Abstract
The effects of simultaneous expression of several efflux pumps on antibiotic resistance were investigated in Escherichia coli and Pseudomonas aeruginosa. Several combinations of efflux pumps have been studied: (i) simultaneous expression of a single-component efflux pump, which exports antibiotics into the periplasm, in combination with a multicomponent efflux pump that accomplishes efflux directly into the external medium; (ii) simultaneous expression of two single-component pumps; and (iii) simultaneous expression of two multicomponent pumps. It was found that when efflux pumps of different structural types were combined in the same cell (the first case), the observed antibiotic resistance was much higher than that conferred by each of the pumps expressed singly. Simultaneous expression of pairs of single-component or multicomponent efflux pumps (the second and third cases) did not produce strong increases in antibiotic resistance.
Efflux of antibiotics out of cells is broadly recognized as a major component of bacterial resistance to many classes of antibiotics (26, 28). This efflux occurs due to the activity of membrane transporter proteins, the so-called drug efflux pumps. Some efflux pumps selectively extrude specific antibiotics, while others, referred to as multidrug resistance (MDR) pumps, expel various structurally diverse antibiotics. While antibiotic-specific efflux pumps are usually encoded on transmissible plasmids and transposons, genes encoding many MDR pumps are normal constituents of bacterial chromosomes. Efflux pumps occur as either single-component or multicomponent systems. In gram-negative bacteria, single-component efflux pumps extrude their substrates into the periplasmic space (40). Examples of such single-component efflux pumps include the transposon-encoded tetracycline- and chloramphenicol-specific pumps, TetA and CmlA, respectively (2, 38), and the MDR pump MdfA, encoded in the chromosome of Escherichia coli (6). Multicomponent efflux pumps (which are found exclusively in gram-negative bacteria) traverse both inner and outer membranes. Examples include the MDR pumps AcrAB-TolC (19) and MexAB-OprM (34) from E. coli and Pseudomonas aeruginosa, respectively. Each pump contains a transporter located in the cytoplasmic membrane (as exemplified by AcrB or MexB), an outer membrane channel (TolC or OprM), and a periplasmic linker protein (AcrA or MexA), which is thought to bring into contact the other two components (42). This structural organization allows extrusion of substrates directly into the external medium, bypassing the periplasm and the outer membrane (27). The outer membrane of gram-negative bacteria serves as an efficient permeability barrier for both hydrophobic and hydrophilic antibiotics (29). Therefore, when antibiotics are extruded directly into the external medium, two independent mechanisms, efflux and low uptake through this permeability barrier, contribute to decreased intracellular accumulation of antibiotics (26).
A single bacterial cell may contain multiple efflux pumps that are capable of extruding the same antibiotic. Cells of E. coli or P. aeruginosa may acquire plasmid-encoded transporters such as TetA or CmlA, even though they already encode in their genomes endogenous MDR pumps (AcrAB-TolC or MexAB-OprM) that also can extrude tetracycline and chloramphenicol. P. aeruginosa contains at least four MDR pumps that can confer resistance to fluoroquinolones (1, 13, 34, 35). We sought to investigate the effect that simultaneous expression of several efflux pumps would have on susceptibility to antibiotics that are the substrates of both pumps. Our results indicate that pairs of efflux pumps may produce either additive effects or much greater than simple additive effects on drug resistance.
MATERIALS AND METHODS
Bacterial strains and media.
The bacterial strains used in this study are listed in Table 1. Bacterial cells were grown in L broth (1% [wt/vol] tryptone, 0.5% [wt/vol] yeast extract, 0.5% [wt/vol] NaCl) or L agar (L broth plus 1.5% agar) at 37°C. Antibiotics were added to the media at the following concentrations: tetracycline, 20 μg/ml for E. coli and 100 to 150 μg/ml for P. aeruginosa; chloramphenicol, 20 μg/ml for E. coli and 100 μg/ml for P. aeruginosa; HgCl2, 15 μg/ml for both E. coli and P. aeruginosa; ampicillin, 100 μg/ml for E. coli; and kanamycin, 50 μg/ml for E. coli. Levofloxacin was synthesized at Daiichi Pharmaceutical Co., Ltd. (Tokyo, Japan). All other antibiotics were purchased from Sigma Chemical Co. (St. Louis, Mo.). MC-207,110 is an efflux pump inhibitor (36) with activity against various RND transporters from several bacterial species (J. Blais, D. Cho, K. Tangen, C. Ford, A. Lee, O. Lomovskaya, and S. Chamberland, Abstr. 39th Intersci. Conf. Antimicrob. Agents Chemother., abstr. F-1266, p. 327, 1999). ABS and EFS compounds are recently discovered inhibitors that are selective for the P. aeruginosa MexAB-OprM and MexEF-OprN pumps, respectively (D. Cho, J. Blais, K. Tangen, K. Ford, A. Lee, O. Lomovskaya, S. Chamberland, and G. Miller, Abstr. 39th Intersci. Conf. Antimicrob. Agents Chemother., abstr. F-1267, p. 327, 1999). All compounds were from Microcide Pharmaceuticals, Inc. or from Daiichi Pharmaceutical Co., Ltd.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Genotypea | Construction, selection, source, or referenceb |
|---|---|---|
| P. aeruginosa strains | ||
| PAM1020 | PAO1 prototroph | 17 |
| PAM1032 | nalB1032 | 17 |
| PAM1033 | nfxB1033 | 17 |
| PAM1034 | nfxC1034 | 17 |
| PAM1262 (K879) | met-9011 amiE200 rpsL pvd-9 mexB::ΩHg | K. Poole |
| PAM1154 | oprM::ΩHg | 17 |
| PAM1014 | nfxC oprM::ΩHg | 17 |
| PAM1610 | nalB1032 ΔmexEF-oprN::ΩHg | 17 |
| PAM1409 | ΔmexCD-oprJ::Gm | 17 |
| PAM1275 | mexB::ΩHg | PAM1020 × (PAM1262); HgCl2c |
| PAM1277 | nfxB1033 mexB::ΩHg | PAM1033 × (PAM1262); HgCl2 |
| PAM1438 | nalB1032 nfxB1438 | Selection on LBA + levofloxacin at 1 μg/ml and ABS at 20 μg/ml from PAM1032 |
| PAM1465 | nfxB1438 mexB::ΩHg | PAM1438 × (PAM1262); HgCl2 |
| PAM1466 | nalB1032 ΔmexCD-oprJ::Gm | PAM1438 × (PAM1409); Gm |
| PAM1278 | nfxC1034 mexB::ΩHg | PAM1034× (PAM1262); HgCl2 |
| PAM2281 | nalB2281 nfxC1034 | Selection on LBA + levofloxacin at 1 μg/ml and EFS at 5 μg/ml from PAM1034 |
| PAM2282 | nalB2282 nfxC1034 | Selection on LBA + levofloxacin at 1 μg/ml and EFS at 5 μg/ml from PAM1034 |
| PAM1610 | ΔmexEF-oprN::ΩHg | 17 |
| PAM2359 | nalB2281 ΔmexEF-oprN::ΩHg | PAM2281 × (PAM1610); HgCl2 |
| PAM2360 | nalB2282 ΔmexEF-oprN::ΩHg | PAM2282 × (PAM1610); HgCl2 |
