Abstract
Colonization of food-producing animals by antimicrobial-resistant Staphylococcus aureus, especially methicillin-resistant S. aureus (MRSA), has become a serious public health problem worldwide. In the current study, clonal diversities of livestock-associated S. aureus isolates collected from broiler farms, slaughterhouses, and retail chicken meat were examined. Two-hundred S. aureus isolates (43 MRSA and 157 methicillin-susceptible S. aureus [MSSA] isolates) were analyzed to determine 1) the genotypes of the isolates (multilocus sequence, agr, and spa types), 2) the methicillin resistance phenotype and staphylococcal cassette chromosome mec (SCCmec) types, 3) the antimicrobial resistance profiles, and 4) the mutational changes in gyrA, gyrB, parC, and parE in fluoroquinolone-resistant isolates. Fifteen different sequence types (STs) of MSSA strains displaying a relatively high degree of genetic diversity were detected in broiler farms, slaughterhouses, and retail chicken meat. In contrast to MSSA, 2 dominant genetic lineages of MRSA (ST692-SCCmecV with t2249 spa type, and ST188-SCCmecIVa with spa type t189) were found in healthy broilers. The high prevalence of ST692 and ST188 in healthy broilers is associated with high levels of multiple antimicrobial-resistance phenotypes, particularly fluoroquinolone resistance. All fluoroquinolone-resistant isolates carried double point mutations in gyrA (S84L) and parC (S80F), regardless of STs or methicillin resistance. Notably, only the ST188 lineage carried an additional third mutation in gyrB (D494N), correlating with enhanced ciprofloxacin minimum inhibitory concentration values versus the strains with double mutations. These results provide important insights into the genetic diversity of antimicrobial-resistant S. aureus strains associated with the chicken meat production chain, including healthy broilers, in Korea.
Key words: Staphylococcus aureus, clonal diversity, broiler farms, slaughterhouses, retail chicken meat
Introduction
The widespread occurrence and persistence of antimicrobial-resistant Staphylococcus aureus in livestock animals is a global public health concern, affecting both humans and animals (van der Mee-Marquet et al., 2014; Cuny et al., 2015; Becker et al., 2017). Recently, zoonotic transmission of livestock-associated (LA) methicillin-susceptible S. aureus (LA-MSSA) and methicillin-resistant S. aureus (LA-MRSA) from food-producing animals to humans has been reported in several studies worldwide (Vanderhaeghen et al., 2010; Fluit, 2012; van Cleef et al., 2014; van der Mee-Marquet et al., 2014). The high prevalence of sequence type (ST) 398 and ST541 LA-MRSA with the staphylococcal cassette chromosome mec V (SCCmec V) in swine farms and pork production systems has been associated with an increased number of MRSA infections in humans with direct or indirect animal contact (Moon et al., 2015; Back et al., 2020). In addition to farm animals and farm environments, frequent detection of LA-type S. aureus, such as clonal complex (CC) 398 (ST398 and ST541) MRSA and MSSA, has been reported in milk and retail meat samples (Eom et al., 2019; Back et al., 2020).
Although a relatively large number of studies have investigated the prevalence and characteristics of LA-MRSA and LA-MSSA in pig and cattle farms (Ho et al., 2012; Nemeghaire et al., 2014; Reynaga et al., 2016; Eom et al., 2019; Moon et al., 2019; Back et al., 2020; Lee et al., 2020a; ), very limited data are available on S. aureus isolates from broilers, the environment, and workers on broiler farms (Friese et al., 2013; Geenen et al., 2013). In a few studies, poultry-associated CC9 and CC398 MRSA strains were identified in healthy and diseased chickens in European countries (Persoons et al., 2009; Kraushaar et al., 2017). In Korea, two recent studies have investigated the prevalence and characteristics of S. aureus and MRSA in retail chicken meat samples produced by integrated broiler operations (Kim et al., 2018) and chicken carcass samples from slaughterhouses (Moon et al., 2015), respectively. Moon et al. detected 5 strains of ST692 MRSA carrying SCCmec III where four strains were from chicken carcasses and one strain was from a slaughterhouse worker (Moon et al., 2015). Although these previous studies provide valuable information on poultry-associated S. aureus in Korea, studies on the prevalence and clonal distribution of MRSA and MSSA strains in chicken meat production systems encompassing broiler farms, slaughterhouses, and retail chicken meats are still lacking. Thus, the present study aimed to analyze both LA-MRSA and LA-MSSA isolates from healthy broilers, carcasses in slaughterhouses, and retail chicken meat samples. In addition, the carriage of MRSA and MSSA in the personnel and environment of broiler farms and slaughterhouses was investigated. To assess the genetic diversity of S. aureus isolates, the multilocus sequence type (MLST), SCCmec type, staphylococcal protein A (spa) type, and accessory gene regulator (agr) type were determined. Moreover, the antimicrobial resistance profiles of the S. aureus isolates and genetic factors associated with the resistance phenotype, especially fluoroquinolone resistance, were analyzed.
Materials and methods
Sample collection
Samples were collected from 20 randomly selected broiler farms located in eight provinces of Korea between February and December 2019. On each broiler farm, approximately 10 different broiler chickens (skin under one wing, throat, and cloacal swabs), farm environment (sewage, floor, fence, ventilator opening, and drinking system), and farm workers (nasal swabs) were sampled. Samples at each slaughterhouse were collected from 20 different chicken carcasses, facility environments (sewage, floor, tables, and ventilators), and facility workers. Retail chicken meat samples (n = 100) were purchased from 20 retail markets and groceries located in eight provinces of Korea during the sampling period. All chicken meat samples and swabs were stored at 4°C in ice-cooled containers and transported to the laboratory within 24 h of sample collection.
S. aureus Isolation and Identification
All swab samples were inoculated into 3 mL of tryptic soy broth (TSB; Becton, Dickinson and Company, Franklin Lakes, NJ) supplemented with 10% sodium chloride (NaCl) and incubated at 37°C for 18 to 24 h with shaking (200 rpm). Chicken carcass and meat samples (25 g) were homogenized in 225 mL of buffered peptone water using HAPS homogenizer (Hukobio, Seoul, Korea), and 1 mL of the solutions was incubated into the 10% NaCl-TSB and enriched for 18 to 24 h at 37°C. After pre-enrichment, 25 μL aliquots of the samples were streaked onto Baird-Parker agar (Difco Laboratories, Detroit) containing potassium tellurite and egg yolk supplements (Difco Laboratories). After 24 to 48 h incubation, the 1 or 2 most dominant colonies of possible S. aureus were selected from each sample and subcultured for subsequent identification. All S. aureus isolates were confirmed by 16S rRNA sequencing (BIONICS, Seoul, Korea) and matrix-assisted laser desorption ionization (MALDI)-Biotyper real-time classification system (Bruker Daltonics GmbH, Bremen, Germany) as described previously (Lee et al., 2020b; Yang et al., 2022). S. aureus strains from the same sample that exhibited distinctive phenotypic and genotypic features were considered to be different isolates.
Determination of antimicrobial susceptibilities
The antimicrobial susceptibility profiles of S. aureus isolates were determined using the standard disc diffusion method according to the Clinical and Laboratory Standards Institute (CLSI) recommendations (CLSI, 2022). The antimicrobial agents used in the susceptibility testing were ampicillin (AMP, 10 μg), cefoxitin (CEF, 30 μg), penicillin (PEN, 10 units), chloramphenicol (CHL, 30 μg), clindamycin (CLI, 2 μg), erythromycin (ERY, 15 μg), fusidic acid (FUS, 10 μg), gentamicin (GEN, 30 μg), mupirocin (MUP, 200 μg), rifampin (RIF, 5 μg), sulfamethoxazole-trimethoprim (SXT, 23.73–1.25 μg), quinupristin-dalfopristin (SYN, 15 μg), tetracycline (TET, 30 μg), ciprofloxacin (CIP, 5 μg), enrofloxacin (ENR, 5 μg), and levofloxacin (LEV, 5 μg) (Oxoid, Hampshire, UK). Susceptibilities to linezolid (LZD), teicoplanin (TEC), vancomycin (VAN), and tigecycline (TGC) were determined using the standard E-test (Bio Mérieux, France) on Mueller–Hinton agar (Difco Laboratories) plates. For evaluation of fluoroquinolone resistance, minimum inhibitory concentrations (MICs) for CIP (ranging from 0.0625 to 256 μg/mL) and ENR (ranging from 0.5 to 64 μg/mL) were determined using the standard broth microdilution assay (Grobbel et al., 2007; CLSI, 2018). The resistance breakpoints for the antimicrobial agents were interpreted based on the CLSI documents of VET01S and M100 (CLSI, 2020; CLSI, 2022). S. aureus ATCC 29213 was included as a reference strain for antimicrobial susceptibility tests.
