Abstract
Pigeon paramyxovirus type 1 (PPMV-1), a variant of Newcastle disease virus (NDV), causes severe Newcastle disease (ND) in pigeons. However, there is no PPMV-1 vaccine available worldwide. In this study, a strain of PPMV-1 was isolated from outbreaks in a vaccinated racing pigeon (Columbia livia) loft in China, namely, PPMV-1/pigeon/Gansu/China/02/2020 (GS02). Experimental infection with GS02 showed mortality rates of 100% and 87.50% in 4- and 12-week-old pigeons, respectively, suggesting that GS02 is virulent and more sensitive to young pigeons. The whole genome of GS02 determined the fusion (F) protein possessing virulence cleavage site 112RRQKRF117. Phylogenetic analysis indicated that GS02 was a subgenotype VI.2.1.1.2.2 (VIk) of Class II NDV and more closely related to the JS/06/20/Pi (MW271791) strain, but it was far from the genetic distance from the commercial vaccine chicken-origin La Sota strain. Using inactivated GS02 as a vaccine candidate and inactivated vaccine La Sota to immunize the pigeons, both of them provided complete protection against GS02 challenge. The GS02 vaccine candidate induced higher antibody titers than the La Sota vaccine, and cross-reactivity testing showed antigenically slight differences between GS02 and La Sota. These results indicated that the GS02 candidate could be a potential pigeon-derived vaccine for the prevention and control of PPMV-1 in pigeons.
Key words: pigeon paramyxovirus type 1, racing pigeon, pathogenicity, inactivated vaccine
INTRODUCTION
Newcastle disease (ND) is one of the most serious viral diseases in poultry worldwide (Alexander, 2001). It is caused by Newcastle disease virus (NDV), which belongs to the genus Orthoavulavirus in the subfamily Avulavirinae of the family Paramyxoviridae (Amarasinghe et al., 2019). The NDV genome is a single-stranded, negative-sense, nonsegmented RNA that encodes 6 structural proteins, including nucleocapsid protein (NP), phosphoprotein (P), matrix protein (M), fusion protein (F), hemagglutinin-neuraminidase (HN), the large protein (L), and 2 nonstructural proteins, V and W (Millar and Emmerson, 1988; Steward et al., 1993). According to the updated unified classification system of NDVs, NDV strains can be grouped into 2 major classes: Class I and Class II. To date, there are 21 identified genotypes of Class II (Dimitrov et al., 2019).
Pigeon paramyxovirus type 1 (PPMV-1) is an antigenic and host variant of NDV, most of which belongs to Genotype VI of Class II (Ujvári et al., 2003). PPMV-1 was first identified in meat pigeons in Iraq in 1978 (Kaleta et al., 1985), and pigeons as a medium caused the third epizootic during the 1980s (Collins et al., 1989). PPMV-1 was introduced into Hong Kong in 1985, subsequently spread throughout China, and now poses a serious threat to the Chinese pigeon breeding industry and wild bird population (Wang et al., 2015). Outbreaks of PPMV-1 infection in racing pigeons are common every year, especially affecting young and unvaccinated pigeons (Teske et al., 2013). Pigeons infected with PPMV-1 show neurological, respiratory, and digestive symptoms, including moderate-to-severe depression with neck twist, ataxia, crouching, paralysis, eyelid edema, diarrhea, and green loose stools (Marlier and Vindevogel, 2006).
According to the definition of the World Organization for Animal Health, avian paramyxovirus serotype I is judged to be strongly virulent when its intracerebral pathogenicity index (ICPI) is higher than or equal to 0.7 (OIE 2021). However, some studies have shown that some pigeon paramyxovirus type I strains have F protein cleavage sites (112R/K-R-Q/K/R-K/R-R-F117) but their ICPI is lower than 0.7 (Panda et al., 2004; Kim et al., 2008). This amino acid composition allows the F protein to be cleaved by intracellular furan-like proteases present in all tissues of the host, resulting in a virus that can cause infection in a wide range of tissues (Nidzworski et al., 2011).
Pigeon racing is an increasingly popular sport, and large international competitions can attract participants from all over China (Celia, 2014). For example, the Golden Island One-loft Race in China has a distance of 310 miles and prize money of 4 million Euros (Jerolmack, 2007; Proskura et al., 2014). However, racing pigeons may be exposed to a variety of wild birds, free-ranging poultry, and contaminated environments during free-flight training and may become infected with PPMV-1 (Celia, 2014). Birds from different breeders and different regions are brought together for long-distance races, which not only promotes the spread of pathogens, but the stress of transport often leads to vaccination failure (Liu et al., 2015a). The most commonly used live vaccines La Sota and Clone-30 were Genotype II of Class II (Liu et al., 2015a). These vaccines do not protect vaccinated individuals from infection and viral shedding in chickens (Kapczynski and King, 2005). The outbreaks of ND in pigeons may be due to significant biological, serological and genetic differences between the prevalent NDV strain and the current vaccine strain (Zhang et al., 2010; Liu et al., 2015a). Numerous studies have shown that the use of vaccines genotypically matched to prevalent strains provides better control of ND by reducing viral shedding in infected birds (Miller et al., 2007; Hu et al., 2009; Xiao et al., 2012; Roohani et al., 2015; Liu et al., 2015b; Sun et al., 2017). The immunogenicity of pigeon-derived Genotype VI of NDV differs from the chicken-origin vaccine strain La Sota (He et al., 2020; Xie et al., 2020). However, the protective properties of PPMV-1 as an inactivated vaccine and commercial vaccine La Sota in pigeons are less known. Therefore, the development of a usable PPMV-1 vaccine has great commercial value to prevent ND in meat pigeons as well as racing pigeons.
In this study, a GS02 strain of PPMV-1 was isolated from a racing pigeon loft in 2020. The pathogenicity and whole genome sequencing of GS02 were determined. An inactivated vaccine candidate of GS02 was prepared and compared with the inactivated vaccine La Sota in immunization and challenge experiments in pigeons. Both vaccines showed effective protection, and the GS02 candidate induced higher antibody titers than the La Sota vaccine. These results suggested that inactivated pigeon-derived Genotype VI could be a potential vaccine for the prevention and control of PPMV-1.
MATERIALS AND METHODS
Ethics Statement
This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the Ministry of Science and Technology of the People's Republic of China. All animal experiments were reviewed and approved by the Animal Ethics and Welfare Committee of Northwest A&F University, and accords with the principles of animal protection, animal welfare and ethics, as well as the relevant provisions of national laboratory animal welfare ethics (Approval number: 2021022).
Cells and Animals
BHK-21 cells (ATCC CCL-10) were cultured in Dulbecco's modified Eagle's medium (DMEM) (Life Technologies, United States) containing 10% fetal bovine serum (FBS) (SeraPro, Germany) and 1% penicillin-streptomycin (P/S). All 9-day-old specific pathogen-free (SPF) embryonated chicken eggs and 1-day-old SPF chickens were supplied from Yangling Green Bio-Engineering Co., Ltd. (Yangling, China). All pigeons were purchased from a pigeon farm in Yangling, China and were housed in isolators throughout the experiments. Adequate food and drinking water were provided. All pigeons were confirmed without antibodies against NDV by performing hemagglutination inhibition (HI) tests on the days before the experiments.
Virus Isolation and Identification
There were outbreaks of suspected ND in a racing pigeon loft in Gansu Province, China, on September 2020. The brains, livers and kidneys of dead pigeons were collected and homogenized. Then, the samples were centrifuged after freeze–thawing 3 times to obtain the supernatants. BHK-21 cells in 6-well plates were infected with the supernatants to observe cytopathic effects (CPEs), including syncytium formation. Total RNA was extracted from the sample supernatants of infected BHK-21 cells and allantoic fluids of 9-day-old embryonated chicken eggs by TRI Gene Reagent (GenStar, China) according to the manufacturer's instructions. The DNA was synthesized using a StarScript II First-strand cDNA Synthesis Kit (GenStar, China). The detection genes were amplified from the cDNA by PCR utilizing Taq DNA Polymerase (GenStar, China), and the primers were designed according to the conserved sequence of PPMV-1. The sequence of the forward primer was 5’-ATGGGCYCCAGAYCTTCTAC-3’, and the reverse primer was 5’-CTGCCACTGCTAGTTGTGATAATCC-3’ (designed in the current study). The program was as follows: 95°C for 5 min; 35 cycles of denaturation at 95°C for 30 s, annealing at 55°C for 30 s, an extension at 72°C for 30 s, and a final extension at 72°C for 10 min. The PCR products were visualized by 1% agarose gel electrophoresis and sequenced.