| PAM2302 | nalB2281 nfxC1034 nfxB2302 | Selection on LBA + levofloxacin at 1 μg/ml, ABS at 20 μg/ml, and EFS at 5 μg/ml from PAM2281 |
| PAM2303 | nalB2281 nfxC1034 nfxB2303 | Selection on LBA + levofloxacin at 1 μg/ml, ABS at 20 μg/ml, and EFS at 5 μg/ml from PAM2282 |
| PAM2387 | oprM::ΩHg nfxC1034 nfxB2303 | PAM2303 × (PAM1014); HgCl2 |
| PAM1064 | mexA-phoA::Tc | 17 |
| PAM1116 | nalB1032 mexA-phoA::Tc | Integration of plasmid pSUP202-mexA-phoA (Tcr Cbr Cmr) in the chromosome of PAM1032; Tcr Cbr |
| PAM2454 | nalB1032 mexA-phoA::Tc oprM::ΩHg | PAM1116 × (PAM1014); HgCl2 |
| PAM2455 | mexA-phoA::Tc oprM::ΩHg | PAM1064 × (PAM1014); HgCl2 |
| PAM1194 | oppA::Tc | Transposon mutagenesis with D171; Tc |
| PAM2386 | nalB1032 oppA::Tc | PAM1032 × (PAM1194); Tc |
| PAM1316 | oprM::ΩHg oppA::Tc | PAM1194 × (PAM1014); HgCl2 |
| PAM2458 | oprM::ΩHg mexA-phoA::Tc oppA::Tc | PAM1194 × (PAM2455); HgCl2 |
| E. coli strains | ||
| ECM1194 (AMS6) | wt | 18 |
| ECM1642 | marR1642 | Selection on LBA + chloramphenicol at 10 μg/ml from ECM1194 |
| ECM1694 | ΔacrAB::Km | ECM1194 × (ECM1343); Km |
| ECM1668 | marR1642 ΔacrAB::Km | ECM1642 × (ECM1343); Km |
| ECM1749 | marR1642 ΔacrAB::Km/pLQ821 | ECM1668/pLQ821; Ap |
| ECM1750 | ΔacrAB::Km/pLQ821 | ECM1694/pLQ821; Ap |
| ECM1735 | marR1642 ΔacrAB::Km/pAL261 | ECM1668/pAL261; Ap |
| ECM1640 | ΔacrAB::Km/pAL261 | ECM1694/pAL261; Ap |
| ECM1748 | wt/pLQ821 | ECM1194/pLQ821; Ap |
| ECM1730 | wt/pAL261 | ECM1194/pAL261; Ap |
| ECM1751 | marR1642/pLQ821 | ECM1642/pLQ821; Ap |
| ECM1754 | marR1642/pAL261 | ECM1642/pAL261; Ap |
| ECM1776 (UTL2) | ΔmdfA::Km | 7 |
| ECM1174 (ZK796) | F− araD139 Δ(argF-lac)205 ptsB5301 ptsF25 relA1 rpsL150 deoC1 flbB tolC::Tn10 | R. Kolter |
| ECM1556 | tolC::Tn10 | ECM1194 × (ECM1174); Tc |
| ECM1816 | tolC::Tn10 ΔmdfA::Km | ECM1556 × (ECM1176); Km |
| ECM1888 | tolC::Tn10 mdfR | Stepwise selection on LBA + chloramphenicol (first at 1 μg/ml, then at 4 μg/ml) from ECM1556 |
| ECM1915 | tolC::Tn10 mdfR ΔmdfA::Km | ECM1888 × (ECM1176); Km |
| ECM1911 | tolC::Tn10 ΔmdfA::Km/pLQ821 | ECM1816/pLQ821; Ap |
| ECM1908 | tolC::Tn10 mdfR/pLQ821 | ECM1888/pLQ821; Ap |
| ECM1955 | tolC::Tn10 mdfR ΔmdfA::Km/pLQ821 | ECM1915/pLQ821; Ap |
| ECM1343 (AG100A) | argE3 thi-1 rpsL xyl mlt Δ(gal uvrB) supE44 ΔacrAB::Km | 31 |
| DH5α | endA hsdR17 supE44 thi-1 recA1 gyrA relA1 Δ(lacZYA-argF)U169 deoR [φ80dlacΔ(lacZ)M15] | 2 |
| S17-1 | thi pro hsdR recA tra+ | 39 |
| Plasmids | ||
| pSUP202-mexA-phoA | pSUP202 (Tcr Cbr CmroriT) carrying the 5′ upstream region of mexA fused to promoterless phoA gene | K. Poole |
| pSE380 | trc promoter expression vector; lacIq Apr | Invitrogen |
| pAL261 | Apr; pSE380 with EcoRI-HindIII PCR fragment containing the mdfA gene | This study |
| pLQ821 | Apr; cmlA | 3 |
| pMarR | Apr; marR | P. Miller |
ΩHg, HgCl2 resistance derivative of interposon Ω; Gm, gentamicin resistance; Ap, ampicillin resistance; Cb, carbenicillin resistance; Cm, chloramphenicol resistance; Km, kanamycin resistance; Tc, tetracycline resistance; oriT, origin of transfer from RP4; wt, wild type.
ABS and EFS, efflux pump inhibitors specific for the MexAB-OprM and the MexEF-OprN pumps, respectively; LBA, L agar.
Description of transduction experiment. The first strain is the recipient; the strain in parentheses is the source of a transducing lysate. Antibiotics used for selection of transductants, transformants, or transconjugants are also shown.
Selection of MDR mutants overexpressing multiple pumps in P. aeruginosa.
Selection was performed using pump-specific efflux pump inhibitors referred to as the ABS and EFS compounds. The structures of these compounds will be presented elsewhere. To isolate mutants simultaneously overexpressing the mexAB-oprM and the mexCD-oprJ operons, 0.1 ml of overnight culture of strain PAM1032 (nalB; mexAB-oprM overexpressed) was plated on L-agar plates containing levofloxacin at 1 μg/ml and the ABS compound at 20 μg/ml to inhibit the activity of MexAB-OprM. Strain PAM1438 was selected under these conditions. To confirm functionality of both pumps, the mexB gene and the mexCD-oprJ operon were disrupted in strain PAM1438, resulting in strains PAM1465 and PAM1466, respectively. The antibiotic susceptibility profiles of PAM1465 and PAM1466 were consistent with overexpression of either mexCD-oprJ or mexAB-oprM, respectively (see Results).
The mutants simultaneously overexpressing the mexAB-oprM and mexEF-oprN efflux operons were selected by plating 0.1 ml of overnight culture of strain PAM1034 (nfxC; mexEF-oprN overexpressed) on L-agar plates containing levofloxacin at 1 μg/ml and the EFS compound at 5 μg/ml to inhibit the activity of MexEF-OprM. Susceptibility testing of the mutants selected under these conditions showed that two strains, PAM2281 and PAM2282, acquired increased resistance to the β-lactams carbenicillin and aztreonam, indicating overexpression of mexAB-oprM (23, 41). Disruption of the mexEF-oprN operon in PAM2281 and PAM2282 rendered PAM2359 and PAM2360, with phenotypes consistent with overexpression of the MexAB-OprM efflux pump. Both PAM2281 and PAM2282 were used for the subsequent selection to isolate mutants overexpressing MexCD-OprJ in addition to two other pumps. To do so, 0.1-ml portions of overnight cultures of PAM2281 and PAM2282 were plated on L-agar plates containing levofloxacin at 1 μg/ml and the EFS and ABS compounds at 5 and 20 μg/ml, respectively. PAM2302 and PAM2303 were selected from PAM2281 and PAM2303, respectively. In all strains, overexpression of the pumps was confirmed using Western analysis with anti-OprM, anti-OprJ, and anti-OprN antibodies (obtained from N. Gotoh) (not shown).
Selection of mutants overexpressing various pumps in E. coli.
Selection of mar (multiple antibiotic resistance) mutants overexpressing the acrAB operon pump was performed by plating 0.1 ml of overnight culture of the wild-type strain ECM1194 on L-agar plates containing chloramphenicol at 10 μg/ml. Strain ECM1642 had the pattern of antibiotic susceptibility consistent with overproduction of the AcrAB efflux pump (26, 31). Deletion of the acrAB operon in strain ECM1642 by transducing in the ΔacrAB::Km construct produced strain ECM1668, which had a hypersensitive phenotype that was indistinguishable from the phenotype of ECM1194 lacking the acrAB operon (PAM1694) (data not shown). Introduction of plasmid pMarR, containing the wild-type marR gene, into strain ECM1642 reversed the drug resistance phenotype, indicating that ECM1642 contained a recessive mutation in this gene (data not shown).