Molecular characterization of S. aureus isolates
To analyze the genetic diversity of S. aureus isolates, MLST analysis was carried out as described previously (Enright et al., 2000). Internal sequences of seven housekeeping genes (arcC, aroE, glpF, gmk, pta, tpi, and yqiL) were amplified and sequenced using polymerase chain reaction (PCR), as described previously (Enright et al., 2000). The allelic profiles, STs, and CC types of each isolate were assigned based on the S. aureus MLST database (http://pubmlst.org/saureus). Genetic relationships between STs and CC types of S. aureus isolates were analyzed using eBURST V3 (http://eburst.mlst.net).
Analysis of spa sequences has been considered a useful typing method due to its great discriminatory power among S. aureus isolates (Koreen et al., 2004). The polymorphic X region of spa was sequenced using a specific primer set (Koreen et al., 2004), and the spa types were determined according to the SpaServer database (http://spa.ridom.de/).
The agr typing (types I-IV) was performed on all S. aureus isolates, as previously described (Gilot et al., 2002). Briefly, agrD sequences were amplified using PCR-amplified using specific primer sets. PCR products of 441, 575, 323, or 659-bp correspond to agr type I, II, III, or IV, respectively.
mecA detection and SCCmec typing
All S. aureus isolates exhibiting resistance phenotype to CEF were examined for the carriage of mecA using PCR method as previously described (Lee, 2003). For mecA-positive isolates, SCCmec types were determined based on the combinations of chromosomal cassette recombinase (ccr) types and mec complexes, as previously described (Kondo et al., 2007; Milheirico et al., 2007).
Sequencing analysis of Quinolone-Resistance Determining Regions (QRDRs)
The bacterial type II topoisomerases, DNA gyrase and topoisomerase IV enzymes, are 2 essential targets of fluoroquinolone (Yoshida et al., 1991; Gonzalez et al., 1998). Quinolone resistance in staphylococci is associated with frequent mutations of QRDRs within the gyr and par genes encoding DNA gyrase and topoisomerase IV, respectively (Ferrero et al., 1994; Ito et al., 1994). To detect mutations in QRDR regions, gyrA, gyrB, parC, and parE genes were PCR-amplified using specific primer sets: gyrA (5’-AATGAACAAGGTATGACACC and 5’-GCGATACCTGATGCACCATT, 368 bp); gyrB (5’-TCGTAAATCAGCGTTAGATGT and 5’-CGATTTTGTGATATCTTGCTTTCG, 570 bp), parC (5’-ACTTGAAGATGTTTTAGGTGAT and 5’-TTAGGAAATCTTGATGGCAA, 459 bp); and parE (5’-CGATTAAAGCACAACAAGCAAG and 5’-GCGCACCATCAGTATCAG, 393 bp) (Schmitz et al., 1998). The amplified PCR products of QRDRs from quinolone-resistant S. aureus isolates were sequenced at BIONICS, Seoul, Korea. Alterations in nucleotide and amino acid sequences within the QRDRs were identified using a multiple sequence alignment program using the BoxShade server (http://embnet.vital-it.ch/software/BOX_form.html). S. aureus NCTC 8325 (GenBank accession no. CP000253) was used as the reference strain for the comparison of QRDR sequences.
Statistical analysis
Statistical analyses were performed using the SPSS statistics software (IBM, Chicago, IL ). Pearson's correlation coefficient was calculated to analyze the relationship between the number of mutations in QRDRs and fluoroquinolone MIC values. P-value < 0.05 was considered to be statistically significant.
Results
S. aureus in the chicken meat production system
As shown in Table 1, 200 S. aureus strains (43 MRSA and 157 MSSA strains) were isolated from samples collected from broiler farms (n = 94 isolates), slaughterhouses (n = 42 isolates), and retail markets (n = 64 isolates) in South Korea during the study period (Table 1). Although the overall rate of methicillin resistance in 200 S. aureus isolates was 21.5% (43/200 isolates), 86% of the MRSA isolates (37/43 isolates) were recovered from broiler farms, 34 isolates from healthy broilers, and 3 isolates from farm environments (Table 1). The rates of methicillin resistance in S. aureus isolates were 39% (37/94), 2.4% (1/42), and 7.8% (5/64) in broiler farms, slaughterhouses, and retail chicken meat samples, respectively. None of the nasal swab samples tested positive for MRSA.
Table 1.
Staphylococcus aureus strains isolated from broiler farms, slaughterhouses, and retail chicken meat samples in Korea.
| Broiler farms |
Slaughterhouses |
Retail markets | |||||
|---|---|---|---|---|---|---|---|
| Broilers (n = 200) | Workers (n = 21) | Environment (n = 71) | Carcasses (n = 120) | Workers (n = 13) | Environment (n = 30) | Chicken meats (n = 100) | |
| MRSA (n = 43) |
34 (17%a) Skin-13 Throat-6 Cloaca-15 |
0 | 3 (4.2%) Floor-2 Fence-1 |
1 (0.83%) | 0 | 0 | 5 (5%) |
| MSSA (n = 157) |
51 (25.5%) Skin-16 Throat-20 Cloaca-15 |
2 (9.5%)a Nasal-2 |
4 (5.6%) Floor-1 Fence-1 Ventilator-1 Toilet-1 |
33 (27.5%) | 4 (30.8%) Nasal-4 |
4 (13.3%) Line-1 Floor-3 |
59 (55%) |
| Total (n = 200) |
85 (42.5%)b | 2 (9.5%) | 7 (9.9%) | 34 (28.3%) | 4 (30.8%) | 4 (13.3%) | 64 (64%)b |
Number of Staphylococcus aureus isolates divided by the number of broiler or facility workers.
Four broiler and four retail chicken meat samples were double-positive for both methicillin-resistant S. aureus (MRSA) and methicillin-susceptible S. aureus (MSSA) strains.
Unlike MRSA, which was mostly associated with broilers, 157 MSSA strains were isolated from all 3 sample sites: broiler farms (n = 57, 36%), slaughterhouses (n = 41, 26%), and retail meat (n = 59, 38%). Six of the 157 MSSA strains were isolated from nasal swabs of farmworkers (n = 2) and slaughterhouse workers (n = 4).
Notably, four broiler and four retail meat samples were double-positive for both MRSA and MSSA strains.