Plaque Purification
To purify the isolated virus, the diluted supernatant was infected with BHK-21 cells and then covered with 1% methylcellulose. After the plaques grew to the appropriate size, selected single plaques were inoculated into the allantoic cavity of 9-day-old SPF embryonated chicken eggs, and the above procedure was repeated 3 times. The isolate was designated PPMV-1/pigeon/Gansu/China/02/2020 and abbreviated as GS02.
Transmission Electron Microscopy (TEM)
Allantoic fluids containing purified virus were centrifuged at 6000 rpm for 20 min, and 20 µl of supernatant was used for TEM imaging. Samples were placed on paraffin film, and 150-mesh carbon-coated copper grids were placed over drops for 3 to 5 min. Then, they were washed 5 times with distilled water and stained with 2% phosphotungstic acid on a copper grid for 1 to 2 min. The samples were covered with filter papers to absorb excess liquids and dried at room temperature. The cuprum grids were observed, and images were taken under TEM (HT7800/HT7700, HITACHI).
Biological Virulence Assessment
The virulence of the isolate was determined by the mean death time (MDT) test on 9-day-old SPF embryonated chicken eggs and ICPI tests on 1-day-old SPF chickens according to standard assay methods (OIE, 2021). Proliferation of this virus strain in chicken embryos was achieved by inoculating 9-day-old embryonated chicken eggs with 1000 PFU. Allantoic fluids were collected every 24 h until 72 h. The viral titers at each time point were determined by a hemagglutination (HA) test.
Whole Genome Sequencing
RNA was extracted from allantoic fluids containing purified virus using TRIGene and prepared for next-generation sequencing. Briefly, reverse transcription used random hexamers. Subsequent DNase treatment and cleanup was followed by second-strand synthesis before library preparation using Nextera XT reagents and sequencing on the NovaSeq 6000 (Illumina, Yangzhou, China). Although originally described as a consensus-level sequencing methodology, the depth of coverage was such that deep sequencing analysis could also be carried out. Bioinformatics analysis of the data was completed using the pipeline previously described.
Read quality trimming was performed using Skewer (https://sourceforge.net/projects/skewer) (Jiang et al., 2014) with an additional trimming filter for unreliable sequences after a user-specified quality score. Host read subtraction by read-mapping was performed by using the BWASW program (Li and Durbin, 2009) against ribosomal RNAs (16, 18, 23, 28, 5S, and internal transcribed rRNA spacers were retrieved from the following ftp site), bacterial genome sequences and the latest host organism genome sequences. We then used SPAdes and MEGAHIT software to de novo assemble the reads obtained after removal of the abovementioned contamination sequence. The de novo assembly followed the A5-miseq pipeline (Coil et al., 2015). The final scaffolds were subjected to bwasw read mapping and a mega blast homology search against the NCBI NT database. The complete genome of the isolate was submitted to GenBank (ID: OM640464).
Phylogenetic Analyses
A total of 44 complete amino acid sequences of the F and HN genes of PPMV-1 were collected from GenBank (Supplementary Table S1). Molecular evolutionary genetic analysis (MEGA 7.0) software was used to conduct multiple amino acid sequence alignment using the ClustalW algorithm. The phylogenetic tree based on the amino acid sequences of the F and HN genes was constructed by using the maximum likelihood method with the JTT + G substitution model (Jones et al., 1992) by using the genotype II NDV strain La Sota (GenBank: AF077761.1) as an outgroup. The tree was drawn to scale with branch lengths measured in the number of substitutions per site. The statistical significance of the phylogenetic tree was assessed with a bootstrap value of 1000. The accession numbers of these NDVs used for phylogenetic analysis are shown in the phylogenetic trees. Evolutionary analyses were conducted in MEGA 7.0 (Kumar et al., 2016).
Challenge Experiment and Viral Load
To further determine the pathogenicity of the virus, 4-week-old pigeons were randomly divided into 2 groups: 8 in the infected group and 3 in the control group. Twelve-week-old pigeons were grouped in the same way as the 4-week-old pigeons. All infected groups were inoculated with 106 PFU of the GS02 virus in a 100-µl volume by intramuscular injection. Additionally, the negative control group received the same volume of phosphate-buffered saline (PBS) solution at pH 7.2. Subsequently, all pigeons were observed daily for clinical signs, and clinical symptoms, mortality, and morbidity were recorded. Oropharyngeal and cloacal swabs were collected at 3, 6, 9, 12, and 15 d postinfection (dpi) and suspended in 800 µl of PBS with 20% glycerol and antibiotics (P/S, 4000 U/ml). Virus titers were determined by tissue culture infectious dose (TCID50) assay in BHK-21 cells. Serum samples from all surviving pigeons at 3, 6, 9, 12, and 15 dpi were collected for serological testing by the HI test with GS02. Tissue samples were collected and observed for pathological changes. A portion of each tissue was fixed with 4% paraformaldehyde fixative and made into paraffin sections for hematoxylin-eosin (HE) staining. The other portion of the tissues was subjected to TCID50 assay.
Preparation of Inactivated Vaccine Candidates
The virus was propagated in 9-day-old SPF chicken embryos. The allantoic fluids were collected and centrifuged at 6000 rpm for 20 min. The supernatants were taken and ultracentrifuged as described below. We added 30% sterile sucrose to the bottom of the centrifuge tube and slowly added the allantoic fluid at a ratio of 1:2. Subsequently, the samples were centrifuged at 20,000 rpm for 2 h at 4°C. The precipitates were resuspended in sterile PBS, diluted tenfold and inoculated with 100-µl of each SPF embryonated egg, with the virus titers were determined by 50% egg infectious dose (EID50). Formaldehyde solution was added to viral suspensions (EID50 = 108.35) to make final formaldehyde concentrations of 0.1% that were incubated at 37°C for 24 h. To test the viability of the inactivated virus, 2 blind passages were performed in 9-day-old SPF embryonated eggs via allantoic cavity inoculation. During each passage, the inactivated virus was inoculated into 5 eggs. Embryonic mortality was monitored for 6 d before harvesting allantoic fluids for the next passage or storage. Under the condition that all eggs survived, the inactivated viral suspensions were filtered and desterilized through a sterile 0.22-µm needle-type filter. The inactivated viral suspensions with 4% were mixed with 96% sterilized Tween-80 solution for preparation of the aqueous phase. The oil phase was prepared by mixing 94% white oil-10 and 6% Span-80, adding aluminum stearate at a final concentration of 2%, and then sterilizing with an autoclave. The 25% water phase and 75% oil phase were fully emulsified by an emulsion machine. At the same time, oil adjuvant without inactivated virus was prepared as a negative control. All of the above formulations were inoculated on different plate cultures for sterilization tests. Stability testing of emulsions involves determination of stability during long-term storage at 4°C and 25°C. The stored vaccine was centrifuged at 3000 rpm for 30 min to observe for stratification.
