Abstract
Establishment and maintenance of a lysogen of the lambdoid bacteriophage 434 require that the 434 repressor both activate transcription from the PRM promoter and repress transcription from the divergent PR promoter. Several lines of evidence indicate that the 434 repressor activates initiation of PRM transcription by occupying a binding site adjacent to the PRM promoter and directly contacting RNA polymerase. The overlapping architecture of the PRM and PR promoters suggests that an RNA polymerase bound at PR may repress PRM transcription initiation. Hence, part of the stimulatory effect of the 434 repressor may be relief of interference between RNA polymerase binding to the PRM promoter and to the PR promoter. Consistent with this proposal, we show that the repressor cannot activate PRM transcription if RNA polymerase binds at PR prior to addition of the 434 repressor. However, unlike the findings with the related λ phage, formation of RNA polymerase promoter complexes at PRM and at PR apparently are mutually exclusive. We find that the RNA polymerase-mediated inhibition of repressor-stimulated PRM transcription requires the presence of an open complex at PR. Taken together, these results indicate that establishment of an open complex at PR directly prevents formation of an RNA polymerase-PRM complex.
Each lambdoid bacteriophage contains a right operator (OR) region on its chromosome that is at the center of a complex regulatory circuit responsible for governing the phage's choice between lytic and lysogenic development. Proper regulation of transcription initiation from the divergently oriented PR and PRM promoters that lie within the OR region is crucial to the lysis-lysogeny decision. In each phage, the activities of these promoters are regulated, in part, by the binding of the bacteriophage repressor to three recognition sites that partially overlap the PR and PRM promoters. In the absence of the repressor, the PRM promoter is virtually inactive and RNA polymerase preferentially initiates transcription at the PR promoter. During an infection or induction of a lysogen, continued activity of the PR promoter drives the phage to develop lytically. If the phage is to develop or maintain the lysogenic state, there must be exclusive expression of PRM over PR.
To perform its role in the lysis-lysogeny decision, the repressor must bind to each of the three binding sites or operators within OR with different affinities and act both as transcriptional activator of PRM and as repressor of PR. In a developing or existing lysogen, the repressor binds with highest affinity to two sites, OR1 and OR2. In this configuration, the repressor molecule bound at OR2 activates transcription from the PRM promoter. This event leads to expression of the cI gene that encodes the repressor, which is the sole protein responsible for maintenance of the lysogenic state. This binding configuration also permits the repressor to concurrently inhibit transcription initiation from PR and in doing so prevents the transcription of genes needed for lytic growth (for a review, see reference 23).
Comparisons among the lambdoid phages have added to our understanding of OR function (1) as well as provided insight into the general mechanisms of transcriptional regulation by DNA binding proteins. In recent years, much has been learned about how the repressors of these phages activate transcription. Evidence suggests that the OR2-bound repressor of each phage activates transcription initiation by directly contacting the ς70 subunit of the PRM-bound RNA polymerase (16, 18). Studies of bacteriophage λ indicate that, in addition to activating PRM transcription by directly contacting RNA polymerase, the λ repressor also activates transcription indirectly by relieving interference with an RNA polymerase bound at PR (6, 10, 11). Interestingly, in λ phage, formation of an open complex at the PR promoter does not prevent RNA polymerase from binding at PRM but rather impairs isomerization of the RNA polymerase-PRM closed complex to an open complex (6, 10). These findings lead to the suggestion that formation of an open complex at the λ PR promoter interferes with formation of a similar complex at PRM. Consistent with this suggestion, on templates bearing a single base deletion in λ OR, which decreases the distance between the transcription start site of PRM and that of PR, open complex formation at PRM is drastically inhibited by RNA polymerase bound at PR (26).
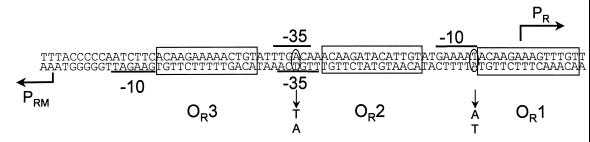
In the OR region of bacteriophage 434, the transcription start sites of the PR and PRM promoters are separated by 65 bp, compared to the 82-bp separation found in λ's OR region. This separation results in a strikingly different placement of the PR and PRM promoter elements with respect to the OR2 sites in 434 phage relative to phage λ. In λ phage, the −35 elements of its PRM and PR promoters overlap the left and right ends of OR2, respectively. In 434 phage, the −35 elements of 434 PR and PRM promoters are located on the left side of OR2 and almost completely overlap (Fig. 1). As a result of this geometry, severe promoter interference between PR and PRM in 434 OR was anticipated (2, 3). Until now, however, this prediction had not been confirmed.
