Abstract
Pseudomonas sp. strain TW3 is able to metabolize 4-nitrotoluene to 4-nitrobenzoate and toluene to benzoate aerobically via a route analogous to the upper pathway of the TOL plasmids. We report the cloning and characterization of a benzyl alcohol dehydrogenase gene (ntnD) which encodes the enzyme for the catabolism of 4-nitrobenzyl alcohol and benzyl alcohol to 4-nitrobenzaldehyde and benzaldehyde, respectively. The gene is located downstream of the previously reported ntn gene cluster. NtnD bears no similarity to the analogous TOL plasmid XylB (benzyl alcohol dehydrogenase) protein either in its biochemistry, being NAD(P)+ independent and requiring assay via dye-linked electron transfer, or in its deduced amino acid sequence. It does, however, have significant similarity in its amino acid sequence to other NAD(P)+-independent alcohol dehydrogenases and contains signature patterns characteristic of type III flavin adenine dinucleotide-dependent alcohol oxidases. Reverse transcription-PCR demonstrated that ntnD is transcribed during growth on 4-nitrotoluene, although apparently not as part of the same transcript as the other ntn genes. The substrate specificity of the enzyme expressed from the cloned and overexpressed gene was similar to the activity expressed from strain TW3 grown on 4-nitrotoluene, providing evidence that ntnD is the previously unidentified gene in the pathway of 4-nitrotoluene catabolism. Examination of the 14.8-kb region around the ntn genes suggests that one or more recombination events have been involved in the formation of their current organization.
Nitrotoluenes, like many xenobiotics, are candidate molecules for understanding the evolution of microbial catabolism, since they have been present in the biosphere as potential substrates for microbes for relatively short periods of time; 2- and 4-nitrotoluene and 2,4- and 2,6-dinitrotoluenes are precursors of TNT and are therefore by-products of the explosives industry. There are also very few naturally occurring nitro compounds upon which bacteria have had the opportunity to evolve a repertoire of catabolic strategies. Pathways by which nitro-substituted xenobiotics are degraded encompass a wide range of different biochemical mechanisms. Some proceed by elimination of nitrogen from the nitro groups as nitrite as a result of oxygenase-catalyzed reactions, as in the degradation of 2,4-dinitrotoluene by Burkholderia sp. strain DNT (28, 30, 31) and 2-nitrotoluene by Pseudomonas sp. strain JS42 (13, 20). Although Pseudomonas putida OU83 reduces most of 3-nitrotoluene to 3-aminotoluene, it appears to mineralize a proportion by first oxidizing its methyl group to form 3-nitrobenzoate which is then converted to 3-nitrophenol, from which nitrite is subsequently eliminated (1). In the case of 4-nitrotoluene catabolism by a Mycobacterium, the nitro group is reduced to an amino group in 6-amino-m-cresol and is eventually released as ammonium by an uncharacterized metabolic reaction (29). Pseudomonas sp. strains TW3 (23) and 4NT (12) use a different route for 4-nitrotoluene catabolism with initial steps similar to those of OU83, i.e., sequential oxidation of the methyl group, to form 4-nitrobenzoate. The nitro group is reduced through 4-nitrosobenzoate and 4-hydroxylaminobenzoate from which it is directly eliminated as ammonia. These reactions from 4-nitrobenzoate were first reported in the 4-nitrobenzoate degrader Comamonas acidovorans NBA10 (10, 11), and similar reactions have since been found in Pseudomonas sp. strain YH102 (L. M. Newman and G. J. Zylstra, unpublished data) and Ralstonia pickettii YH105 (35), but none of these utilize 4-nitrotoluene. The genes encoding the initial enzymes of the 4-nitrotoluene pathway (from 4-nitrotoluene to 4-nitrobenzoate) have been cloned from Pseudomonas sp. strain TW3 (17) and are very similar in sequence and organization to the TOL plasmid-encoded upper pathway genes which catalyze the analogous reactions on unsubstituted and methyl-substituted substrates (15). Pseudomonas sp. strain 4NT also contains a similar set of TOL plasmid-like genes (K. D. James and P. A. Williams, unpublished data). Strains TW3 and 4NT differ in the 4-nitrobenzyl alcohol dehydrogenase reaction which, in strain TW3, is NAD(P)+ independent (17, 23), whereas the activity in 4NT is NAD+ dependent like the TOL plasmid benzyl alcohol dehydrogenase XylB (26). The ntn operon in strain TW3 does contain a xylB homolog, but it is only a pseudogene containing a stop codon within the reading frame and is interrupted by the insertion of a transposable element fragment (17).
Here, we report the cloning and sequencing of a region downstream of the previously reported ntn gene cluster that includes a gene, ntnD, which encodes the missing NAD(P)+-independent alcohol dehydrogenase of the pathway.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains and plasmids used in this study are listed in Table 1. Pseudomonas sp. strain TW3 utilizes 4-nitrotoluene as its sole carbon and nitrogen source (23) and will also grow on toluene.
TABLE 1.