Clonal linages and genetic characteristic of MRSA and MSSA isolates
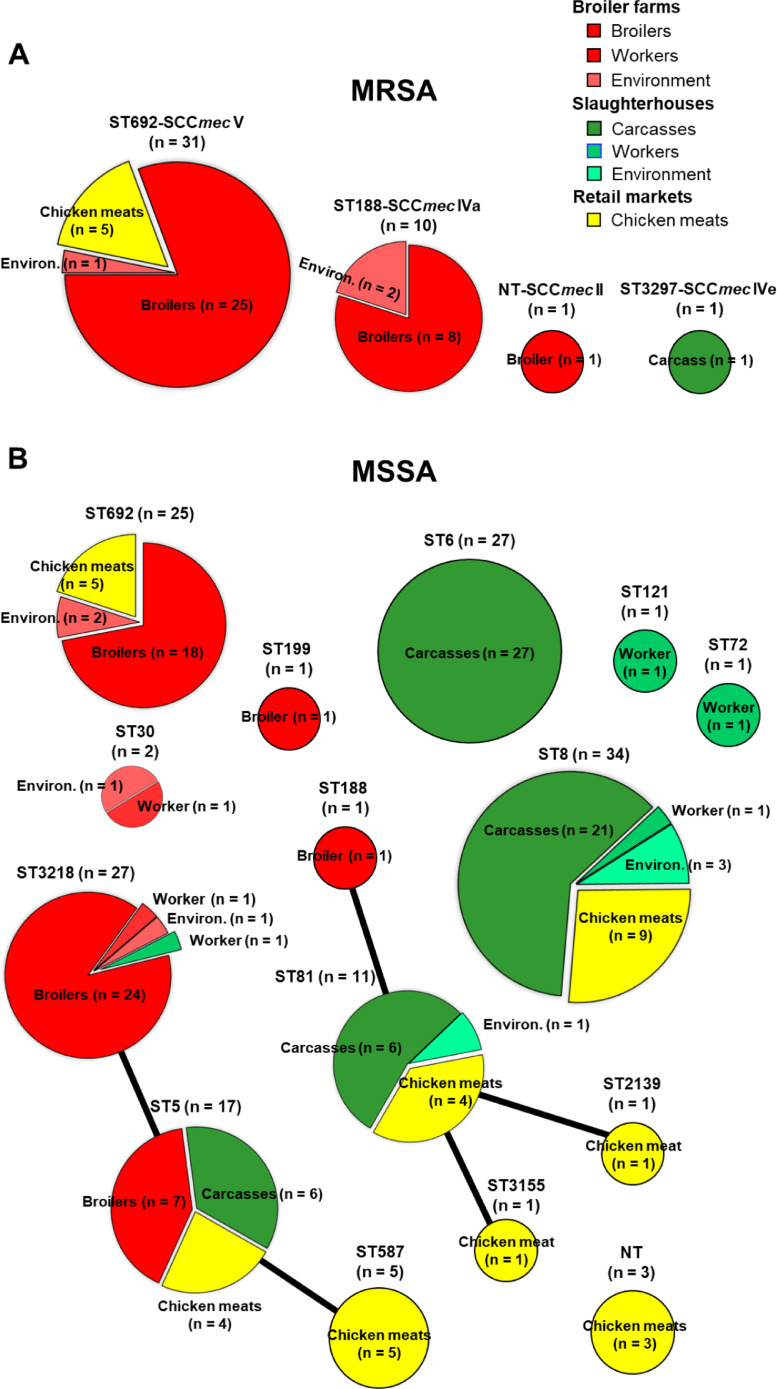
As presented in Figure 1A and B, MLST analyses of 200 S. aureus isolates revealed 15 different STs. Except for two MRSA strains (one ST3297 and one nontypeable strain), of 41/43 MRSA isolates were either ST692-SCCmec V (n = 31) or ST188-SCCmec IVa (n = 10). In contrast to the 2 dominant genotypes of MRSA strains (ST692 and ST188), the MSSA strains exhibited 14 different STs. Of the 12 STs, 4 major STs were most frequently observed in 157 MSSA isolates: ST8 (n = 34, 21.7%), ST6 (n = 27, 17.2%), ST3218 (n = 27, 17.2%), and ST692 (n = 25, 15.9%). The ST8 and ST6 MSSA strains were mainly associated with chicken carcasses and retail chicken meat samples. Except for one ST3218 strain from a worker in a slaughterhouse and five ST692 MSSA strains from retail chicken meat samples, all ST3218 (n = 26) and ST692 MSSA (n = 20) strains were isolated from broiler chickens and broiler farm environments. Two human MSSA isolates obtained from broiler farms were ST3218 and ST30, whereas each one of 4 MSSA isolates from slaughterhouse workers was ST3218, ST8, ST72, and ST121, respectively.
Figure 1.
Genetic diversity of methicillin-resistant Staphylococcus aureus (MRSA) (A) and methicillin-susceptible S. aureus (MSSA) (B) isolates collected from broiler farms, slaughterhouses, and retail chicken meat samples. eBURST analyses display genetic relatedness among 200 S. aureus strains depending on the allelic profiles of the multilocus sequence type (MLST). Clusters consisting of multiple sequence types (STs) within single-locus variants (SLVs) were linked with clonal ancestor STs using solid lines. MRSA, methicillin-resistant S. aureus; MSSA, methicillin-susceptible S. aureus; NT, nontypeable.
As shown in Table 2, agr type analyses revealed that all 41 MRSA strains were agr type I (agr I) regardless of their STs. In the analyses of spa types, all 31 ST692 MRSA strains were spa type t2247 (spa t2247), while 10 ST188 MRSA strains were spa t189. Analyses of agr types in 157 MSSA isolates also revealed that agr I (95/157, 60.5%) was the most prevalent in the MSSA strain group, followed by agr II (55/157, 35%). Interestingly, in contrast to the MRSA isolates, which showed a single type of spa for each STs (t2247 for ST692 and t189 for ST188), multiple spa types were identified in ST5, ST6, ST3218, ST692, and ST81 MSSA strains.
Table 2.
Clonal lineages of MRSA and MSSA strains isolated from the chicken meat production system in Korea.
| No. of isolates | |||||
|---|---|---|---|---|---|
| MRSA/MSSA | Clonal complex | ST | SCCmec | agr | spa |
| MRSA (43) | CC385 (31) | ST692 | V | I (31) | t2247 |
| CC1 (10) | ST188 | IVa | I (10) | t189 | |
| ND (2) | ST3297 (1) | IVe | IV (1) | t2247 | |
| NT (1) | II | II (1) | t2431 | ||
| MSSA (157) | CC5 (76) | ST5 (17) | - | I (2) II (13) NT (2) |
t002 (1); t2247 (1) t548 (4); t062 (4); t2247(2), t002 (1); t3478 (1); NT (2) |
| ST6 (27) | - | I (27) | t304 (17), t190 (9), t701 (1) | ||
| ST3218 (27) | - | II (27) | t002 (22), t045 (3), t3478 (1), t548 (1) | ||
| ST587 (5) | - | II (5) | t002 | ||
| CC8 (34) | ST8 (34) | - | I (34) | t008 | |
| CC385 (25) | ST692 (25) | - | I (24) | t2247 | |
| - | II (1) | t3478 | |||
| CC1 (14) | ST81 (11) | - | I (2) II (7) NT (2) |
NT t548 (2), NT (5) NT (2) |
|
| ST2139 (1) | - | III (1) | t286 | ||
| ST3155 (1) | - | I (1) | t189 | ||
| ST188 (1) | - | I (1) | t189 | ||
| CC30 (2) | ST30 (2) | - | I (2) | t840 | |
| Singletons (6) | ST72 (1) | - | I (1) | t17433 | |
| ST199 (1) | - | II (1) | t2036 | ||
| ST121 (1) | - | IV (1) | t4956 | ||
| NT (3) | - | I (1) | t190 | ||
| II (1) | t548 | ||||
| NT (1) | NT | ||||
Abbreviations: NT; nontypeable, ND; not determined.