FIG. 1.
The sequence of 434 OR region. OR1, OR2, and OR3 are enclosed in boxes; the transcription start sites of PRM and PR are indicated by bent arrows. The −35 and −10 regions of PR and PRM are underlined. The positions and sequences of the promoter mutants used in this study are indicated.
The potential for simultaneous occupancy of PR by RNA polymerase and the repressor at OR leads to a question about the precise mechanism that the repressor uses in inhibiting transcription initiation from PR. Jacob and Monod (13) advanced the idea that gene regulation could occur by preventing or repressing the expression of genes. Their classical model of repressor function proposes that repressors block access of RNA polymerase to the promoter by occluding the RNA polymerase binding site. More recent studies indicate, however, that repressors of transcription often act subsequent to the formation of the initial RNA polymerase-promoter complex (4, 9, 17, 20).
MATERIALS AND METHODS
Enzyme and reagents.
Wild-type and mutant 434 repressors were prepared as described in reference 24. Sigma-saturated wild-type Escherichia coli RNA polymerase was obtained from Epicentre Technologies. [α-32P]UTP and [α-32P]dATP (3,000 Ci/mmol) were obtained from New England Nuclear. Unlabeled nucleoside triphosphates were purchased from Boehringer Mannheim.
DNA templates.
Transcription reactions were programmed with DNA fragments isolated from variants of the plasmid pJX (28). The 450-bp transcription templates were prepared by isolating a 450-bp PvuII fragment from this plasmid or its derivatives. Point mutations in the template (Fig. 1) were introduced by PCR mutagenesis as described previously (19).
Transcription in vitro.
Transcription reactions were performed essentially as described previously (28). Briefly, 5 nM (each) DNA template was separately incubated with or without varying amounts of the 434 repressor for 10 min at 23°C in transcription buffer containing 100 mM KCl, 40 mM Tris (pH 7.9), 10 mM MgCl2, and 10 mM dithiothreitol. RNA polymerase was added to a final concentration of 50 nM, and incubation was continued for another 15 min at 37°C to allow the formation of open complexes. The transcription reaction was started by the addition of 0.25 mM ATP, GTP, or CTP; 0.04 mM UTP; 10 μCi of [α-32P]UTP; and 0.1 mg of heparin per ml. After 10 min of further incubation, the reactions were stopped by addition of formamide dye mix (90% formamide) and fractionated on 6% denaturing gels. The amounts of RNA transcripts resulting from initiation at PR and PRM were quantified by PhosphorImager analysis.
DNase I footprinting.
DNase I footprinting assays were performed essentially as described previously (28). Briefly, a 400-bp PvuII-HindIII DNA fragment derived from the desired pJX derivative was 3′ end labeled using the Klenow fragment and [α-32P]dATP. The DNA was mixed with increasing amounts of the 434 repressor in transcription buffer. After 10 min of incubation at 23°C, sufficient DNase I was added to give, on average, one cleavage per DNA molecule in 5 min of further incubation. The reaction products were precipitated with ethanol and sec-butanol, dissolved in a formamide dye, and resolved on 6% denaturing gels.
KMnO4 footprinting.
KMnO4 footprinting was performed essentially as described previously (27). Briefly, the 400-bp PvuII-HindIII DNA fragment was 3′ end labeled as described above. This fragment was incubated with or without varying concentrations of the 434 repressor at 23°C for 10 min, followed by the addition of RNA polymerase. After an additional 10-min incubation at 37°C, the DNA was exposed to 10 mM KMnO4 for 1 min. The oxidation reaction was stopped by adding 1.3 M 2-mercaptoethanol, and the DNA was purified by two ethanol precipitations. The precipitated DNA fragments were dissolved in 100 μl of 1 M piperidine and incubated for 15 min at 90°C to induce cleavage at the modified bases. The DNA was then diluted in the same volume of ddH2O, lyophilized twice, dissolved in formamide dye mix, and fractionated on a 6% denaturing polyacrylamide gel. The products were visualized by PhosphorImager analysis.