The Vaccination-Challenge Trial
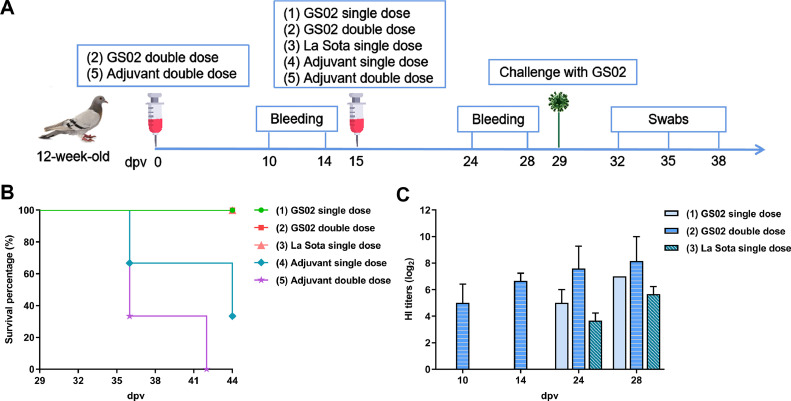
A total of 24 pigeons at 12-week-old were randomly divided into 5 groups: Groups 1 to 3 were vaccinated with 6 pigeons each, and Groups 4 and 5 were negative control groups injected with oil adjuvant of 3 pigeons each. On Day 0, Group 2 was vaccinated with the inactivated GS02 vaccine candidate, and Group 5 was vaccinated with virus-free oil adjuvant. At 15 d postvaccination (dpv), Groups 1 to 2 received inactivated GS02 vaccine candidates, and Group 3 received inactivated commercial NDV (La Sota) vaccine (EID50 = 108.5, purchased from Yangzhou Weike Biological Engineering Co., Ltd.), and Groups 4 and 5 received virus-free oil adjuvant. All pigeons were vaccinated with 0.5 ml by subcutaneous injection in the neck (He, 2012; Aljumaili et al., 2020). Groups 2 and 5 received 2 immunizations, and the remaining groups received only a single immunization. Group 5 is the control of Group 2. The pigeons were later challenged with 106 PFU of GS02 by intramuscular injection at 29 dpv. The timeline of immunization and challenge is shown in Figure 7A. All pigeons were observed daily for clinical signs, and clinical symptoms, mortality, and morbidity were recorded. Serum samples were collected at 10, 14, 24, and 28 dpv and analyzed using the HI test with 4 HA units of antigens GS02 and La Sota. Oral and cloacal swabs were collected at 32, 35, and 38 dpv. Viral shedding was detected by TCID50 and HA assays of allantoic fluids after inoculation of embryonated chicken eggs.
Figure 7.
Protective effects of inactivated GS02 vaccination on pigeons. (A) The timeline of immunization with different vaccines and challenges with the GS02 strain. (B) Survival curve of immunized pigeons after challenge with the GS02 virus. (C) HI titers of pigeons immunized with different vaccines (with 4 HA units of antigens GS02). GS02, Gansu/China/02/2020; HA, hemagglutination; HI, hemagglutination inhibition.
Cross-Inhibition Testing
The R-value of serum cross-inhibition was calculated using the formula from a previous report (Archetti and Horsfall, 1950). An R-value closer to 1 indicates a greater correlation between serotypes. An R-value greater than 0.8 indicates the same serotype, whereas an R-value lower than 0.1 indicates a different serotype. An R-value between 0.8 and 0.1 indicates a different subserotype.
Statistical Analysis
All experimental data were analyzed by an unpaired Student's t test using Prism 8.0 software (GraphPad Inc., San Diego, CA). The values were expressed as the mean ± standard deviation (SD), and the significant differences were assigned to P values < 0.05.
RESULTS
Outbreak and Virus Isolation
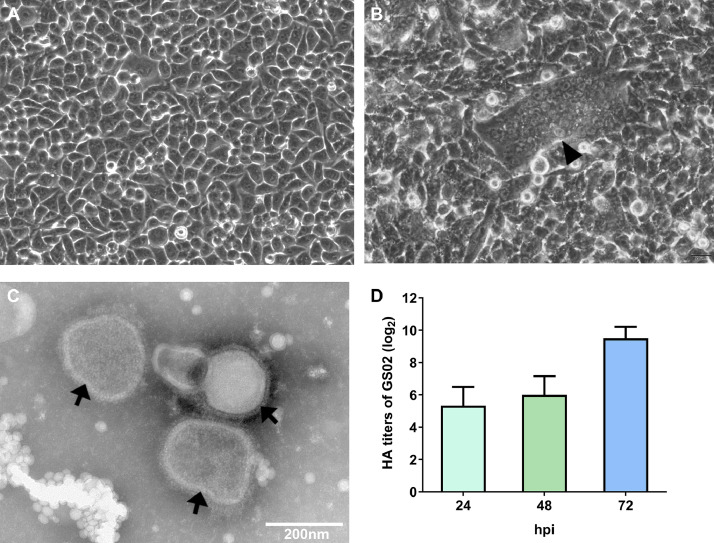
In September 2020, some racing pigeons with sour liquid in the mouth, green feces, crooked neck, paralyzed legs and even death in serious cases appeared in a racing pigeon loft in Gansu, China. The mortality rate was 10%, and morbidity was 20%, with young pigeons accounting for most of the cases. The main pathological changes found after the dissection of the sick and dead pigeons were inflammation of the intestinal mucosa, bleeding of the glandular gastric mucosa, redness and swelling of the lungs and bleeding spots in the trachea. Based on the clinical symptoms and pathological changes, the disease was suspected to be ND. Then, the observation was confirmed for NDV infection by RT-PCR. Before this outbreak, the racing pigeons in the loft were vaccinated with a chicken conventional inactivated-vaccine La Sota, which was originally isolated from chickens. The preliminary serological test of several sick racing pigeons revealed that HI titers of 21 to 23 units showed low levels of antibody. Following infection of BHK-21 cells with tissue homogenates from the dead pigeons, we observed a CPE. The syncytia were obviously observed (Figures 1A and 1B). After plaque purification 3 times on BHK-21 cells and inoculation into the allantoic cavity of SPF embryonated chicken eggs, one PPMV-1 strain was identified by RT–PCR assays. The morphology of viral particles by TEM showed that GS02 was a spherical or irregular shape with a diameter of 100 to 400 nm and with a vesicle membrane and spikes on the surface (Figure 1C), which was a typical paramyxovirus particle. The proliferation of this strain in chicken embryos reached 29.5 HA units at 72 h postinfection (hpi), as shown in Figure 1D, indicating that this virus has the ability to effectively grow.
Figure 1.
Isolation and proliferation of the PPMV-1 GS02 virus. (A) Negative control of BHK-21 cells. (B) Syncytium formation (black solid arrowhead) in brain tissue homogenates infected with BHK-21 cells 48 hpi. (C) Transmission electron microscope images of allantoic fluids of GS02 (black solid arrows). (D) HA titers of GS02 proliferation in embryonated chicken eggs. GS02, Gansu/China/02/2020; HA, hemagglutination; hpi, hours postinfection; PPMV-1, pigeon paramyxovirus type 1.
Sequence Analysis of the GS02 Strain
Next-generation sequencing of GS02 yielded a total of 7,621,672 raw reads, of which 7,209,420 reads passed quality filtering. After removing the host, bacterial and rRNA reads and performing genome assembly, 2,508,679 reads were mapped. We used SPAdes and MEGAHIT software to de novo assemble the reads to obtain the whole genome sequence of the isolate, which was designated PPMV-1/pigeon/Gansu/China/02/2020 (GS02) under GenBank accession number OM640464. The full-length genome of GS02 was 15,192 bp, and its gene order was 5’-NP-P-M-F-HN-L-3’. The amino acid analysis was deduced at the F protein cleavage site of GS02 with multiple basic amino acids 112RRQKRF117, which is typical for virulent NDV.