Antimicrobial resistance phenotypes
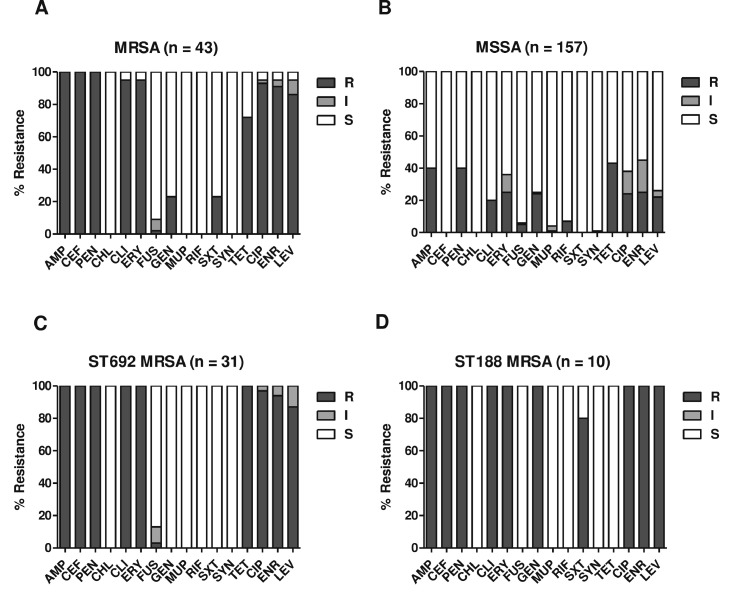
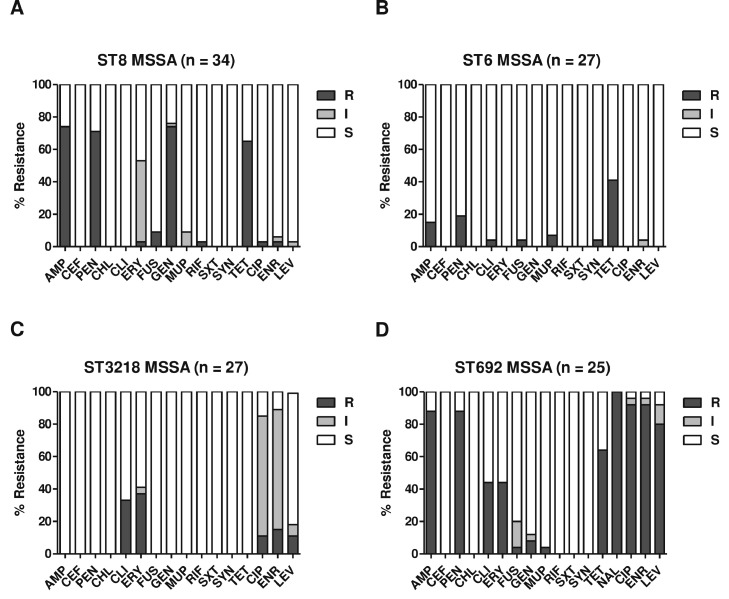
The antimicrobial resistance profiles of the MRSA and MSSA isolates are shown in Figure 2A and B. All 200 S. aureus strains were susceptible to CHL, LZD, TEC, VAN, and TGC (data not shown). As expected, the MRSA strains showed significantly higher levels of resistance to β-lactams (AMP, CEF, and PEN), CLI, ERY, and fluoroquinolones (CIP, ENR, and LEV) than the MSSA isolates. Interestingly, the 2 major ST groups of MRSA isolates (ST692-SCCmec V and ST188-SCCmec IVa) exhibited similar levels of MDR phenotypes (Figure 2C and D) despite the opposite resistance phenotypes to GEN, TET, and SXT. All ST692 MRSA strains displayed resistance to TET, with 100% susceptibility to GEN and SXT. The ST188 MRSA strains exhibited opposite resistance phenotypes to these three antibiotics. When the 4 major STs of MSSA strain groups, ST8 (n = 34), ST6 (n = 27), ST3218 (n = 27), and ST692 (n = 25), were compared for antimicrobial resistance, the ST692 MSSA strain displayed higher levels of multidrug resistance (MDR) phenotype (≥3 different classes of antimicrobial agents) and fluoroquinolones than the other 3 ST groups (Figure 3).
Figure 2.
Antimicrobial resistance profiles of S. aureus strains isolated from broiler farms, slaughterhouses, and retail chicken meat samples. Antimicrobial resistance phenotypes of MRSA (A), MSSA (B), ST692 MRSA (C), and ST188 MRSA (D) strain groups are shown. Antimicrobial susceptibility assays and interpretation criteria were implemented according to the Clinical and Laboratory Standards Institute (CLSI) guidelines of M100 and VET01S (CLSI, 2022; CLSI, 2020). AMP, ampicillin; CEF, cefoxitin; CHL, chloramphenicol; CIP, ciprofloxacin; CLI, clindamycin; ENR, enrofloxacin; ERY, erythromycin; FUS, fusidic acid; GEN, gentamicin; LEV, levofloxacin; MUP, mupirocin; PEN, penicillin; RIF, rifampicin; SXT, trimethoprim-sulfamethoxazole; SYN, quinupristin-dalfopristin; TET, tetracycline.
Figure 3.
Antimicrobial resistance profiles of major ST groups of MSSA strains. Antimicrobial susceptibilities of ST8 (A), ST6 (B), ST3218 (C), and ST692 (D). AMP, ampicillin; CEF, cefoxitin; CHL, chloramphenicol; CIP, ciprofloxacin; CLI, clindamycin; ENR, enrofloxacin; ERY, erythromycin; FUS, fusidic acid; GEN, gentamicin; LEV, levofloxacin; MUP, mupirocin; PEN, penicillin; RIF, rifampicin; SXT, trimethoprim-sulfamethoxazole; SYN, quinupristin-dalfopristin; TET, tetracycline.
Mutations in the QRDRs of fluoroquinolone-resistant S. aureus
Forty-one MRSA (41/43, 95.3%) and 39 MSSA (39/157, 24.8%) isolates were resistant to fluoroquinolones (Figure 2A and B). Fluoroquinolone resistance phenotype in S. aureus is caused by point mutations in the QRDRs encoding the two essential enzymes, DNA gyrase and topoisomerase IV (Ito et al., 1994; Schmitz et al., 1998). As shown in Table 3, S84L and S80F mutations were identified in gyrA and parC genes of all 80 fluoroquinolone-resistant S. aureus strains, except for one ST3218 MSSA strain that only had the S80F mutation in parC. In addition to mutations in gyrA and parC, a mutation of D494N in parE was detected in 10 fluoroquinolone-resistant ST188 MRSA strains and one ST188 MSSA strain. The ST188 MRSA strains carrying the triple point mutations in gyrA, parC, and parE displayed significantly higher CIP MIC values than ST692 MRSA strains (P = 0.008) and ST692 MSSA strains (P = 0.006) carrying dual mutations in gyrA and parC (Supplementary Figure 1). No statistically significant difference was observed in the ENR MICs between the triple (gyrA, parC, and parE) and dual mutations (gyrA and parC) (data not shown).
Table 3.
Mutational changes detected in the QRDRs of gyrA, gyrB, parC, and parE in fluoroquinolone-resistant MRSA and MSSA strains.
| Mutational changes in QRDRs |
|||||
|---|---|---|---|---|---|
| STs | gyrA | gyrB | parC | parE | |
| MRSA (n = 41) | ST692 (n = 31) | S84L (100%) | S80F (100%) | ||
| ST188 (n = 10) | S84L (100%) | D494N (100%) | S80F (100%) | ||
| MSSA (n = 39) | ST692 (n = 24) | S84L (100%) | S80F (100%) | ||
| ST5 (n = 9) | S84L (100%) | S80F (100%) | |||
| ST3218 (n = 3) | S84L (67%) | S80F (100%) | |||
| ST188 (n = 1) | S84L (100%) | D494N (100%) | S80F (100%) | ||
| ST8 (n = 1) | S84L (100%) | S80F (100%) | |||
| ST199 (n = 1) | S84L (100%) | S80F (100%) | |||
Abbreviations: STs, sequence types; QRDRs, quinolone resistance-determining regions.
Discussion
The present study describes the cross-sectional occurrence and genetic relatedness of antimicrobial-resistant S. aureus strains isolated from chicken meat production systems, including broiler farms, slaughterhouses, and retail chicken meat.
The overall proportion of MRSA among S. aureus isolates in the chicken meat production chain was 21.5% (43/200 isolates), and 79% of MRSA isolates (34/43 isolates) were recovered from healthy broilers (Table 1). High rates of MRSA carriage, ranging from 1.1% to 6.9%, in healthy broilers have been reported in several previous studies in European countries (Mulders et al., 2010; Geenen et al., 2013; Nemeghaire et al., 2013; Krupa et al., 2014). The carriage rates of MRSA in carcass samples from slaughterhouses and retail chicken meat were only 0.83% and 5%, respectively. Previous studies have reported the detection rates of MRSA in chicken carcass samples (1.5%) (Okorie-Kanu et al., 2020) and retail chicken meat samples (0.5%–6.9%) (Mulders et al., 2010; Wang et al., 2013; Krupa et al., 2014; Tang et al., 2017; Bernier-Lachance et al., 2020). Consistent with previous studies in Korea (Kim et al., 2018) and other countries (Hanning et al., 2012; Wang et al., 2013; Krupa et al., 2014; Tang et al., 2017; Okorie-Kanu et al., 2020), the detection rates of MSSA were significantly higher than those of MRSA at all three sampling sites: broiler farms, slaughterhouses, and retail chicken meat (Table 1). In particular, the isolation rates of MSSA were >10 times higher than those of MRSA in slaughterhouse carcass samples (1 MRSA isolate vs. 33 MSSA isolates) and retail chicken meat samples (5 MRSA isolates vs. 59 MSSA isolates). Although previous studies have mainly focused on the prevalence of MRSA in livestock and foods of animal origin, our results indicate that the carriage of MSSA and MRSA in broilers and chicken carcass/meat samples should be investigated as a potential source of food contamination in future studies.