Bacterial strains and plasmids used in this study
| Bacterial strain or plasmid | Genotype or phenotype | Reference or source |
|---|---|---|
| Pseudomonas sp. strain TW3 | 4NT+ 4NBA+ 4NBZate+a | 23 |
| E. coli XL1-Blue MRA | Δ(mcrA)183 Δ(mcrCB-hsdSMR-mrr)173 endA1 supE44 thi-1 gyrA96 relA1 lac | Stratagene |
| E. coli XL1-Blue MRA (P2) | XL1-Blue MRA (P2 lysogen) | Stratagene |
| E. coli TOP 10 | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZM15 ΔlacX74 deoR recA1 araD139 Δ(ara-leu)7679 galU galK rpsL(Strr) endA1 nupG | Invitrogen |
| E. coli BL21(DE3)pLysS | F−ampT hsdSB(rB− mB−) dcm gal λ(DE3) pLysS(Cmr) | Promega |
| pTW3.6 | 6-kbp EcoRI clone containing ntnCMAB in pUC19 | 17 |
| pTW3.10 | 8.5-kbp XbaI/XhoI clone containing partial ntnA, ntnB*, ntnD, and downstream sequences in pBluescript SK | This study |
| pETntnD | NdeI/BamHI fragment containing ntnD in pET5a | This study |
4NT, 4-nitrotoluene; 4NBA, 4-nitrobenzyl alcohol; 4NBZate, 4-nitrobenzoate.
Chemicals and growth media.
Aromatic and aliphatic substrates were obtained from Aldrich Chemical Co. Pseudomonas strain TW3 was grown on minimal salts medium (5) supplemented with either solid 4-nitrotoluene (0.5 g/liter) or sodium succinate added at 10 mM. Escherichia coli strains were grown on Luria-Bertani medium (25). Where appropriate, ampicillin was added at 100 μg/ml and kanamycin was added at 50 μg/ml. Bacteriophage λ (FIX II; Stratagene) was propagated according to the supplier's instructions.
DNA manipulations.
Unless otherwise stated, standard methods for DNA manipulation were used (25). Total DNA was prepared from Pseudomonas sp. strain TW3 by the method of Ausubel et al. (3). Plasmid DNA was prepared from E. coli strains by using Qiaprep columns (QIAGEN). DNA fragments were recovered from agarose gels by using Qiaquick columns (QIAGEN). Southern blotting and plaque lifts were carried out as described by Sambrook et al. (25). Hybridizations were carried out with ECL direct labelling (Amersham) according to the manufacturer's instructions.
Preparation and screening of Pseudomonas sp. strain TW3 genomic library.
Genomic DNA was partially digested with Sau3AI, and the ends were filled in by incubation with DNA polymerase Klenow fragment and the appropriate deoxynucleogide triphosphates (dNTP) (dATP and dGTP), leaving a 2-bp overhang. The genomic DNA fragments were ligated into phage λ FIX II arms (Stratagene) previously digested with XhoI and partially filled in. The library was screened by hybridization to plaque lifts, and lambda DNA was prepared from positive clones as described by Sambrook et al. (25).
DNA sequencing and sequence analysis methods.
DNA sequences were determined by MWG-Biotech, Ltd. (Ebersberg, Germany). PCR primers were designed with the aid of the Lasergene software package (DNAStar, Inc., Madison, Wis.). Sequence databases were searched by using FASTA (21), BLASTN, and BLASTX programs (2). The PROSITE and Pfam motif databases were searched with custom Perl scripts. Frame plot analysis (6) and determination of percentage G+C content and open reading frame location were carried out with Artemis (Pathogen Sequencing Unit, The Sanger Centre). Multiple sequence alignments were carried out by using ClustalW.
Expression of ntnD in E. coli.
The ntnD gene was amplified by PCR from plasmid pTW3.10 with Taq polymerase (Promega). Primers incorporating an NdeI site in the forward primer and a BglII site in the reverse primer were designed. The NdeI site was positioned at the start codon of the ntnD open reading frame. Primer sequences (with restriction sites underlined) were as follows: forward, CATATGAATAATAATAACTTTGACGTG; reverse, AGATCTAACTCCTCGGTAGGAAGAGC (bp 8084 to 8110 and 9741 to 9716, respectively [GenBank accession no. AF043544]). PCR amplifications were carried out in a 100-μl reaction volume containing 10 ng of template DNA, 100 pmol of each primer, 200 μM each dNTP, 2 mM MgCl2, and 1 U of Taq polymerase. After a 2-min hot start at 94°C, the reaction mixtures were given 25 cycles of 1 min at 94°C, 1 min at 53°C, and 2 min at 72°C. The PCR product was cloned into pCR-blunt (Invitrogen) in E. coli TOP10 with selection on 50 μg of kanamycin per ml. The amplified gene was cut from the cloning vector with NdeI and BglII and was ligated into pET5a cut with NdeI and BamHI, placing the ntnD gene in frame with the T7 promoter to create pETntnD. The NtnD protein was expressed in E. coli BL21(DE3)/pLysS (Promega) grown in Luria-Bertani broth to an optical density at 600 nm of 0.3 and was induced with 0.2 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 3 h prior to harvesting.
SDS-PAGE.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was carried out by using the method of Laemmli (18) on a Mini-PROTEAN II Electrophoresis Cell (Bio-Rad, Hemel Hempstead, United Kingdom).
Enzyme assays.