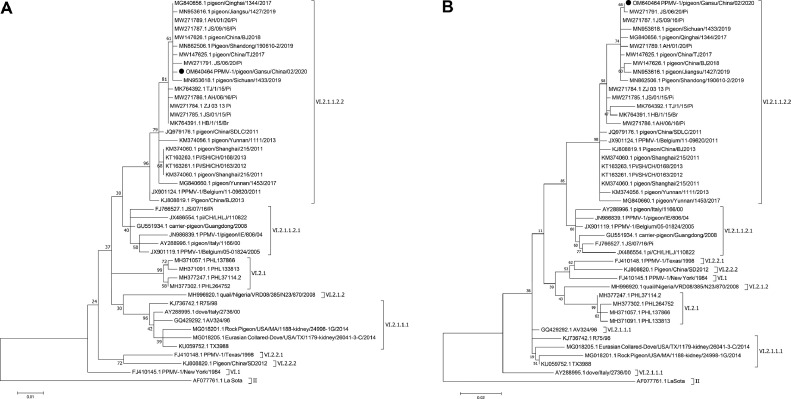
To investigate the evolutionary relationships between isolate GS02 and other PPMV-1 strains, we performed a genetic analysis. The phylogenetic trees were constructed based on the amino acid sequences of the complete F and HN genes using MEGA 7.0 software. According to the recently reported proposed NDV classification nomenclature system by Dimitrov et al. (Dimitrov et al., 2019), as shown in Figure 2A, GS02 was classified into subgenotype VI.2.1.1.2.2 of Class II. It was also classified into VIk based on another classification system by Diel et al. (Diel et al., 2012; He et al., 2020). Although NDV is evolving and the classification system of Diel et al. is no longer perfect, Dimitrov et al. updated the NDV classification system by identifying 3 new Class II genotypes and reducing the number of subgenotypes. According to the 2 classification systems, the remaining 44 PPMV-1 strains in the phylogenetic trees were shown in Supplementary Table S1. The majority of PPMV-1 strains isolated from China after 2011 were identified as VI.2.1.1.2.2. The evolutionary tree constructed from the amino acid sequences of F and HN showed that GS02 was most closely related to the JS/06/20/Pi (MW271791) strain (Figures 2A and 2B). The F protein has 3 different amino acid sites, T20A, L28S and V121I, and HN has only one amino acid site, A4V. By comparing the amino acid sequence identities of F and HN between GS02 and La Sota, we found 88% and 87.9%, respectively, and the GS02 strain is more genetically distant from the vaccine strain La Sota commonly used for chickens.
Figure 2.
Phylogenetic analysis of GS02. The phylogenetic tree of the amino acid sequences of F (553 aa) (A) and HN (571 aa) (B). The trees were constructed by the JTT + G substitution model and 1000 bootstrap replicates using the maximum likelihood method in MEGA 7.0 software. The isolate in this study is indicated by a black solid circle. F, fusion, GS02, Gansu/China/02/2020; HN, hemagglutinin-neuraminidase.
Virulence and Pathogenicity of GS02
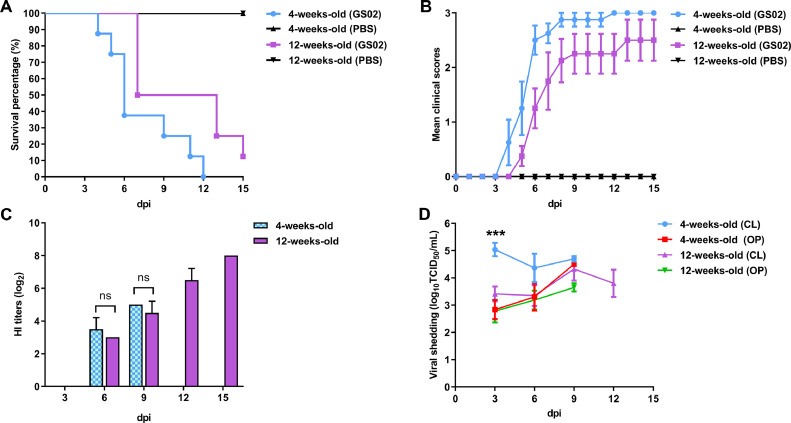
To determine the virulence of GS02, the MDT and ICPI of the GS02 strain were 72 h and 1.36, respectively, indicating that GS02 was moderately virulent. Furthermore, we infected pigeons of different ages with GS02 to evaluate pathogenicity at a dose of 106 PFU with daily observations, which revealed that all eight 4-week-old pigeons died within 12 dpi, but one 12-week-old pigeon still survived until 21 dpi, i.e., the mortality rate was 100% for young pigeons and 87.5% for adults (Figure 3A). In the infected group of 4-week-old pigeons, one pigeon suddenly died, and one pigeon with severe symptoms, such as paralysis, was observed at 4 dpi; 2 pigeons died, and 2 pigeons exhibited severe clinical signs, including head tremors, crooked necks, and unilateral or bilateral wing paralysis, at 5 dpi. Twelve-week-old pigeons were first observed clinically at 5 dpi. Some pigeons in 2 age groups showed swollen eyelids, mild conjunctivitis, and sunken eyeballs. Seven out of eight 12-week-old pigeons died during the whole experiment, but all 4-week-old pigeons died (Figure 3B). As expected, no clinical signs were observed in the control group. The results indicated that GS02 was virulent in pigeons. To evaluate the immune responses of the infected pigeons, serum samples were collected from each surviving pigeon at 3, 6, 9, 12, and 15 dpi. All serum samples were negative at 3 dpi. As shown in Figure 3C, the HI titers of the surviving 4-week-old and 12-week-old pigeons at 6 and 9 dpi were almost the same, after which the antibody titers of the surviving 12-week-old pigeons gradually increased, reaching 9.50 (log2) HI units in the 2 surviving 12-week-old pigeons at 15 dpi.
Figure 3.
Pathogenicity of GS02 in pigeons. (A) Survival curves of 4-week-old and 12-week-old pigeons. (B) Clinical scores of the infected pigeons. The clinical score was calculated as follows: 0, normal; 1, sick; 2, paralysis/torticollis/wing drop/incoordination; 3, death. The mean scores per group per day are shown. (C) Serum antibody HI titers of GS02-infected pigeons. (D) Viral shedding of the GS02 virus in cloacal and oral swabs of pigeons. Swabs collected every 3 d were used for virus titration in BHK-21 cells. CL, cloacal swabs; HI, hemagglutination inhibition; GS02, Gansu/China/02/2020; OP, oropharyngeal swabs. ns P > 0.05, *** P < 0.001.
To determine viral shedding in inoculated pigeons, oropharyngeal and cloacal swabs were collected from all surviving pigeons. Viral shedding could be detected in both oropharyngeal and cloacal swabs of inoculated 4- and 12-week-old pigeons during the early experimentation period (Table 1). At 3 dpi, the cloacal swab virus shedding rate was significantly higher in 4-week-old pigeons than in oral swabs in 4-week-old pigeons (P < 0.001) and significantly higher than cloacal and oral swabs in 12-week-old pigeons (P < 0.001; Figure 3D).
Table 1.
Viral shedding in GS02-infected pigeons.
| Group | 3 dpi |
6 dpi |
9 dpi |
12 dpi |
15 dpi |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| CL | OP | CL | OP | CL | OP | CL | OP | CL | OP | |
| (1) 4-week-old with GS02 | 8/8 | 6/8 | 3/3 | 2/3 | 2/2 | 1/2 | - | - | - | - |
| (2) 4-week-old with PBS | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 |
| (3) 12-week-old with GS02 | 8/8 | 4/8 | 8/8 | 5/8 | 2/4 | 1/4 | 1/4 | 0/4 | 0/1 | 0/1 |
| (4) 12-week-old with PBS | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 |
Abbreviations: CL, cloacal swabs; dpi, day postinfection; GS02, Gansu/China/02/2020; OP, oropharyngeal swabs; -, all pigeons died.
