Abstract
The uvrD gene in Escherichia coli encodes a 720-amino-acid 3′-5′ DNA helicase which, although nonessential for viability, is required for methyl-directed mismatch repair and nucleotide excision repair and furthermore is believed to participate in recombination and DNA replication. We have shown in this study that null mutations in uvrD are incompatible with lon, the incompatibility being a consequence of the chronic induction of SOS in uvrD strains and the resultant accumulation of the cell septation inhibitor SulA (which is a normal target for degradation by Lon protease). uvrD-lon incompatibility was suppressed by sulA, lexA3(Ind−), or recA (Def) mutations. Other mutations, such as priA, dam, polA, and dnaQ (mutD) mutations, which lead to persistent SOS induction, were also lon incompatible. SOS induction was not observed in uvrC and mutH (or mutS) mutants defective, respectively, in excision repair and mismatch repair. Nor was uvrD-mediated SOS induction abolished by mutations in genes that affect mismatch repair (mutH), excision repair (uvrC), or recombination (recB and recF). These data suggest that SOS induction in uvrD mutants is not a consequence of defects in these three pathways. We propose that the UvrD helicase participates in DNA replication to unwind secondary structures on the lagging strand immediately behind the progressing replication fork, and that it is the absence of this function which contributes to SOS induction in uvrD strains.
DNA helicases are enzymes involved in a variety of processes such as DNA replication, repair, and recombination, and at least 12 have been identified in Escherichia coli (29, 30). Of these, only DnaB helicase (with 5′-3′ polarity) is essential for cell viability, being associated with movement of the chromosome replication fork (27). Three other helicases, Rep, PriA, and UvrD, each with 3′-5′ polarity, have also been suggested to participate in chromosome replication, but their roles are not as well established.
The UvrD helicase (also called helicase II) is the product of the uvrD gene and is a polypeptide of 720 amino acids. UvrD is required in the pathways of both methyl-directed mismatch repair (34, 35) and UvrABC-mediated excision repair (46, 47), and uvrD mutants exhibit a spontaneous mutator phenotype as well as increased sensitivity to UV irradiation. Genetic evidence has implicated UvrD in homologous recombination (21, 23, 26, 32). The suggestion has also been made that UvrD participates in DNA replication, based on (i) experiments with cell-free systems (17, 22), (ii) the observation that uvrD null mutations are incompatible with rep (encoding the Rep helicase) or polA (encoding DNA polymerase I) mutations (56), and (iii) identification of dominant-lethal uvrD mutations (8, 59).
In the present study, we discovered that lon (encoding the Lon protease) represents a third locus, mutations in which are incompatible with uvrD null alleles. Chronic induction of the SOS response (reviewed in references 23 and 55) in the uvrD null strains was shown to be the basis for lon incompatibility; other mutations (in dam, priA, mutD, and polA) that exhibited persistent SOS induction were also lon incompatible. We propose a model suggesting a role for the UvrD helicase in removing regions of secondary structure in the lagging strand during replication fork progression, which could account for chronic generation of an SOS-inducing signal in uvrD mutants.
MATERIALS AND METHODS
Growth media and conditions.
The defined and nutrient media were, respectively, glucose-minimal A and Luria-Bertani (LB) medium (33). cpsB-lac expression was monitored on MacConkey-lactose agar (Difco). Unless otherwise indicated, the growth temperature was 37°C. Isopropyl β-d-thiogalactopyranoside (IPTG) was added at a final concentration of 1 mM. Concentrations of antibiotics and 5-bromo-4-chloro 3-indolyl-β-d-galactoside (X-Gal) used were as described previously (43, 44).
Bacterial strains and phages.
The E. coli strains used are listed in Tables 1 and 2. Phage P1kc was from our laboratory stock. The transposon vehicle phages λ1105, λ1323, and λ1324 for transposition, respectively, of Tn10dKan, Tn10dTet, and Tn10dCm, have been described elsewhere (16). The ordered λ phage library of the E. coli genome (19) was obtained from K. Isono.
TABLE 1.
List of E. coli strainsa
| Strain | Genotypeb | Source or reference |
|---|---|---|
| BL21 | hsdS gal | 33 |
| JL2301c | thr leu arg his ΔlexA300::(Smr Spr) sulA (λsulA::lac) | J. Little via M. Radman |
| MG1655 | Wild type | Lab stock |
| SG20780 | araD139 Δ(argF-lac)U169 rpsL150 relA1 flbB5301 Δlon-510 cpsB10::lac-Mu-λ-imm | 57 |
| SG20781 | SG20780 lon+ | 57 |
| GJ1823 | MG1655 zbh-900::Tn10dKan(Ts)1 lacZ4525::Tn10dKan lon-103::IS186 | 44 |
| GJ1901 | ara zbh-900::Tn10dKan(Ts)1 lacZ(Am) lacI3098::Tn10Kan | From CSH142 (33), in several steps |
| GJ1904 | GJ1901 uvrD400::Tn10dTet | This study |
| GJ1907 | MG1655 zbh-900::Tn10dKan(Ts)1 lon-103::IS186 | This study |
| GJ1912 | MG1655 lon-103::IS186 zba-901::Tn10dKan | This study |
| GJ1913 | MG1655 zba-901::Tn10dKan | This study |
| GJ1916 | MG1655 lon-104::IS186::Tn10dCm zba-901::Tn10dKan | This study |
| GJ1922 | JL2301 lexA+ malB::Tn9 | This study |
| GJ1925 | JL2301 lexA+ malF3180::Tn10Kan | This study |
| GJ1988 | GJ1907 sulA::Tn5 uvrD400::Tn10dTet | This study |
| GJ1989 | GJ1907 lexA3 malB::Tn9 uvrD400::Tn10dTet | This study |
| GJ1992 | JL2301 lexA+ | This study |
| GJ1997 | GJ1907 sulA::Tn5 dam::Tn9 | This study |
| GJ1998 | GJ1907 sulA::Tn5 dnaQ (mutD5) zae-502::Tn10 | This study |
| GJ1999 | GJ1907 lexA3 malB::Tn9 priA1::Kan | This study |
| GJ2101 | SG20780 ΔuvrD288::Kan/pHYD141 | This study |
| GJ2102 | SG20780 ΔuvrD288::Kan recA56 srl-300::Tn10 | This study |
| GJ2122 | MG1655 ΔpolA::Kan lon-146::Tn10dTet/F′[polA(Klenow+)Cmr] | This study |
| GJ2126 | SG20780 uvrD3 ilv-500::Tn10 | This study |
| GJ2127 | SG20780 Δrep::Cm | This study |
Strain BL21 is an E. coli B derivative. All other strains are derivatives of E. coli K-12. Genotypes of additional sulA-lac strains used in this study are described in Table 2.