Cells were harvested by centrifugation, washed with 50 mM Na2HPO4 (pH 7.5), and resuspended in the same buffer at approximately 0.2 g (wet weight) per ml. Cells were disrupted by passing them through a precooled French pressure cell (SLM Instruments Inc., Urbana, Ill.), and particulates were removed by centrifugation at 45,000 × g and 4°C for 30 min. Assays were carried out spectrophotometrically by following the reduction of 2,6-dichlorophenol indophenol (DCPIP) at 600 nm. Each 3-ml assay contained 50 μl of 20 mM phenazine methosulfate, 25 μl of 6.7 mM DCPIP, 30 μl of substrate (100 mM in dimethyl sulfoxide), and 50 μl of cell extract and was buffered with 2,845 μl of 80 mM Tris-HCl (pH 8.7). The Tris-HCl buffer was replaced with Bicine (pH 7.5 to 9.5) or CAPS (3-[cyclohexylamino]-1 propanesulfonic acid) (pH 9.5 to 11) to determine the pH profile. Assays were carried out at 28°C. The assay mixture was preincubated without substrate for 2 min at 28°C, and the reaction was started by the addition of substrate. The molar extinction coefficient for DCPIP at 600 nm was taken to be 21,000 M−1 cm−1.
RNA isolation and RT-PCR.
Cells were grown on minimal media supplemented with either 4-nitrotoluene or succinate until they reached an optical density at 600 nm of 0.3. Total RNA was prepared from 109 cells with RNeasy Mini columns (QIAGEN), with elution in 30 μl of water. The RNA was treated with DNase I to remove any genomic DNA contamination by incubation for 30 min at 37°C with 1 U of RNase-free DNase (Promega) and 1 U of RNasin (Promega) in 40 mM Tris-HCl (pH 7.5) containing 10 mM NaCl, 10 mM CaCl2, and 6 mM MgSO4. The RNA was cleaned by passage through an RNeasy Mini column before reverse transcription-PCR (RT-PCR) was carried out with total RNA by using an Access RT-PCR kit (Promega). Primer sequences for ntnD were (Dforward) 5′-CGTGATCGTAGTTGGTAGCGGTGC and (Dreverse) 5′-GGGTTGGTGCGTTGGTGTTGC (bp 8107 to 8130 and 9629 to 9609, respectively [GenBank accession no. AF043544]). The primer sequences for the region spanning ntnAB*D were (AB*Dforward) 5′-GCAACTCGATTGGGGTGGGC and (AB*Dreverse) 5′-GCCCAACACCTTGCCAGAGCG (bp 6739 to 6758 and 8338 to 8318, respectively). PCRs were carried out in a 50-μl volume containing 0.1 μg of template RNA, 50 pmol of each primer, 50 μM each dNTP, 1 mM MgSO4, 5 U of avian myeloblastosis virus reverse transcriptase, and 5 U of Tfl DNA polymerase in the reaction buffer supplied by the manufacturer. After reverse transcription at 48°C for 45 min, the reaction mixtures were heated to 94°C for 2 min and given 40 cycles of 30 s at 94°C, 1 min at 52°C, and 2 min at 68°C followed by a final extension at 68°C for 10 min. Negative control reactions were performed in the same way without reverse transcriptase to eliminate the possibility of amplifying residual genomic DNA.
Nucleotide sequence accession number.
The nucleotide sequence of 14,861 bp presented in Fig. 1 is available in GenBank under accession no. AF043544.
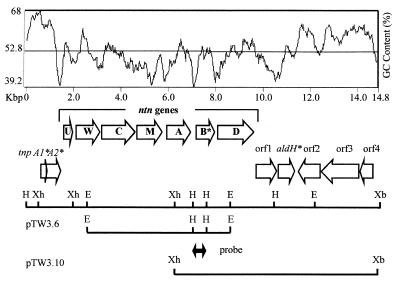
FIG. 1.
Map of the ntn gene cluster and flanking genes of TW3 with its guanine-plus-cytosine content profile. Open reading frames are marked by the open arrows, the direction of the arrowheads indicating the direction of transcription. The solid lines below the open reading frame arrows represent the inserts of recombinant plasmids pTW3.6 from which the 579-bp HindIII probe was isolated and pTW3.10 from which the nucleotide sequence was derived. Only restriction sites relevant to the clones and construct described in this figure are shown.
RESULTS
Screening of Pseudomonas sp. strain TW3 genomic library.
The TW3 genomic library was screened by hybridization by using a probe containing part of the ntn operon previously characterized (17). The probe was obtained by digesting plasmid pTW3.6 with HindIII, yielding a 579-bp HindIII fragment which included part of ntnB*, downstream of ntnA (Fig. 1). Fifty positive clones were obtained from screening 3.2 × 105 λ phage. One positive clone was selected, from which an 8,626-bp XhoI/XbaI fragment was subcloned into pBluescript (Stratagene) to create plasmid pTW3.10. The DNA sequence of the insert of pTW3.10 was determined.
Analysis of the nucleotide sequence of pTW3.10.