A mutant overexpressing the gene mdfA (carrying a mutation in a gene tentatively called mdfR) was selected from strain ECM1556 (ECM1194 tolC::Tn10) by stepwise selection on chloramphenicol. ECM1556 lacks the functional AcrAB-TolC pump and is hypersensitive to multiple antibiotics. It has been previously demonstrated that TolC was not required for the MdfA activity (7). Chloramphenicol and ethidium bromide MICs are higher for strain ECM1888 (mdfR) (16 μg/ml for both agents) than for the parent strain (MIC of 1 μg/ml for both agents). Disruption of the mdfA gene in ECM1888 (to give ECM1915) by transducing the mdfA::Km insertion from strain UTL2 (obtained from E. Bibi) decreased the MICs of chloramphenicol and ethidium bromide to 1 μg/ml, implicating MdfA in increased resistance observed for the ECM1888. The nature of the mdfR mutation(s) in ECM1888 is presently unknown.
Transductions.
Transductions in P. aeruginosa were performed using phage F116L according to a previously described protocol (14). Transductions in E. coli were performed using phage P1 as previously described (18).
MIC determinations.
MIC determinations were carried out in 96-well microtiter plates using a standard broth microdilution method (25) in Muller-Hinton broth (Difco). In the case of E. coli strains containing various plasmids, ampicillin was added to Muller-Hinton broth to a final concentration of 50 μg/ml. In the case of pAL261-containing strains, IPTG (isopropyl-β-d-thiogalactopyranoside) was added to a final concentration of 0.5 mM to induce expression of the gene mdfA (see below). Inocula were 104 to 105 cells/ml.
Construction of tetA- and tetC-containing strains of P. aeruginosa.
The plasmid pSUP202-mexA-phoA (a gift from K. Poole) contains the mexA-phoA transcriptional fusion inserted into the vector pSUP202 (Tcr Cbr Cmr) (39), which can replicate in E. coli but cannot replicate in P. aeruginosa. The plasmid also carries the mob (mobilization) site, which allows conjugal transfer. This plasmid was transformed into E. coli strain S-17 (39) and mobilized into P. aeruginosa PAM1020 and PAM1032 via conjugation (33). Transconjugants were selected on tetracycline at 150 μg/ml and were expected to contain the entire plasmid pSUP202-mexA-phoA integrated into the mexA locus. Indeed, PAM1064 and PAM1116 were confirmed by PCR and antibiotic susceptibility profiles to contain chromosomal mexA-phoA fusions, the closely linked plasmid-encoded Tcr and Cbr markers, and functional mexAB-oprM operons at either the wild-type or overexpressed level, respectively. The tetracycline resistance gene in pSUP202 originates from pBR325 and belongs to the TetC class (15). To construct the TetC-containing strains with nonfunctional MexAB-OprM, we have transduced oprM::ΩHg from PAM1014 into PAM1116 and PAM1064. PAM2454 and PAM2455 were among the rare transductants that retained the mexA-phoA fusion and were still resistant to carbenicillin (MIC of 128 μg/ml for PAM2454 and PAM2455 versus 0.5 μg/ml for PAM1154). Note that PAM2454 also contained the nalB mutation.
To construct TetA-containing strains, we first performed transposon mutagenesis of the wild-type P. aeruginosa with the phage mini-D171Tc (TetA) (5). In one of the strains (PAM1194) the mini-D171 insertion was mapped to the gene oppA (A. Mistry and O. Lomovskaya, unpublished data). The oprM::ΩHg locus was then transduced into PAM1194 from PAM1014 to give PAM1316, and the oppA::Tet insertion was transduced from PAM1194 into PAM1032 (nalB; overexpression of mexAB-oprM) using the phage F116L to give PAM2386.
To construct the strain with both Tet pumps, the entire mexA-phoA-Tcr-Cbr-oprM::Hg locus was transduced from PAM2455 into PAM1194 to give PAM2458.
DNA manipulations.
Plasmid DNA was purified using an RPM Spin Kit (BIO 101 Inc., Vista, Calif.). Chromosomal DNA was prepared by using a Blood and Cell Culture Mini Kit (Qiagen Inc., Valencea, Calif.). DNA fragments were gel purified and extracted using a Qiagen Gel Purification Kit. Restriction enzymes were obtained from New England Biolabs (Beverly, Mass.), and AmpliTaq was obtained from Perkin-Elmer (Branchburg, N.J.). Plasmid DNA was introduced into E. coli strains by electroporation (Bio-Rad Laboratories, Mississauga, Ontario, Canada). All molecular biology techniques were performed according to the manufacturer's instructions or as described by Sambrook et al. (37). PCR was carried out in a Perkin-Elmer GeneAmp 9600 thermal cycler. Typically, 30 cycles of denaturing (30 s at 95°C), annealing (30 s at 55°C), and extending (1 min at 72°C) were used to amplify the chromosomally carried genes. The chromosome of the strain DH5α was used as a template. The gene mdfA was amplified with primers MdfA-EcoRI (forward) GGAATTCATGCAAAATAAATTAGCTTCCGGTGCC and MdfA-HindIII (reverse) CCCAAGCTTGGCTTACCCTTCGTGAGAATTT (6). The PCR fragment was subsequently ligated into the EcoRI and HindIII sites of pSE380 (Invitrogen) to create pAL261. In pAL261 mdfA was expressed from the trc promoter of pSE380; expression required addition of IPTG (0.5 mM). No MdfA-mediated drug resistance was seen without induction.
SDS-PAGE and Western immunoblotting.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed according to a previously described protocol (8) with 10% (wt/vol) acrylamide in the running gel. SDS-PAGE-separated proteins were electrophoretically transferred to a nitrocellulose membrane (BA85; Schleicher & Schuell) as described previously (9) with the exception that SDS (0.1% [wt/vol]) was included in the buffer and transfer was carried out at 100 mA for 90 min. Membranes were processed as described previously (8) with the murine monoclonal antibodies specific to the OprM, OprJ, and OprN protein (obtained from N. Gotoh) as the primary antibodies and alkaline phosphatase-conjugated goat antibodies to mouse immunoglobulin G (Bio-Rad) as the secondary antibodies, respectively. Blots were developed using the AP Conjugate Substrate Kit (Bio-Rad) according to the manufacturer's protocol.
RESULTS
Effect of simultaneous expression of multicomponent and single-component efflux pumps on resistance to chloramphenicol in E. coli.
In the case of E. coli, we have determined the resistance to chloramphenicol in a series of strains that carried either plasmid pLQ821, encoding the chloramphenicol-specific CmlA transporter (2), or plasmid pAL261, encoding the MDR transporter MdfA (6), and that additionally either lacked (ECM1750 and ECM1740), contained the wild-type (ECM1748 and ECM1730), or overexpressed (ECM1751 and ECM1754) the AcrAB-TolC efflux pump. It is noteworthy that while mdfA is a normal constituent of the E. coli chromosome, it appears to be nonexpressed in the wild-type strains, since deleting mdfA did not result in increased susceptibility to chloramphenicol (7).
The results are presented in Table 2. Overexpression of AcrAB (ECM1642) and CmlA (ECM1750) in E. coli conferred comparable fold increases (calculated as ratios of MICs) in resistance to chloramphenicol, yielding MICs of 16 and 32 μg/ml, respectively. Overexpression of MdfA from the trc promoter of pSE380 (induced with 0.5 mM IPTG [see Materials and Methods]) resulted in the MIC of 8 μg/ml (ECM1740). Interestingly, the effect on drug resistance conferred by CmlA and MdfA appears to be independent of the level of AcrAB-TolC expression. CmlA and MdfA conferred almost equal fold increases in drug resistance, regardless of whether the strain lacked, contained the wild-type level of, or overexpressed the AcrAB pump. Consequently, the E. coli strains ECM1751 and ECM1754, each overexpressing pairs of efflux pumps, had MICs of >128 and 128 μg/ml, respectively.
