Abstract
The products of three genes named CARGRI, CARGRII, and CARGRIII were shown to repress the expression of CAR1 and CAR2 genes, involved in arginine catabolism. CARGRI is identical to UME6 and encodes a regulator of early meiotic genes. In this work we identify CARGRII as SIN3 and CARGRIII as RPD3. The associated gene products are components of a high-molecular-weight complex with histone deacetylase activity and are recruited by Ume6 to promoters containing a URS1 sequence. Sap30, another component of this complex, is also required to repress CAR1 expression. This histone deacetylase complex prevents the synthesis of the two arginine catabolic enzymes, arginase (CAR1) and ornithine transaminase (CAR2), as long as exogenous nitrogen is available. Upon nitrogen depletion, repression at URS1 is released and Ume6 interacts with ArgRI and ArgRII, two proteins involved in arginine-dependent activation of CAR1 and CAR2, leading to high levels of the two catabolic enzymes despite a low cytosolic arginine pool. Our data also show that the deletion of the UME6 gene impairs cell growth more strongly than the deletion of the SIN3 or RPD3 gene, especially in the Σ1278b background.
The first mutations affecting the expression of the arginine catabolic genes CAR1 and CAR2, encoding arginase and ornithine transaminase, respectively, were located in three unlinked genes, ARGRI (ARG80), ARGRII (ARG81), and ARGRIII (ARG82). The products of these genes were required for the induction of arginase and ornithine transaminase synthesis, and their loss impaired cell ability to use arginine and ornithine as nitrogen sources. The same proteins repressed the synthesis of five arginine anabolic enzymes when exogenous arginine was present in the growth medium (1, 42). Later, it was shown that the pleiotropic factor Mcm1 also participated, with the ArgR proteins, in the arginine-specific regulation (10, 28). The growth defect of an argR mutant allowed the selection of suppressor mutations falling into three complementation groups containing the CARGRI (CAR80), CARGRII (CAR81) and CARGRIII (CAR82) genes. Mutations in any of these genes led to overproduction of arginase and ornithine transaminase, even in an argR background (7, 9). The CARGRI gene was identical to UME6, a gene whose product is involved in controlling the expression of early meiotic genes (34, 41). Kadosh and Struhl (20) showed that repression by Ume6 at URS1, a sequence present in a wide variety of yeast promoters (24), involved recruitment of a Sin3-Rpd3 complex and targeted histone deacetylation.
Although CAR1 and CAR2 genes are coordinately induced by arginine and the ArgR-Mcm1 complex (29) and repressed by the three CargR proteins, only CAR2 is induced by Dal82 and allophanate, the last intermediate of the allantoin-degrading pathway (17, 35), whereas CAR1 is activated by Gln3 and Nil1 through multiple GATAA sequences in the absence of optimal nitrogen sources (ammonia, glutamine) (11, 40). In addition CAR1 expression is controlled not only by the quality of the nitrogen source (NCR) but also by the quantity of nitrogen available to the cell (8). Derepression of CAR1 upon nitrogen deficiency requires the integrity of both the ArgR and Ume6 proteins and their target sequences (arginine boxes and URS1) but does not require the integrity of the GATAA sequences, the targets of Gln3 and Nil1 (11, 41). It was also shown that arginase derepression upon severe nitrogen starvation required the presence of arginine or a nonmetabolizable inducer, such as homoarginine (46).
This work aimed at characterizing the CARGRII and CARGRIII genes and at determining their role in the response of CAR1 and CAR2 genes to exogenous nitrogen availability. In this paper, we identify CARGRII as SIN3 and CARGRIII as RPD3 and show that Ume6, Sin3, and Rpd3 proteins repress the expression of CAR1 and CAR2 genes, as long as nitrogen is available in the growth medium. We also identify a physiological interaction between Ume6 and ArgRI or ArgRII upon nitrogen depletion.
(A preliminary report of this work has been published as an abstract [12].)
MATERIALS AND METHODS
Strains and media.
Saccharomyces cerevisiae strains 12T7c (ura3), 27061b (ura3 trp1), and 27029c (ura3 leu2) were derived from the wild-type strain Σ1278b. Strain BY4709 (ura3) was derived from the wild-type strain S288c (3). The in vivo-selected cargRII (11S52a) and cargRIII (02451c) mutants (7) were ura3 recombinants from crosses between derivatives of wild-type strain Σ1278b and a ura3 mutant strain isogenic to FL100. Strain HY (22) was used as the recipient strain for two-hybrid experiments.
Escherichia coli strain XL1-Blue (Stratagene) was used for plasmid amplification, and HB101 (Life Technologies) was used for plasmid recovery from yeast.
All yeast strains were grown on minimal medium containing 3% glucose, vitamins, mineral traces, and 0.02 M (NH4)2SO4 (M.ammonia medium) (27). Nitrogen starvation was achieved by filtering the cells grown on M.ammonia medium and cultivating them on fresh minimal medium without nitrogen for 2 h. The lithium acetate procedure (19) was used to transform the recipient yeast strains.
Construction of yeast deletant strains.
The long flanking homology strategy was used to perform deletion of the following genes: SIN3, RPD3, SAP30, UME6, GCN5, HDA1, and HOS2 (45). Long flanking homology replacement cassettes were synthesized using a two-step PCR, leading to the kanMX4 cassette flanked by about 500 bp, corresponding to the promoter and terminator regions, respectively, of the target genes. The DNA fragments containing the different cassettes were used to transform yeast strains 12T7c (ura3) and BY4709 (ura3) on rich-medium plates containing 200 μg of Geneticin/ml. The correct targeting of the deletions in G418r transformants was verified by PCR, using whole cells as a source of DNA and appropriate primers. In the Σ1278b background, the deletion of UME6 had to be performed in the diploid strain obtained by crossing strain 27061b with 27029c. To construct strains with multiple deletions, we have used the gene disruption cassette loxP-kanMX-loxP (16). To eliminate the kanMX marker from the disrupted gene, the mutated strain was transformed with the cre expression plasmid pSH47, which carries the URA3 marker gene and the cre gene under the control of the inducible GAL1 promoter. Expression of the cre recombinase was induced by shifting cells from YPD (glucose) to YPG (galactose) medium for 2 h. The loss of the kanMX cassette was detected by plating cells on YPD and replica plating the colonies onto YPD-G418. The cre expression plasmid was removed from the strains by streaking cells on plates containing 5-fluoroorotic acid to counterselect for the loss of the plasmid.