Genotype designations are as in the work of Berlyn (2). All strains are F− unless otherwise indicated. Allele numbers are given where they are known. Listed below in parentheses are references or sources for various mutations introduced into the GJ strains: ilv-500::Tn10, lacI3098::Tn10Kan, and malF3180::Tn10Kan (52); lacZ(Am), dnaQ, and zae-502::Tn10 (33); ΔpolA::Kan and F′[polA(Klenow+)Cmr] (12); lon-146::Tn10dTet (31); dam::Tn9 (N. Kleckner); ΔuvrD288::Kan (56); priA1::Kan (36); lexA3, sulA::Tn5, and malB::Tn9 (M. Radman); Δrep::Cm (B. Michel); recA56 and srl-300::Tn10 (44); and uvrD3 (E. coli Genetic Stock Center).
Genotype markers of JL2301 relevant to this study are listed.
TABLE 2.
Effects of mutations on sulA::lac expressiona
| Strain | Mutationb | Enzyme sp act |
|---|---|---|
| GJ1922 derivatives | ||
| GJ1922 | None | 49 |
| With pHYD136 | 44 | |
| GJ1923 | ΔlexA300::(Smr Spr) | 3,574 |
| GJ1927 | dnaQ (mutD5) zae-502::Tn10 | 138 |
| GJ1930 | uvrD400::Tn10dTet | 264 |
| Grown at 42°C | 236 | |
| GJ1931 | priA1::Kan | 397 |
| GJ1965 | ΔuvrD288::Kan | 258 |
| GJ1966 | uvrD3 ilv-500::Tn10 | 36 |
| GJ1991 | lexA3 malF3180::Tn10Kan uvrD400::Tn10dTet | 23 |
| GJ2103 | mutH471::Kan | 51 |
| GJ2104 | mutS::Tn10dTet | 62 |
| GJ2105 | uvrC279::Tn10 | 59 |
| GJ2106 | mutH471::Kan uvrC279::Tn10 | 65 |
| GJ2107 | mutS::Tn10dTet ΔuvrD288::Kan | 239 |
| GJ2108 | uvrC279::Tn10 ΔuvrD288::Kan | 271 |
| GJ1925 derivatives | ||
| GJ1925 | None | 52 |
| GJ1929 | dam::Tn9 | 237 |
| GJ1992 derivatives | ||
| GJ1992 | None | 52 |
| GJ1993 | ΔpolA::Kan | 2,050 |
| GJ1994 | ΔpolA::Kan/F′ [polA+ Cmr] | 37 |
| GJ1995 | ΔpolA::Kan/F′ [polA(Klenow+)Cmr] | 1,488 |
| With IPTG | 41 | |
| GJ1996 | ΔpolA::Kan/F′ [polA(5′ Exo+)Cmr] | 197 |
The specific activity of β-galactosidase was determined in each of the cultures grown to exponential phase in glucose-minimal medium (for strains GJ1993 and GJ1995) or LB (for all other strains), and values (representing the means of at least three independent experiments) are given in Miller units (33).
Mutations in addition to those present in the parental strain GJ1922, GJ1925, or GJ1992 are listed for each derivative. The reference or source for each of the mutations not described in Table 1 is given below in parentheses: mutH471::Kan (E. coli Genetic Stock Center); mutS::Tn10dTet (33); uvrC279::Tn10 (52); F′[polA+ Cmr] and F′[polA (5′ Exo+)Cmr] (12).
Plasmids.
The plasmid vectors pCL1920 (pSC101 based, encoding Smr Spr, for general cloning [24]) and pMAK705 (pSC101 based, encoding Cmr, temperature-sensitive replicon [10]) have been described previously. Plasmids pHYD130 and pHYD141 were constructed in this study and carry the lon+ gene subcloned from the λ phage 148 of the ordered library of Kohara et al. (19): the former bears a 6-kb insert comprising two contiguous PstI chromosomal segments in the PstI site of vector pCL1920, and the latter bears a 4.1-kb BglII-KpnI chromosomal segment in the BamHI-KpnI sites of vector pMAK705 (see Fig. 1). Plasmids pHYD136 and pHYD138 were also constructed in this study and are described below.
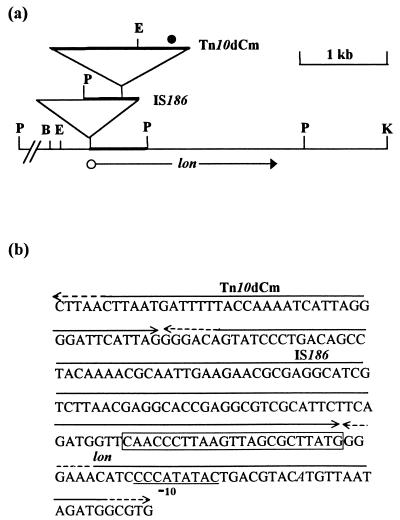
FIG. 1.
Molecular characterization of lon-103 and lon-104 mutations. (a) The line at the bottom depicts the physical map of the wild-type lon locus (4, 19) for the enzymes BglII (B), EcoRI (E), KpnI (K), and PstI (P); the open circle and arrow beneath the line denote, respectively, the promoter and transcribed region of the lon gene. Horizontal line segments of the lower and upper inverted triangles represent, respectively, IS186 and Tn10dCm; the vertices denote sites of insertion of these two elements in the lon-103 mutant and the lon-104 suppressor-bearing strain, respectively. The figure is drawn to the scale marked; the distance between the PstI site at the left and the BglII site is 2.42 kb. The contiguous region of DNA (delimited by PstI sites) corresponding to the insert cloned in plasmid pHYD138 is shown by heavy line segments. The recognition site for the Tn10dCm-specific sequencing primer (see the text) is marked by the solid circle. (b) Nucleotide sequence determined with a Tn10dCm-specific sequencing primer using pHYD138 DNA as a template. The extents of Tn10dCm, IS186, and lon sequences are indicated by double-headed arrows above the sequence; dashed lines have been used where the delimited ends do not represent the natural ends of the sequence in question. The right end of IS186 is as specified in the work of Sengstag et al. (50), and the sequence corresponding to the 23-bp right inverted repeat is boxed. The −10 region of the lon promoter (4, 11) is underlined, and the base representing the transcription start site is italicized.