As shown in Figure 1A, MLST analyses of all 43 MRSA isolates identified 2 dominant STs, ST692 and ST188, each harboring SCCmec V and SCCmec IVa for methicillin resistance. Except for five ST692-SCCmec V strains from retail chicken meat samples, 26 ST692-SCCmec V and 10 ST188-SCCmec IVa strains were recovered from broilers (n = 33) and farm environments (n = 3). Further analyses of spa types of these 2 dominant MRSA lineages revealed ST-specific spa types of MRSA strains, ST692-SCCmec V-t2247 and ST188-SCCmec IVa-t189 (Table 2). Unlike the ST-specific spa types, all ST692 and ST188 MRSA strains were determined to be agr I. Furthermore, the clonal lineage of ST692 LA-MRSA with spa t2247 is uniquely associated with the chicken carcass and retail chicken meat in Korea (Moon et al., 2015; Kim et al., 2020). Although Moon et al. reported ST692-SCCmec III-t2247 in chicken carcass samples in 2015 (Moon et al., 2015; Moon et al., 2016), all 31 ST692 MRSA-t2247 strains used in this study carried SCCmec V (Figure 2A; Table 2). Recent studies in Korea have also reported that SCCmec V is frequently identified in MRSA strains and methicillin-resistant non-aureus staphylococci isolated from livestock and companion animals (Lee et al., 2019; Back et al., 2020; Lee and Yang, 2020). In contrast to ST692 MRSA, which has emerged as a poultry-specific clonal lineage in Korea (Moon et al., 2015; Kim et al., 2020), ST188-SCCmec IVa-t189 MRSA is frequently detected in clinical settings (Wang et al., 2018; Huang et al., 2019), livestock (Wang et al., 2018; Moon et al., 2019), meat products (Wu et al., 2018; Kim et al., 2020), and even in primates (Roberts et al., 2018) in European and Asian countries, implying that the ST188 clonal lineage has a broader host range than the ST692 strain.
In contrast to MRSA strains, diverse genetic lineages have been identified in the MSSA strains. ST692 and ST3218 MSSA strains are usually associated with broiler farms, whereas ST6, ST8, and ST81 were identified in MSSA strains from slaughterhouse carcasses and retail chicken meat samples (Figure 1B). In line with previous studies from other countries (Tang et al., 2017; Li et al., 2019; Okorie-Kanu et al., 2020), which indicated that CC5 (ST5, ST6, ST3218, and ST587) was the most prominent genotype in MSSA found in chicken meat production systems, 48.4% (n = 76) of the MSSA strains used in this study were found to have the CC5 genotype. In particular, the ST5 clonal lineage has evolved from a major human pathogen to a poultry pandemic strain owing to the acquisition of avian-specific prophages and novel virulence genes (Lowder et al., 2009). None of the six MSSA strains collected from human workers belonged to the ST5 lineage (Table 1; Figure 1B). Although ST5 and ST3218, a single locus variant of ST5, have been found in chicken meat samples carrying avian-specific mobile genetic elements (Muller et al., 2016), only 24 ST3218 MSSA strains were associated with healthy broilers in this study (Figure 1B). In contrast to the ST692 and ST3218 MSSA strains in broiler farms, slaughterhouse carcass samples are usually associated with ST6, ST8, and ST81. ST6 and ST81 have been reported in food poisoning outbreaks caused by retail meat and dairy products in Japan, China, Algeria, and the USA (Waters et al., 2011; Yan et al., 2012; Li et al., 2015; Sato et al., 2017; Titouche et al., 2019). ST8 MSSA-t008, which was one of the most prevalent genetic lineages of MSSA strains in this study, is known to be a global community-associated MRSA lineage of USA300 (Nimmo, 2012). Overall, our results suggest that MSSA strains from healthy broilers and farm environments are associated with poultry- or LA-type clonal lineages of S. aureus, whereas MSSA strains from slaughterhouses and retail chicken meat are linked to human-type clonal lineages. Currently, whole-genome sequencing analyses of broiler-associated MRSA and MSSA strains are in progress in our laboratory.
In this study, MRSA strains displayed higher levels of antimicrobial resistance to most of the tested antibiotics than the MSSA strains (Figure 2A and B). Although the two major ST groups of MRSA isolates, ST692 and ST188, showed high levels of MDR and fluoroquinolone resistance, opposite resistance phenotypes to GEN, TET, and SXT were identified (Figures 2C and D). These opposite resistance phenotypes have also been identified in previous studies (Lim et al., 2010; Wang et al., 2014; Moon et al., 2015; Kim et al., 2020), which showed the acquisition of TET resistance genes, such as tet(L) and tet(S), in ST692 (Moon et al., 2015) and aminoglycoside/trimethoprim resistance genes in ST188 strains (Wang et al., 2014; Kim et al., 2020). When the 4 major ST groups of MSSA strains (ST8, ST6, ST3218, and ST692) were compared for antimicrobial resistance, ST692 MSSA strains exhibited the highest rate of MDR, including all three fluoroquinolones (Figure 3A). High fluoroquinolone resistance in MRSA and MSSA strains was most frequently associated with clonal lineages isolated from broiler farms, especially from healthy broilers (Figures 1, 2A, 3C and D). Fluoroquinolones are very effective therapeutic and preventive options for pathogenic bacteria that cause gastroenteritis in poultry because of their broad-spectrum bactericidal activity (Endtz et al., 1991; Engberg et al., 2004; Waters et al., 2011). The frequent occurrence of fluoroquinolone resistance in Staphylococci has been attributed to mutations in QRDRs of gyrA and parC genes encoding DNA gyrase and topoisomerase IV, respectively (de Oliveira et al., 2019; Ostrer et al., 2019). As shown in Table 3, point mutations in gyrA (S84L) and parC (S80F) were identified in all 80 fluoroquinolone-resistant S. aureus isolates (41 MRSA and 39 MSSA isolates), regardless of methicillin resistance or STs. In addition to the point mutations in gyrA and parC, the ST188 lineage of MRSA (n = 10) and MSSA (n =1) strains had a point mutation in gyrB (D494N) for the observed fluoroquinolone resistance (Table 3). The triple mutations of ST188 S. aureus strains correlated with significantly higher CIP MIC values than those of non-ST692 S. aureus strains with dual mutations (Supplementary Figure 1). However, there was no significant difference in the MIC values of ENR between the triple- and double-mutant strains (data not shown). Nakaminami et al. reported that a novel point mutation (E477D) in gyrB of MRSA (Nakaminami et al., 2014) confers a high level of resistance to third-generation quinolones. Further investigations are required to elucidate the molecular mechanisms involved in fluoroquinolone resistance caused by point mutations in gyrB.
It should be recognized that the present study has several limitations. The relatively low number of MRSA isolates may have contributed to the lower degree of clonal diversity compared with the MSSA isolates. Moreover, future studies need to confirm the clonality of the MRSA and MSSA isolates with additional phylogenetic methods, including whole genome sequence analysis and pulsed-field gel electrophoresis. Furthermore, composite profiles of antimicrobial resistance genes associated with the observed resistance phenotype have not been assessed in this study.