Construction of wild-type UME6, RPD3, and SIN3 wild-type cognate clones.
To clone these wild-type genes, the gap repair procedure was used (36). The UME6, RPD3, and SIN3 open reading frame replacement cassettes were cloned into the vector pRS416 (39). The kanMX4 modules were excised by restriction with EcoRI and BamHI. The linearized plasmids were used to transform the standard FY1679 strain. Plasmids bearing the wild-type alleles pFV111 (CEN6 ARS4 URA3 UME6), pFV33 (CEN6 ARS4 URA3 SIN3) and pFV36 (CEN6 ARS4 URA3 RPD3) from URA+ transformants were isolated and amplified in E. coli. The presence of the genes of interest was verified by restriction analysis.
Fusion of CAR2 promoter to the lacZ gene.
A fragment of about 1,000 bp containing the CAR2 promoter and its first two codons was produced by PCR using appropriate oligonucleotides based on S. cerevisiae genomic data and was extended with BamHI restriction sites. This fragment was fused in frame to the lacZ coding sequence by insertion in the BamHI site of plasmid pMC310 (pFL1 containing the E. coli lacZ gene inserted at the BamHI site [5]); gift from M. Crabeel), yielding plasmid pFV118 (CAR2-lacZ). We determined the nucleotide sequence of the junction between the promoter and the lacZ gene to make sure that the fusion was in frame.
Construction of GBD and GAD fusions.
The DNA-binding domain of the Gal4 activator, Gal4(1-147), is referred to as GBD, and its activation domain, Gal4(768-881), is referred to as GAD. GBD-ARGRI and GBD-ARGRII fusions were described previously (14). To produce the GAD-UME6 fusion, we used PCR to synthesize a BamHI-BamHI DNA fragment containing the UME6 coding sequence (from nucleotide +4 to 2505). The primers used were based on the published UME6 sequence (41), each flanked by a BamHI restriction site. The BamHI-BamHI fragment was inserted into the BamHI site of plasmid pACTII (13), yielding plasmid pFV124 (GAD-Ume6). In this GAD gene fusion, we determined the nucleotide sequence of the junction between the GAD-encoding region and the UME6 gene to make sure that the fusions were in frame.
Enzyme assay.
β-Galactosidase activity was assayed as described by Miller (31). Protein contents were determined by the Folin method (23).
Activity of arginase was assayed as described previously (30).
Measurement of amino acid pools.
Differential extractions of cytosolic and vacuolar amino acid pools were performed as described by Ohsumi et al. (33).
The intracellular concentrations of glutamate, glutamine, arginine, ornithine, and lysine were determined by the PICOTag method. This system employs phenylisothiocyanate to rapidly and quantitavely derivatize both primary and secondary amino acids in a simple, one-step reaction. This derivatization is the first step of the well known Edman degradation. The stable phenylthiocarbamyl derivatives were easily separated by reverse-phase high-pressure liquid chromatography (2).
After peak identification, the amount of each amino acid was calculated by integration of the peak surface on the chromatogram and comparison to calibration curves established with standard amino acid solutions. These amounts, taking into account the different dilution factors and the dry weight of each sample, were used to establish pool concentrations. The dry weights were estimated by precisely measuring absorbance of the cells just before filtration using a conversion curve establishing the relation between the absorbance and the dry weight of the cells.
RESULTS
Identification of CARGRII as SIN3 and CARGRIII as RPD3.
Mutations in CARGRII and CARGRIII genes led to the derepression of two arginine catabolic genes, which was one of the phenotypes observed in cargRI (ume6) mutants. One role of Ume6 is to recruit to target promoters the Rpd3 histone deacetylase, through its interaction with Sin3. We have therefore tested the effect on CAR1 and CAR2 expression of deletions in the SIN3 and RPD3 genes. These deletions were created in strain 12T7c (ura3) by replacing the complete coding sequence of each gene by the kanMX4 cassette, confering resistance to Geneticin. The levels of arginase, the product of the CAR1 gene, from strains 12T7cII (ura3 sin3::kanMX4; Table 1, experiment 3) and 12T7cIII (ura3 rpd3::kanMX4; Table 1 experiment 4) were comparable to those of cargRII (11S52a; Table 1, experiment 2) and cargRIII (02451c; Table 2, experiment 2) mutants. The similar phenotypes produced by the cargRII and cargRIII mutations and the sin3 and rpd3 deletions prompted the question of their allelism. We crossed cargRII or cargRIII point mutant haploid strains to sin3 or rpd3 deletion mutants and sporulated the resulting diploids. The arginase levels presented in Table 1 showed that the cargRII mutant allele was incapable of complementing the sin3 deletion allele (Table 1 experiment 5) but fully complemented the rpd3 deletion allele (Table 1, experiment 6). In contrast, the cargRIII mutant allele complemented the sin3 deletion allele (Table 2, experiment 6) and not the rpd3 deletion allele (Table 2, experiment 5). These data suggest that CARGRII is identical to SIN3 and that CARGRIII is identical to RPD3. We confirmed these results by analysis of the tetrads issued from the sporulation of the cargRII-Δsin3 diploid. All the spores from 10 tetrads had derepressed arginase levels on M.ammonia medium. This analysis could not be performed with the cargRIII-Δrpd3 diploid, which did not sporulate. It was reported that homozygous sin3 or rpd3 diploid strains were sporulation defective (43, 44). Our cargRII point mutant is thus not impaired in sporulation, in contrast to our cargRIII point mutant. The SIN3 and RPD3 genes were cloned by gap repair (see Materials and Methods) and introduced into cargRII (11S52a) and cargRIII (02451c) mutant strains. The arginase-specific activities in the cargRII strain transformed with plasmid pFV33 (pRS416 ARS CEN URA3 SIN3) (Table 1, experiment 7) and the cargRIII strain transformed with pFV36 (pRS416 ARS CEN URA3 RPD3) (Table 2, experiment 7) were comparable to that of the wild-type strain, as expected.