Isolation and characterization of the uvrD400::Tn10dTet mutant.
Strain GJ1901, carrying a lacZ(Am) mutation, was subjected to random insertion mutagenesis with the Tn10dTet transposon following infection with λ1323, as described elsewhere (16). Tetr colonies exhibiting increased Lac+ papillation frequency were identified on LB agar plates supplemented with 0.1% lactose and X-Gal, as described earlier (43). One such mutant clone was designated GJ1904, and the following characteristics led to the conclusion that it carried a uvrD insertion, designated uvrD400::Tn10dTet. (i) Strain GJ1904 both was UV sensitive and exhibited a high frequency of spontaneous mutations to Lac+, nalidixic acid resistance, or rifampin resistance (data not shown). (ii) The Tn10dTet insertion in GJ1904 was linked 30% to the ilv locus in P1 transduction. (iii) Lastly, the uvrD400::Tn10dTet insertion was cloned from the chromosome of GJ1904 as part of a 6.6-kb PstI fragment (size inclusive of the 2.9-kb Tn10dTet element) in the plasmid vector pCL1920, and the resulting plasmid was designated pHYD136. The nucleotide sequence at either junction of Tn10dTet with the E. coli chromosomal DNA was determined by using pHYD136 DNA as a template and the oligonucleotides 5′-TGGTCACCAACGCTTTTCCCGAG-3′ and 5′-CTGTTGACAAAGGGAATCATAG-3′ as primers (designed so as to read outward from sites immediately internal to the inverted terminal repeat at either end of Tn10dTet); comparison of our results with the E. coli genome sequence (4) permitted the conclusion that the insertion had disrupted the (720-residue) uvrD open reading frame immediately after the second base of codon 476 (data not shown).
Transposon tagging and genetic mapping of uvrD-incompatible mutation in GJ1823.
Strain GJ1907 (a spontaneous Lac+ revertant of GJ1823) is Kanr at 30°C but Kans at 42°C, because of the presence in GJ1823 (and its retention in GJ1907) of a Kanr(Ts) insertion near the gal locus (44). GJ1907 was initially transduced to Kanr at 42°C with a P1 lysate prepared on a population of random Tn10dKan insertion clones of the wild-type strain MG1655. The pool of GJ1907 Kanr cells was then used as a recipient for transduction to Tetr with a P1 lysate prepared on the uvrD400::Tn10dTet strain GJ1904. Since the uvrD mutation is ordinarily lethal in GJ1823 and its derivatives (see below), only a small number of Tetr transductants were recovered in this cross, and these were presumed to represent clones in which the locus conferring incompatibility in GJ1823 had been replaced with the wild-type allele from MG1655 in the first transduction to Kanr. By this approach, a Kanr insertion (designated zba-901::Tn10dKan) 83% linked to the locus governing uvrD incompatibility was identified, and it was mapped to the 10-min region (data not shown). Subsequently, the mutation conferring incompatibility with uvrD was itself shown to be 92% cotransducible with a hupB::Kan insertion (data not shown).
Transposon-generated mutation that suppresses uvrD incompatibility in GJ1912.
Strain GJ1912 (which carries the uvrD-incompatible mutation) was subjected to random insertion mutagenesis with Tn10dCm following infection with λ1324, as described previously (16). The introduction of an uvrD400::Tn10dTet insertion into the pooled culture of Cmr clones, by P1 transduction, was then attempted. One viable Tetr transductant colony (GJ1916) so recovered was shown to have a Cmr insertion that (i) was located in the same 10-min chromosomal region as the locus conferring uvrD incompatibility in GJ1912 (and GJ1823) and (ii) was 100% cotransducible with the phenotype of suppression of uvrD incompatibility. The chromosomal region bearing the Tn10dCm insertion was cloned from GJ1916 on a 3.2-kb PstI fragment in the plasmid vector pCL1920, and the resultant plasmid was designated pHYD138. The oligonucleotide primer 5′-ATTCTGCCTCCCAGAGCCTGA-3′, designed to read outward from a site immediately internal to the inverted terminal repeat at one end of Tn10dCm, was used to determine the sequence at the junction of the Tn10dCm insertion in the insert DNA of pHYD138.
Tests for lon incompatibility.
Incompatibility of different mutations with lon was established by transduction experiments, wherein it was demonstrated that no colonies were recovered when an attempt was made to introduce an insertion mutation in the concerned locus into a lon strain, or vice versa, unless the recipient carried a plasmid such as pHYD130 bearing the lon+ gene. The demonstration in this study (see below) that the lon incompatibility of different mutations could be partially suppressed by growth at 42°C also allowed us to recover transductants from various crosses at the elevated temperature and then test them for a lethality phenotype at 37 or 30°C.
DNA methods.
The standard protocols of Sambrook et al. (45) were followed for experiments involving recombinant DNA. Nucleotide sequence determinations were done with the aid of an automated DNA sequencer, using double-stranded plasmid DNAs as templates. The BLAST programs were employed for comparison of DNA sequences with the GenBank sequence database.
Other techniques.
Procedures for conjugation (33), P1kc transduction (9), measurement of spontaneous mutation frequency to nalidixic acid or rifampin resistance (43), assay of cell survival following UV radiation (33), and measurement of β-galactosidase activity in cultures (33) were as described previously.
RESULTS
Identification of a cryptic uvrD-incompatible mutation in strain GJ1823 as lon.