Gel mobility shift assay.
The 400-bp PvuII-HindIII DNA fragment isolated from pJX or its derivatives was 3′ end labeled as described above. The labeled DNA was mixed with increasing amounts of RNA polymerase in transcription buffer at 37°C and incubated for 15 min to allow the open complex formation. Subsequently, 0.1 mg of heparin per ml was added to remove nonspecifically bound RNA polymerases and/or closed promoter complexes before 5% glycerol was added prior to loading the sample onto a nondenaturing 3.5% polyacrylamide gel. The gels were run at 4°C with 0.5× Tris-borate-EDTA at 160 V for approximately 4 h. The gels were then dried, and the reaction products were visualized by PhosphorImager analysis.
RESULTS
Transcription from PRM cannot be activated by the 434 repressor if RNA polymerase is added before the 434 repressor.
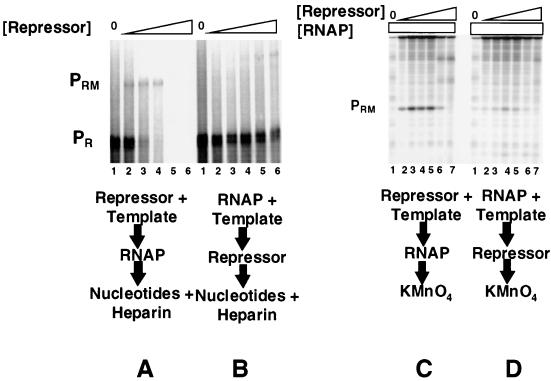
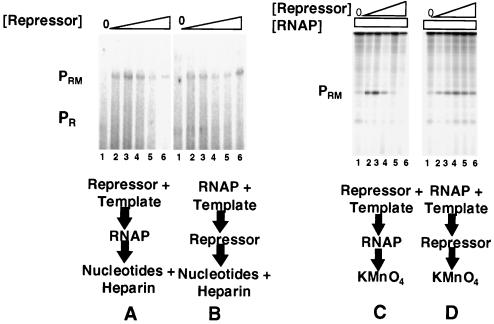
The distance between the PR and PRM promoters in bacteriophage 434 is 17 bp less than in the related bacteriophage λ. Since, in both phages, the PR promoter is substantially stronger than PRM and an RNA polymerase-PR complex interferes with open complex formation at PRM in bacteriophage λ, we were interested in characterizing the predicted (1) promoter interference mechanism in bacteriophage 434. To begin this study, we compared the abilities of the 434 repressor to activate transcription from 434 PRM when it is incubated with DNA prior to or after addition of RNA polymerase on a template that contains both the wild-type PR and the PRM promoters (Fig. 1). Consistent with previous reports (1, 2, 28), incubating the 434 repressor with the DNA template prior to the addition of RNA polymerase allows repressor-mediated activation of PRM transcription and repression of PR transcription (Fig. 2A). In contrast, activation of PRM transcription by the 434 repressor is significantly reduced when the DNA template is first incubated with RNA polymerase at 37°C prior to adding the 434 repressor (Fig. 2B). Under the conditions of this experiment, RNA polymerase forms an open complex at PR (1, 2). Apparently, the 434 repressor's ability to activate transcription from PRM is inhibited by the formation of stable RNA polymerase open complexes at PR.
FIG. 2.
Prior addition of RNA polymerase inhibits repressor-activated PRM transcription (A and B) or open complex formation (C and D). For panels A and B, DNA fragments containing wild-type PR and PRM were transcribed in vitro in the absence of repressor (lanes 1) and at various increasing repressor concentrations. Repressor concentrations were increased in 2.5-fold steps starting at 250 nM protein (lanes 2 to 6). Positions of transcripts resulting from initiation of transcription from PRM and PR are indicated. (A) The 434 repressor was incubated with DNA template at 23°C for 10 min, followed by addition of RNA polymerase. The reaction mixture was transferred to 37°C for 10 min before the transcription reaction was initiated by the addition of nucleotides and heparin. (B) RNA polymerase was incubated with DNA at 37°C for 10 min before addition of the 434 repressor. After an additional 10-min incubation at 37°C, the transcription reaction was initiated by the addition of nucleotides and heparin. (C and D) Shown are the open complexes formed at PRM in the absence of repressor (lanes 1) and the presence of increasing concentrations of the 434 repressor as detected by KMnO4 footprinting. Repressor concentrations were increased in 2.5-fold steps starting at 250 nM protein. Footprinting conditions are given in Materials and Methods. Positions of PR and PRM open complexes are indicated. (C) Incubation conditions were as described for panel A. (D) Incubation conditions were as described for panel B. RNAP, RNA polymerase.