TABLE 1.
Identification of CARGRII gene as SIN3
| Expt | Strain | Plasmid | Genotype | Arginase specific activity on M.ammoniaa |
|---|---|---|---|---|
| 1 | 12T7c | ura3 | 8* | |
| 2 | 11S52a | ura3 cargRII | 40* | |
| 3 | 12T7cII | ura3 sin3::kanMX4 | 49* | |
| 4 | 12T7cIII | ura3 rpd3::kanMX4 | 33* | |
| 5 | 11S52a × 12T7cII | ura3 cargRII × ura3 sin3::kanMX4 | 46* | |
| 6 | 11S52a × 12T7cIII | ura3 cargRII × ura3 rpd3::kanMX4 | 9* | |
| 7 | 11S52a | pFV33b | ura3 cargRII | 16 |
| 8 | 11S52a | pFV36c | ura3 cargRII | 40 |
Arginase specific activity was measured at 30°C after growth of the different strains on M.ammonia–25 μg of uracil per ml where indicated (∗). The specific activity is expressed in micromoles of urea produced per hour and per milligram of protein. The values are the means of two independent assays, and the standard error was 10 to 15%.
Corresponds to pRS416 CEN6 ARS4 URA3 SIN3.
Corresponds to pRS416 CEN6 ARS4 URA3 RPD3.
TABLE 2.
Identification of CARGRIII gene as RPD3
| Expt | Strain | Plasmid | Genotype | Arginase specific activity on M.ammoniaa |
|---|---|---|---|---|
| 1 | 12T7c | ura3 | 8* | |
| 2 | 02451c | ura3 cargRIII | 50* | |
| 3 | 12T7cIII | ura3 rpd3::kanMX4 | 33* | |
| 4 | 12T7cII | ura3 sin3::kanMX4 | 49* | |
| 5 | 02451c × 12T7cIII | ura3 cargRIII × ura3 rpd3::kanMX4 | 30* | |
| 6 | 02451c × 12T7cII | ura3 cargRIII × ura3 sin3::kanMX4 | 12* | |
| 7 | 02451c | pFV36c | ura3 cargRIII | 13 |
| 8 | 02451c | pFV33b | ura3 cargRIII | 38 |
Arginase specific activity was measured at 30°C after growth of the different strains on M.ammonia–25 μg of uracil per ml where indicated (∗). The specific activity is expressed in micromoles of urea produced per hour per milligram of protein. The values are the means of two independent assays, and the standard error was 10 to 15%.
Corresponds to pRS416 CEN6 ARS4 URA3 SIN3.
Corresponds to pRS416 CEN6 ARS4 URA3 RPD3.
Sap30, but not Hda1, Hos2, or Gcn5, controls CAR1 expression.
In S. cerevisiae as in humans, histone acetylation and deacetylation are catalyzed by structurally distinct multisubunit complexes. In yeast, Gcn5, present in Ada and SAGA (Spt/Ada) complexes (4, 15, 26), and TAF145/130 (the yeast equivalent of mammalian TAF250), present in TFIID (32), were shown to have histone acetylation activity. Hda1, a component of histone deacetylase A, has identity to Rpd3, Hos1, Hos2, and Hos3, and it was shown that Hda1 and Rpd3 are members of distinct histone deacetylase complexes (38). Sap30 was recently shown to be part of the Rpd3-Sin3 complex in S. cerevisiae (47). To test the role of these different proteins involved in histone acetylation and deacetylation in CAR1 expression, we have created strains with deletions of the HDA1, HOS2, SAP30, and GCN5 genes by insertion of the kanMX4 cassette (see Materials and Methods). As shown in Table 3, the arginase levels on minimal medium in strains with the HDA1 (12T7cV), HOS2 (12T7cVI), or GCN5 (12T7cVII) gene deleted were comparable to the level in the wild-type strain, whereas deletion of the SAP30 gene (strain 12T7cIV) led to a derepression of CAR1 expression, as in SIN3- and RPD3-deleted strains. It is noteworthy that the activation of CAR1 in a gcn5::kanMX4 strain in response to the presence of arginine or a poor nitrogen source was not impaired (data not shown). Only the Rpd3-Sin3-Sap30 complex seems to regulate the expression of the CAR1 gene, probably by deacetylation of histones. In contrast, the histone acetyltransferase activity of Gcn5 or TAF145/130 (according to global analysis by Holstege et al. [18]) does not seem to be required for CAR1 expression.
TABLE 3.
Effect of mutations impairing histone acetylation and deacetylation activities on CAR1 expression
| Strain | Genotype | Arginase specific activity on M.ammoniaa |
|---|---|---|
| 12T7c | ura3 | 8 |
| 12T7cII | ura3 sin3::kanMX4 | 49 |
| 12T7cIII | ura3 rpd3::kanMX4 | 33 |
| 12T7cIV | ura3 sap30::kanMX4 | 27 |
| 12T7cV | ura3 hda1::kanMX4 | 5 |
| 12T7cVI | ura3 hos2::kanMX4 | 4 |
| 12T7cVII | ura3 gcn5::kanMX4 | 9 |
Arginase specific activity was measured at 30°C after growth of the different strains on M.ammonia–25 μg of uracil per ml. The specific activity is expressed in micromoles of urea produced per hour per milligram of protein. The values are the means of two independent assays, and the standard error was 10 to 15%.
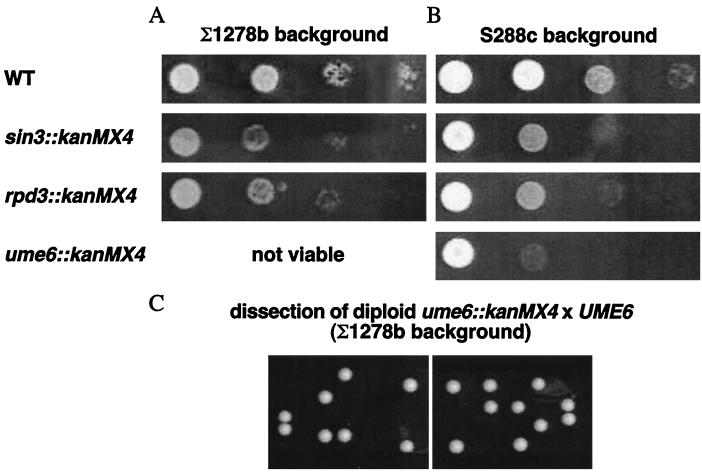
Effect of deletions in UME6, SIN3, and RPD3 genes on cellular growth in the Σ1278b background.
