular characterization of a novel serotype infectious bronchitis virus (GI-28) in China. Vet. Microbiol. 2017;198:108–115. doi: 10.1016/j.vetmic.2016.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domanska-Blicharz K., Lisowska A., Pikula A., Sajewicz-Krukowska J. Specific detection of GII-1 lineage of infectious bronchitis virus. Lett. Appl. Microbiol. 2017;65:141–146. doi: 10.1111/lam.12753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y., Zhang L., Ren M., Han Z., Sun J., Zhao Y., Liu S. A highly pathogenic GI-19 lineage infectious bronchitis virus originated from multiple recombination events with broad tissue tropism. Virus Res. 2020;285 doi: 10.1016/j.virusres.2020.198002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houta M.H., Hassan K.E., El-Sawah A.A., Elkady M.F., Kilany W.H., Ali A., Abdel-Moneim A.S. The emergence, evolution and spread of infectious bronchitis virus genotype GI-23. Arch. Virol. 2021;166:9–26. doi: 10.1007/s00705-020-04920-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M., Zou C., Liu Y., Han Z., Xue C., Cao Y. A novel low virulent respiratory infectious bronchitis virus originating from the recombination of QX, TW and 4/91 genotype strains in China. Vet. Microbiol. 2020;242 doi: 10.1016/j.vetmic.2020.108579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackwood M.W. Review of infectious bronchitis virus around the world. Avian Dis. 2012;56:634–641. doi: 10.1637/10227-043012-Review.1. [DOI] [PubMed] [Google Scholar]
- Jiang L., Zhao W., Han Z., Chen Y., Zhao Y., Sun J., Li H., Shao Y., Liu L., Liu S. Genome characterization, antigenicity and pathogenicity of a novel infectious bronchitis virus type isolated from South China. Infect Genet. Evol. 2017;54:437–446. doi: 10.1016/j.meegid.2017.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian J., Wang Z., Xu Z., Chen T., Shao G., Zhang X., Qin J., Xie Q., Lin W. Distribution and molecular characterization of avian infectious bronchitis virus in southern China. Poult. Sci. 2021;100 doi: 10.1016/j.psj.2021.101169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S.Y., Chen H.W. Infectious Bronchitis virus variants: molecular analysis and pathogenicity investigation. Int. J. Mol. Sci. 2017;18 doi: 10.3390/ijms18102030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma T., Xu L., Ren M., Shen J., Han Z., Sun J., Zhao Y., Liu S. Novel genotype of infectious bronchitis virus isolated in China. Vet. Microbiol. 2019;230:178–186. doi: 10.1016/j.vetmic.2019.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molenaar R.J., Dijkman R., de Wit J.J. Characterization of infectious bronchitis virus D181, a new serotype (GII-2) Avian Pathol. 2020;49:243–250. doi: 10.1080/03079457.2020.1713987. [DOI] [PubMed] [Google Scholar]
- Niesters H.G., Lenstra J.A., Spaan W.J., Zijderveld A.J., Bleumink-Pluym N.M., Hong F., van Scharrenburg G.J., Horzinek M.C., van der Zeijst B.A. The peplomer protein sequence of the M41 strain of coronavirus IBV and its comparison with Beaudette strains. Virus Res. 1986;5:253–263. doi: 10.1016/0168-1702(86)90022-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjaak de Wit J.J., Cook J.K., van der Heijden H.M. Infectious bronchitis virus variants: a review of the history, current situation and control measures. Avian Pathol. 2011;40:223–235. doi: 10.1080/03079457.2011.566260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomsen M.C., Nielsen M. Seq2Logo: a method for construction and visualization of amino acid binding motifs and sequence profiles including sequence weighting, pseudo counts and two-sided representation of amino acid enrichment and depletion. Nucleic Acids Res. 2012;40:W281–W287. doi: 10.1093/nar/gks469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valastro V., Holmes E.C., Britton P., Fusaro A., Jackwood M.W., Cattoli G., Monne I. S1 gene-based phylogeny of infectious bronchitis virus: an attempt to harmonize virus classification. Infect. Genet Evol. 2016;39:349–364. doi: 10.1016/j.meegid.2016.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen F., Li W., Guo J., Yang J., Zhang X., Mei K., Liu H., El-Ashram S., Luo K., Yuan S., Chi S., Huang S. Genetic characterization of a novel genotype H9N2 avian influenza virus from chicken in South China. J. Infect. 2020;81:816–846. doi: 10.1016/j.jinf.2020.09.011. [DOI] [PubMed] [Google Scholar]
- Wen F., Yang J., Li A., Gong Z., Yang L., Cheng Q., Wang C., Zhao M., Yuan S., Chen Y., El-Ashram S., Li Y., Yu H., Guo J., Huang S. Genetic characterization and phylogenetic analysis of porcine epidemic diarrhea virus in Guangdong, China, between 2018 and 2019. PLoS One. 2021;16 doi: 10.1371/journal.pone.0253622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickramasinghe I.N., de Vries R.P., Grone A., de Haan C.A., Verheije M.H. Binding of avian coronavirus spike proteins to host factors reflects virus tropism and pathogenicity. J. Virol. 2011;85:8903–8912. doi: 10.1128/JVI.05112-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Li Y., Guo J., Luo K., Yu H., Chen Y., Li A., Yuan S., El-Ashram S., Huang S., Wen F. Genetic characterization of an H5N6 avian influenza virus from chickens in Guangdong, China. J. Infect. 2021;82:414–451. doi: 10.1016/j.jinf.2020.10.014. [DOI] [PubMed] [Google Scholar]
- Zhang X., Deng T., Lu J., Zhao P., Chen L., Qian M., Guo Y., Qiao H., Xu Y., Wang Y., Li X., Zhang G., Wang Z., Bian C. Molecular characterization of variant infectious bronchitis virus in China, 2019: implications for control programmes. Transbound Emerg. Dis. 2020;67:1349–1355. doi: 10.1111/tbed.13477. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. The nucleotide (A) and amino acid (B) homology of the S1 gene between the emerging IBV strain identified in this study and the reference strain. The multiple sequence alignment and genetic distance matrix were performed using the UGENE software (Version 36.0).