The starting point of this study was our observation that a Tn10dTet insertion in codon 476 of the uvrD gene (uvrD400), obtained as described above, could not be transduced into GJ1823, a strain that had been derived (44) by transduction from wild-type MG1655 and that was not expected to carry a uvrD-incompatible mutation. We also confirmed that GJ1823 had not acquired a rep or polA mutation (data not shown); these are the two loci known earlier to be uvrD incompatible.
A transposon-tagging strategy was employed to localize the cryptic mutation in GJ1823 to the 10-min chromosomal region, and the following lines of evidence indicated that the mutation is in the lon locus. (i) Colonies of strains carrying the GJ1823 mutation were mucoid, and expression of cpsB-lac in strain SG20781 was markedly increased following introduction of the mutation by P1 transduction (data not shown); these are characteristic Lon phenotypes, since one of the substrates of the Lon protease is RcsA, an activator of the cps operon involved in capsular polysaccharide biosynthesis (31, 57). (ii) The uvrD::Tn10dTet insertion could not be introduced by transduction into strain SG20780, the isogenic Δlon derivative of SG20781. (iii) Strains GJ1823 and SG20780 could both be complemented for the uvrD incompatibility phenotype by plasmid pHYD130 carrying the cloned lon+ gene. (iv) Finally, DNA sequence determination of the insert cloned in plasmid pHYD138 (whose construction has been described above), and its comparison with the E. coli genome sequence (4), indicated, first, that the mutation in GJ1823 is an IS186 insertion (6, 20) within a run of G's in the spacer sequence between the −10 and −35 regions of the lon promoter and, second, that a Tn10dCm insertion which suppressed the UvrD incompatibility phenotype of GJ1823 was situated within, and 116 bp from the right end of, IS186 (as depicted in Fig. 1). The same Cmr insertion was also able to suppress the phenotypes of mucoidy and cpsB-lac derepression associated with the GJ1823 mutation (data not shown). The simplest hypothesis to explain these observations is that the IS186 insertion in GJ1823 disrupts the lon promoter and that the Tn10dCm suppressor restores lon+ expression by providing a new promoter (see Fig. 1). The original cryptic mutation in GJ1823 and the suppressor have been designated lon-103::IS186 and lon-104::IS186::Tn10dCm, respectively.
These results therefore established lon as an additional locus in which mutations are inviable in combination with uvrD.
SOS induction in uvrD400::Tn10dTet as basis for lon incompatibility.
Another substrate of the Lon protease is the cell septation inhibitor SulA, whose synthesis is induced during the SOS response; as a consequence, lon mutants are known to be sensitive to SOS-inducing agents (reviewed in references 23 and 55). At least some uvrD mutations have been shown earlier to induce the SOS response (3, 25, 38), and we therefore tested the hypothesis that the uvrD-lon combination is rendered inviable because Lon becomes essential to degrade the increased concentration of SulA in the uvrD mutant background. (i) Using sulA-lac expression as a measure of SOS induction, we found that the uvrD400::Tn10dTet insertion (which is lon incompatible) caused a moderate (5-fold) induction of the SOS regulon, as against the 70-fold increase observed in a fully derepressed (ΔlexA) strain (Table 2). The expression value for a merodiploid strain bearing uvrD+ on the chromosome and uvrD400 on plasmid pHYD136 was the same as that for haploid uvrD+ (Table 2; compare the first and second rows), indicating that the mutation is recessive to the wild-type gene. (ii) Contrarily, the uvrD3 mutation, which does not induce the SOS response (38) (see Table 2), was lon compatible (strain GJ2126). (iii) Finally, the uvrD400::Tn10dTet incompatibility with lon could be suppressed by sulA and lexA3 (Ind−) mutations in strains GJ1988 and GJ1989, respectively, in which the inhibitory accumulation of SulA is expected to be abolished (21, 23, 55); predictably, the uvrD400-mediated derepression of sulA-lac was not observed in the lexA3 background (Table 2, GJ1991).
By these experiments, therefore, we could conclude (i) that the lon incompatibility of uvrD400 is correlated with chronic SOS induction and the consequent accumulation of SulA in the latter strain and (ii) that such induction is not essential for the viability of the uvrD mutant (as the uvrD lexA3 strains are viable).
SOS induction is associated with null mutations in uvrD.
In order to address the question of whether SOS induction in uvrD strains is the null phenotype, we tested two deletion-insertion uvrD mutations, constructed by Kushner and coworkers (56) and recessive to uvrD+, for their effects on reporter gene expression from SOS-responsive promoters and for lon incompatibility. The ΔuvrD288::Kan allele (in which more than two-thirds of the gene has been deleted from its 3′ end) was associated with sulA-lac derepression (Table 2). Furthermore, both ΔuvrD288 and the ΔuvrD290::Tet mutation (which represents a deletion of the uvrD promoter and the 5′ region of the gene) were lon-103::IS186 incompatible (data not shown). We therefore conclude that SOS induction by uvrD is the null phenotype.
We also employed the uvrD288::Kan mutation to construct a viable uvrD lon recA (Def) triple mutant (GJ2102), as follows. The uvrD allele was first introduced by P1 transduction at 30°C into SG20780/pHYD141, a chromosomal lon mutant strain carrying lon+ on a Ts plasmid replicon. The resulting strain (GJ2101) could be successfully cured of the plasmid after it was made recA(Def). This experiment demonstrated that recA (which also serves to abrogate the SOS response [21, 23, 55]) is an additional suppressor of lon-uvrD incompatibility.
dam, mutD, polA, and priA mutations are also lon incompatible.
Mutations in dam, dnaQ (mutD), polA, and priA have previously been shown (or suggested) to lead to chronic SOS induction (1, 36, 39, 40, 48, 53), and we examined whether they are also lon incompatible. We initially confirmed that each of the mutations did lead to a 3- to 40-fold derepression of sulA-lac (Table 2). We then showed, by the criteria listed above, that all four mutations were lon incompatible (dam, dnaQ, and priA were tested with lon-103::IS186; ΔpolA was tested with lon-146::Tn10dTet) (data not shown). Suppression of lon incompatibility by sulA was demonstrated for dam and dnaQ (strains GJ1997 and GJ1998, respectively), and suppression of lon incompatibility by lexA3 was demonstrated for priA (strain GJ1999).