Deletions of SIN3 and RPD3 genes from strain 12T7c, which is a ura3 derivative of Σ1278b, were performed. These two strains resulting from the deletions (12T7cII and 12T7cIII) were affected in their growth on minimal medium (Fig. 1A). In the same background, we could not obtain a viable ume6::kanMX4 strain. We thus performed the ume6 deletion in the diploid strain 27061b (ura3 trp1) × 27029c (ura3 leu2) by insertion of the kanMX4 cassette and obtained viable diploids resistant to Geneticin (see Materials and Methods). After sporulation of these diploids, tetrad analysis revealed that only two spores were viable (Fig. 1C) and that the viable spores were Geneticin sensitive. The UME6 gene product is thus essential in the Σ1278b background, whereas it only reduces the growth rate in other backgrounds. We have indeed created UME6, SIN3, and RPD3 deletions in strain BY4709 (ura3; gift from J. Boeke), and, as shown in Fig. 1B, deletion of the three genes also affects the cellular growth rate in this background, especially for the strain with the UME6 gene deleted.
FIG. 1.
Effect of deletions in SIN3, RPD3, and UME6 genes on cell growth. (A and B) Tenfold serial dilutions of cells were plated and incubated at 30°C for 3 days on M.ammonia–25 μg of uracil. Strains 12T7c (ura3), 12T7cII (ura3 sin3::kanMX4), and 12T7cIII (ura3 rpd3::kanMX4) are isogenic to strain Σ1278b (A). Strains BY4709 (ura3), BY4709II (ura3 sin3::kanMX4), BY4709III (ura3 rpd3::kanMX4), and BY4709I (ura3 ume6::kanMX4) are isogenic to S288c (B). (C) Tetrad analysis of the Σ1278b isogenic diploid strain 27061b (ura3 trp1) × 27029c (ura3 leu2) in which one copy of the UME6 gene was replaced by the kanMX4 cassette. Spores were isolated on YPD medium. WT, wild type.
CAR1 expression in strains bearing different combinations of mutations impairing histone deacetylation.
To test whether simultaneous deletion of RPD3, SIN3, and UME6 genes had a cumulative effect on CAR1 expression, we have created in strain BY4709 single, double, and triple deletions. Each gene was deleted by the kanMX4 cassette flanked by lox sequences, which allowed the removal of the kanMX4 cassette by recombination using the cre recombinase (16) (see Materials and Methods). As shown in Table 4, the arginase level was higher in the ume6::kanMX4 strain (BY4709I) than in the rpd3::kanMX4 strain (BY4709III), in the sin3::kanMX4 strain (BY4709II), or in the rpd3::kanMX4 sin3::kanMX4 strain (BY4709II-III). The derepression of arginase synthesis resulting from the UME6 deletion was not increased by additional deletion of SIN3 (BY4709I-II), RPD3 (BY4709I-III), or SIN3 plus RPD3 (BY4709I-II-III). These results confirm the interdependence of these three proteins to repress a gene containing a URS1 sequence. The deletion of Ume6, which is the DNA binding protein, abolishes the repression of CAR1 expression, whereas the deletion of Sin3 and/or Rpd3, two components of the histone deacetylase complex, relieves only partially the repression. This suggests that other deacetylases and deacetylase-associated proteins could fulfil the Rpd3 or Sin3 functions.
TABLE 4.
CAR1 expression in strains bearing single, double, and triple deletions of UME6, SIN3, and RPD3
| Strain | Genotype | Arginase specific activity on M.ammoniaa |
|---|---|---|
| BY4709 | ura3 | 7 |
| BY4709I | ura3 ume6::kanMX4 | 66 |
| BY4709II | ura3 sin3::kanMX4 | 38 |
| BY4709III | ura3 rpd3::kanMX4 | 34 |
| BY4709I-II | ura3 ume6::kanMX4 sin3::lox | 63 |
| BY4709I-III | ura3 ume6::kanMX4 rpd3::lox | 60 |
| BY4709II-III | ura3 sin3::kanMX4 rpd3::lox | 41 |
| BY4709I-II-III | ura3 ume6::kanMX4 sin3::lox rpd3::lox | 60 |
Arginase specific activity was measured at 30°C after growth of the different strains on M.ammonia–25 μg of uracil per ml. The specific activity is expressed in micromoles of urea produced per hour per milligram of protein. The values are the means of two independent assays, and the standard error was 10 to 15%.
The Ume6-Sin3-Rpd3 complex represses arginine catabolic genes in response to exogenous nitrogen availability.
We have previously shown that the response of the CAR1 gene to nitrogen starvation is impaired in a ume6 mutant strain and in a strain in which the Ume6 target sequence, namely, URS1, was deleted from the CAR1 promoter (11, 41). Since Ume6 works in conjunction with Sin3 and Rpd3, we tested the role of the last two proteins in the control of CAR1 by nitrogen availability. The results showed that, upon nitrogen starvation, the arginase levels in strains with sin3::kanMX4 (BY4709II) or rpd3::kanMX4 (BY4709III) deleted were comparable to that in the wild-type strain (Table 5), indicating that, besides Ume6, Sin3 and Rpd3 were required for mediating repression of CAR1 expression as long as nitrogen was available. Since cargRI (ume6), cargRII (sin3), and cargRIII (rpd3) mutations also led to derepression of the CAR2 gene (7, 9), we analyzed the response of the CAR2 promoter to nitrogen starvation in these strains with deletions. Since the basal level of the CAR2 gene product (ornithine transaminase) was very low, we measured CAR2 expression using a CAR2-lacZ fusion (pFV118). The deletion of the UME6, SIN3, or RPD3 gene led to a strong increase of β-galactosidase synthesis, and, similar to what was found for CAR1, CAR2 derepression was significantly higher in the ume6-deleted strain. When the three strains with deletions were starved completely for exogenous nitrogen, further derepression of CAR2 was abolished in the ume6 strain and strongly reduced in the sin3 and rpd3 strains.