TABLE 2.
Effect of simultaneous expression of a single-component pump, CmlA or MdfA, and a multicomponent pump, AcrAB-TolC, on susceptibility to chloramphenicol in E. coli
| Single-component pump | MIC of chloramphenicol (μg/ml) for the indicated strain with the following AcrAB pump status:
|
||
|---|---|---|---|
| ΔacrAB::Km | Wild type | AcrAB overexpressed | |
| None | 1 (ECM1694) | 4 (ECM1194) | 16 (ECM1642) |
| CmlA | 32 (ECM1750) | 64 (ECM1748) | >128 (ECM1751) |
| MdfA | 8 (ECM1740) | 16 (ECM1730) | 128 (ECM1754) |
It was important to clarify whether simultaneous overexpression of two pumps affected the activity of each individual pump. To assess the activity of AcrAB-TolC, we took advantage of the fact that AcrAB-TolC is an MDR pump with an extremely broad spectrum of substrates (16, 31), including such antibiotics as tetracycline, erythromycin, levofloxacin, and trimethoprim that are not extruded by either the CmlA or the MdfA transporter. The level of resistance to these antibiotics in strain PAM1642 overexpressing AcrAB-TolC was the same whether or not CmlA or MdfA was present (data not shown), indicating that activity (and expression) of AcrAB-TolC was independent of CmlA or MdfA. It has previously been reported that MdfA confers some resistance to aminoglycosides (6), the rare class of antibiotics that are not extruded by AcrAB. Unfortunately, we have not detected MdfA-mediated resistance to apramycin, tobramycin, or gentamicin in any of our strains, possibly because of the difference in strain backgrounds. Therefore, due to the lack of antibiotics that are extruded exclusively by CmlA and MdfA but not by AcrAB, we were unable to assess if an active AcrAB-TolC somehow affected the activity of CmlA and/or MdfA. However, we were able to demonstrate, with the efflux pump inhibitor MC-207,110 (36), that the AcrAB pump did not affect the activity of the MdfA protein. MC-207,110 at 20 μg/ml decreased the MIC of chloramphenicol for PAM1642 (16 μg/ml) to the level seen for ECM1694 (acrAB deleted) (1 μg/ml), indicating complete inhibition of the AcrAB pump. MC-207,110 did not affect the MIC of chloramphenicol for ECM1740 (lacking AcrAB but containing MdfA), indicating that this compound did not inhibit MdfA. For the strain ECM1754 (ECM1642/pAL261) the chloramphenicol MIC in the presence of MC-207,110 dropped from 128 to 8 μg/ml, i.e., the level of resistance was exactly that seen in the ΔacrAB strain ECM1740 (Table 2). Thus, MdfA appeared to function with the same activity whether or not AcrAB was present in the same strain. Since CmlA itself is partially inhibited by MC-207,110, this useful tool could not be applied in this case.
It was also important to investigate whether the marR mutation, which was present in ECM1642, had a pleiotropic effect on expression of plasmid-carried cmlA and/or mdfA. To do so we have introduced the cmlA- and mdfA-containing plasmids in strain ECM1668, which contained the marR1642 (see Materials and Methods) mutation but lacked the acrAB operon. The level of resistance to chloramphenicol conferred by pLQ821 and pAL261 was independent of the mar1642 mutation (data not shown), ruling out the possibility that the marR mutation influences antibiotic resistance by altering the expression of the cmlA or mdfA gene.
Effect of simultaneous expression of multicomponent and single-component efflux pumps on resistance to tetracycline in P. aeruginosa.
In the case of P. aeruginosa, we have determined resistance to tetracycline in the strains that carried the chromosomally encoded TetA or TetC transporters and additionally either lacked (PAM1316 and PAM2455), contained the wild-type (PAM1194 and PAM1064), or overexpressed (PAM2386 and PAM1116) the MexAB-OprM efflux pump.
The resistance to tetracycline was then measured (Table 3). Each singly overexpressed pump conferred similar levels of resistance to tetracycline. MICs ranged from 16 to 32 μg/ml for MexAB-OprM (PAM1032), TetA (PAM1316), or TetC (PAM2455). Similar to the case described above for E. coli, it also appeared that the effect on drug resistance conferred by TetA or TetC was independent of the level of expression of MexAB-OprM: the Tet pumps conferred similar fold increases in drug resistance, whether or not the strain lacked, contained the wild-type level of, or overexpressed the MexAB-OprM pump. Consequently, strains PAM2386 and PAM1116, overexpressing both pumps, had MICs of >512 μg/ml. Activity of the MexAB-OprM was unaffected by the Tet transporters. This conclusion was based on the fact that the level of resistance to levofloxacin (the substrate of the MexAB-OprM but not the Tet pumps), provided by the overexpressed MexAB-OprM pump, remained the same regardless of the presence of the Tet pumps. It also appeared that the nalB mutation did not affect expression of at least the tetC gene: the tetracycline MICs for strains PAM2454 (nalB) and PAM2455 (not nalB), both of which had tetC while lacking functional MexAB-OprM, were still the same.
TABLE 3.
Effect of simultaneous expression of a single-component pump, TetA or TetC, and a multicomponent pump, MexAB-OprM, on susceptibility to tetracycline in P. aeruginosa
| Single-component Tet pump | MIC of tetracycline (μg/ml) for the indicated strain with the following MexAB-OprM pump status:
|
||
|---|---|---|---|
| oprM::ΩHg (MexAB-OprM nonfunctional) | Wild type | nalB (MexAB-OprM overproduced) | |
| None | 0.5 (PAM1154) | 4 (PAM1020) | 32 (PAM1032) |
| TetA | 32 (PAM1316) | 512 (PAM1194) | >512 (PAM2386) |
| TetC | 16 (PAM2455) | 512 (PAM1064) | >1,024 (PAM1116) |
Our data obtained in these two groups of experiments indicate that simultaneous overexpression of pairs of efflux pumps can result in much higher levels of drug resistance than are provided by each of the singly overexpressed pumps. In fact, the fold increase in drug resistance that was seen for the strain overexpressing both multicomponent and single-component efflux pumps appeared to be close to the product of the fold increases produced by each of the individually expressed pumps.
In the studies described above, the pumps under investigation exhibited different efflux mechanisms due to differences in structural organization, extruding their substrates either into the periplasm or into the external medium. In the following experiments, we investigated interplay between pumps with similar structural types.
Effect of simultaneous expression of two single-component efflux pumps on the resistance to chloramphenicol in E. coli and to tetracycline in P. aeruginosa.
In the case of E. coli, we investigated interplay between the CmlA and MdfA transporters. All E. coli strains used for these studies lacked the gene tolC, encoding the essential outer membrane component of the multicomponent efflux pump AcrAB-TolC. It was necessary to inactivate AcrAB-TolC since the basal level of expression of this pump contributes to the intrinsic resistance to the antibiotics studied. It is noteworthy that other known multicomponent E. coli pumps either are not expressed without acquisition of regulatory mutations, do not confer chloramphenicol or tetracycline resistance, or require the TolC protein for activity like AcrAB (27). Importantly, TolC is not required for the activity of the single-component pump MdfA or CmlA: the MICs of chloramphenicol, conferred by pLQ821 (cmlA) or by pAL261 (mdfA), were the same in strains ECM1694 (ΔacrAB::Km) and ECM1556 (tolC::Tn10), lacking acrAB and tolC, respectively (data not shown). These data also indicate that deletion of tolC does not impair the efflux activity of other pumps.