From the nucleotide sequence of pTW3.10, the presence of six complete open reading frames was deduced (Fig. 1). Immediately downstream of ntnB* is a 1,599-nucleotide gene (designated ntnD) corresponding to a protein of 532 amino acids with a molecular mass of 57.4 kDa. NtnD is similar to Gluconobacter oxydans l-sorbose dehydrogenase which catalyzes the conversion of l-sorbose to l-sorbosone (24) and to the alcohol dehydrogenase (AlkJ) from Pseudomonas oleovorans (32) and from P. putida (27) which converts aliphatic medium-chain-length alcohols to aldhehydes during the metabolism of alkanes (Table 2). The second and fourth open reading frames (orf1 and -2, respectively) correspond to proteins of unknown function and the 5′ end of the fifth open reading frame (orf3) shows some similarity to a putative transposase from Deinococcus radiodurans (Table 2). The third open reading frame (designated aldH*) is similar to an aldehyde dehydrogenase from P. putida but is interrupted by a stop codon and therefore appears to be a pseudogene like ntnB*. This interruption coincides with an abrupt increase in G+C content (Fig. 1). The final open reading frame (orf4) corresponds to part of a transposase from Xanthomonas campestris.
TABLE 2.
Pseudomonas sp. strain TW3 genes and gene products
| Gene designationa | Putative function of gene product or gene | Position in sequence | Amino acid identity/similarity of open reading frame (%) | Most-similar gene products (species) (accession no.) |
|---|---|---|---|---|
| tnpA1* | Transposase pseudogene | 632–895 | 56/72 | Part of TnpA (Bordetella parapertussis) (X66858) |
| tnpA2* | Transposase pseudogene | 864–1505 | 55/69 | Part of TnpA (B. parapertussis) (X66858) |
| ntnU | Unknown | 1590–1985 | 100 | XylU (P. putida) (U20269) |
| ntnW | Unknown | 2111–3157 | 99/99 | XylW (P. putida) (U20269) |
| ntnC | Nitrobenzaldehyde dehydrogenase | 3189–4652 | 96/98 | XylC (P. putida) (AF019635) |
| ntnM | Nitrotoluene monooxygenase (hydroxylase component) | 4677–5786 | 86/94 | XylM (P. putida) (AF019635) |
| ntnA | Nitrotoluene monooxygenase (reductase component) | 5936–6988 | 99/99 | XylA (P. putida) (AF019635) |
| ntnB* | Benzyl alcohol dehydrogenase pseudogene containing stop codon and insertion | 7178–7981 | 97/98 | XylB (P. putida) (M94184)b; insertion related to transposase from pEST1226 (M57500)c |
| ntnD | Nitrobenzyl alcohol dehydrogenase | 8087–9685 | 34/54 | l-Sorbose dehydrogenase (Gluconobacter oxydans) (D86622) |
| orf1 | Unknown | 9711–10625 | 33/50 | Unknown orf3 (Pseudomonas sp. strain YH102) (AF187880) |
| aldH* | Aldehyde dehydrogenase pseudogene | 10642–11367 | 42/58 | Part of aldehyde dehydrogenase (Synechocystis sp.) (D64004) |
| orf2 | Unknown | 12451–11480 | 96/99 | Protein of unknown function (Pseudomonas syringae) (AF141883) |
| orf3 | Transposase | 14102–12441 | 24/40 | N terminus homologous to putative transpose (D. radiodurans) (AE001862) |
| orf4 | Transposase | 14658–14083 | 62/76 | TnpR (X. campestris) (Z73593) |
Genes with asterisks are proposed to be nonfunctional pseudogenes either because the length of the open reading frame differs significantly from that of the closest gene in the data bank or because the region of homology to the closest gene in the data bank is interrupted by a stop codon or an insertion of unrelated DNA.
Bases 7178 to 7830.
Bases 7883 to 7981.
Upstream of ntnU we have also sequenced a region which carries two overlapping and incomplete transposase-like pseudogenes which we have designated tnpA1* and tnpA2*.
Expression of ntnD in E. coli and alcohol dehydrogenase assays.
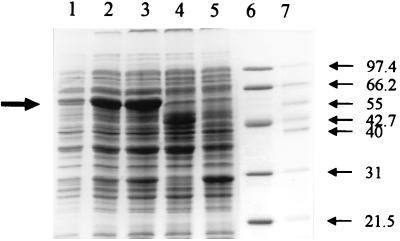
A 4-nitrobenzyl alcohol dehydrogenase specific activity of 0.28 U/mg with 4-nitrobenzyl alcohol as the substrate was obtained at pH 8.7 from cell extracts of the cloned ntnD gene overexpressed in E. coli BL21(DE3). SDS-PAGE showed high levels of a polypeptide of ∼57 kDa (Fig. 2). No activity or enhanced 57-kDa protein band was detectable in controls where the expression vector contained no insert. Activity was found only by the dye-linked assay, and no reduction of NAD+, NADP+, or flavin adenine dinucleotide (FAD) was detected in the presence of 4-nitrobenzyl alcohol, confirming earlier data obtained by using cell extracts of 4-nitrotoluene-grown Pseudomonas sp. strain TW3 (17, 23).
FIG. 2.
SDS-PAGE of overexpressed nitrobenzyl alcohol dehydrogenase (NtnD) in E. coli on a 12% gel. Lanes 1 to 3, cell extracts obtained at 1, 2, and 3 h, respectively, after induction with IPTG; lane 4, uninduced pETntnD; lane 5, pET5a alone; lanes 6 and 7, low-range marker (Bio-Rad) and mid-range marker (Promega), respectively. Marker masses are indicated on the right in kilodaltons. The molecular mass of the overexpressed protein (indicated by the large arrow) is ∼55 kDa.