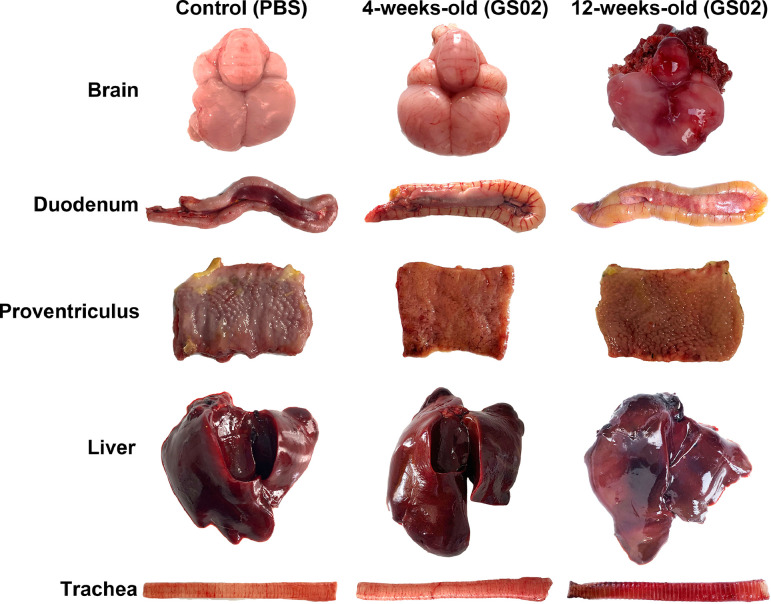
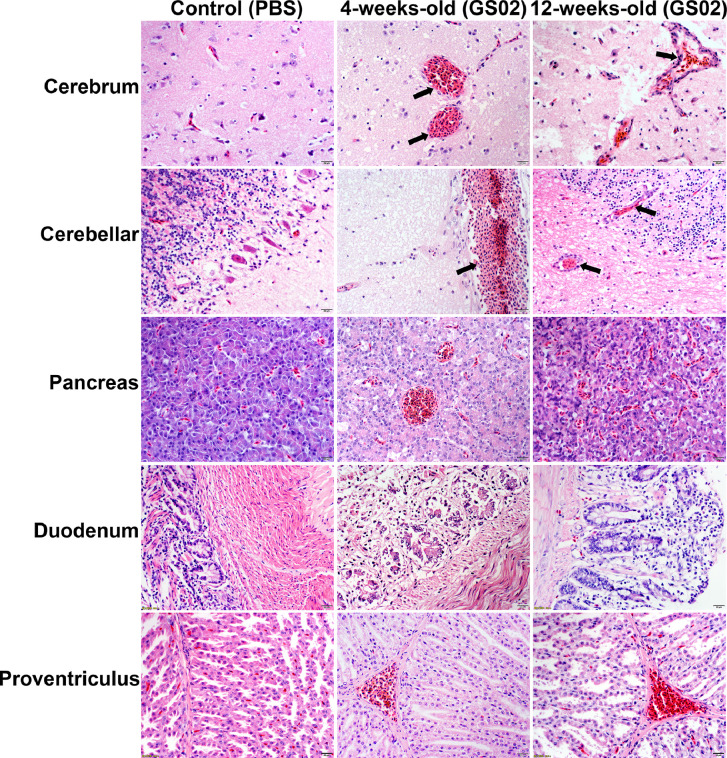
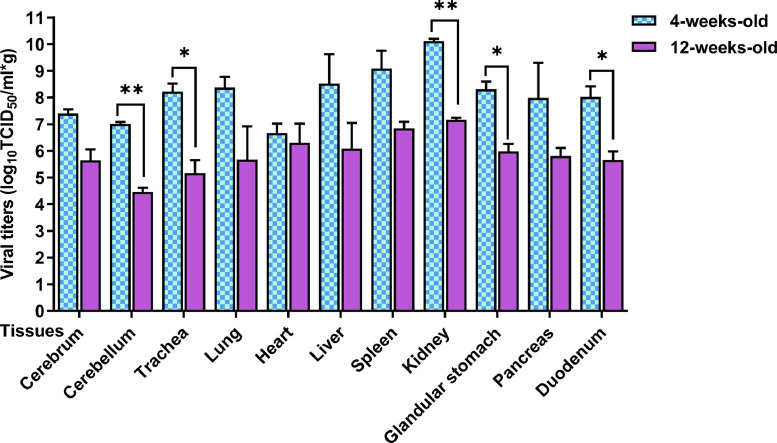
Dead pigeons were used for gross pathology observation, and tissue samples were collected for histopathological analysis. On gross examination, there were foci of hemorrhage in the brain; congestion, hemorrhage and necrosis in the pancreas; hemorrhage from the glandular gastric papillae; swollen and friable liver with patchy hemorrhage; and reddening and swelling of the trachea (Figure 4). The pigeons in the control group did not exhibit gross lesions. Histopathological results indicated that GS02 caused the following moderate to severe tissue pathological changes in 5 sampled tissues: perivascular lymphocytic cuffing in the cerebral and cerebellum cortex; degeneration and necrosis of pancreatic cells; vacuolar degeneration of intestinal villous epithelial cells; and hemorrhage, degeneration and necrosis of glandular gastric connective tissue (Figure 5). The age of the pigeon did not seem to be strongly associated with the types of tissue lesions. The tissue samples of 4- and 12-week-old pigeons from the cerebrum, cerebellum, trachea, lungs, heart, liver, spleen, kidney, glandular stomach, pancreas and duodenum were used to determine the virus titers (Figure 6). Tissue virus titers in the cerebellum (P < 0.01), trachea (P < 0.05), kidneys (P < 0.01), glandular stomach (P < 0.05), and duodenum (P < 0.05) were significantly higher in 4-week-old pigeons than in 12-week-old pigeons. The results indicated that the virus could effectively replicate in different tissues of pigeons, and the virus proliferates faster in young pigeons.
Figure 4.
Gross lesions of the organs in the 4-week-old and 12-week-old pigeons. Lesions are as follows: Foci of hemorrhage in the brain. Congestion, hemorrhage and necrosis in the pancreas. Hemorrhage from the glandular gastric papillae. Swollen and friable liver with patchy hemorrhage. Reddening and swelling of the trachea.
Figure 5.
Histopathology of GS02-infected pigeons. Pigeon tissues were fixed with 4% paraformaldehyde fixative, sectioned and stained with hematoxylin and eosin. The histological lesions were as follows: perivascular lymphocytic cuffing in the cerebral and cerebellar cortex (black arrows). Degeneration and necrosis of pancreatic cells. Vacuolar degeneration of intestinal villous epithelial cells. Hemorrhage, degeneration and necrosis of glandular gastric connective tissues. GS02, Gansu/China/02/2020.
Figure 6.
Viral loads in different organs of GS02-infected pigeons. Virus titers of different tissues from dead pigeons (log10 median TCID50/ml) were quantified by serial end-point dilution in 96-well plates using BHK-21 cells. The asterisk indicates a significant difference in tissue virus titers between 4- and 12-week-old pigeons. * P < 0.05, ** P < 0.01. GS02, Gansu/China/02/2020; TCID50, tissue culture infectious dose.
Protective Effects of the Inactivated GS02 Vaccine Candidate
To prepare the inactivated GS02 vaccine, the virus was incubated with 0.1% formaldehyde for 24 h. The inactivated virus was monitored by inoculated embryonic mortality. The inactivated vaccine fully emulsified the water phase with the oil phase and passed the sterility test. The water/oil vaccine preparation formed an emulsion that remained intact for more than 3 months at 25℃ or 4℃. These results indicate the stability of the inactivated vaccine emulsified in the oil phase.
Groups 1 and 2 were vaccinated with the GS02 vaccine candidate, Group 3 was vaccinated with inactivated La Sota vaccine, and Groups 4 and 5 were vaccinated with oil adjuvant. Groups 2 and 5 were vaccinated twice, and the remaining groups were vaccinated once. After vaccination of pigeons by subcutaneous injection in the neck, no local or systemic reactions were observed in Groups 1 to 5 of the pigeons. Importantly, all pigeons vaccinated with inactivated vaccine (Groups 1–3) were healthy after the challenge (Figure 7B), and none were found to shed the virus, as shown in Table 2. These results indicated that the inactivated vaccine provided full immune protection. The pigeons in Groups 4 and 5 infected with virus-free oil adjuvant showed obvious clinical signs, and the mortality rates were 66.7% and 100%, respectively, after the challenge (Figure 7B). High levels of antibody titers in Groups 1 to 3 were induced according to results of HI test using GS02 antigen after vaccination (Figure 7C). Importantly, inactivated GS02 vaccination induced higher antibody titers than the commercial inactivated La Sota vaccine, suggesting that the PPMV-1-matched vaccine could elicit a better humoral response. Pigeons in Group 2 showed an antibody titer of 28.20 after 2 immunizations with the GS02 candidate, showing a higher antibody level compared to Group 1, but all had a 100% survival rate after the challenge, which indicated that our candidate vaccine was able to induce a positive immune response. The serum cross-inhibition test confirmed a modest difference in antigenicity between GS02 and La Sota, as the R-value was 0.5, indicating an antigenic difference between GS02 and La Sota viruses within one serotype.