To investigate the interplay between the CmlA and MdfA transporters, we first selected a mutant that overexpressed mdfA due to a presently uncharacterized chromosomal mutation. The mutant strain ECM1888, carrying a mutation in a gene tentatively called mdfR, was selected from strain ECM1556 (tolC::Tn10) (see Materials and Methods). It is noteworthy that there are no genes with homology to transcriptional regulators immediately upstream or downstream of mdfA (3). Therefore, the mdfR mutation occurred either in the promoter region of mdfA or in a gene that was unlinked to the mdfA locus.
Next, we compared the MICs of chloramphenicol for strains overexpressing the chromosomally encoded MdfA and the plasmid-encoded CmlA singly (ECM1888 and ECM1911) and in combination (ECM1908). The results are shown in Table 4. Each singly expressed pump conferred a similar level of chloramphenicol resistance (MIC of 16 μg/ml for MdfA and 32 μg/ml for CmlA). However, combining MdfA and CmlA in the same strain did not produce any multiplicative effect, such as that seen in the case of the CmlA-AcrAB or MdfA-AcrAB pair: the MIC for strain ECM1908, which contained both pumps, was still 32 μg/ml. One of the possibilities was that CmlA and MdfA do not function independently and decreased each other's activity. Since the MdfA protein confers resistance to ethidium bromide, while CmlA does not, it was possible to assess MdfA activity (and expression) in the presence or absence of CmlA. The MIC of ethidium bromide was not affected by the CmlA protein, thus ruling out the possibility of an inhibitory effect of CmlA. Due to the lack of CmlA-specific substrates we were not able to clarify if MdfA affected the activity of CmlA. To learn whether the mdfR mutation (assuming that it is not a cis mutation but a mutation in an mdfA-unlinked locus) represses expression of CmlA, pLQ821 was introduced in strain ECM1915 (mdfR ΔmdfA::Km). The chloramphenicol MIC for the resulting strain, ECM1955, was the same as that for ECM1911 (ΔmdfA::Km/pLQ821), making this possibility very unlikely.
TABLE 4.
Effect of simultaneous expression of single-component pumps CmlA and MdfA on susceptibility to chloramphenicol in E. coli
| CmlA | MIC of chloramphenicol (μg/ml) for the indicated strain with the following MdfA status:
|
|
|---|---|---|
| Absent | Present | |
| Absent | 1 (ECM1816) | 16 (ECM1888) |
| Present | 32 (ECM1911) | 32 (ECM1908) |
In the case of the two single-component pumps in P. aeruginosa, we have studied the effects of simultaneous expression of the TetA and TetC transporters on resistance to tetracycline. All studied P. aeruginosa strains lacked the functional pump MexAB-OprM, which is constitutively expressed in wild-type cells of P. aeruginosa and contributes to intrinsic antibiotic resistance (13, 32, 35).
Measurements of susceptibility for tetracycline demonstrated that combining two different Tet transporters in the same cell did not produce an increase in drug resistance much stronger than that conferred by each pump expressed singly (Table 5). The results with Tet transporters in P. aeruginosa were similar to the results described above for the two single-component pumps in E. coli. However, it still remains to be clarified whether the Tet transporters are being expressed and are working independently from each other.
TABLE 5.
Effect of simultaneous expression of single-component pumps TetA and TetC on susceptibility to tetracycline in P. aeruginosa
| TetA | MIC of tetracycline (μg/ml) for the indicated strain with the following TetC status:
|
|
|---|---|---|
| Absent | Present | |
| Absent | 0.25–0.5 (PAM1154) | 16 (PAM2455) |
| Present | 32 (PAM1316) | 32 (PAM2458) |
Effect of simultaneous expression of multicomponent efflux pumps on the resistance to antibiotics in P. aeruginosa.
We have studied the interplay between the three multicomponent pumps, MexAB-OprM, MexCD-OprJ, and MexEF-OprN, in P. aeruginosa. All three pumps confer resistance to multiple antibiotics and, importantly, have several overlapping substrates (fluoroquinolones, tetracycline, and chloramphenicol, etc.).
We have constructed strains of P. aeruginosa overexpressing each of the mentioned Mex pumps singly or in combination. We have also selected a strain which simultaneously overexpressed all three of these efflux pumps. Mutants simultaneously overexpressing multiple Mex pumps were selected using pump-specific efflux pump inhibitors (see Materials and Methods) (Table 1). Expression of multiple efflux pumps did not affect the growth rate of P. aeruginosa (data not shown). Western analysis with antibodies against the outer membrane components of the pumps has confirmed overexpression of each of the pumps in double and triple overexpressors (data not shown). This was also confirmed in the gene disruption experiments: the resistance to antibiotics in the double overexpressors was not reversed when only genes encoding a single pump were inactivated (Table 6).
TABLE 6.
Effect of simultaneous expression of pairs of multicomponent pumps on susceptibility to antibiotics in P. aeruginosa
| Strain | Genotype | Pump Status
|
MIC (μg/ml)
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MexAB-OprM | MexCD-OprJ | MexEF-OprN | Levofloxacin | Tetracycline | Chloramphenicol | Cefpirome | Cefepime | Carbenicillin | Aztreonam | Rifampin | ||
| PAM1020 | Wild type | + | − | − | 0.125 | 4 | 128 | 1 | 0.5 | 64 | 2 | 16 |
| PAM1275 | mexB::Hg | − | − | − | 0.03 | 0.5 | 4 | 0.25 | 0.125 | 0.5 | 0.25 | 16 |
| PAM1032 | nalB1032 | +++ | − | − | 1 | 32 | 512 | 4 | 4 | 256 | 16 | 16 |
| PAM1277 | mexB::Hg nfxB1033 | − | +++ | − | 2 | 16 | 512 | 16 | 4 | 0.5 | 0.125 | NDa |
| PAM1465 | mexB::Hg nfxB1438 | − | +++ | − | 1 | 16 | 256 | 8 | 2 | 0.5 | 0.125 | ND |
| PAM1438 | nalB1032 nfxB1438 | +++ | +++ | − | 2 | 64 | 512 | 16 | 4 | 256 | 16 | 16 |
| PAM2359 | nalB2281 ΔmexEF::Gm | +++ | − | − | 1 | 32 | 512 | ND | ND | 256 | 16 | ND |
| PAM2360 | nalB2282 ΔmexEF::Gm | +++ | − | − | 1 | 16 | 256 | 2 | 2 | 256 | 16 | ND |
| PAM1278 | mexB::Hg nfxC1034 | − | − | +++ | 2 | 8 | 2048 | 0.25 | 0.25 | 0.5 | 0.125 | ND |
| PAM2281 | nalB2281 nfxC1034 | +++ | − | +++ | 4–8 | 64 | 2048 | 4 | 4 | 256 | 16 | 16 |
| PAM2282 | nalB2282 nfxC1034 | +++ | − | +++ | 4 | 32 | 2048 | 2 | 2 | 256 | 8 | 16 |
| PAM2302 | nalB2281 nfxC1034 nfxB2302 | +++ | +++ | +++ | 8 | 128 | >512 | 8 | 4 | 256 | 16 | 16 |
| PAM2303 | nalB2281 nfxC1034 nfxB2303 | +++ | +++ | +++ | 4–8 | 64 | 2048 | 8 | 4 | 128 | 8 | 16 |
| PAM2387 | oprM::Hg nfxC1034 nfxB2303 | − | +++ | +++ | 4–8 | 32 | 2048 | 8 | 2 | 0.5 | 0.125 | ND |
ND, not determined.