The pH of the assays was varied by using Bicine at pHs 7.5, 8.0, 8.5, 9.0, and 9.5 and CAPS at pHs 9.5, 10.0, and 10.5 with 4-nitrobenzyl alcohol as substrate; maximum activity was obtained at pH 10, although the pH profile is very broad and the activity at pH 10 is less than twice the activity at pH 7.5 (data not shown).
The relative activities on different substrates of the wild-type 4-nitrobenzyl alcohol dehydrogenase expressed during growth on 4-nitrotoluene and of the overexpressed enzyme were compared. Cell extracts were assayed against all three nitrobenzyl alcohols and all the hydroxy-, methyl-, and dimethyl-substituted analogs. Two independent cultures of each were grown, and triplicate assays were performed with cell extracts against each of the substrates. The activity against 4-nitrobenzyl alcohol was taken to be 100% in each case, and activities against the other substrates were expressed relative to this (Table 3). The activity against position 2-substituted alcohols is generally low whereas that against position 3- and 4-substituted substrates is high. The exception to this is the low activity against 4-hydroxybenzyl alcohol, for which we have no explanation.
TABLE 3.
Relative activities of 4-nitrobenzyl alcohol dehydrogenasea
| Substrate | Source of cell extract
|
|
|---|---|---|
| TW3 grown on 4-nitrotoluene | E. coli BL21(DE3) expressing ntnD | |
| 4-Nitrobenzyl alcohol | 100 | 100 |
| 3-Nitrobenzyl alcohol | 30 | 33 |
| 2-Nitrobenzyl alcohol | 3 | 3 |
| 4-Methylbenzyl alcohol | 117 | 126 |
| 3-Methylbenzyl alcohol | 98 | 120 |
| 2-Methylbenzyl alcohol | 14 | 24 |
| 4-Hydroxybenzyl alcohol | 3 | 5 |
| 3-Hydroxybenzyl alcohol | 104 | 110 |
| 2-Hydroxybenzyl alcohol | 5 | 18 |
| 2,4-Dimethylbenzyl alcohol | 28 | 36 |
| 2,5-Dimethylbenzyl alcohol | 2 | 2 |
| 3,4-Dimethylbenzyl alcohol | 140 | 148 |
| 3,5-Dimethylbenzyl alcohol | 70 | 63 |
| 4-Ethylbenzyl alcohol | 114 | 113 |
| Benzyl alcohol | 121 | 108 |
| l-Sorbose, decanol, octanol, hexanol, butanol, ethanolb | 2 | 2 |
Activities are expressed as percentages relative to that observed with 4-nitrobenzyl alcohol, each value being the mean from two different cultures. No individual value was more than ± 3% from the mean.
All of these aliphatic alcohols stimulated the same low activities.
RT-PCR analysis of transcripts present in TW3.
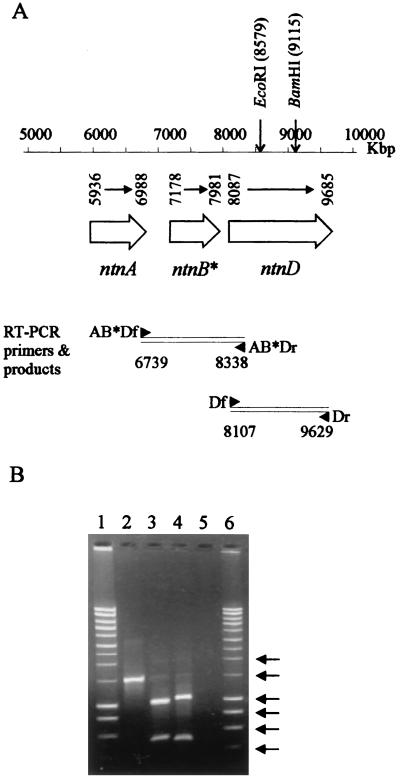
In order to show that the ntnD gene encodes an enzyme involved in the catabolism of 4-nitrotoluene, we examined transcripts from cells grown on 4-nitrotoluene and on succinate as a control. Two primer sets were constructed, one spanning from ntnA, across ntnB*, and through to ntnD and the other spanning ntnD alone (Fig. 3A). The expected RT-PCR size for ntnAB*D was 1,599 bp, and for ntnD alone it was 1,522 bp. The PCR products obtained, together with restriction digests of the products chosen to confirm the presence of expected restriction sites, were analyzed by agarose gel electrophoresis. Figure 3B shows that products of the expected size were obtained from total RNA of cells grown on 4-nitrotoluene by using the ntnD primers, and the presence of restriction sites in the expected positions was confirmed by digestion with BamHI and EcoRI. No products were obtained from total RNA of succinate-grown cells (data not shown) or from reaction mixtures from which the reverse transcriptase had been omitted. No products were obtained across ntnAB*D by using RNA prepared from 4-nitrotoluene-grown cells, although the primers did amplify a product of the correct size by using genomic DNA as the template (data not shown).
FIG. 3.