In conclusion, the present study aimed to gain insight into the clonal diversity of both MRSA and MSSA isolates collected from broiler farms, slaughterhouses, and retail chicken meat samples. To the best of our knowledge, this is the first study to systematically analyze the clonal lineages and antimicrobial resistance profiles of MRSA and MSSA strains isolated from the 3 major sectors of the chicken meat production system in Korea. Taken together, our results suggest that 1) colonization or contamination with 2 dominant genetic lineages of MRSA, ST692-SCCmec V-t2247 and ST188-SCCmec IVa-t189, is mainly observed in broiler farms, especially in healthy broilers; 2) there is a higher degree of genetic diversity (MLST, agr, and spa types) in MSSA versus MRSA strains; 3) the prevalence of ST692 MRSA/MSSA and ST188 MRSA in broilers appears to be correlated with higher levels of antimicrobial resistance phenotypes, especially fluoroquinolone resistance; 4) most fluoroquinolone resistant MRSA/MSSA strains carry double point mutations in gyrA (S84L) and parC (S80F) regardless of their genotypes and sampling sites; and 5) in addition to the double point mutations, ST188 lineage of strains tend to have a third point mutation in gyrB (D494N), which correlates with higher CIP MIC values vs. double point-mutated strains.
ACKNOWLEDGMENTS
This study was supported by funding from the Research of Korea Centers for Disease Control and Prevention (Project No. 2020ER540500).
DISCLOSURES
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.psj.2022.102070.
Appendix. Supplementary materials
REFERENCES
- Back S.H., Eom H.S., Lee H.H., Lee G.Y., Park K.T., Yang S.J. Livestock-associated methicillin-resistant Staphylococcus aureus in Korea: antimicrobial resistance and molecular characteristics of LA-MRSA strains isolated from pigs, pig farmers, and farm environment. J. Vet. Sci. 2020;21:e2. doi: 10.4142/jvs.2020.21.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker K., Ballhausen B., Kahl B.C., Kock R. The clinical impact of livestock-associated methicillin-resistant Staphylococcus aureus of the clonal complex 398 for humans. Vet. Microbiol. 2017;200:33–38. doi: 10.1016/j.vetmic.2015.11.013. [DOI] [PubMed] [Google Scholar]
- Bernier-Lachance J., Arsenault J., Usongo V., Parent E., Labrie J., Jacques M., Malouin F., Archambault M. Prevalence and characteristics of livestock-associated methicillin-resistant Staphylococcus aureus (LA-MRSA) isolated from chicken meat in the province of Quebec, Canada. PLoS One. 2020;15 doi: 10.1371/journal.pone.0227183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clinical and Laboratory Standards Institute (CLSI) 5th Edition. Clinical and Laboratory Standards Institute; Wayne, PA: 2020. Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated From Animals. VET01S. [Google Scholar]
- Clinical and Laboratory Standards Institute (CLSI). 2022. Performance Standards for Antimicrobial Susceptibility Testing. M100 32nd Edition. Clinical and Laboratory Standards Institute, Wayne, PA.
- Cuny C., Wieler L.H., Witte W. Livestock-associated MRSA: the impact on humans. Antibiotics (Basel) 2015;4:521–543. doi: 10.3390/antibiotics4040521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Oliveira T.L.R., Cavalcante F.S., Chamon R.C., Ferreira R.B.R., Dos Santos K.R.N. Genetic mutations in the quinolone resistance-determining region are related to changes in the epidemiological profile of methicillin-resistant Staphylococcus aureus isolates. J. Glob. Antimicrob. Resist. 2019;19:236–240. doi: 10.1016/j.jgar.2019.05.026. [DOI] [PubMed] [Google Scholar]
- Endtz H.P., Ruijs G.J., van Klingeren B., Jansen W.H., van der Reyden T., Mouton R.P. Quinolone resistance in Campylobacter isolated from man and poultry following the introduction of fluoroquinolones in veterinary medicine. J. Antimicrob. Chemother. 1991;27:199–208. doi: 10.1093/jac/27.2.199. [DOI] [PubMed] [Google Scholar]
- Engberg J., Neimann J., Nielsen E.M., Aerestrup F.M., Fussing V. Quinolone-resistant Campylobacter infections: risk factors and clinical consequences. Emerg. Infect. Dis. 2004;10:1056–1063. doi: 10.3201/eid1006.030669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enright M.C., Day N.P., Davies C.E., Peacock S.J., Spratt B.G. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 2000;38:1008–1015. doi: 10.1128/jcm.38.3.1008-1015.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eom H.S., Back S.H., Lee H.H., Lee G.Y., Yang S.J. Prevalence and characteristics of livestock-associated methicillin-susceptible Staphylococcus aureus in the pork production chain in Korea. J. Vet. Sci. 2019;20:e69. doi: 10.4142/jvs.2019.20.e69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrero L., Cameron B., Manse B., Lagneaux D., Crouzet J., Famechon A., Blanche F. Cloning and primary structure of Staphylococcus aureus DNA topoisomerase IV: a primary target of fluoroquinolones. Mol. Microbiol. 1994;13:641–653. doi: 10.1111/j.1365-2958.1994.tb00458.x. [DOI] [PubMed] [Google Scholar]
- Fluit A.C. Livestock-associated Staphylococcus aureus. Clin. Microbiol. Infect. 2012;18:735–744. doi: 10.1111/j.1469-0691.2012.03846.x. [DOI] [PubMed] [Google Scholar]
- Friese A., Schulz J., Zimmermann K., Tenhagen B.A., Fetsch A., Hartung J., Rosler U. Occurrence of livestock-associated methicillin-resistant Staphylococcus aureus in turkey and broiler barns and contamination of air and soil surfaces in their vicinity. Appl. Environ. Microb. 2013;79:2759–2766. doi: 10.1128/AEM.03939-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geenen P.L., Graat E.A., Haenen A., Hengeveld P.D., Van Hoek A.H., Huijsdens X.W., Kappert C.C., Lammers G.A., Van Duijkeren E., Van De Giessen A.W. Prevalence of livestock-associated MRSA on Dutch broiler farms and in people living and/or working on these farms. Epidemiol. Infect. 2013;141:1099–1108. doi: 10.1017/S0950268812001616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilot P., Lina G., Cochard T., Poutrel B. Analysis of the genetic variability of genes encoding the RNA III-activating components Agr and TRAP in a population of Staphylococcus aureus strains isolated from cows with mastitis. J. Clin. Microbiol. 2002;40:4060–4067. doi: 10.1128/JCM.40.11.4060-4067.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez I., Georgiou M., Alcaide F., Balas D., Linares J., de la Campa A.G. Fluoroquinolone resistance mutations in the parC, parE, and gyrA genes of clinical isolates of viridans group streptococci. Antimicrob. Agents. Chemother. 1998;42:2792–2798. doi: 10.1128/aac.42.11.2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grobbel M., Lubke-Becker A., Wieler L.H., Froyman R., Friederichs S., Filios S. Comparative quantification of the in vitro activity of veterinary fluoroquinolones. Vet. Microbiol. 2007;124:73–81. doi: 10.1016/j.vetmic.2007.03.017. [DOI] [PubMed] [Google Scholar]
- Hanning I., Gilmore D., Pendleton S., Fleck S., Clement A., Park S.H., Scott E., Ricke S.C. Characterization of Staphylococcus aureus isolates from retail chicken carcasses and pet workers in Northwest Arkansas. J. Food. Prot. 2012;75:174–178. doi: 10.4315/0362-028X.JFP-11-251. [DOI] [PubMed] [Google Scholar]