To assess the effect of simultaneously overexpressing multiple pumps, we compared the levels of resistance to the common substrates levofloxacin, chloramphenicol, tetracycline, and cefepime or cefpirome (which are shared by MexAB-OprM and MexCD-OprJ) in the strains overexpressing single and multiple efflux pumps (Table 6). For all common antibiotic substrates tested, each of the efflux pumps when individually overexpressed conferred a comparable level of resistance. However, the effect on antibiotic resistance (for all common antibiotic substrates, without exception) conferred by each of the Mex pumps was dependent on the level of expression of other Mex pumps present in the same cell. As an example, MexCD-OprJ and MexEF-OprN conferred 32- to 64-fold, 16- to 32-fold, and 64- to 512-fold increases in levofloxacin, tetracycline, and chloramphenicol resistance, respectively, in strains lacking MexAB-OprM (compare PAM1465 and PAM1278 with PAM1275). However, the same pumps conferred only a two- to fourfold increase in resistance to these antibiotics in strains overexpressing the MexAB-OprM efflux pump (compare PAM1438 and PAM2281/PAM2282 with PAM1032). The data from Table 6 show that regardless of the nature of the antibiotic or of the absolute level of antibiotic resistance, simultaneous overexpression of pairs of multicomponent efflux pumps only results in additive effects on drug resistance similar to that seen for pairs of single-component pumps.
As for the previously described cases, we addressed the possibility that pumps that are simultaneously present in the same strain are less active than when they are expressed singly. The MexAB-OprM pump is the only one which confers resistance to the β-lactams carbenicillin and aztreonam (22). The level of resistance to these antibiotics in strains PAM1438 (MexAB-OprM and MexCD-OprJ overexpressed) and PAM2281 and PAM2282 (MexAB-OprM and MexEF-OprN overexpressed) or in strains PAM2302 and PAM2303 (overexpressing all three pumps) was the same as in strain PAM1032 (overexpressing MexAB-OprM alone) (256 and 16 μg/ml for carbenicillin and aztreonam, respectively), indicating that the MexAB-OprM pump is just as functional in the strains overexpressing other pumps as it is in the strain expressing this pump alone (Table 6). The MexCD-OprJ pump confers resistance to the cephalosporins cefepime and cefpirome (21). The level of resistance to these antibiotics in strain PAM2387 (MexAB-OprM nonfunctional, MexCD-OprJ and MexEF-OprN overexpressed) was the same as in PAM1465 (MexCD-OprJ overexpressed, MexAB-OprM nonfunctional), indicating that the MexCD-OprJ pump is as active in the strain overexpressing MexEF-OprN as it is in the strain expressing only MexCD-OprJ. Finally, overexpression of known Mex pumps did not appear to affect activity of other efflux pumps present in P. aeruginosa. This conclusion was based on the following observation. The antibiotic rifampin is not a substrate of the known Mex pumps (Table 6). At the same time, the MIC of rifampin is significantly decreased in the presence of the broad-spectrum efflux pump inhibitor MC-207,110 (A. Lee and O. Lomovskaya, unpublished data), implying that rifampin is extruded by a pump that has not yet been identified. We have demonstrated that the MIC of rifampin remained unchanged in the strains overexpressing various efflux pumps. Thus, our data indicate that in the strains overexpressing combinations of efflux pumps, at least some of these pumps were fully functional.
DISCUSSION
Multiple mechanisms of resistance to a particular antibiotic can coexist in the same bacterial strain. Effects on antibiotic resistance due to interplay of different resistance mechanisms have been previously studied in some detail. Examples include interplay between efflux-based and target-mediated resistance to fluoroquinolones in P. aeruginosa (17) and E. coli (30), between increased efflux and decreased permeability in resistance to carbapenems (12), and between efflux pumps and β-lactamases in resistance to a variety of β-lactams in P. aeruginosa (20, 24). For both of the first two cases it was demonstrated that the combined presence conferred a multiplicative effect on drug resistance; i.e., the fold increase in MIC due to the combined presence was close to the product of the individual fold increases produced by individual mechanisms. However, in the third case, interplay between different mechanisms produced only a smaller, additive, effect (the fold increase in MIC was a sum of the individual fold increases). We sought to investigate the effect on drug resistance due to the interplay between various efflux pumps. Several types of pump combinations have been studied: (i) simultaneous expression of a single-component efflux pump, which exports antibiotics into the periplasm, in combination with a multicomponent efflux pump that accomplishes efflux directly into the external medium; (ii) simultaneous expression of two single-component pumps; and (iii) simultaneous expression of two multicomponent pumps. It was found that when efflux pumps with different structural types were combined in the same cell (the first case), the observed antibiotic resistance was much higher than that conferred by each of the pumps expressed singly, and the fold increase in drug resistance was close to the product of the fold increases due to the individual pumps. Simultaneous expression of either two single-component or two multicomponent efflux pumps (the second and third cases) did not produce similar large increases in antibiotic resistance. It appears that in the latter two cases, simultaneous expression of efflux pumps resulted in only an additive effect on drug resistance.
What are the reasons that such different effects on drug resistance are seen for various combinations of the pumps? One possibility is that multiplicative versus additive effects on drug resistance could be attributed to factors specific to each particular pair of the pumps used in experiments. For example, depending on whether any particular efflux pump is present singly or in combination with another pump, it may have a different level of expression and/or activity. Moreover, one can imagine that some pumps may increase the activity while other pumps inhibit the activity and/or expression of another pump present in the same cell. In several instances we specifically addressed this possibility without obtaining any confirmatory data. Still, this possibility shall not be ruled out completely. For example, we did not have tools to assess the efflux activity of the MexEF-OprN efflux pump in the presence of active MexAB-OprM or MexCD-OprJ efflux pumps. Therefore, we cannot exclude the possibility that the lack of a multiplicative effect on drug resistance in case of the MexEF-MexAB combination was due to inhibition of activity of the MexEF-OprN pump. Similarly, due to the lack of tools to assess the independence of CmlA or MdfA from AcrAB-TolC and of TetA or TetC from MexAB-OprM, we shall not exclude the possibility that the multiplicative effect on drug resistance in the case of these combinations could be explained by a stimulatory activity of the multicomponent pumps.
It is noteworthy, however, that in several cases we could prove that at least one pump from the pair was expressed and worked independently of the other pump. These facts indicate that it is feasible to assume that the studied efflux pumps are independent of each other (when overexpressed). Based on this assumption, a very simple and general explanation of our results could be offered. We hypothesize that multiplicative or additive effects are due to differences in the structural organizations of the pumps and, consequently, different modes of efflux. Our conjecture is that pumps of the same structural type are organized in parallel, while pumps of different structural types function in series.
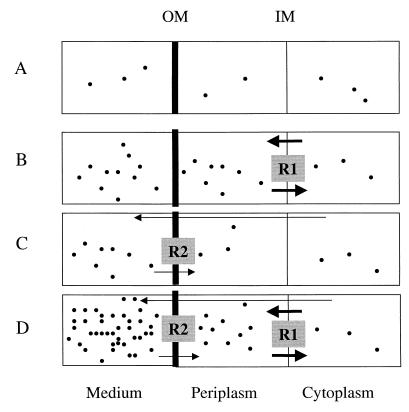
This explanation is justified by considering a simple model, presented in Fig. 1. Consider the steady state at which the concentration of an inhibitor in the cytoplasm, Cin, corresponds to the concentration sufficient to inhibit the antibiotic's target and always remains the same regardless of whether or not a strain has an efflux pump(s). The concentration in the external medium, Cout, is equal to the MIC and will be different depending on the presence or absence of an efflux pump(s). For the pumpless strain, Cin = Cout. In the range of concentrations where neither of the pumps is saturated, each pump maintains a concentration gradient, R, between the external medium and the cytoplasm (R = Cout/Cin). Accordingly, R approximately corresponds to the fold increase in drug resistance in the presence of a pump. In the case of a single-component efflux pump, whose substrates are extruded into the periplasm, the gradient is maintained across the inner membrane (R1) (Fig. 1B). As a result, the MIC afforded by this pump is equal to Cout ≈ Cin · R1. For a multicomponent pump, the gradient exists across the outer membrane (R2) (Fig. 1C), and MIC ≈ Cout ≈ Cin · R2. When both pumps are simultaneously engaged, a gradient of concentrations is maintained across both membranes. As a result, the concentration in the periplasm is now Cin · R1, and the concentration in the external medium, which corresponds approximately to the new MIC, is Cin · R1 · R2. Therefore, the fold increase in resistance for the case of series pumps is equal to R1 · R2, or the product of the corresponding fold increases.