RT-PCR of ntn genes. (A) The positions of ntnA, -B*, and -D relative to the positions shown in Fig. 1, the primers used for RT-PCR and the EcoRI and BamHI restriction sites. (B) The agarose gel electrophoresis of RT-PCR products amplified from TW3 grown on 4-nitrotoluene. Molecular markers in lanes 1 and 6 (HyperLadder I; Bioline, London, United Kingdom) are indicated by arrows (from top to bottom, 2,000, 1,500, 1,000, 800, 600, and 400 bp). Lane 2, ntnD, 4-nitrotoluene-grown cells (expected size, 1,522 bp); lane 3, ntnD, 4-nitrotoluene-grown cells cut with BamHI (1,008 and 514 bp); lane 4, ntnD, 4-nitrotoluene-grown cells cut with EcoRI (1,050 and 472 bp); lane 5, ntnD, 4-nitrotoluene-grown cells with reverse transcriptase omitted from the reaction. No detectable products were obtained in reactions carried out on succinate-grown cells and in reactions with ntnAB*D primers (data not shown).
DISCUSSION
We have cloned and sequenced a genomic DNA fragment from Pseudomonas sp. strain TW3 downstream of the genes of the ntn operon (17). We have located ntnD directly adjacent to and downstream of the insertionally inactivated XylB homologue ntnB*. The NtnD protein catalyzes the catabolism of 4-nitrobenzyl alcohol to 4-nitrobenzaldehyde, and the location and identification of its gene complete the characterization of the genes encoding the pathway from 4-nitrotoluene to 4-nitrobenzoate. Previous biochemical analysis of this pathway demonstrated that the enzyme catalyzing this step in 4-nitrotoluene- or toluene-grown cells was anomalous in being NAD(P)+ independent (17, 23). This anomaly was highlighted by the reports that, in the other 4-nitrotoluene-degrading Pseudomonas sp. strain, 4NT, the analogous enzyme was NAD+ dependent (12) and that the other genes for the catabolism to 4-nitrobenzoate in TW3 were highly homologous to the genes on the TOL plasmid pWW0 (15, 17), in which the benzyl alcohol dehydrogenase XylB is a Zn2+-containing, NAD+- dependent alcohol dehydrogenase (26). There is in TW3 a considerable region of homology with the TOL plasmid xylB, but this appears to be a pseudogene with the potential reading frame interrupted by an insertion and has been designated ntnB* (17).
The NtnD amino acid sequence shares clear homology with similar NAD(P)+-independent enzymes, i.e., the alcohol dehydrogenase (AlkJ) from P. oleovorans and P. putida (27, 32) and l-sorbose dehydrogenase from G. oxydans (24). NtnD, like its NAD(P)+-independent counterparts, possesses a possible glycine box (GXGXXG) close to the amino terminus at residues 12 to 17 (GSGAAG) which is typical of the binding site of the ADP moiety of FAD (19, 33). Examination of the amino acid sequence in PROSITE (4) shows that at residues 80 to 103 (GKVLGGGTSVNAMCYVRGQKRDFD) and at residues 253 to 267 (GAVHSPKILMHSGIG), NtnD has signature patterns characteristic of FAD oxidoreductases (GA)-(RKN)-X- (LIV)-G(2)-(GST)(2)-X-(LIVM)-N-X(3)-(FYWA)-X(2)- (PAG)-X(5)-(DNESH) and (GS)-(PSTA)-X(2)-(ST)-P-X-(LIVM)(2)-X(2)-S-G-(LIVM)-G, respectively, but the function of these domains is not yet known. These data suggest that NtnD is also a flavoprotein.
There are three major classes of alcohol dehydrogenases (22): (i) the NAD(P)+-dependent alcohol dehydrogenases which are subdivided into three subgroups according to their metal dependence, the medium-chain Zn-dependent enzymes, the short-chain Zn-independent enzymes, and the Fe-activated enzymes; (ii) the NAD(P)+-independent enzymes which use pyrroloquinoline quinone, a heme group in association with pyrroloquinoline quinone (7–9), or cofactor F420 as a cofactor; and (iii) the FAD-dependent alcohol oxidases, which catalyze an essential irreversible oxidation of alcohols. The presence of a FAD signature sequence in NtnD suggests that it is a FAD-dependent alcohol dehydrogenase. Although the in vivo electron acceptor of NtnD, like that of the homologous AlkJ, is unknown, Van Beilen et al. (32) have shown that AlkJ transfers electrons from the substrate onto molecular O2, possibly through the electron transfer chain.
In this study, NtnD has been highly expressed from the vector pET5a. The cloned NtnD alcohol dehydrogenase activity shows the same relative substrate specificity as that of the wild-type TW3 grown on 4-nitrotoluene, providing evidence that the cloned gene is indeed the one expressed during growth on 4-nitrotoluene or toluene. Similarly, the RT-PCR provides further evidence that ntnD is being transcribed during growth on 4-nitrotoluene. However, it appears that the gene is not being cotranscribed with the other ntn genes. Because of its location, downstream of ntnB*, this implies that within the insertion in that pseudogene there is some termination signal for transcription. However, the inducibility of NtnD during growth on toluene and 4-nitrotoluene also implies that upstream on ntnD there is also a regulatory element controlling its growth substrate-dependent expression separately from that of the other ntn genes.