TABLE 5.
Effect of mutations in UME6, SIN3, and RPD3 on the starvation response of CAR1 and CAR2 genes
| Strain | Genotype | Mediuma | Expression of:
|
|
|---|---|---|---|---|
| CAR1c | CAR2-lacZb | |||
| BY4709 | ura3 | M.ammonia | 7 | 176 |
| −Nd | 60 | 1,184 | ||
| BY4709I | ura3 ume6::kanMX4 | M.ammonia | 66 | 1,644 |
| −N | 76 | 1,743 | ||
| BY4709II | ura3 sin3::kanMX4 | M.ammonia | 38 | 900 |
| −N | 63 | 1,165 | ||
| BY4709III | ura3 rpd3::kanMX4 | M.ammonia | 34 | 653 |
| −N | 65 | 1,045 | ||
When no plasmid was present in the strains, 25 μg of uracil was added.
β-Galactosidase activity was measured in extracts from different strains transformed with plasmid pFV118 (CAR2-lacZ). The β-galactosidase specific activity is expressed in nanomoles of o-nitrophenyl-β-d-galactopyranoside hydrolyzed per minute per milligram of protein.
Arginase-specific activity is expressed in micromoles of urea produced per hour per milligram of protein. The standard error was 10 to 15%.
−N, medium with no nitrogen source (see Materials and Methods).
Induction of arginase upon nitrogen starvation does not result from a burst of arginine in the cytoplasm.
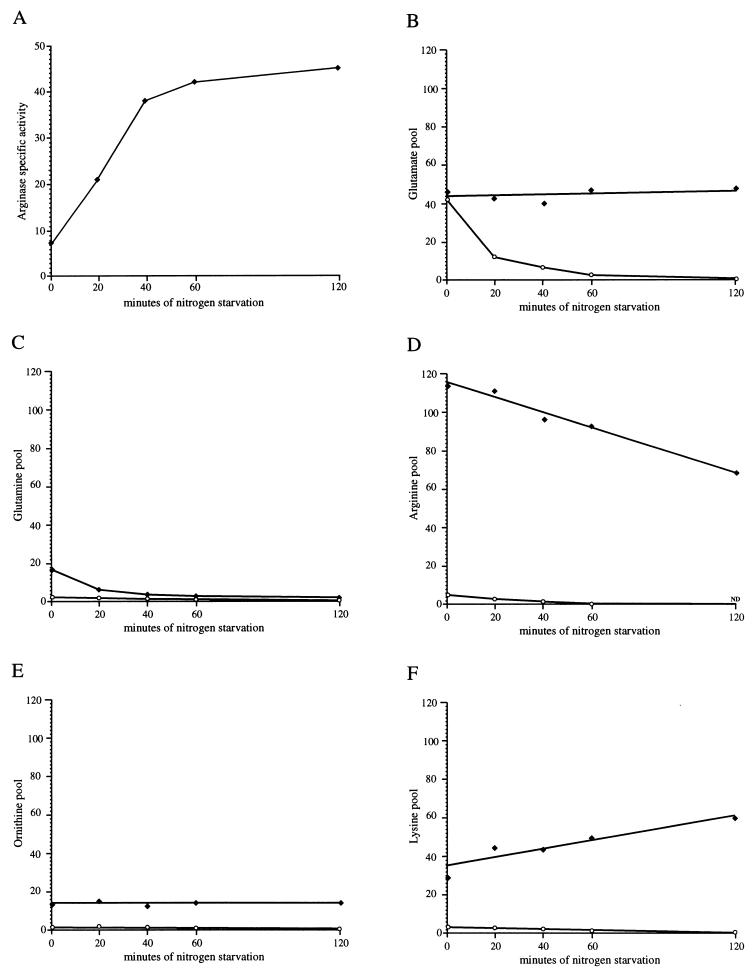
It was previously reported that starvation of an arginine auxotroph for both arginine and nitrogen did not result in the synthesis of arginase unless homoarginine, a nonmetabolizable inducer, was added, suggesting that production of arginase under conditions of nitrogen starvation was the result of induction and was contingent upon the presence of the inducer arginine in the amino acid pools of the cells (46). Later it was proposed that the internal induction of arginase in cells deprived of any nitrogen source resulted from the release of arginine from the vacuole (6). The data shown in Fig. 2 indicate that maximal arginase derepression was reached after 40 min of nitrogen starvation (Fig. 2A). To test whether this rapid increase of arginase level was a consequence of arginine accumulation in the cytosol, we have measured the evolution of amino acid pools in the vacuole and the cytoplasm after a shift from M.ammonia medium to a medium devoid of nitrogen. Using the standard Cu22+ method (33), we have determined the differential pools of glutamate, glutamine, arginine, ornithine, and lysine. As shown in Fig. 2B, the glutamate pool was equally distributed between the cytoplasm and the vacuole. Interestingly, nitrogen deprivation led to a rapid consumption of cytosolic glutamate without release from the vacuole. Glutamine was 90% vacuolar when the cells were grown on M.ammonia but was quickly released in the cytoplasm and immediately utilized as a nitrogen source upon nitrogen starvation (Fig. 2C). In contrast, the vacuolar arginine was released more slowly. After 40 min of starvation, only 15% of the vacuolar arginine was consumed without accumulation in the cytosol (Fig. 2D), while the production of arginase was maximal. These results did not contradict those of Kitamoto et al. (21). In their experiments, nitrogen starvation was performed after growth on arginine as the sole nitrogen source. In that condition at time zero, the arginase level was about 50-fold higher than that on M.ammonia, which explained why the vacuolar arginine pool decreased more rapidly but no burst of arginine was detected in the cytosol. During the nitrogen starvation, the ornithine pool remained constant (Fig. 2E), probably because the ornithine transaminase level was not sufficient to degrade it, and the lysine vacuolar pool accumulated (Fig. 2F), since lysine was not used as a nitrogen source and was less incorporated into proteins. These results indicate that the production of arginase after transfer to a nitrogen-free medium does not result from an induction in response to a higher cytosolic arginine concentration.