FIG. 1.
Simplified model of antibiotic fluxes in bacterial cells without efflux pumps (A), cells expressing a single-component pump (B), cells expressing a multicomponent efflux pump (C), and cells simultaneously expressing a multicomponent and a single-component pump (D). The models show the fluxes of antibiotics at steady state. External concentrations of antibiotics are equal to MICs and the density of dots corresponds approximately to the concentration of antibiotic in each compartment. In each case, the concentration of antibiotics in the cytoplasm is the same and is sufficient to inhibit the target for antibiotics. Arrows indicate directions, and their thicknesses indicate rates of flux. The thick line separating the periplasm and the external medium indicates the low permeability of the outer membrane (OM) compared to the inner membrane (IM). In cells without efflux pumps (A), all compartments are essentially in equilibrium. In cells expressing a single-component pump (B), which extrudes substrates into the periplasm, thus balancing the rapid influx across the inner membrane, the external medium and the periplasm are in equilibrium, and the gradient of concentrations (R1) exists at the inner membrane. In cells expressing a multicomponent efflux pump (C), which extrudes antibiotics in the external medium, bypassing the outer membrane barrier, the cytoplasm and the periplasm are in equilibrium, and the gradient of concentrations (R2) emerges at the outer membrane. In the case of simultaneous expression of both efflux pumps (D), respective concentration gradients are maintained at both the inner and the outer membranes. This results in a multiplicative effect on drug resistance.
If two independent pumps are of the same type, the concentration in the external medium for pairs of multicomponent pumps or in the periplasm for pairs of the single-component pumps (which is equal to the MIC) will be equal to Cin · R1 + Cin · R2 (where R1 and R2 are the concentration gradients maintained by each of the pumps). Therefore, the fold increase in resistance for the case of parallel pumps is equal to R1 + R2, or the sum of the corresponding fold increases.
This explanation is substantiated by the fact that the same results were obtained for both P. aeruginosa and E. coli, for several pump combinations belonging to each of the cases, and that these results were independent of the antibiotics used in the study. Moreover, chloramphenicol and tetracycline were antibiotics that were common for each of the three cases studied.
Both cases investigated in this study could be relevant in clinical settings. In some strains of E. coli, which are highly resistant to chloramphenicol (MIC > 128 μg/ml), the resistance observed is the result of simultaneous overexpression of both the multicomponent AcrAB-TolC pump and the single-component specific efflux pump, Flo (Lee and Lomovskaya, unpublished data). This pump was first discovered in Pasteurella piscicida (11) but recently has also been detected in clinical strains of Salmonella typhimurium (4) and E. coli (10). Strains simultaneously overexpressing Mex pumps have also been reported among clinical isolates of P. aeruginosa (Cho et al., 39th ICAAC). The difference between series and parallel pairs of pumps should be taken into consideration when devising strategies to combat efflux pump-mediated drug resistance. In the case of a series pair, inhibition of either pump will effectively decrease drug resistance. In the parallel case, inhibition of both pumps is required to achieve a substantial effect.
ACKNOWLEDGMENTS
We thank Paul Roy, Hiroshi Nikaido, Paul Miller, and Eitan Bibi for providing various strains and plasmids. We are grateful to Naomasa Gotoh for monoclonal antibodies against OprM, OprJ, and OprN. We are indebted to Kim Lewis for his ideas regarding the mechanism of interactions between efflux pumps. We are grateful to Don Biek, Will Watkins, Molly Schmid, and George Miller for critical reading of the manuscript.
REFERENCES
- 1.Aires J R, Kohler T, Nikaido H, Plesiat P. Involvement of an active efflux system in the natural resistance of Pseudomonas aeruginosa to aminoglycosides. Antimicrob Agents Chemother. 1999;43:2624–2628. doi: 10.1128/aac.43.11.2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bissonnette L, Champetier S, Buisson J P, Roy P H. Characterization of the nonenzymatic chloramphenicol resistance (cmlA) gene of the In4 integron of Tn1696: similarity of the product to transmembrane transport proteins. J Bacteriol. 1991;173:4493–4502. doi: 10.1128/jb.173.14.4493-4502.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blattner F R, Plunkett III G, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 4.Bolton L F, Kelley L C, Lee M D, Fedorka-Cray P J, Maurer J J. Detection of multidrug-resistant Salmonella enterica serotype Typhimurium DT104 based on a gene which confers cross-resistance to florfenicol and chloramphenicol. J Clin Microbiol. 1999;37:1348–1351. doi: 10.1128/jcm.37.5.1348-1351.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Darzins A, Casadaban M J. Mini-D3112 bacteriophage transposable elements for genetic analysis of Pseudomonas aeruginosa. J Bacteriol. 1989;171:3909–3916. doi: 10.1128/jb.171.7.3909-3916.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Edgar R, Bibi E. MdfA, an Escherichia coli multidrug resistance protein with an extraordinarily broad spectrum of drug recognition. J Bacteriol. 1997;179:2274–2280. doi: 10.1128/jb.179.7.2274-2280.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Edgar R, Bibi E. A single membrane-embedded negative charge is critical for recognizing positively charged drugs by the Escherichia coli multidrug resistance protein MdfA. EMBO J. 1999;18:822–832. doi: 10.1093/emboj/18.4.822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gotoh N, Itoh N, Tsujimoto H, Yamagishi J, Oyamada Y, Nishino T. Isolation of OprM-deficient mutants of Pseudomonas aeruginosa by transposon insertion mutagenesis: evidence of involvement in multiple antibiotic resistance. FEMS Microbiol Lett. 1994;122:267–273. doi: 10.1111/j.1574-6968.1994.tb07179.x. [DOI] [PubMed] [Google Scholar]
- 9.Gotoh N, Tsujimoto H, Tsuda M, Okamoto K, Nomura A, Wada T, Nakahashi M, Nishino T. Characterization of the MexC-MexD-OprJ multidrug efflux system in Δ(mexA-mexB-oprM) mutants of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1998;42:1938–1943. doi: 10.1128/aac.42.8.1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Keyes K, Hudson C, Maurer J J, Thayer S, White D G, Lee M D. Detection of florfenicol resistance genes in Escherichia coli isolated from sick chickens. Antimicrob Agents Chemother. 2000;44:421–424. doi: 10.1128/aac.44.2.421-424.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim E, Aoki T. Sequence analysis of the florfenicol resistance gene encoded in the transferable R-plasmid of a fish pathogen, Pasteurella piscicida. Microbiol Immunol. 1996;40:665–9. doi: 10.1111/j.1348-0421.1996.tb01125.x. [DOI] [PubMed] [Google Scholar]
- 12.Kohler T, Michea-Hamzehpour M, Epp S F, Pechere J C. Carbapenem activities against Pseudomonas aeruginosa: respective contributions of OprD and efflux systems. Antimicrob Agents Chemother. 1999;43:424–427. doi: 10.1128/aac.43.2.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kohler T, Michea-Hamzehpour M, Henze U, Gotoh N, Curty L K, Pechere J C. Characterization of MexE-MexF-OprN, a positively regulated multidrug efflux system of Pseudomonas aeruginosa. Mol Microbiol. 1997;23:345–354. doi: 10.1046/j.1365-2958.1997.2281594.x. [DOI] [PubMed] [Google Scholar]