By introducing F′ plasmids carrying the appropriate polA variants into a chromosomal ΔpolA strain, we also tested the effects of the polA (5′ Exo+) and polA (Klenow+) mutations on SOS induction and lon incompatibility. The former encodes the isolated 5′-3′ exonuclease activity, and the latter encodes the 5′-3′ polymerase with associated 3′-5′ exonuclease activities of DNA polymerase I (12). We found that the polA (5′ Exo+) mutant exhibited fourfold chronic SOS induction (Table 2) and was also lon incompatible; the suggestion that polA (5′ Exo+) is associated with SOS induction has also been made earlier (1). On the other hand, the polA (Klenow+) allele (which is known to require IPTG for its expression [12]) was able to cause sulA-lac expression in a ΔpolA strain to revert to basal levels (Table 2); correspondingly, the lon polA (Klenow+) strain GJ2122 was viable on IPTG-supplemented media.
Finally, mutations in rep were shown earlier to be associated with a low level of chronic SOS induction (38, 49), and we also observed that a rep lon mutant strain (GJ2127) is very sick. Taken together, these data serve to establish the general validity of the notion that lon mutations are lethal in strain backgrounds which exhibit chronic SOS induction and that the lethality is sulA and lexA3 suppressible.
lon incompatibility of SOS-inducing mutations is partially suppressed at 42°C.
We noticed that the various SOS-inducing mutations such as uvrD400::Tn10dTet, uvrD288, dam, priA, and mutD could be introduced by transduction into a lon strain to give small colonies on plates incubated at 42°C and that the transductants could grow, albeit poorly, upon restreaking at 42°C but not at 30 or 37°C. The degree of sulA-lac derepression by uvrD400::Tn10dTet was unaffected at 42°C (Table 2). On the other hand, cpsB-lac expression in the lon mutant was less pronounced at 42°C (data not shown). Our results support the findings by other workers (13, 15, 51, 57) that alternative proteases such as HslVU (ClpYQ) whose synthesis is induced upon heat shock are able to substitute for Lon in degrading the substrates RcsA and SulA at the elevated temperature.
Dissociation of the uvrD role in SOS induction from its functions in mismatch repair, excision repair, and recombination.
The following experiments indicated that SOS induction in uvrD strains is not the consequence of the absence of mismatch repair or nucleotide excision repair. Mutations in mutH or mutS that interfere with mismatch repair did not induce the SOS response, nor did a mutation in uvrC that renders defective the pathway of nucleotide excision repair (Table 2); similar results with mutH, mutL, mutS, and uvrA have been reported earlier (28, 38). A mutH uvrC double mutant in which both pathways are simultaneously nonfunctional (as is the case in uvrD) also was not derepressed for sulA-lac expression (Table 2).
We considered the possibility that SOS induction in the uvrD mutant is the result of accumulation of a trapped intermediate in either the mismatch repair or the excision repair pathway. For example, UvrD has been shown to be necessary for turnover of UvrBC during excision repair (37). However, the observation that strains that were uvrD mutS or uvrD uvrC continued to exhibit SOS induction to the same extent as the uvrD single mutant (Table 2) served to exclude this explanation.
In view of the possibility that UvrD participates in recombination, we tested the effects of mutations in several recombination genes (other than recA) on SOS induction in uvrD mutants. The expression of sulA-lac in a uvrD strain remained elevated despite introduction of mutations in recB, recF, recQ, or ruvABC (data not shown). Substantially similar results have also been described earlier (3).
uvrD derivatives of BL21 are very sick.
It is known that E. coli B strains are naturally Lon protease deficient. Consistent with the results described above was our observation that uvrD400::Tn10dTet transductants of the E. coli B strain derivative BL21 grow at 37°C as tiny colonies and that they are somewhat more healthy at 42°C. Other workers (8) have earlier reported the construction of UvrD-deficient derivatives of BL21(DE3), but it is possible that these derivatives may inadvertently also have been selected for suppressors of uvrD-lon incompatibility such as sulA.
DISCUSSION
We have identified in this study a new set of incompatible pairs of mutations involving the lon gene on the one hand and uvrD, priA, mutD, dam, or polA on the other. The common features that characterize this set are as follows: (i) each of the latter partners of the set results in moderate SOS induction, and (ii) the incompatibilities are suppressed by sulA, lexA3 (Ind−), or recA mutations and are partially suppressed by growth at 42°C. Included in the category of lon-incompatible SOS-inducing mutations are both those (such as uvrD, priA, and mutD) for which SOS induction itself is not essential for viability and others (such as dam and polA) for which it is. The simplest interpretation to explain the incompatibility is that persistent SOS induction causes a moderate elevation in levels of SulA protein and that the Lon protease becomes essential for inactivating SulA under these conditions.
The demonstration of lon incompatibility for mutations such as uvrD and priA permits the inference that the latter mutations lead to moderate SOS induction in all (or a majority of) cells in the culture rather than to extreme SOS induction in a smaller subpopulation. The rep mutation appears to be associated with an even lower level of SOS induction than uvrD or priA, since a lon rep mutant is sick yet viable. Chronic SOS induction has also been reported in wild-type cells in the stationary-growth phase (54) but is probably mild enough not to be lethal in a lon mutant. Likewise, mutations in recF (40), ruv (40), or recBC sbcBC (14, 28, 39) have been reported to cause SOS induction, but we found each of them to be lon compatible (data not shown).
It may be noted that the IS186 insertion mutation in the promoter region of lon in strain GJ1823 had arisen spontaneously, in the absence of any imposition of ostensible selection. Ignatov and Chistyakova (11) have also independently reported the identification of a spontaneous IS186 insertion at the same site in the lon promoter. Combined with the fact that wild-type E. coli B isolates are lon negative, our observations raise the question whether natural selection may operate for lon-defective strains under certain conditions.
SOS induction in uvrD mutants probably occurs through a defect in DNA replication.
It has been established from earlier studies that the signal for SOS induction in vivo is single-stranded DNA (reviewed in references 23 and 55). Our results indicate that chronic SOS induction in uvrD mutants is the null phenotype. Nevertheless, identification of the precise mechanism is still complicated by the fact that UvrD is involved in several discrete functions and pathways, loss of any one of which could plausibly be modeled to account for SOS induction.