Open complex formation on PRM is inhibited if RNA polymerase is added before the repressor.
The RNA polymerase-mediated “inhibition” of activated PRM transcription shown in Fig. 2B could occur at any of the steps of the transcription pathway including closed complex and open complex formation at PRM or transcription initiation and elongation processes. To distinguish between these possibilities, we observed open complex formation at PRM by monitoring the KMnO4 reactivity of the bases in the −10 region of this promoter. Since this method detects only unpaired thymines under the conditions used here, and since there are no accessible thymines in the labeled strand at PR, this experiment monitors only open complex formation at PRM. Incubating the 434 repressor with DNA template prior to adding RNA polymerase leads to a repressor-dependent activation of PRM open complex formation. This finding is consistent with the repressor's ability to activate transcription from PRM under these conditions (Fig. 2C). However, the ability of the 434 repressor to stimulate open complex formation at PRM is almost completely eliminated if the template is incubated with RNA polymerase at 37°C prior to addition of the 434 repressor (Fig. 2D). Thus, RNA polymerase inhibition of repressor-mediated activation of PRM transcription occurs prior to formation of an open complex. We note, however, that the inhibition of open complex formation is incomplete, in that some PRM open complexes are formed under these conditions. Since we do not observe any transcripts initiating at PRM when the repressor is added to DNA after RNA polymerase (Fig. 2B), the observation of residual open complexes may indicate that the subsequent addition of the repressor also blocks PRM promoter clearance.
The repressor binds to OR2 in the presence of RNA polymerase bound at PR.
Since transcription initiation at PRM requires a repressor-OR2 complex (Fig. 2A), a simple explanation for the inhibitory effect of RNA polymerase on repressor activation of PRM transcription would be that an RNA polymerase molecule at PR prevents repressor binding. To test this idea, we examined the ability of the repressor to bind the sites in OR in the absence or presence of RNA polymerase by DNase I footprinting. We first characterized the DNase I footprinting pattern of the repressor in the absence of RNA polymerase. Figure 3A shows that, in the absence of RNA polymerase, increasing 434 repressor concentrations result in progressive occupancy of the operator sites. The occupancy of these sites as a function of repressor concentration (OR1 ≈ OR2 > OR3) reflects their relative affinity for the repressor in intact OR (25).
FIG. 3.
The 434 repressor binds to OR in the presence of RNA polymerase at PR. DNA templates containing wild-type PR and PRM promoters were partially digested by DNase I in the presence of various amounts of the 434 repressor (A) or the 434 repressor and RNA polymerase (B and C). (A) Increasing concentrations of the repressor were incubated with DNA at 23°C for 10 min prior to addition of DNase I and heparin. Lane 1 shows the DNase I cleavage pattern of the DNA in the absence of added repressor. In lanes 2 to 6, repressor concentrations were increased in 2.5-fold steps starting at 250 nM protein. (B and C) DNA and 50 nM RNA polymerase were incubated in the absence (lanes 1) or the presence of RNA polymerase (lanes 2) and the 434 repressor (lanes 3 to 7). The repressor concentrations were increased in 2.5-fold steps starting at 250 nM protein. (B) In the lanes containing the repressor, the repressor was incubated with DNA template at 23°C for 10 min, followed by addition of RNA polymerase. The reaction mixture was transferred to 37°C for 10 min before the addition of DNase I and heparin. (C) RNA polymerase was incubated with DNA at 37°C for 10 min before addition of the 434 repressor (lanes 2 to 7). After an additional 10-min incubation at 37°C, cleavage was initiated by addition of DNase I and heparin. Positions of protection and enhancements resulting from protein binding to the sites indicated are denoted as described in the text. RNAP, RNA polymerase.