FIG. 2.
Determination of arginase-specific activity and cytosolic and vacuolar glutamate, glutamine, arginine, ornithine, and lysine pools upon nitrogen starvation. Arginase-specific activity was measured in extracts from wild-type strain Σ1278b after growth on M.ammonia (time zero) and shifted to minimal medium without a nitrogen source (times 20, 40, 60, and 120 min) (A). The specific activity is expressed in micromoles of urea produced per hour per milligram of protein. (B to F) Solid diamonds, vacuolar amino acid pool; open circles, cytosolic amino acid pool. Amino acid intracellular concentrations are expressed in nanomoles per milligram of dry weight.
Arginase induction upon nitrogen starvation could result from the interaction between Ume6 and components of the ArgR-Mcm1 complex.
We have previously shown that the control of CAR1 expression by nitrogen availability required URS1, the arginine boxes (arginine upstream activation sequence [UASarg]), and the integrity of Ume6 and the ArgR-Mcm1 complex (11). We had proposed that when cells were starved for nitrogen, the release of URS1 facilitated the accessibility of the ArgR-Mcm1 complex to UASarg in spite of the weak arginine internal pool. Using the two-hybrid system we identified an interaction between Ume6 and ArgRI or ArgRII, but only under nitrogen starvation conditions. As shown in Table 6, in strain HY transformed with plasmid pME46 (GBD-ArgRI) or plasmid pNA33 (GBD-ArgRII) and plasmid pFV124 (GAD-Ume6), the level of β-galactosidase was increased in the absence of nitrogen. No interaction between Ume6 and the pleiotropic regulators ArgRIII and Mcm1, which are also regulators of the arginine genes (data not shown), was detected. The interaction between Ume6 and ArgRI or ArgRII seems thus specific and physiological.
TABLE 6.
Interaction between Ume6 and ArgRI or ArgRII under nitrogen starvation conditionsa
| Protein | Medium | β-Galactosidase specific activityb for:
|
|
|---|---|---|---|
| GAD | GAD-Ume6 | ||
| GBD | M.ammonia | <1 | <1 |
| −Nc | <1 | <1 | |
| GBD-ArgRI | M.ammonia | 2 | 2 |
| −N | 2 | 21 | |
| GBD-ArgRII | M.ammonia | 33 | 37 |
| −N | 35 | 162 | |
The full-length Ume6 was fused in frame with the activation domain of Gal4 (GAD-Ume6), and full-length ArgRI and ArgRII were fused in frame with the DNA binding domain of Gal4 (GBD-ArgRI and GBD-ArgRII).
Transcription activation of the lacZ reporter gene was determined by β-galactosidase activity assays performed at 30°C on extracts of at least two independent transformants containing both plasmids. The standard error was 15% of the mean. The β-galactosidase specific activity is expressed in nanomoles of o-nitrophenyl-β-d-galactopyranoside hydrolyzed per minute per milligram of protein.
−N, medium with no nitrogen source.
DISCUSSION
CARGRI, CARGRII, and CARGRIII gene products were identified as repressors of CAR1 and CAR2 genes. CargRI turned out to be Ume6, and we show here that CargRII is identical to Sin3 and that CargRIII is identical to Rpd3. These three proteins are part of a high-molecular-weight complex regulating the expression of a large set of genes, which explains the wide variety of names attributed to these three genes. Ume6 is able to interact with DNA in vitro at a sequence called URS1 and recruits Sin3, which then binds to Rpd3 causing repression of transcription through core histone deacetylation (20). Sap30, which is part of this protein complex, is also required for CAR1 repression, whereas two other histone deacetylases, Hda1 and Hos2, belonging to other histone deacetylase complexes, do not control CAR1 expression. In contrast, neither Gcn5 nor TAF145/130, both of which show histone acetyltransferase activity (4, 32), is involved in the derepression of CAR1. Formation of a more active chromatin state at the CAR1 promoter should thus depend on another histone acetyltransferase. As for other genes controlled by Ume6, the repression at CAR1 results mainly from the action of the Sin3-Rpd3 complex, since deletion of SIN3 and/or RPD3 does not increase the derepression of arginase observed in a ume6-deleted strain. However, this derepression is always higher in a ume6-deleted strain than in a sin3- or rpd3-deleted strain. This could result from a repressing activity of Ume6 independent of histone deacetylase recruitment or from a partial effect of other histone deacetylases in the absence of Sin3 or Rpd3. The fact that Ume6 could play a role independent of Sin3 and Rpd3 is sustained by our observation that the growth of a ume6 deletant is more impaired than the growth of sin3 or rpd3 deletants. This difference in behavior is increased in the Σ1278b background, in which deletion of UME6 leads to lethality. Such a phenotype suggests that Ume6 could recruit positive transcription factors involved in the control of essential genes. It has already been reported that one of the positive effects of Ume6 is to recruit the transcriptional activator Ime1 to activate the expression of early meiotic genes upon nitrogen starvation (37). This Ime1-Ume6 complex formation requires an interaction between Rim11 and Ume6, resulting in a carbon source-dependent phosphorylation of Ume6 (25).
In the control of arginine catabolic genes, the role of the Ume6-Sin3-Rpd3 complex is to block the expression of CAR1 and CAR2 promoters as long as exogenous nitrogen is available. Indeed, a mutation in UME6 abolishes completely the response of these two promoters to nitrogen depletion. Arginase and ornithine transaminase production under nitrogen starvation conditions also requires the integrity of the ArgR-Mcm1 complex (11). However, this enzyme synthesis does not result from a burst of arginine stored in the vacuole toward the cytosol, as shown by our differential arginine pool measurements. Under these growth conditions, arginine leaks slowly out of the vacuole to the cytosol, where it is used as a nitrogen source, without sufficient accumulation of arginine to allow the interaction between the ArgR-Mcm1 complex and the arginine boxes (11). This induction could rather result from an interaction between Ume6 and the components of the ArgR-Mcm1 complex, leading to a more efficient binding at the arginine boxes at low arginine concentration. Such a hypothesis is supported by our two-hybrid experiments showing an interaction between ArgRI or ArgRII and Ume6 only under nitrogen starvation conditions.