- Ho P.L., Chow K.H., Lai E.L., Law P.Y., Chan P.Y., Ho A.Y., Ng T.K., Yam W.C. Clonality and antimicrobial susceptibility of Staphylococcus aureus and methicillin-resistant S. aureus isolates from food animals and other animals. J. Clin. Microbiol. 2012;50:3735–3737. doi: 10.1128/JCM.02053-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y.C., Su L.H., Wu T.L., Lin T.Y. Methicillin-resistant Staphylococcus aureus nasal carriage in international medical conference attendees. J. Microbiol. Immunol. Infect. 2019;52:242–247. doi: 10.1016/j.jmii.2018.08.004. [DOI] [PubMed] [Google Scholar]
- Ito H., Yoshida H., Bogaki-Shonai M., Niga T., Hattori H., Nakamura S. Quinolone resistance mutations in the DNA gyrase gyrA and gyrB genes of Staphylococcus aureus. Antimicrob. Agents. Chemother. 1994;38:2014–2023. doi: 10.1128/aac.38.9.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.B., Seo K.W., Jeon H.Y., Lim S.K., Lee Y.J. Characteristics of the antimicrobial resistance of Staphylococcus aureus isolated from chicken meat produced by different integrated broiler operations in Korea. Poult. Sci. 2018;97:962–969. doi: 10.3382/ps/pex357. [DOI] [PubMed] [Google Scholar]
- Kim Y.H., Kim H.S., Kim S., Kim M., Kwak H.S. Prevalence and characteristics of antimicrobial-resistant Staphylococcus aureus and methicillin-resistant Staphylococcus aureus from retail meat in Korea. Food. Sci. Anim. Resour. 2020;40:758–771. doi: 10.5851/kosfa.2020.e50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo Y., Ito T., Ma X.X., Watanabe S., Kreiswirth B.N., Etienne J., Hiramatsu K. Combination of multiplex PCRs for staphylococcal cassette chromosome mec type assignment: rapid identification system for mec, ccr, and major differences in junkyard regions. Antimicrob. Agents. Chemother. 2007;51:264–274. doi: 10.1128/AAC.00165-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koreen L., Ramaswamy S.V., Graviss E.A., Naidich S., Musser J.M., Kreiswirth B.N. spa typing method for discriminating among Staphylococcus aureus isolates: implications for use of a single marker to detect genetic micro- and macrovariation. J. Clin. Microbiol. 2004;42:792–799. doi: 10.1128/JCM.42.2.792-799.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraushaar B., Ballhausen B., Leeser D., Tenhagen B.A., Kasbohrer A., Fetsch A. Antimicrobial resistances and virulence markers in methicillin-resistant Staphylococcus aureus from broiler and turkey: a molecular view from farm to fork. Vet. Microbiol. 2017;200:25–32. doi: 10.1016/j.vetmic.2016.05.022. [DOI] [PubMed] [Google Scholar]
- Krupa P., Bystron J., Bania J., Podkowik M., Empel J., Mroczkowska A. Genotypes and oxacillin resistance of Staphylococcus aureus from chicken and chicken meat in Poland. Poult. Sci. 2014;93:3179–3186. doi: 10.3382/ps.2014-04321. [DOI] [PubMed] [Google Scholar]
- Lee G.Y., Lee H.H., Hwang S.Y., Hong J., Lyoo K.S., Yang S.J. Carriage of Staphylococcus schleiferi from canine otitis externa: antimicrobial resistance profiles and virulence factors associated with skin infection. J. Vet. Sci. 2019;20:e6. doi: 10.4142/jvs.2019.20.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee G.Y., Yang S.J. Comparative assessment of genotypic and phenotypic correlates of Staphylococcus pseudintermedius strains isolated from dogs with otitis externa and healthy dogs. Comp. Immunol. Microbiol. Infect. Dis. 2020;70 doi: 10.1016/j.cimid.2019.101376. [DOI] [PubMed] [Google Scholar]
- Lee H.H., Lee G.Y., Eom H.S., Yang S.J. Occurrence and characteristics of methicillin-resistant and -susceptible Staphylococcus aureus isolated from the beef production chain in Korea. Food. Sci. Anim. Resour. 2020;40:401–414. doi: 10.5851/kosfa.2020.e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.H. Methicillin (Oxacillin)-resistant Staphylococcus aureus strains isolated from major food animals and their potential transmission to humans. Appl. Environ. Microbiol. 2003;69:6489–6494. doi: 10.1128/AEM.69.11.6489-6494.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.I., Kim S.D., Park J.H., Yang S.J. Species distribution, antimicrobial resistance, and enterotoxigenicity of non-aureus staphylococci in retail chicken meat. Antibiotics (Basel) 2020;13:809. doi: 10.3390/antibiotics9110809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., Wu S., Luo W., Su Y., Luan Y., Wang X. Staphylococcus aureus ST6-t701 isolates from food-poisoning outbreaks (2006-2013) in Xi'an, China. Foodborne. Pathog. Dis. 2015;12:203–206. doi: 10.1089/fpd.2014.1850. [DOI] [PubMed] [Google Scholar]
- Li H., Andersen P.S., Stegger M., Sieber R.N., Ingmer H., Staubrand N., Dalsgaard A., Leisner J.J. Antimicrobial resistance and virulence gene profiles of methicillin-resistant and -susceptible Staphylococcus aureus from food products in Denmark. Front. Microbiol. 2019;10:2681. doi: 10.3389/fmicb.2019.02681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim S.K., Nam H.M., Park H.J., Lee H.S., Choi M.J., Jung S.C., Lee J.Y., Kim Y.C., Song S.W., Wee S.H. Prevalence and characterization of methicillin-resistant Staphylococcus aureus in raw meat in Korea. J Microbiol. Biotechnol. 2010;20:775–778. [PubMed] [Google Scholar]
- Lowder B.V., Guinane C.M., Ben Zakour N.L., Weinert L.A., Conway-Morris A., Cartwright R.A., Simpson A.J., Rambaut A., Nubel U., Fitzgerald J.R. Recent human-to-poultry host jump, adaptation, and pandemic spread of Staphylococcus aureus. Proc. Natl. Acad. Sci. U S A. 2009;106:19545–19550. doi: 10.1073/pnas.0909285106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milheirico C., Oliveira D.C., de Lencastre H. Multiplex PCR strategy for subtyping the staphylococcal cassette chromosome mec type IV in methicillin-resistant Staphylococcus aureus: 'SCCmec IV multiplex'. J. Antimicrob. Chemother. 2007;60:42–48. doi: 10.1093/jac/dkm112. [DOI] [PubMed] [Google Scholar]
- Moon D.C., Jeong S.K., Hyun B.H., Lim S.K. Prevalence and characteristics of methicillin-resistant Staphylococcus aureus isolates in pigs and pig farmers in Korea. Foodborne. Pathog. Dis. 2019;16:256–261. doi: 10.1089/fpd.2018.2509. [DOI] [PubMed] [Google Scholar]
- Moon D.C., Kim B.Y., Tamang M.D., Nam H.M., Jang G.C., Jung S.C., Lee H.S., Park Y.H., Lim S.K. Genome sequence of a unique t2247-ST692-III livestock-associated methicillin-resistant Staphylococcus aureus strain from chicken carcass. Genome. Announc. 2016;25 doi: 10.1128/genomeA.00026-16. e00026-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon D.C., Tamang M.D., Nam H.M., Jeong J.H., Jang G.C., Jung S.C., Park Y.H., Lim S.K. Identification of livestock-associated methicillin-resistant Staphylococcus aureus isolates in Korea and molecular comparison between isolates from animal carcasses and slaughterhouse workers. Foodborne. Pathog. Dis. 2015;12:327–334. doi: 10.1089/fpd.2014.1868. [DOI] [PubMed] [Google Scholar]
- Mulders M.N., Haenen A.P., Geenen P.L., Vesseur P.C., Poldervaart E.S., Bosch T., Huijsdens X.W., Hengeveld P.D., Dam-Deisz W.D., Graat E.A., Mevius D., Voss A., Van De Giessen A.W. Prevalence of livestock-associated MRSA in broiler flocks and risk factors for slaughterhouse personnel in the Netherlands. Epidemiol. Infect. 2010;138:743–755. doi: 10.1017/S0950268810000075. [DOI] [PubMed] [Google Scholar]
- Muller A., Seinige D., Jansen W., Klein G., Ehricht R., Monecke S., Kehrenberg C. Variety of antimicrobial resistances and virulence factors in Staphylococcus aureus isolates from meat products legally and illegally introduced to Germany. PLoS One. 2016;11 doi: 10.1371/journal.pone.0167864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaminami H., Sato-Nakaminami K., Noguchi N. A novel GyrB mutation in meticillin-resistant Staphylococcus aureus (MRSA) confers a high level of resistance to third-generation quinolones. Int. J. Antimicrob. Agents. 2014;43:478–479. doi: 10.1016/j.ijantimicag.2014.02.002. [DOI] [PubMed] [Google Scholar]