Table 2.
Viral shedding in GS02 postchallenged pigeons.
| 32 dpi |
35 dpi |
38 dpi |
||||
|---|---|---|---|---|---|---|
| Group | CL | OP | CL | OP | CL | OP |
| (1) GS02 single dose | 0/6 | 0/6 | 0/6 | 0/6 | 0/6 | 0/6 |
| (2) GS02 double dose | 0/6 | 0/6 | 0/6 | 0/6 | 0/6 | 0/6 |
| (3) La Sota single dose | 0/6 | 0/6 | 0/6 | 0/6 | 0/6 | 0/6 |
| (4) Adjuvant single dose | 3/3 | 1/3 | 3/3 | 3/3 | 1/2 | 0/2 |
| (5) Adjuvant double dose | 3/3 | 2/3 | 3/3 | 1/3 | - | - |
Abbreviations: CL, cloacal swabs; dpi, day postinfection; GS02, Gansu/China/02/2020; OP, oropharyngeal swabs; -, all pigeons died.
DISCUSSION
In recent years, outbreaks of PPMV-1 have commonly occurred in racing pigeon lofts in China. We isolated the PPMV-1 strain GS02 from an NDV-vaccinated loft in Gansu Province and GS02 showed pathogenicity to pigeons. GS02 belonged to subgenotype VIk and subgenotype VI.2.1.1.2.2 of Class II classified by phylogenetic analysis that differed from the commercial vaccine strain La Sota of Genotype II. Although the prepared inactivated vaccine candidate GS02 and commercial vaccine La Sota effectively protected against GS02 challenge in pigeons, the antibody level induced by the GS02 candidate was higher than that induced by the La Sota vaccine. The difference in the antigenicity of GS02 and La Sota implied that the development of a genotype-matched vaccine has potential for use in pigeons.
Despite the La Sota vaccine used in the outbreak of ND in a racing pigeon loft in Gansu Province, PPMV-1 still caused morbidity and mortality in this study. The reasons might be due to low antibody levels against PPMV-1. The low antibody level (21–23 HI units) could be influenced in practice by individual differences in pigeons, local inoculation operation, vaccine validity, dosage, etc. The commonly used vaccine La Sota is genotype II, but PPMV-1 belongs to Genotype VI. Comparing the immune efficacies of the La Sota vaccine and GS02 vaccine candidates, the GS02 candidate induced a higher level of antibody in pigeons than the La Sota vaccine. The serum cross-reactivity assay showed an R-value of 0.5, indicating slight antigenic differences between GS02 and La Sota. Previous studies showed insufficient protection rates of the commercial La Sota inactivated vaccine against PPMV-1 ND32 strain with 50% (Ye et al., 2012) and PL strain with 65% (He, 2012) in 5-week-old young pigeons, but the protection rates of PPMV-1 candidate vaccines were 100% (He, 2012; Ye et al., 2012). In addition, numerous studies have shown that antigenic matching between vaccine strains and prevalent strains can increase the effectiveness of the vaccine in chickens (Kim et al., 2008; Hu et al., 2009; Liu et al., 2015a). Thus, using a genotype-matched vaccine is a potent choice for the prevention and control of PPMV-1 in pigeons. Here having a conflict that we found the La Sota vaccine providing fully protection in pigeons comparing to previous reports (He, 2012; Ye et al., 2012). Several factors might account for the differences in protection rate of La Sota vaccine between our study and previous reports: age of pigeon, challenge route, challenge dose and the strain. We speculated that the age of pigeons might be an important role in the protection rate. The challenge experiments in 4-week-old young pigeons or various age pigeons need to be investigated to clarify this hypothesis in future.
The assessment of NDV virulence usually follows the OIE protocol in chickens. PPMV-1, a pigeon-derived pathogen, may not be suitable for assessment in chickens. The MDT and ICPI of the GS02 strain were 72 h and 1.36, respectively, indicating that it is moderately virulent to chickens. However, the GS02 strain was found to be virulent to pigeons based on challenge tests. This suggests that the virulence of NDV strains is determined by MDT and ICPI, and clinical studies of hosts susceptible to the genotype to accurately characterize viral pathogenicity. This is consistent with previous studies showing that the pathogenicity of PPMV-1 varies between pigeons and chickens. Highly virulent PPMV-1 isolated from pigeons often shows weak or even nonvirulence in chickens (Guo et al., 2014; Heiden et al., 2014; Wang et al., 2017). However, the PPMV-1 strain became more virulent when tested for pathogenicity and clinical symptoms after successive passages in chickens that caused ND (Alexander, 2011; Dortmans et al., 2011). These studies, including our study, concluded that pigeons should be used for pathogenicity testing of PPMV-1 to assess real virulence. Moreover, the pathogenicity of PPMV-1 could be age-dependent in pigeons (Guo et al., 2014; Ren et al., 2017; Xiang et al., 2019). Our study analyzed the differences in PPMV-1 pathogenicity in different-aged pigeons, and the mortality rates of young pigeons and adults were 100% and 87.5%, respectively, indicating that the GS02 strain is more susceptible and pathogenic for young pigeons, which is consistent with the clinical cases appearing in racing pigeon lofts in Gansu.
In past decades, the genotypes of the known transmitted NDV strains have undergone major shifts, although they remain as a single serotype (Kapczynski et al., 2013). The main subgenotypes of PPMV-1 affecting the pigeon breeding industry in China are VI.1 and VI.2.1.1.2.2. These viruses were originally identified in China in 2005 and have been isolated in at least 20 provinces (He et al., 2020). These 2 subgenotypes of PPMV-1 could have been introduced into China from Europe (Cai et al., 2011; Kapczynski et al., 2013; He et al., 2020). According to Figures 2A and 2B, the GS02 strain was genetically closest to the strains isolated in East China, suggesting that PPMV-1 may have spread from East China to Northwest China. Migration of birds, phylogeographic analysis, and monitoring of PPMV-1 in wild birds will contribute to a more detailed study of PPMV-1 epidemiology.
In conclusion, we isolated a VIk subgenotype of the PPMV-1 strain from a racing pigeon loft in Gansu Province that was virulent in pigeons. Four-week-old young pigeons could be susceptible to GS02 infection. The immunization experiments of a prepared inactivated GS02 vaccine candidate and La Sota vaccine in pigeons showed effective protection, and the GS02 candidate could induce high levels of antibodies. This study provides a potential application for the development of a genotype-matched vaccine to prevent PPMV-1 in pigeons.
ACKNOWLEDGMENTS
This work was partially supported by the National Natural Science Foundation of China (31972662 to SX), and the Foundation of Yangling Vocational and Technical College (A2019071 to JB). We thank Man Zhang of Yangling Vocational and Technical College for primary survey in racing pigeon lofts.
DISCLOSURES
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper. Sa Xiao reports financial support was provided by Northwest Agriculture and Forestry University. Jun Bai reports financial support was provided by Yangling Vocational and Technical College.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.psj.2022.102097.