Next, we determined the DNase I footprinting pattern of the repressor-RNA polymerase-DNA ternary complex under the condition where RNA polymerase is added to DNA subsequent to the formation of the repressor-DNA complex. A difficulty with analyzing these footprinting results is that the repressor is able to both repress transcription of PR and activate transcription of PRM under these conditions (1, 2) (Fig. 2). Hence, the footprinting patterns reflect not only repressor binding but also the repressor's redirection of RNA polymerase binding from PR to PRM. Comparison of lanes 1 and 2 of Fig. 3B reveals that RNA polymerase fully or partially inhibits the DNAse I-mediated cleavage of numerous bases in the region of OR that comprises the PR promoter, from about +20 through −50 relative to the start site of PR transcription (protected bases are labeled with solid circles in lane 2 of Fig. 3B). Addition of twofold-more RNA polymerase does not appreciably change the observed pattern of protection and enhancements (data not shown). Together with control KMnO4 probe experiments (data not shown), these findings suggest that this pattern represents that of the RNA polymerase-PR complex.
The observation that adding RNA polymerase to the repressor-DNA complex results in activation of PRM transcription (Fig. 2A) suggests that these conditions should allow us to examine the DNase I footprint pattern of the repressor-RNA polymerase-PRM ternary complex. Comparing lanes 1 and 2 with lanes 3 to 7 of Fig. 3B shows that adding increasing concentrations of the repressor followed by subsequent addition of RNA polymerase results in protection of several bases at the center of OR2 and at either end of the OR1 site (asterisk-marked bases in lane 3 in Fig. 3B) that are not protected by RNA polymerase alone. These additional protections result from occupancy of OR1 and OR2 by the repressor (see Fig. 3A for comparison). Inspection of the repressor-alone footprinting results in Fig. 3A shows that the repressor protects a region of DNA upstream of OR1 that extends to position +20 of PR from DNase I cleavage. This region is similarly protected in the presence of RNA polymerase bound at PR (Fig. 3B, lane 2). However, when RNA polymerase is added to the repressor-DNA complex, this region is not protected but instead shows hyperreactive DNase I cleavage (positions marked with open arrowheads in Fig. 3B, lanes 3 to 7). These hyperreactive cleavages are not observed when RNA polymerase is added to DNA prior to adding the repressor (Fig. 3C, lanes 3 to 7). Adding RNA polymerase to the repressor-DNA complex also results in the appearance of a weak hypersensitive cleavage at position −15 of PR (denoted by a caret), which is not seen when the repressor is added alone (compare lane 4 of Fig. 3A with that of 3B) or subsequent to RNA polymerase addition (Fig. 3C; see below). A stronger hypersensitive cleavage site is observed at position −30 of PRM. In addition, several protections are observed in the −10 region of PRM that are not well resolved on this gel (Fig. 3B, lanes 4 to 6, and data not shown). As supported by a similar analysis of template bearing a strong mutation in PR (data not shown), we assert that this pattern of DNase I protections and enhancements represents that of the repressor-RNA polymerase-PRM open complex (also Fig. 2C).