- Nemeghaire S., Argudin M.A., Haesebrouck F., Butaye P. Epidemiology and molecular characterization of methicillin-resistant Staphylococcus aureus nasal carriage isolates from bovines. BMC. Vet. Res. 2014;10:153. doi: 10.1186/1746-6148-10-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemeghaire S., Roelandt S., Argudin M.A., Haesebrouck F., Butaye P. Characterization of methicillin-resistant Staphylococcus aureus from healthy carrier chickens. Avian. Pathol. 2013;42:342–346. doi: 10.1080/03079457.2013.805183. [DOI] [PubMed] [Google Scholar]
- Nimmo G.R. USA300 abroad: global spread of a virulent strain of community-associated methicillin-resistant Staphylococcus aureus. Clin. Microbiol. Infect. 2012;18:725–734. doi: 10.1111/j.1469-0691.2012.03822.x. [DOI] [PubMed] [Google Scholar]
- Okorie-Kanu O.J., Anyanwu M.U., Ezenduka E.V., Mgbeahuruike A.C., Thapaliya D., Gerbig G., Ugwuijem E.E., Okorie-Kanu C.O., Agbowo P., Olorunleke S., Nwanta J.A., Chah K.F., Smith T.C. Molecular epidemiology, genetic diversity and antimicrobial resistance of Staphylococcus aureus isolated from chicken and pig carcasses, and carcass handlers. PLoS One. 2020;15 doi: 10.1371/journal.pone.0232913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrer L., Khodursky R.F., Johnson J.R., Hiasa H., Khodursky A. Analysis of mutational patterns in quinolone resistance-determining regions of GyrA and ParC of clinical isolates. Int. J. Antimicrob. Agents. 2019;53:318–324. doi: 10.1016/j.ijantimicag.2018.12.004. [DOI] [PubMed] [Google Scholar]
- Persoons D., Van Hoorebeke S., Hermans K., Butaye P., de Kruif A., Haesebrouck F., Dewulf J. Methicillin-resistant Staphylococcus aureus in poultry. Emerg. Infect. Dis. 2009;15:452–453. doi: 10.3201/eid1503.080696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynaga E., Navarro M., Vilamala A., Roure P., Quintana M., Garcia-Nunez M., Figueras R., Torres C., Lucchetti G., Sabria M. Prevalence of colonization by methicillin-resistant Staphylococcus aureus ST398 in pigs and pig farm workers in an area of Catalonia, Spain. BMC. Infect. Dis. 2016;16:716. doi: 10.1186/s12879-016-2050-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts M.C., Feßler A.T., Monecke S., Ehricht R., No D., Schwarz S. Molecular analysis of two different MRSA clones ST188 and ST3268 from primates (Macaca spp.) in a united states primate center. Front. Microbiol. 2018;9:2199. doi: 10.3389/fmicb.2018.02199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T., Usui M., Konishi N., Kai A., Matsui H., Hanaki H., Tamura Y. Closely related methicillin-resistant Staphylococcus aureus isolates from retail meat, cows with mastitis, and humans in Japan. PLoS One. 2017;12 doi: 10.1371/journal.pone.0187319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz F.J., Jones M.E., Hofmann B., Hansen B., Scheuring S., Luckefahr M., Fluit A., Verhoef J., Hadding U., Heinz H.P., Kohrer K. Characterization of grlA, grlB, gyrA, and gyrB mutations in 116 unrelated isolates of Staphylococcus aureus and effects of mutations on ciprofloxacin MIC. Antimicrob. Agents. Chemother. 1998;42:1249–1252. doi: 10.1128/aac.42.5.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y., Larsen J., Kjeldgaard J., Andersen P.S., Skov R., Ingmer H. Methicillin-resistant and -susceptible Staphylococcus aureus from retail meat in Denmark. Int. J. Food. Microbiol. 2017;249:72–76. doi: 10.1016/j.ijfoodmicro.2017.03.001. [DOI] [PubMed] [Google Scholar]
- Titouche Y., Hakem A., Houali K., Meheut T., Vingadassalon N., Ruiz-Ripa L., Salmi D., Chergui A., Chenouf N., Hennekinne J.A., Torres C., Auvray F. Emergence of methicillin-resistant Staphylococcus aureus (MRSA) ST8 in raw milk and traditional dairy products in the Tizi Ouzou area of Algeria. J. Dairy. Sci. 2019;102:6876–6884. doi: 10.3168/jds.2018-16208. [DOI] [PubMed] [Google Scholar]
- van Cleef B.A., van Benthem B.H., Verkade E.J., van Rijen M., Kluytmans-van den Bergh M.F., Schouls L.M., Duim B., Wagenaar J.A., Graveland H., Bos M.E., Heederik D., Kluytmans J.A. Dynamics of methicillin-resistant Staphylococcus aureus and methicillin-susceptible Staphylococcus aureus carriage in pig farmers: a prospective cohort study. Clin. Microbiol. Infect. 2014;20:O764–O771. doi: 10.1111/1469-0691.12582. [DOI] [PubMed] [Google Scholar]
- Vanderhaeghen W., Hermans K., Haesebrouck F., Butaye P. Methicillin-resistant Staphylococcus aureus (MRSA) in food production animals. Epidemiol. Infect. 2010;138:606–625. doi: 10.1017/S0950268809991567. [DOI] [PubMed] [Google Scholar]
- van der Mee-Marquet N.L., Corvaglia A., Haenni M., Bertrand X., Franck J.B., Kluytmans J., Girard M., Quentin R., Francois P. Emergence of a novel subpopulation of CC398 Staphylococcus aureus infecting animals is a serious hazard for humans. Front. Microbiol. 2014;5:652. doi: 10.3389/fmicb.2014.00652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Li G., Xia X., Yang B., Xi M., Meng J. Antimicrobial susceptibility and molecular typing of methicillin-resistant Staphylococcus aureus in retail foods in Shaanxi, China. Foodborne. Pathog. Dis. 2014;11:281–286. doi: 10.1089/fpd.2013.1643. [DOI] [PubMed] [Google Scholar]
- Wang X., Tao X.Y., Xia X.D., Yang B.W., Xi M.L., Meng J.H., Zhang J., Xu B.J. Staphylococcus aureus and methicillin-resistant Staphylococcus aureus in retail raw chicken in China. Food. Control. 2013;29:103–106. [Google Scholar]
- Wang Y., Liu Q., Liu Q., Gao Q., Lu H., Meng H., Xie Y., Huang Q., Ma X., Wang H., Qin J., Li Q., Li T., Xia Q., Li M. Phylogenetic analysis and virulence determinant of the host-adapted Staphylococcus aureus lineage ST188 in China. Emerg. Microbes. Infect. 2018;7:45. doi: 10.1038/s41426-018-0048-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters A.E., Contente-Cuomo T., Buchhagen J., Liu C.M., Watson L., Pearce K., Foster J.T., Bowers J., Driebe E.M., Engelthaler D.M., Keim P.S., Price L.B. Multidrug-resistant Staphylococcus aureus in US meat and poultry. Clin. Infect. Dis. 2011;52:1227–1230. doi: 10.1093/cid/cir181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S., Huang J., Wu Q., Zhang J., Zhang F., Yang X., Wu H., Zeng H., Chen M., Ding Y., Wang J., Lei T., Zhang S., Xue L. Staphylococcus aureus isolated from retail meat and meat products in China: incidence, antibiotic resistance and genetic diversity. Front. Microbiol. 2018;9:2767. doi: 10.3389/fmicb.2018.02767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X., Wang B., Tao X., Hu Q., Cui Z., Zhang J., Lin Y., You Y., Shi X., Grundmann H. Characterization of Staphylococcus aureus strains associated with food poisoning in Shenzhen, China. Appl. Environ. Microbiol. 2012;78:6637–6642. doi: 10.1128/AEM.01165-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y.J., Lee G.Y., Kim S.D., Park J.H., Lee S.I., Kim G.B., Yang S.J. Profiles of non-aureus staphylococci in retail pork and slaughterhouse carcasses: prevalence, antimicrobial resistance, and genetic determinant of fusidic acid resistance. Food. Sci. Anim. Resour. 2022;42:225–239. doi: 10.5851/kosfa.2021.e74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H., Bogaki M., Nakamura M., Yamanaka L.M., Nakamura S. Quinolone resistance-determining region in the DNA gyrase gyrB gene of Escherichia coli. Antimicrob. Agents. Chemother. 1991;35:1647–1650. doi: 10.1128/aac.35.8.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.