Appendix. Supplementary materials
REFERENCES
- Alexander D.J. Gordon memorial lecture. Newcastle disease. Brit. Poult. Sci. 2001;42:5–22. doi: 10.1080/713655022. [DOI] [PubMed] [Google Scholar]
- Alexander D.J. Newcastle disease in the European Union 2000 to 2009. Avian Pathol. 2011;40:547–558. doi: 10.1080/03079457.2011.618823. [DOI] [PubMed] [Google Scholar]
- Aljumaili O.A., Bello M.B., Yeap S.K., Omar A.R., Ideris A. Protective efficacy of inactivated Newcastle disease virus vaccines prepared in two different oil-based adjuvants. Onderstepoort J. Vet. Res. 2020;87:e1–e7. doi: 10.4102/ojvr.v87i1.1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amarasinghe G.K., Ayllón M.A., Bào Y., Basler C.F., Bavari S., Blasdell K.R., Briese T., Brown P.A., Bukreyev A., Balkema-Buschmann A., Buchholz U.J., Chabi-Jesus C., Chandran K., Chiapponi C., Crozier I., de Swart R.L., Dietzgen R.G., Dolnik O., Drexler J.F., Dürrwald R., Dundon W.G., Duprex W.P., Dye J.M., Easton A.J., Fooks A.R., Formenty P.B.H., Fouchier R.A.M., Freitas-Astúa J., Griffiths A., Hewson R., Horie M., Hyndman T.H., Jiāng D., Kitajima E.W., Kobinger G.P., Kondō H., Kurath G., Kuzmin I.V., Lamb R.A., Lavazza A., Lee B., Lelli D., Leroy E.M., Lǐ J., Maes P., Marzano S.L., Moreno A., Mühlberger E., Netesov S.V., Nowotny N., Nylund A., Økland A.L., Palacios G., Pályi B., Pawęska J.T., Payne S.L., Prosperi A., Ramos-González P.L., Rima B.K., Rota P., Rubbenstroth D., Shī M., Simmonds P., Smither S.J., Sozzi E., Spann K., Stenglein M.D., Stone D.M., Takada A., Tesh R.B., Tomonaga K., Tordo N., Towner J.S., van den Hoogen B., Vasilakis N., Wahl V., Walker P.J., Wang L.F., Whitfield A.E., Williams J.V., Zerbini F.M., Zhāng T., Zhang Y.Z., Kuhn J.H. Taxonomy of the order mononegavirales: update 2019. Arch. Virol. 2019;164:1967–1980. doi: 10.1007/s00705-019-04247-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archetti I., Horsfall F.L., Jr Persistent antigenic variation of influenza A viruses after incomplete neutralization in ovo with heterologous immune serum. J. Exp. Med. 1950;92:441–462. doi: 10.1084/jem.92.5.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai S., Li J., Wong M.T., Jiao P., Fan H., Liu D., Liao M., Jiang J., Shi M., Lam T.T., Ren T., Leung F.C. Genetic characterization and evolutionary analysis of 4 Newcastle disease virus isolate full genomes from waterbirds in South China during 2003-2007. Vet. Microbiol. 2011;152:46–54. doi: 10.1016/j.vetmic.2011.04.014. [DOI] [PubMed] [Google Scholar]
- Celia A. A current review of avian influenza in pigeons and doves (Columbidae) Vet. Microbiol. 2014;170:181–196. doi: 10.1016/j.vetmic.2014.02.042. [DOI] [PubMed] [Google Scholar]
- Coil D., Jospin G., Darling A.E. A5-miseq: an updated pipeline to assemble microbial genomes from Illumina MiSeq data. Bioinformatics. 2015;31:587–589. doi: 10.1093/bioinformatics/btu661. [DOI] [PubMed] [Google Scholar]
- Collins M.S., Alexander D.J., Brockman S., Kemp P.A., Manvell R.J. Evaluation of mouse monoclonal antibodies raised against an isolate of the variant avian paramyxovirus type 1 responsible for the current panzootic in pigeons. Arch. Virol. 1989;104:53–61. doi: 10.1007/BF01313807. [DOI] [PubMed] [Google Scholar]
- Diel D.G., Susta L., Cardenas Garcia S., Killian M.L., Brown C.C., Miller P.J., Afonso C.L. Complete genome and clinicopathological characterization of a virulent Newcastle disease virus isolate from South America. J. Clin. Microbiol. 2012;50:378–387. doi: 10.1128/JCM.06018-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrov K.M., Abolnik C., Afonso C.L., Albina E., Bahl J., Berg M., Briand F.X., Brown I.H., Choi K.S., Chvala I., Diel D.G., Durr P.A., Ferreira H.L., Fusaro A., Gil P., Goujgoulova G.V., Grund C., Hicks J.T., Joannis T.M., Torchetti M.K., Kolosov S., Lambrecht B., Lewis N.S., Liu H., Liu H., McCullough S., Miller P.J., Monne I., Muller C.P., Munir M., Reischak D., Sabra M., Samal S.K., Servan de Almeida R., Shittu I., Snoeck C.J., Suarez D.L., Van Borm S., Wang Z., Wong F.Y.K. Updated unified phylogenetic classification system and revised nomenclature for Newcastle disease virus. Infect. Genet. Evol. 2019;74 doi: 10.1016/j.meegid.2019.103917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dortmans J.C., Koch G., Rottier P.J., Peeters B.P. A comparative infection study of pigeon and avian paramyxovirus type 1 viruses in pigeons: evaluation of clinical signs, virus shedding and seroconversion. Avian Pathol. 2011;40:125–130. doi: 10.1080/03079457.2010.542131. [DOI] [PubMed] [Google Scholar]
- Guo H., Liu X., Xu Y., Han Z., Shao Y., Kong X., Liu S. A comparative study of pigeons and chickens experimentally infected with PPMV-1 to determine antigenic relationships between PPMV-1 and NDV strains. Vet. Microbiol. 2014;168:88–97. doi: 10.1016/j.vetmic.2013.11.002. [DOI] [PubMed] [Google Scholar]
- He F. Gansu Agricultural University; 2012. Studies on the vaccine of PPMV-1 and cloning, expression and the biological activity of F gene segments of PPMV-1. (Thesis, in Chinese) [Google Scholar]
- He Y., Lu B., Dimitrov K.M., Liang J., Chen Z., Zhao W., Qin Y., Duan Q., Zhou Y., Liu L., Li B., Yu L., Duan Z., Liu Q. Complete genome sequencing, molecular epidemiological, and pathogenicity analysis of pigeon paramyxoviruses type 1 isolated in Guangxi, China during 2012-2018. Viruses. 2020;12:366. doi: 10.3390/v12040366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiden S., Grund C., Höper D., Mettenleiter T.C., Römer-Oberdörfer A. Pigeon paramyxovirus type 1 variants with polybasic F protein cleavage site but strikingly different pathogenicity. Virus Genes. 2014;49:502–506. doi: 10.1007/s11262-014-1111-7. [DOI] [PubMed] [Google Scholar]
- Hu S., Ma H., Wu Y., Liu W., Wang X., Liu Y., Liu X. A vaccine candidate of attenuated genotype VII Newcastle disease virus generated by reverse genetics. Vaccine. 2009;27:904–910. doi: 10.1016/j.vaccine.2008.11.091. [DOI] [PubMed] [Google Scholar]
- Jerolmack C. Animal archeology: domestic pigeons and the nature-culture dialectic. Qual. Sociol. Rev. 2007;3:74–95. [Google Scholar]
- Jiang H., Lei R., Ding S.W., Zhu S. Skewer: a fast and accurate adapter trimmer for next-generation sequencing paired-end reads. BMC Bioinform. 2014;15:182. doi: 10.1186/1471-2105-15-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones D.T., Taylor W.R., Thornton J.M. The Rapid generation of mutation data matrices from protein sequences. CABIOS, Comput. Appl. Biosci. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- Kaleta E.F., Alexander D.J., Russell P.H. The first isolation of the avian PMV-1 virus responsible for the current panzootic in pigeons ? Avian Pathol. 1985;14:553–557. doi: 10.1080/03079458508436258. [DOI] [PubMed] [Google Scholar]
- Kapczynski D.R., Afonso C.L., Miller P.J. Immune responses of poultry to Newcastle disease virus. Dev. Comp. Immunol. 2013;41:447–453. doi: 10.1016/j.dci.2013.04.012. [DOI] [PubMed] [Google Scholar]