The average G+C content of the DNA from ntnU through to the stop codon in aldH*, being only 49.7%, is unusually low for Pseudomonas. This contrasts with the DNA at either end, which is 59.4% (at the 5′ end) and 58.2% (at the 3′ end) (Fig. 1), both corresponding much more closely to the norm for Pseudomonas. This suggests that it was incorporated into the genome of strain TW3 by some (relatively) recent recombination event. It also shows its close relationship with the homologous xylUWCMABN operon from the TOL plasmid, which is also unusually low in G+C content (50.2%) compared with the norm for Pseudomonas and which contrasts with the TOL lower operon xylXYZLTEGFJKIH (61.8%). The presence of DNA within and around the ntn gene cluster of sequences homologous to transposase-like genes (or partial genes), including the insertion within pseudogene ntnB*, adds credence to the likelihood that this cluster of genes was incorporated into the TW3 genome by one or more transposition events. It is possible that at some stage during, or after, acquisition of these genes, the xylB homolog became inactivated by insertion, and this event may have also been accompanied by the deletion of a xylN homolog which is present at the 3′ end of the TOL operon but absent from the ntn gene cluster. To compensate for the inactivation of ntnB, the gene for NtnD may have been recruited from elsewhere within the genome or from incoming heterologous DNA in order to carry out the conversion of benzyl alcohols to benzaldehydes. This scenario, based upon the structure of the DNA around these ntn genes, adds additional support for hypotheses (14, 16, 34) that strains have evolved novel catabolic pathways through the acquisition of genetic modules encoding operons or parts of operons.
ACKNOWLEDGMENT
This research was funded under the auspices of the Biotechnology Research program of the European Commission.
REFERENCES
- 1.Alisadat S, Mohan K S, Walia S K. A novel pathway for the biodegradation of 3-nitrotoluene in Pseudomonas putida. FEMS Microbiol Ecol. 1995;17:169–176. [Google Scholar]
- 2.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel F M, Brent R, Kingston R E, Moore D E, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1987. [Google Scholar]
- 4.Bairoch A, Bucher P, Hofman K. The PROSITE database, its status in 1997. Nucleic Acids Res. 1997;25:217–221. doi: 10.1093/nar/25.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bauchop T, Elsden S R. The growth of microorganisms in relation to energy supply. J Gen Microbiol. 1960;23:457–469. doi: 10.1099/00221287-23-3-457. [DOI] [PubMed] [Google Scholar]
- 6.Bibb M J, Findlay P R, Johnson M W. The relationship between base composition and codon usage in bacterial genes and its use for the simple and reliable identification of protein-coding sequences. Gene. 1984;30:157–166. doi: 10.1016/0378-1119(84)90116-1. [DOI] [PubMed] [Google Scholar]
- 7.Duine J A. Quinoproteins: enzymes containing the quinoid cofactor pyrroloquinoline quinone, topaquinone or tryptophan-tryptophan quinone. Eur J Biochem. 1991;200:271–284. doi: 10.1111/j.1432-1033.1991.tb16183.x. [DOI] [PubMed] [Google Scholar]
- 8.Groen B W, Duine J A. Quinoprotein alcohol dehydrogenase from Pseudomonas aeruginosa and quinohemoprotein alcohol dehydrogenase from Pseudomonas testosteroni. Methods Enzymol. 1990;188:33–39. doi: 10.1016/0076-6879(90)88009-y. [DOI] [PubMed] [Google Scholar]
- 9.Groen B W, van Kleef M A G, Duine J A. Quinohemoprotein alcohol-dehydrogenase apoenzyme from Pseudomonas testosteroni. Biochem J. 1986;234:611–615. doi: 10.1042/bj2340611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Groenewegen P E J, deBont J A M. Degradation of 4-nitrobenzoate via 4-hydroxylaminobenzoate and 3,4-dihydroxybenzoate in Comamonas acidovorans NBA-10. Arch Microbiol. 1992;158:381–386. [Google Scholar]
- 11.Groenewegen P E J, Breeuwer P, Vanhelvoort J M L M, Langenhoff A A M. Novel degradative pathway of 4-nitrobenzoate in Comamonas acidovorans NBA-10. J Gen Microbiol. 1992;138:1599–1605. doi: 10.1099/00221287-138-8-1599. [DOI] [PubMed] [Google Scholar]
- 12.Haigler B E, Spain J C. Biodegradation of 4-nitrotoluene by Pseudomonas sp. strain 4NT. Appl Environ Microbiol. 1993;59:2239–2243. doi: 10.1128/aem.59.7.2239-2243.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haigler B E, Wallace W H, Spain J C. Biodegradation of 2-nitrotoluene by Pseudomonas sp. strain JS42. Appl Environ Microbiol. 1994;60:3466–3469. doi: 10.1128/aem.60.9.3466-3469.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harayama S. Codon usage patterns suggest independent evolution of two catabolic operons on toluene degradative plasmid TOL pWW0 of Pseudomonas putida. J Mol Evol. 1994;39:328–335. doi: 10.1007/BF00163150. [DOI] [PubMed] [Google Scholar]