To confirm the importance of chromatin structure in the regulation of CAR1 and CAR2 expression, mapping nucleosomes along the promoters of these two genes under different growth conditions will be our next goal.
ACKNOWLEDGMENTS
We are grateful to Eric Joris for his excellent technical assistance and to J. Boeke for providing strains.
REFERENCES
- 1.Béchet J, Grenson M, Wiame J M. Mutations affecting the repressibility of arginine biosynthetic enzymes in Saccharomyces cerevisiae. Eur J Biochem. 1970;12:31–39. doi: 10.1111/j.1432-1033.1970.tb00817.x. [DOI] [PubMed] [Google Scholar]
- 2.Bidlingmeyer B A, Cohen S A, Tarvin T L. Rapid analysis of amino acids using pre-column derivatization. J Chromatogr. 1984;336:93–104. doi: 10.1016/s0378-4347(00)85133-6. [DOI] [PubMed] [Google Scholar]
- 3.Brachmann C B, Davies A, Cost G J, Caputo E, Li J, Hieter P, Boeke J D. Designer deletion strains derived from S. cerevisiae S288c: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 4.Candau R, Zhou J X, Allis C D, Berger S L. Histone acetyltransferase activity and interaction with Ada2 are critical for Gcn5 function in vivo. EMBO J. 1997;16:555–565. doi: 10.1093/emboj/16.3.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Casadaban M J, Chou J, Cohen S. In vitro gene fusions that join an enzymatically active β-galactosidase segment to amino-terminal fragments of exogenous proteins: E. coli plasmid vectors for the detection and cloning of translation initiation signals. J Bacteriol. 1980;143:971–980. doi: 10.1128/jb.143.2.971-980.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Courchesne W E. Nitrogen regulation in S. cerevisiae. Ph.D. thesis. Cambridge: Massachusetts Institute of Technology; 1985. [Google Scholar]
- 7.Deschamps J, Dubois E, Wiame J M. l-Ornithine transaminase synthesis in Saccharomyces cerevisiae: regulation by inducer exclusion. Mol Gen Genet. 1979;174:225–232. doi: 10.1007/BF00267794. [DOI] [PubMed] [Google Scholar]
- 8.Dubois E, Grenson M, Wiame J M. The participation of the anabolic glutamate dehydrogenase in the nitrogen catabolite repression of arginase in Saccharomyces cerevisiae. Eur J Biochem. 1974;48:603–616. doi: 10.1111/j.1432-1033.1974.tb03803.x. [DOI] [PubMed] [Google Scholar]
- 9.Dubois E, Hiernaux D, Grenson M, Wiame J M. Specific induction of catabolism and its relation to repression of biosynthesis in the arginine metabolism of Saccharomyces cerevisiae. J Mol Biol. 1978;122:383–406. doi: 10.1016/0022-2836(78)90417-5. [DOI] [PubMed] [Google Scholar]
- 10.Dubois E, Messenguy F. In vitro studies of the binding of the ARGR proteins to the ARG5,6 promoter. Mol Cell Biol. 1991;11:2162–2168. doi: 10.1128/mcb.11.4.2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dubois E, Messenguy F. Integration of the multiple controls regulating the expression of the arginase gene CAR1 of Saccharomyces cerevisiae in response to different nitrogen signals: role of Gln3p, ArgRp-Mcm1p and Ume6p. Mol Gen Genet. 1997;253:568–580. doi: 10.1007/s004380050359. [DOI] [PubMed] [Google Scholar]
- 12.Dubois E, Vierendeels F, Messenguy F. Role of histone deacetylase Rpd3 in the control of expression of arginine catabolic genes. Curr Genet. 1999;35:243. [Google Scholar]
- 13.Durfee T, Becherer K, Chen R-L, Yeh S H, Yang Y, Kilburn A E, Lee W H, Elledge S J. The retinoblastoma protein associates with the protein phosphatase type 1 catalytic subunit. Genes Dev. 1993;7:555–569. doi: 10.1101/gad.7.4.555. [DOI] [PubMed] [Google Scholar]
- 14.El Bakkoury M, Dubois E, Messenguy F. Recruitment of the yeast MADS-box proteins, ArgRI and Mcm1, by the pleiotropic factor ArgRIII is required for their stability. Mol Microbiol. 2000;35:15–31. doi: 10.1046/j.1365-2958.2000.01665.x. [DOI] [PubMed] [Google Scholar]
- 15.Georgakopoulos T, Gounalaki N, Thireos G. Genetic evidence for the interaction of the yeast transcriptional co-activator proteins Gcn5 and Ada2. Mol Gen Genet. 1995;246:723–728. doi: 10.1007/BF00290718. [DOI] [PubMed] [Google Scholar]
- 16.Guldener U, Heck S, Fiedler T, Beinhauwer J, Hegemann J H. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 1996;24:2519–2524. doi: 10.1093/nar/24.13.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hennaut C. Ornithine transaminase synthesis in Saccharomyces cerevisiae: induction by allophanate, intermediate and inducer of the urea degradative pathway, adds to arginine induction. Curr Genet. 1981;4:69–72. doi: 10.1007/BF00376788. [DOI] [PubMed] [Google Scholar]
- 18.Holstege F C, Jennings E G, Wyrick J J, Lee T I, Hengartner C J, Green M R, Golub T R, Lander E S, Young R A. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- 19.Ito H, Fukura Y, Murata K, Timura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kadosh D, Struhl K. Repression by Ume6 involves recruitment of a complex containing Sin3 corepressor and Rpd3 histone deacetylase to target promoters. Cell. 1997;89:365–371. doi: 10.1016/s0092-8674(00)80217-2. [DOI] [PubMed] [Google Scholar]
- 21.Kitamoto K, Yoshizawa K, Ohsumi Y, Anraku Y. Dynamic aspects of vacuolar and cytosolic amino acid pools of Saccharomyces cerevisiae. J Bacteriol. 1988;170:2683–2686. doi: 10.1128/jb.170.6.2683-2686.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Louvet O, Doignon F, Crouzet M. Stable DNA-binding yeast vector allowing high-bait expression for use in the two-hybrid system. BioTechniques. 1997;23:816–817. doi: 10.2144/97235bm11. [DOI] [PubMed] [Google Scholar]