The findings that mutH, mutS, or uvrC strains are not SOS induced and that SOS induction continues to occur in mutS uvrD and uvrC uvrD double mutants indicate that the induction is not due to the loss of mismatch repair or excision repair pathways in the uvrD mutant, nor is it due to a trapped intermediate in either pathway. Likewise, our results render unlikely the possibility that it is the loss of a recombination function of UvrD which is responsible for SOS induction. One could therefore consider some defect in DNA replication to be the cause for spontaneous SOS induction in uvrD strains.
Role for UvrD in lagging-strand replication?
In order to account for generation of a moderate SOS-inducing signal in all cells of a culture of a uvrD strain, we propose that UvrD functions as a 3′-5′ helicase during lagging-strand replication. The chromosomal replication fork is expected to contain a loop of unreplicated lagging strand that is approximately 1 kb long (27). We suggest that the 3′-5′ helicase activity of UvrD helps maintain this single-stranded DNA region free of secondary structure and that in the absence of UvrD, synthesis of some fraction of Okazaki fragments by DNA polymerase III is impeded so that a chronic, low-level SOS-inducing signal is generated by the unreplicated DNA segments on the lagging strand. A similar function was in fact postulated earlier for PriA (29); however, more recent findings have established that the preeminent role of this protein is in the reassembly of collapsed replication forks (7, 18, 58) and that loss of its helicase activity is not sufficient for generation of an SOS-inducing signal in vivo (18, 48).
Our model for UvrD's role in replication may explain the synthetic lethality of null uvrD mutants with polA (reference 56 and this study). Both the polymerase and 5′-3′ exonuclease activities of DNA polymerase I have been implicated in efficient lagging-strand replication (27). Our own observations (data not shown) using the polA (Klenow+) or polA (5′ Exo+) alleles in combination with uvrD400::Tn10dTet are consistent with earlier reports that null uvrD strains are rendered inviable upon loss of either catalytic activity of DNA polymerase I (56). It is also possible that Rep becomes essential for lagging-strand replication in a uvrD mutant, thereby accounting for rep-uvrD incompatibility (56).
Finally, we speculate that the availability of unreplicated stretches of DNA on the lagging strand might contribute to the hyperrecombinational phenotype of uvrD strains described earlier (21, 23), through the mechanism that has been referred to as daughter strand gap repair (7, 23, 55). Previous studies have shown that uvrD mutants exhibit hyperrecombination as a consequence of the loss of mismatch repair (41, 42), and we suggest that the two mechanisms (loss of mismatch repair and daughter strand gap repair) operate independently. For example, the latter mechanism may explain the RecA- and RecF-dependent increase in the frequency of tandem repeat deletions (3) and of transposon precise excisions (5) in uvrD mutants, since such dependence is the hallmark of daughter strand gap repair. Lloyd (25) also suggested earlier that the hyperrecombinational phenotype associated with uvrD operates through the RecF pathway.
In summary, therefore, we propose that several phenotypes associated with uvrD null mutations, including chronic SOS induction and the consequent lon incompatibility described in this study, may be explained by a model in which UvrD participates as a helicase in lagging-strand DNA replication in E. coli.
ACKNOWLEDGMENTS
We thank Susan Gottesman, Carol Gross, Masayori Inouye, Nancy Kleckner, Sidney Kushner, Bénédicte Michel, Josette Rouvière-Yaniv, Miroslav Radman, and the E. coli Genetic Stock Center for making available various strains and plasmids that were used in this study, and we thank Andrei Kuzminov and Imran Siddiqi for suggestions on the manuscript. We also acknowledge the assistance of Mehar Sultana and N. Nagesh with oligonucleotide synthesis and automated DNA sequencing, respectively. Kundan Sengupta and M. V. L. S. Narasimha Rao contributed to the project as summer trainees, by identifying uvrD incompatibility in GJ1823 and constructing plasmid pHYD141, respectively.
This study was supported in part by funds from the Department of Science and Technology, Government of India. J.G. is an honorary faculty member of the Jawaharlal Nehru Centre for Advanced Scientific Research.
REFERENCES
- 1.Bates H, Randall S K, Rayssiguier C, Bridges B A, Goodman M F, Radman M. Spontaneous and UV-induced mutations in Escherichia coli K-12 strains with altered or absent DNA polymerase I. J Bacteriol. 1989;171:2480–2484. doi: 10.1128/jb.171.5.2480-2484.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berlyn M K B. Linkage map of Escherichia coli K-12, edition 10: the traditional map. Microbiol Mol Biol Rev. 1998;62:814–984. doi: 10.1128/mmbr.62.3.814-984.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bierne H, Seigneur M, Ehrlich S D, Michel B. uvrD mutations enhance tandem repeat deletion in the Escherichia coli chromosome via SOS induction of the RecF recombination pathway. Mol Microbiol. 1997;26:557–567. doi: 10.1046/j.1365-2958.1997.6011973.x. [DOI] [PubMed] [Google Scholar]
- 4.Blattner F R, Plunkett III G, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 5.Chan A, Nagel R. Involvement of recA and recF in the induced precise excision of Tn10 in Escherichia coli. Mutat Res. 1997;381:111–115. doi: 10.1016/s0027-5107(97)00157-7. [DOI] [PubMed] [Google Scholar]
- 6.Chong P, Hui I, Loo T, Gillam S. Structural analysis of a new GC-specific insertion element, IS186. FEBS Lett. 1985;192:47–52. doi: 10.1016/0014-5793(85)80040-5. [DOI] [PubMed] [Google Scholar]
- 7.Cox M M. A broadening view of recombinational DNA repair in bacteria. Genes Cells. 1998;3:65–78. doi: 10.1046/j.1365-2443.1998.00175.x. [DOI] [PubMed] [Google Scholar]