- 14.Krishnapillai V. A novel transducing phage. Its role in recognition of a possible new host-controlled modification system in Pseudomonas aeruginosa. Mol Gen Genet. 1972;114:134–143. doi: 10.1007/BF00332784. [DOI] [PubMed] [Google Scholar]
- 15.Levy S B, McMurry L M, Barbosa T M, Burdett V, Courvalin P, Hillen W, Roberts M C, Rood J I, Taylor D E. Nomenclature for new tetracycline resistance determinants. Antimicrob Agents Chemother. 1999;43:1523–1524. doi: 10.1128/aac.43.6.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li X Z, Nikaido H, Poole K. Role of mexA-mexB-oprM in antibiotic efflux in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1995;39:1948–1953. doi: 10.1128/aac.39.9.1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lomovskaya O, Lee A, Hoshino K, Ishida H, Mistry A, Warren M S, Boyer E, Chamberland S, Lee V J. Use of a genetic approach to evaluate the consequences of inhibition of efflux pumps in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1999;43:1340–1346. doi: 10.1128/aac.43.6.1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lomovskaya O, Lewis K, Matin A. EmrR is a negative regulator of the Escherichia coli multidrug resistance pump EmrAB. J Bacteriol. 1995;177:2328–2334. doi: 10.1128/jb.177.9.2328-2334.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ma D, Cook D N, Alberti M, Pon N G, Nikaido H, Hearst J E. Molecular cloning and characterization of acrA and acrE genes of Escherichia coli. J Bacteriol. 1993;175:6299–6313. doi: 10.1128/jb.175.19.6299-6313.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Masuda N, Gotoh N, Ishii C, Sakagawa E, Ohya S, Nishino T. Interplay between chromosomal beta-lactamase and the MexAB-OprM efflux system in intrinsic resistance to beta-lactams in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1999;43:400–402. doi: 10.1128/aac.43.2.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Masuda N, Gotoh N, Ohya S, Nishino T. Quantitative correlation between susceptibility and OprJ production in nfxB mutants of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1996;40:909–913. doi: 10.1128/aac.40.4.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Masuda N, Ohya S. Cross-resistance to meropenem, cephems, and quinolones in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1992;36:1847–1851. doi: 10.1128/aac.36.9.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morshed S R, Lei Y, Yoneyama H, Nakae T. Expression of genes associated with antibiotic extrusion in Pseudomonas aeruginosa. Biochem Biophys Res Commun. 1995;210:356–362. doi: 10.1006/bbrc.1995.1669. [DOI] [PubMed] [Google Scholar]
- 24.Nakae T, Nakajima A, Ono T, Saito K, Yoneyama H. Resistance to beta-lactam antibiotics in Pseudomonas aeruginosa due to interplay between the MexAB-OprM efflux pump and beta-lactamase. Antimicrob Agents Chemother. 1999;43:1301–1303. doi: 10.1128/aac.43.5.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.National Committee for Clinical Laboratory Standards. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. 4th ed. 1997. Approved standards. NCCLS document M7-A4. National Committee for Clinical Laboratory Standards, Wayne, Pa. [Google Scholar]
- 26.Nikaido H. Antibiotic resistance caused by gram-negative multidrug efflux pumps. Clin Infect Dis. 1998;27(Suppl. 1):S32–S41. doi: 10.1086/514920. [DOI] [PubMed] [Google Scholar]
- 27.Nikaido H. Multidrug efflux pumps of gram-negative bacteria. J Bacteriol. 1996;178:5853–5859. doi: 10.1128/jb.178.20.5853-5859.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nikaido H. Multiple antibiotic resistance and efflux. Curr Opin Microbiol. 1998;1:516–523. doi: 10.1016/s1369-5274(98)80083-0. [DOI] [PubMed] [Google Scholar]
- 29.Nikaido H, Vaara M. Molecular basis of bacterial outer membrane permeability. Microbiol Rev. 1985;49:1–32. doi: 10.1128/mr.49.1.1-32.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oethinger M, Kern W V, Jellen-Ritter A S, McMurry L M, Levy S B. Ineffectiveness of topoisomerase mutations in mediating clinically significant fluoroquinolone resistance in Escherichia coli in the absence of the AcrAB efflux pump. Antimicrob Agents Chemother. 2000;44:10–13. doi: 10.1128/aac.44.1.10-13.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Okusu H, Ma D, Nikaido H. AcrAB efflux pump plays a major role in the antibiotic resistance phenotype of Escherichia coli multiple-antibiotic-resistance (Mar) mutants. J Bacteriol. 1996;178:306–308. doi: 10.1128/jb.178.1.306-308.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Poole K, Gotoh N, Tsujimoto H, Zhao Q, Wada A, Yamasaki T, Neshat S, Yamagishi J, Li X Z, Nishino T. Overexpression of the mexC-mexD-oprJ efflux operon in nfxB-type multidrug-resistant strains of Pseudomonas aeruginosa. Mol Microbiol. 1996;21:713–724. doi: 10.1046/j.1365-2958.1996.281397.x. [DOI] [PubMed] [Google Scholar]
- 33.Poole K, Heinrichs D E, Neshat S. Cloning and sequence analysis of an EnvCD homologue in Pseudomonas aeruginosa: regulation by iron and possible involvement in the secretion of the siderophore pyoverdine. Mol Microbiol. 1993;10:529–544. doi: 10.1111/j.1365-2958.1993.tb00925.x. [DOI] [PubMed] [Google Scholar]
- 34.Poole K, Krebes K, McNally C, Neshat S. Multiple antibiotic resistance in Pseudomonas aeruginosa: evidence for involvement of an efflux operon. J Bacteriol. 1993;175:7363–7372. doi: 10.1128/jb.175.22.7363-7372.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Poole K, Tetro K, Zhao Q, Neshat S, Heinrichs D E, Bianco N. Expression of the multidrug resistance operon mexA-mexB-oprM in Pseudomonas aeruginosa: mexR encodes a regulator of operon expression. Antimicrob Agents Chemother. 1996;40:2021–2028. doi: 10.1128/aac.40.9.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Renau T E, Leger R, Flamme E M, Sangalang J, She M W, Yen R, Gannon C L, Griffith D, Chamberland S, Lomovskaya O, Hecker S J, Lee V J, Ohta T, Nakayama K. Inhibitors of efflux pumps in Pseudomonas aeruginosa potentiate the activity of the fluoroquinolone antibacterial levofloxacin. J Med Chem. 1999;42:4928–4931. doi: 10.1021/jm9904598. [DOI] [PubMed] [Google Scholar]
- 37.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 38.Schnappinger D, Hillen W. Tetracyclines: antibiotic action, uptake, and resistance mechanisms. Arch Microbiol. 1996;165:359–369. doi: 10.1007/s002030050339. [DOI] [PubMed] [Google Scholar]
- 39.Simon R, Priefer Y, Puehler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram-negative bacteria. Biotechnology. 1983;1:784–791. [Google Scholar]
- 40.Thanassi D G, Suh G S, Nikaido H. Role of outer membrane barrier in efflux-mediated tetracycline resistance of Escherichia coli. J Bacteriol. 1995;177:998–1007. doi: 10.1128/jb.177.4.998-1007.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yoneyama H, Ocaktan A, Tsuda M, Nakae T. The role of mex-gene products in antibiotic extrusion in Pseudomonas aeruginosa. Biochem Biophys Res Commun. 1997;233:611–618. doi: 10.1006/bbrc.1997.6506. [DOI] [PubMed] [Google Scholar]
- 42.Zgurskaya H I, Nikaido H. AcrA is a highly asymmetric protein capable of spanning the periplasm. J Mol Biol. 1999;285:409–420. doi: 10.1006/jmbi.1998.2313. [DOI] [PubMed] [Google Scholar]