- Kapczynski D.R., King D.J. Protection of chickens against overt clinical disease and determination of viral shedding following vaccination with commercially available Newcastle disease virus vaccines upon challenge with highly virulent virus from the California 2002 exotic Newcastle disease outbreak. Vaccine. 2005;23:3424–3433. doi: 10.1016/j.vaccine.2005.01.140. [DOI] [PubMed] [Google Scholar]
- Kim L.M., King D.J., Guzman H., Tesh R.B., Travassos da Rosa A.P., Bueno R., Jr., Dennett J.A., Afonso C.L. Biological and phylogenetic characterization of pigeon paramyxovirus serotype 1 circulating in wild North American pigeons and doves. J. Clin. Microbiol. 2008;46:3303–3310. doi: 10.1128/JCM.00644-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M., Qu Y., Wang F., Liu S., Sun H. Genotypic and pathotypic characterization of Newcastle disease virus isolated from racing pigeons in China. Poult. Sci. 2015;94:1476–1482. doi: 10.3382/ps/pev106. [DOI] [PubMed] [Google Scholar]
- Liu M.M., Cheng J.L., Yu X.H., Qin Z.M., Tian F.L., Zhang G.Z. Generation by reverse genetics of an effective attenuated Newcastle disease virus vaccine based on a prevalent highly virulent Chinese strain. Biotechnol. Lett. 2015;37:1287–1296. doi: 10.1007/s10529-015-1799-z. [DOI] [PubMed] [Google Scholar]
- Marlier D., Vindevogel H. Viral infections in pigeons. Vet. J. 2006;172:40–51. doi: 10.1016/j.tvjl.2005.02.026. [DOI] [PubMed] [Google Scholar]
- Millar N.S., Emmerson P.T. In: Newcastle Disease. Alexander D.J., editor. Springer; Boston, MA: 1988. Molecular cloning and nucleotide sequencing of Newcastle disease virus; pp. 79–97. Pages. [Google Scholar]
- Miller P.J., King D.J., Afonso C.L., Suarez D.L. Antigenic differences among Newcastle disease virus strains of different genotypes used in vaccine formulation affect viral shedding after a virulent challenge. Vaccine. 2007;25:7238–7246. doi: 10.1016/j.vaccine.2007.07.017. [DOI] [PubMed] [Google Scholar]
- Nidzworski D., Rabalski L., Gromadzka B. Detection and differentiation of virulent and avirulent strains of Newcastle disease virus by real-time PCR. J. Virol. Methods. 2011;173:144–149. doi: 10.1016/j.jviromet.2010.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OIE. 2021. Newcastle disease, chapter 3.3.14. Accessed May 2021. https://www.oie.int/en/what-we-do/standards/codes-and-manuals/terrestrial-manual-online-access/
- Panda A., Huang Z., Elankumaran S., Rockemann D.D., Samal S.K. Role of fusion protein cleavage site in the virulence of Newcastle disease virus. Microb. Pathog. 2004;36:1–10. doi: 10.1016/j.micpath.2003.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proskura W.S., Cichoń D., Grzesiak W., Zaborski D., Sell-Kubiak E., Cheng Y.-H., Dybus A. Single nucleotide polymorphism in the LDHA gene as a potential marker for the racing performance of pigeons. Poult. Sci. 2014;51:364–368. [Google Scholar]
- Ren S., Wang C., Zhang X., Zhao L., Wang X., Yao W., Han Q., Wang Y., Fan M., Gao X., Xiao S., Wang X., Yang Z. Phylogenetic and pathogenic characterization of a pigeon paramyxovirus type 1 isolate reveals cross-species transmission and potential outbreak risks in the northwest region of China. Arch. Virol. 2017;162:2755–2767. doi: 10.1007/s00705-017-3422-1. [DOI] [PubMed] [Google Scholar]
- Roohani K., Tan S.W., Yeap S.K., Ideris A., Bejo M.H., Omar A.R. Characterisation of genotype VII Newcastle disease virus (NDV) isolated from NDV vaccinated chickens, and the efficacy of LaSota and recombinant genotype VII vaccines against challenge with velogenic NDV. J. Vet. Sci. 2015;16:447–457. doi: 10.4142/jvs.2015.16.4.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steward M., Vipond I.B., Millar N.S., Emmerson P.T. RNA editing in Newcastle disease virus. J. Gen. Virol. 1993;74:2539–2547. doi: 10.1099/0022-1317-74-12-2539. [DOI] [PubMed] [Google Scholar]
- Sun M., Xiang B., Li Y., Xie P., Gao S., Kang Y., Gao P., Li Y., Wang Z., Liang J., Yu D., Ren T. Generation and evaluation of a genetically attenuated Newcastle disease virus rGM-VIIm as a genotype-matched vaccine. Virus Genes. 2017;53:35–43. doi: 10.1007/s11262-016-1397-8. [DOI] [PubMed] [Google Scholar]
- Teske L., Ryll M., Rautenschlein S. Epidemiological investigations on the role of clinically healthy racing pigeons as a reservoir for avian paramyxovirus-1 and avian influenza virus. Avian Pathol. 2013;42:557–565. doi: 10.1080/03079457.2013.852157. [DOI] [PubMed] [Google Scholar]
- Ujvári D., Wehmann E., Kaleta E.F., Werner O., Savić V., Nagy E., Czifra G., Lomniczi B. Phylogenetic analysis reveals extensive evolution of avian paramyxovirus type 1 strains of pigeons (Columba livia) and suggests multiple species transmission. Virus Res. 2003;96:63–73. doi: 10.1016/s0168-1702(03)00173-4. [DOI] [PubMed] [Google Scholar]
- Wang J., Liu H., Liu W., Zheng D., Zhao Y., Li Y., Wang Y., Ge S., Lv Y., Zuo Y., Yu S., Wang Z. Genomic characterizations of six pigeon paramyxovirus type 1 viruses isolated from live bird markets in China during 2011 to 2013. PLoS One. 2015;10 doi: 10.1371/journal.pone.0124261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Ren S., Wang X., Wang C.Y., Fan M., Jia Y., Gao X., Liu H., Xiao S., Yang Z. Genomic characterization of a wild-bird-origin pigeon paramyxovirus type 1 (PPMV-1) first isolated in the northwest region of China. Arch. Virol. 2017;162:749–761. doi: 10.1007/s00705-016-3156-5. [DOI] [PubMed] [Google Scholar]
- Xiang B., You R., Kang Y., Xie P., Zhu W., Sun M., Gao P., Li Y., Ren T. Host immune responses of pigeons infected with Newcastle disease viruses isolated from pigeons. Microb. Pathog. 2019;127:131–137. doi: 10.1016/j.micpath.2018.11.049. [DOI] [PubMed] [Google Scholar]
- Xiao S., Nayak B., Samuel A., Paldurai A., Kanabagattebasavarajappa M., Prajitno T.Y., Bharoto E.E., Collins P.L., Samal S.K. Generation by reverse genetics of an effective, stable, live-attenuated Newcastle disease virus vaccine based on a currently circulating, highly virulent Indonesian strain. PLoS One. 2012;7:e52751. doi: 10.1371/journal.pone.0052751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie P., Chen L., Zhang Y., Lin Q., Ding C., Liao M., Xu C., Xiang B., Ren T. Evolutionary dynamics and age-dependent pathogenesis of sub-genotype VI.2.1.1.2.2 PPMV-1 in pigeons. Viruses. 2020;12:433. doi: 10.3390/v12040433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye H., Qiu W., Li M., Liang Z., Wang X., Luo K. Development and immune efficacy of Newcastle disease virus vaccine for pigeon. China Poult. 2012;34:20–24. (In Chinese) [Google Scholar]
- Zhang R., Pu J., Su J., Zhao J., Wang X., Zhang S., Li X., Zhang G. Phylogenetic characterization of Newcastle disease virus isolated in the mainland of China during 2001-2009. Vet. Microbiol. 2010;141:246–257. doi: 10.1016/j.vetmic.2009.09.020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.