In light of the observation that the repressor is unable to activate transcription of PRM if it is added to DNA after RNA polymerase (Fig. 2), a significant question is whether prior binding of RNA polymerase at PR blocks repressor binding to its binding sites in OR. Comparison of lanes 2 and 3 in Fig. 3C shows that adding the repressor to RNA polymerase bound at PR results in the protection of bases at either end of OR1, bases between OR1 and OR2 (denoted by asterisks in Fig. 3C), and bases at the center of OR2 from DNase I digestion. These protections result from repressor binding and, moreover, occur at repressor concentrations that are very similar to those needed to occupy these sites in the absence of RNA polymerase. As a result of the overlap between the DNase I footprints of RNA polymerase-PR complex and the complex of the repressor with OR1 and OR2, it is difficult to assess whether RNA polymerase remains bound to the template at the PR promoter upon addition of the repressor. To answer this question, we compared the patterns of DNase I cleavage that are diagnostic for full repressor occupancy of OR1 and OR2 (Fig. 3A, lane 4) and of a repressor-RNA polymerase-PRM open complex (Fig. 3B, lane 4) with the patterns present in Fig. 3C, lanes 3 to 6. The DNase I patterns of the samples in the latter lanes sample the structure of the complex under conditions where RNA polymerase is capable of transcribing PR in the presence of the repressor (Fig. 2). The DNase I cleavage pattern of RNA polymerase alone (Fig. 3B and C, lanes 2) and complexes formed when RNA polymerase is added prior to the repressor (Fig. 3C, lanes 3 to 7) consistently display hypersensitive cleavages at positions near −50 of PR (lanes 2 of Fig. 3B and C, denoted by plus signs). These cleavages are absent in the repressor-only and the repressor-RNA polymerase-PRM open complex lanes. This finding suggests that these cleavages are diagnostic for an RNA polymerase-PR complex. Significantly, these hypersensitive cleavages are observed when the repressor is added and binds to OR1 and OR2 subsequent to the addition of RNA polymerase (Fig. 3C, lanes 3 to 6). This finding indicates that RNA polymerase remains bound at PR when the repressor is added to DNA subsequent to RNA polymerase-PR promoter complex formation. Moreover, the failure to detect protections in the −10 region of PRM under these conditions suggests that RNA polymerase is unable to form a complex at PRM under these conditions, even though the repressor is bound at OR1 and OR2.
In addition to the footprinting results shown in Fig. 3C, two additional lines of evidence also indicate that RNA polymerase is bound to PR under the conditions of the experiments in Fig. 3C. First, under the same conditions, open complex formation at PR is detected by KMnO4 footprinting (see Fig. 5, below). Second, footprinting experiments performed with Cu(II) phenanthroline suggest that under these conditions RNA polymerase forms an open complex at PR (data not shown). Third, RNA polymerase is capable of initiating transcription from PR under these conditions (data not shown).
FIG. 5.
Damaging PR increases PRM transcription initiation (A) and open complex formation at PRM (B) in the absence of the 434 repressor. DNA containing a wild-type PRM and a wild-type (lanes 1) or defective (lanes 2) PR promoter was transcribed by RNA polymerase (A) or incubated with RNA polymerase and footprinted using KMnO4 (B). The positions of the PR and PRM transcripts are indicated, as is the position of the PRM open complex.
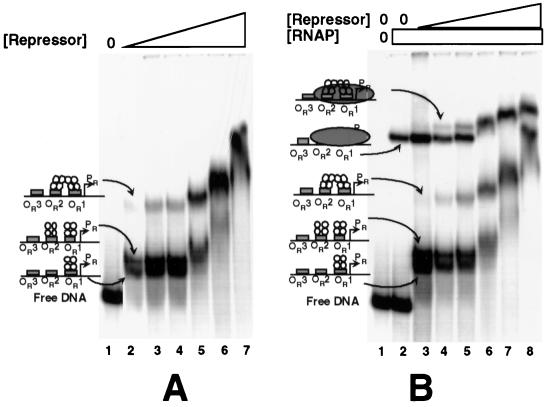
To further explore the potential for an effect of RNA polymerase-PR complex formation on DNA binding by the 434 repressor, we monitored the formation of an RNA polymerase-repressor-DNA ternary complex by gel mobility shift assay. The results in Fig. 4A monitor the formation of 434 repressor-DNA complexes. At all repressor concentrations, we observe the formation of two sets of bands (Fig. 4A). DNase I footprinting studies and gel mobility shift experiments performed with repressor mutants that are unable to cooperatively bind OR1 and OR2 (data not shown) have allowed us to identify the nature of each of these species. The band with the lowest mobility represents a protein-DNA complex in which the repressor is bound at OR1 and OR2 and the repressors at the two sites are cooperatively interacting (Fig. 4A, lanes 2 to 7). The complex with the highest mobility represents the repressor dimer bound at OR1 alone (Fig. 4A, lanes 2 to 4). The other, slightly lower mobility complex represents a DNA fragment on which the repressor is bound at OR1 and OR2 but the two repressors are not interacting, presumably because this interaction is disrupted during entry into the gel or during progression of the complex through the gel matrix (Fig. 4A, lanes 2 to 4). As a result of the very high protein concentrations in the samples in Fig. 4A, lanes 5 to 7, the mobilities of all these protein-DNA complexes decrease.
FIG. 4.