- 23.Lowry O H, Rosebrough N J, Farr A L, Randall R J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 24.Luche R M, Sumrada R, Cooper T G. A cis-acting element present in multiple genes serves as a repressor protein binding site for the yeast CAR1 gene. Mol Cell Biol. 1990;10:3884–3895. doi: 10.1128/mcb.10.8.3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Malathi K, Xiao Y, Mitchell A P. Interaction of yeast repressor-activator protein Ume6p with glycogen synthase kinase 3 homolog Rim11p. Mol Cell Biol. 1997;17:7230–7236. doi: 10.1128/mcb.17.12.7230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marcus G A, Silverman N, Berger S L, Horiuchi J, Guarente L. Functional similarity and physical association between Gcn5 and Ada2: putative transcriptional adaptors. EMBO J. 1994;13:4807–4815. doi: 10.1002/j.1460-2075.1994.tb06806.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Messenguy F. Regulation of arginine biosynthesis in Saccharomyces cerevisiae: isolation of a cis-dominant constitutive mutant for ornithine carbamoyltransferase synthesis. J Bacteriol. 1976;128:49–55. doi: 10.1128/jb.128.1.49-55.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Messenguy F, Dubois E. Genetic evidence for a role for Mcm1 in the regulation of arginine metabolism in Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:2586–2592. doi: 10.1128/mcb.13.4.2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Messenguy F, Dubois E, Boonchird C. Determination of the DNA binding sequences of ARGR proteins to arginine anabolic and catabolic promoters. Mol Cell Biol. 1991;11:2852–2863. doi: 10.1128/mcb.11.5.2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Messenguy F, Penninckx M, Wiame J-M. Interaction between arginase and ornithine carbamoyltransferase in Saccharomyces cerevisiae. Eur J Biochem. 1971;22:277–286. doi: 10.1111/j.1432-1033.1971.tb01542.x. [DOI] [PubMed] [Google Scholar]
- 31.Miller J. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 32.Mizzen C A, Yang X-Y, Kokubo T, Brownell J E, Bannister A J, Owen-Hughes T, Workman J, Wang L, Berger S L, Kouzarides T, Nakatani Y, Allis C D. The TAFII250 subunit of TFIID has histone acetyltransferase activity. Cell. 1996;87:1261–1270. doi: 10.1016/s0092-8674(00)81821-8. [DOI] [PubMed] [Google Scholar]
- 33.Ohsumi Y, Kitamoto K, Anraku Y. Changes induced in the permeability barrier of the yeast plasma membrane by cupric ion. J Bacteriol. 1988;170:2676–2682. doi: 10.1128/jb.170.6.2676-2682.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Park H-D, Luche R M, Cooper T G. The yeast UME6 gene product is required for transcriptional repression mediated by the CAR1 URS1 repressor binding site. Nucleic Acids Res. 1992;20:1909–1915. doi: 10.1093/nar/20.8.1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park H-D, Scott S, Rai R, Dorrington R, Cooper T G. Synergistic operation of the CAR2 (ornithine transaminase) promoter elements in Saccharomyces cerevisiae. J Bacteriol. 1999;181:7052–7064. doi: 10.1128/jb.181.22.7052-7064.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rothstein R J. Targeting, disruption, replacement and allele rescue: integrative DNA transformation in yeast. Methods Enzymol. 1991;194:281–301. doi: 10.1016/0076-6879(91)94022-5. [DOI] [PubMed] [Google Scholar]
- 37.Rubin-Bejerano I, Mandel S, Robzyk K, Kassir Y. Induction of meiosis in Saccharomyces cerevisiae depends on conversion of the transcriptional repressor Ume6 to a positive regulator by its regulated association with the transcriptional activator Ime1. Mol Cell Biol. 1996;16:2518–2526. doi: 10.1128/mcb.16.5.2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rundlett S E, Carmen A A, Kobayashi R, Bavykin S, Turner B M, Grunstein M. Hda1 and Rpd3 are members of distinct yeast histone deacetylase complexes. Proc Natl Acad Sci USA. 1996;93:14503–14508. doi: 10.1073/pnas.93.25.14503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in S. cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smart W C, Coffman J A, Cooper T G. Combinatorial regulation of the Saccharomyces cerevisiae CAR1 (arginase) gene promoter in response to multiple environmental signals. Mol Cell Biol. 1996;16:5876–5887. doi: 10.1128/mcb.16.10.5876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Strich R, Surosky R T, Steber C, Dubois E, Messenguy F, Esposito R E. UME6 is a key regulator of nitrogen repression and meiotic development. Genes Dev. 1994;8:796–810. doi: 10.1101/gad.8.7.796. [DOI] [PubMed] [Google Scholar]
- 42.Thuriaux P. Ph.D. thesis. Brussels, Belgium: Brussels University; 1969. [Google Scholar]
- 43.Vidal M, Gaber R F. RPD3 encodes a second factor required to achieve maximum positive and negative transcriptional states in Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:6317–6327. doi: 10.1128/mcb.11.12.6317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vidal M, Strich R, Esposito R E, Gaber R F. RPD1 (SIN3/UME4) is required for maximal activation and repression of diverse yeast genes. Mol Cell Biol. 1991;11:6306–6316. doi: 10.1128/mcb.11.12.6306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wach A. PCR-synthesis of marker cassettes with long flanking homology regions for gene disruptions in Saccharomyces cerevisiae. Yeast. 1996;12:259–265. doi: 10.1002/(SICI)1097-0061(19960315)12:3%3C259::AID-YEA901%3E3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 46.Withney P A, Magasanik B. The induction of arginase in Saccharomyces cerevisiae. J Biol Chem. 1973;248:6197–6202. [PubMed] [Google Scholar]
- 47.Zhang Y, Sun Z-W, Iratni R, Erdjument-Bromage H, Tempst P, Hampsey M, Reinberg D. Sap30, a novel protein conserved between human and yeast, is a component of an histone deacetylase complex. Mol Cell. 1998;1:1021–1031. doi: 10.1016/s1097-2765(00)80102-1. [DOI] [PubMed] [Google Scholar]