- 8.George J W, Brosh R M, Jr, Matson S W. A dominant negative allele of the Escherichia coli uvrD gene encoding DNA helicase II: a biochemical and genetic characterization. J Mol Biol. 1994;235:424–435. doi: 10.1006/jmbi.1994.1003. [DOI] [PubMed] [Google Scholar]
- 9.Gowrishankar J. Identification of osmoresponsive genes in Escherichia coli: evidence for participation of potassium and proline transport systems in osmoregulation. J Bacteriol. 1985;164:434–445. doi: 10.1128/jb.164.1.434-445.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hamilton C M, Aldea M, Washburn B K, Babitzke P, Kushner S R. New method for generating deletions and gene replacements in Escherichia coli. J Bacteriol. 1989;171:4617–4622. doi: 10.1128/jb.171.9.4617-4622.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ignatov K B, Chistyakova L G. Integration of the insertion element IS186 into the −10 region of the heat shock promoter of the Escherichia coli lon gene. Bioorg Khim. 1997;23:18–20. . (In Russian.) [PubMed] [Google Scholar]
- 12.Joyce C M, Grindley N D F. Method for determining whether a gene of Escherichia coli is essential: application to the polA gene. J Bacteriol. 1984;158:636–643. doi: 10.1128/jb.158.2.636-643.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kanemori M, Yanagi H, Yura T. The ATP-dependent HsIVU/ClpQY protease participates in turnover of cell division inhibitor SulA in Escherichia coli. J Bacteriol. 1999;181:3674–3680. doi: 10.1128/jb.181.12.3674-3680.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karu A E, Belk E D. Induction of E. coli RecA protein via recBC and alternate pathways: quantitation by enzyme-linked immunosorbent assay (ELISA) Mol Gen Genet. 1982;185:275–282. doi: 10.1007/BF00330798. [DOI] [PubMed] [Google Scholar]
- 15.Khattar M M. Overexpression of the hslVU operon suppresses SOS-mediated inhibition of cell division in Escherichia coli. FEBS Lett. 1997;414:402–404. doi: 10.1016/s0014-5793(97)01024-7. [DOI] [PubMed] [Google Scholar]
- 16.Kleckner N, Bender J, Gottesman S. Uses of transposons with emphasis on Tn10. Methods Enzymol. 1991;204:139–180. doi: 10.1016/0076-6879(91)04009-d. [DOI] [PubMed] [Google Scholar]
- 17.Klinkert M-Q, Klein A, Abdel-Monem M. Studies on the functions of DNA helicase I and DNA helicase II of Escherichia coli. J Biol Chem. 1980;255:9746–9752. [PubMed] [Google Scholar]
- 18.Kogoma T, Cadwell G W, Barnard K G, Asai T. The DNA replication priming protein, PriA, is required for homologous recombination and double-strand break repair. J Bacteriol. 1996;178:1258–1264. doi: 10.1128/jb.178.5.1258-1264.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kohara Y, Akiyama K, Isono K. The physical map of the whole E. coli chromosome: application of a new strategy for rapid analysis and sorting of a large genomic library. Cell. 1987;50:495–508. doi: 10.1016/0092-8674(87)90503-4. [DOI] [PubMed] [Google Scholar]
- 20.Kothary R K, Jones D, Candido E P M. IS186: an Escherichia coli insertion element isolated from a cDNA library. J Bacteriol. 1985;164:957–959. doi: 10.1128/jb.164.2.957-959.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kowalczykowski S C, Dixon D A, Eggleston A K, Lauder S D, Rehrauer W M. Biochemistry of homologous recombination in Escherichia coli. Microbiol Rev. 1994;58:401–465. doi: 10.1128/mr.58.3.401-465.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuhn B, Abdel-Monem M. DNA synthesis at a fork in the presence of DNA helicases. Eur J Biochem. 1982;125:63–68. doi: 10.1111/j.1432-1033.1982.tb06651.x. [DOI] [PubMed] [Google Scholar]
- 23.Kuzminov A. Recombinational repair of DNA damage in Escherichia coli and bacteriophage λ. Microbiol Mol Biol Rev. 1999;63:751–813. doi: 10.1128/mmbr.63.4.751-813.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lerner C G, Inouye M. Low copy number plasmids for regulated low-level expression of cloned genes in Escherichia coli with blue/white insert screening capability. Nucleic Acids Res. 1990;18:4631. doi: 10.1093/nar/18.15.4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lloyd R G. lexA dependent recombination in uvrD strains of Escherichia coli. Mol Gen Genet. 1983;189:157–161. doi: 10.1007/BF00326069. [DOI] [PubMed] [Google Scholar]
- 26.Lovett S T, Sutera V A., Jr Suppression of RecJ exonuclease mutants of Escherichia coli by alterations in DNA helicases II (uvrD) and IV (helD) Genetics. 1995;140:27–45. doi: 10.1093/genetics/140.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marians K J. Replication fork propagation. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: American Society for Microbiology; 1996. pp. 749–763. [Google Scholar]
- 28.Matic I, Rayssiguier C, Radman M. Interspecies gene exchange in bacteria: the role of SOS and mismatch repair systems in evolution of species. Cell. 1995;80:507–515. doi: 10.1016/0092-8674(95)90501-4. [DOI] [PubMed] [Google Scholar]
- 29.Matson S W. DNA helicases of Escherichia coli. Prog Nucleic Acids Res Mol Biol. 1991;40:289–326. doi: 10.1016/s0079-6603(08)60845-4. [DOI] [PubMed] [Google Scholar]
- 30.Matson S W, Bean D W, George J W. DNA helicases: enzymes with essential roles in all aspects of DNA metabolism. Bioessays. 1994;16:13–22. doi: 10.1002/bies.950160103. [DOI] [PubMed] [Google Scholar]
- 31.Maurizi M R, Trisler P, Gottesman S. Insertional mutagenesis of the lon gene in Escherichia coli: lon is dispensable. J Bacteriol. 1985;164:1124–1135. doi: 10.1128/jb.164.3.1124-1135.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mendonca V M, Matson S W. Genetic analysis of ΔhelD and ΔuvrD mutations in combination with other genes in the RecF recombination pathway in Escherichia coli: suppression of a ruvB mutation by a uvrD deletion. Genetics. 1995;141:443–452. doi: 10.1093/genetics/141.2.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miller J H. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1992. [Google Scholar]