- 15.Harayama S, Rekik M, Wubbolts M, Rose K, Leppik R A, Timmis K N. Characterization of five genes in the upper-pathway operon of TOL plasmid pWW0 from Pseudomonas putida and identification of the gene products. J Bacteriol. 1989;171:5048–5055. doi: 10.1128/jb.171.9.5048-5055.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horn J M, Harayama S, Timmis K N. DNA sequence determination of the TOL plasmid (pWW0) xylGFJ genes of Pseudomonas putida: implications for the evolution of aromatic catabolism. Mol Microbiol. 1991;5:2459–2474. doi: 10.1111/j.1365-2958.1991.tb02091.x. [DOI] [PubMed] [Google Scholar]
- 17.James K D, Williams P A. ntn genes determining the early steps in the divergent catabolism of 4-nitrotoluene and toluene in Pseudomonas sp. strain TW3. J Bacteriol. 1998;180:2043–2049. doi: 10.1128/jb.180.8.2043-2049.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 19.Lamark T, Kaasen I, Eshoo M W, Falkenberg P, McDougal J, Strom A R. DNA sequence and analysis of the bet genes encoding the osmoregulatory choline-glycine betaine pathway of Escherichia coli. Mol Microbiol. 1991;5:1049–1064. doi: 10.1111/j.1365-2958.1991.tb01877.x. [DOI] [PubMed] [Google Scholar]
- 20.Parales J V, Kumar A, Parales R E, Gibson D T. Cloning and sequencing of the genes encoding 2-nitrotoluene dioxygenase from Pseudomonas sp. JS42. Gene. 1996;181:57–61. doi: 10.1016/s0378-1119(96)00462-3. [DOI] [PubMed] [Google Scholar]
- 21.Pearson W R, Lipman D J. Improved tools for biological sequence comparison. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reid M F, Fewson C A. Molecular characterization of microbial alcohol dehydrogenases. Crit Rev Microbiol. 1994;20:13–56. doi: 10.3109/10408419409113545. [DOI] [PubMed] [Google Scholar]
- 23.Rhys-Williams W, Taylor S C, Williams P A. A novel pathway for the catabolism of 4-nitrotoluene by Pseudomonas. J Gen Microbiol. 1993;139:1967–1972. doi: 10.1099/00221287-139-9-1967. [DOI] [PubMed] [Google Scholar]
- 24.Saito Y, Ishii Y, Hayashi H, Imao Y, Akashi T, Yoshikawa K, Noguchi Y, Soeda S, Yoshida M, Niwa M, Hosoda J, Shimomura K. Cloning of genes coding for l-sorbose and l-sorbosone dehydrogenases from Gluconobacter oxydans and microbial production of 2-keto-l-gulonate, a precursor of l-ascorbic acid, in a recombinant G. oxydans strain. Appl Environ Microbiol. 1997;63:454–460. doi: 10.1128/aem.63.2.454-460.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 26.Shaw J P, Rekik M, Schwager F, Harayama S. Kinetic studies on benzyl alcohol dehydrogenase encoded by TOL plasmid pWW0. J Biol Chem. 1993;265:10842–10850. [PubMed] [Google Scholar]
- 27.Smits T H M, Röthlisberger M, Witholt B, van Beilen J B. Molecular screening for alkane hydroxylase genes in Gram-negative and Gram-positive strains. Environ Microbiol. 1999;1:307–317. doi: 10.1046/j.1462-2920.1999.00037.x. [DOI] [PubMed] [Google Scholar]
- 28.Spanggord R J, Spain J C, Nishino S F, Mortelmans K E. Biodegradation of 2,4-dinitrotoluene by a Pseudomonas sp. Appl Environ Microbiol. 1991;57:3200–3205. doi: 10.1128/aem.57.11.3200-3205.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Spiess T, Desiere F, Fischer P, Spain J C, Knackmuss H J, Lenke H. A new 4-nitrotoluene degradation pathway in a Mycobacterium strain. Appl Environ Microbiol. 1998;64:446–452. doi: 10.1128/aem.64.2.446-452.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Suen W-C, Haigler B E, Spain J C. 2,4-Dinitrotoluene dioxygenase from Burkholderia sp. strain DNT: similarity to naphthalene dioxygenase. J Bacteriol. 1996;178:4926–4934. doi: 10.1128/jb.178.16.4926-4934.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suen W C, Spain J C. Cloning and characterization of Pseudomonas sp. strain DNT genes for 2,4-dinitrotoluene degradation. J Bacteriol. 1993;175:1831–1837. doi: 10.1128/jb.175.6.1831-1837.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Van Beilen J B, Eggink G, Enequist H, Bos R, Witholt B. DNA sequence determination and functional characterization of the OCT-plasmid-encoded alkJKL genes of Pseudomonas oleovorans. Mol Microbiol. 1992;6:3121–3136. doi: 10.1111/j.1365-2958.1992.tb01769.x. [DOI] [PubMed] [Google Scholar]
- 33.Wierenga R K, Terpstra P, Hol W G J. Prediction of the occurrence of the ADP-binding βαβ-fold in proteins, using an amino acid sequence fingerprint. J Mol Biol. 1986;187:101–107. doi: 10.1016/0022-2836(86)90409-2. [DOI] [PubMed] [Google Scholar]
- 34.Williams P A, Sayers J R. The evolution of pathways for aromatic hydrocarbon oxidation in Pseudomonas. Biodegradation. 1994;5:195–217. doi: 10.1007/BF00696460. [DOI] [PubMed] [Google Scholar]
- 35.Yabannavar A V, Zylstra G J. Cloning and characterization of the genes for p-nitrobenzoate degradation from Pseudomonas pickettii YH105. Appl Environ Microbiol. 1995;61:4284–4290. doi: 10.1128/aem.61.12.4284-4290.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]