- 34.Modrich P. Mechanisms and biological effects of mismatch repair. Annu Rev Genet. 1991;25:229–253. doi: 10.1146/annurev.ge.25.120191.001305. [DOI] [PubMed] [Google Scholar]
- 35.Modrich P, Lahue R. Mismatch repair in replication fidelity, genetic recombination, and cancer biology. Annu Rev Biochem. 1996;65:101–133. doi: 10.1146/annurev.bi.65.070196.000533. [DOI] [PubMed] [Google Scholar]
- 36.Nurse P, Zavitz K H, Marians K J. Inactivation of the Escherichia coli PriA DNA replication protein induces the SOS response. J Bacteriol. 1991;173:6686–6693. doi: 10.1128/jb.173.21.6686-6693.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Orren D K, Selby C P, Hearst J E, Sancar A. Post-incision steps of nucleotide excision repair in Escherichia coli. J Biol Chem. 1992;267:780–788. [PubMed] [Google Scholar]
- 38.Ossanna N, Mount D W. Mutations in uvrD induce the SOS response in Escherichia coli. J Bacteriol. 1989;171:303–307. doi: 10.1128/jb.171.1.303-307.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peterson K R, Mount D W. Analysis of the genetic requirements for viability of Escherichia coli K-12 DNA adenine methylase (dam) mutants. J Bacteriol. 1993;175:7505–7508. doi: 10.1128/jb.175.22.7505-7508.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peterson K R, Ossanna N, Thliveris A T, Ennis D G, Mount D W. Derepression of specific genes promotes DNA repair and mutagenesis in Escherichia coli. J Bacteriol. 1988;170:1–4. doi: 10.1128/jb.170.1.1-4.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Radman M. Mismatch repair and genetic recombination. In: Kucherlapati R, Smith G R, editors. Genetic recombination. Washington, D.C.: American Society for Microbiology; 1988. pp. 169–192. [Google Scholar]
- 42.Rayssiguier C, Thaler D S, Radman M. The barrier to recombination between Escherichia coli and Salmonella typhimurium is disrupted in mismatch-repair mutants. Nature. 1989;342:396–401. doi: 10.1038/342396a0. [DOI] [PubMed] [Google Scholar]
- 43.Reddy M, Gowrishankar J. Identification and characterization of ssb and uup mutants with increased frequency of precise excision of transposon Tn10 derivatives: nucleotide sequence of uup in Escherichia coli. J Bacteriol. 1997;179:2892–2899. doi: 10.1128/jb.179.9.2892-2899.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reddy M, Gowrishankar J. A genetic strategy to demonstrate the occurrence of spontaneous mutations in nondividing cells within colonies of Escherichia coli. Genetics. 1997;147:991–1001. doi: 10.1093/genetics/147.3.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 46.Sancar A. Mechanisms of DNA excision repair. Science. 1994;266:1954–1956. doi: 10.1126/science.7801120. [DOI] [PubMed] [Google Scholar]
- 47.Sancar A. DNA excision repair. Annu Rev Biochem. 1996;65:43–81. doi: 10.1146/annurev.bi.65.070196.000355. [DOI] [PubMed] [Google Scholar]
- 48.Sandler S J, Samra H S, Clark A J. Differential suppression of priA2::kan phenotypes in Escherichia coli K-12 by mutations in priA, lexA, and dnaC. Genetics. 1996;143:5–13. doi: 10.1093/genetics/143.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Seigneur M, Bidnenko V, Ehrlich S D, Michel B. RuvAB acts at arrested replication forks. Cell. 1998;95:419–430. doi: 10.1016/s0092-8674(00)81772-9. [DOI] [PubMed] [Google Scholar]
- 50.Sengstag C, Iida S, Hiestand-Nauer R, Arber W. Terminal inverted repeats of prokaryotic transposable element IS186 which can generate duplications of variable length at an identical target sequence. Gene. 1986;49:153–156. doi: 10.1016/0378-1119(86)90395-1. [DOI] [PubMed] [Google Scholar]
- 51.Seong I S, Oh J Y, Yoo S J, Seol J H, Chung C H. ATP-dependent degradation of SulA, a cell division inhibitor, by the HslVU protease in Escherichia coli. FEBS Lett. 1999;456:211–214. doi: 10.1016/s0014-5793(99)00935-7. [DOI] [PubMed] [Google Scholar]
- 52.Singer M, Baker T A, Schnitzler G, Deischel S M, Goel M, Dove W, Jaacks K J, Grossman A D, Erickson J W, Gross C A. A collection of strains containing genetically linked alternating antibiotic resistance elements for genetic mapping of Escherichia coli. Microbiol Rev. 1989;53:1–24. doi: 10.1128/mr.53.1.1-24.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Slater S C, Lifsics M R, O'Donnell M, Maurer R. holE, the gene coding for the θ subunit of DNA polymerase III of Escherichia coli: characterization of a holE mutant and comparison with a dnaQ (ɛ subunit) mutant. J Bacteriol. 1994;176:815–821. doi: 10.1128/jb.176.3.815-821.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Taddei F, Matic I, Radman M. cAMP-dependent SOS induction and mutagenesis in resting bacterial populations. Proc Natl Acad Sci USA. 1995;92:11736–11740. doi: 10.1073/pnas.92.25.11736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Walker G C. The SOS response of Escherichia coli. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: American Society for Microbiology; 1996. pp. 1400–1416. [Google Scholar]
- 56.Washburn B K, Kushner S R. Construction and analysis of deletions in the structural gene (uvrD) for DNA helicase II of Escherichia coli. J Bacteriol. 1991;173:2569–2575. doi: 10.1128/jb.173.8.2569-2575.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu W-F, Zhou Y, Gottesman S. Redundant in vivo proteolytic activities of Escherichia coli Lon and the ClpYQ (HslUV) protease. J Bacteriol. 1999;181:3681–3687. doi: 10.1128/jb.181.12.3681-3687.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zavitz K H, Marians K J. Dissecting the functional role of PriA protein-catalysed primosome assembly in Escherichia coli DNA replication. Mol Microbiol. 1991;5:2869–2873. doi: 10.1111/j.1365-2958.1991.tb01846.x. [DOI] [PubMed] [Google Scholar]
- 59.Zhang G, Deng E, Baugh L R, Hamilton C M, Maples V F, Kushner S R. Conserved motifs II to VI of DNA helicase II from Escherichia coli are all required for biological activity. J Bacteriol. 1997;179:7544–7550. doi: 10.1128/jb.179.23.7544-7550.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]