The 434 repressor binds DNA in the presence of RNA polymerase at PR. A DNA fragment containing wild-type OR including the PR and PRM promoters was incubated with increasing concentrations of the 434 repressor in the absence (A) or the presence (B) of 50 nM RNA polymerase. Shown is a native gel of the resulting complexes. (A) OR-containing DNA incubated in the absence (lane 1) or the presence (lanes 2 to 7) of increasing concentrations of the 434 repressor. The concentration of the repressor was increased in 2.5-fold steps starting at 100 nM. (B) OR-containing DNA incubated in the absence (lane 1) or the presence of RNA polymerase (lane 2) or RNA polymerase and increasing concentrations of the 434 repressor (lanes 3 to 8). In lanes 3 to 8, RNA polymerase was added to the labeled DNA fragment, and this allowed the formation of an open complex at PR by incubation for 10 min at 37°C. Subsequently, the repressor was added and the mixture was incubated at 37°C for an additional 10 min before addition of heparin and loading on the gel. RNAP, RNA polymerase.
For the experiment in Fig. 4B, we first added RNA polymerase to the labeled DNA fragment. Closed complexes and nonspecifically bound RNA polymerase molecules were removed by heparin addition. This reaction results in the formation of a single band that corresponds to an RNA polymerase-PR promoter complex (Fig. 4B, lane 2). Control experiments using a template bearing mutations in PRM that prevent RNA polymerase from forming any complexes with PRM formed an identical species, confirming that RNA polymerase is bound only at PR under these conditions (data not shown; also Fig. 3B and C). Additionally, using KMnO4 footprinting we confirmed that the only open complex formed under these conditions was at PR (data not shown). Adding the 434 repressor to this mixture results in the formation of the same complexes identified in Fig. 4A. More importantly, the added repressor supershifts the band corresponding to the RNA polymerase-PR promoter complex. Identical results were obtained with the template that bears mutations in PRM that prevent RNA polymerase from forming a complex with PRM (data not shown). These findings confirm that the 434 repressor is capable of binding to its operators even in the presence of RNA polymerase at PR. Together, the results in Fig. 3 and 4 show that, in the presence of RNA polymerase bound at PR, the 434 repressor is able to bind within OR and that the repressor likely occupies both OR1 and OR2. This finding is somewhat surprising, considering that the PR promoter sequence overlaps OR1 and OR2 (Fig. 1).
Only one open complex is allowed to form on the 434 OR region.
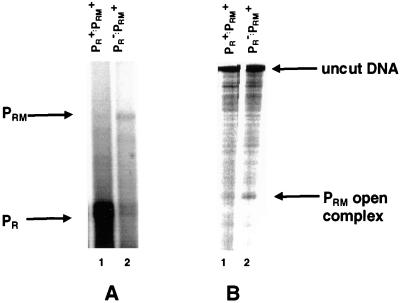
As discussed above, the −35 regions of the PR and PRM promoters substantially overlap (Fig. 1). This observation implies that the binding of RNA polymerase to the strong PR promoter may directly interfere with RNA polymerase binding to the weaker PRM promoter and that this direct interference may result in the repressor's inability to activate PRM transcription initiation. To test this hypothesis, we compared the basal level of PRM transcription and the amount of open complexes formed on a template bearing a defective PR promoter with that found on a template bearing wild-type PR. For this experiment, we mutated PR by substituting a base pair at the −10 consensus region (see Fig. 1 for sequence). This mutation dramatically decreases the basal level of PR transcription (Fig. 5A, compare lanes 1 and 2; see also Fig. 7 below). We find that, on the template bearing the mutant PR promoter, approximately three times as many transcripts initiate at PRM as on the template bearing the wild-type PR promoter (Fig. 5A). Similarly, two- to threefold as many open complexes are formed at PRM when PR is defective as when it has wild-type activity (Fig. 5B). These data indicate that RNA polymerase bound at the PR promoter interferes with open complex formation at PRM.
FIG. 7.
Damaging PR obviates RNA polymerase inhibition of repressor-activated PRM transcription (A and B) or open complex formation (C and D). For panels A and B, DNA fragments containing wild-type PRM and −10 mutant PR (see Fig. 1 for sequence) were transcribed in vitro in the absence of repressor (lanes 1) and at various increasing repressor concentrations. Repressor concentrations were increased in 2.5-fold steps starting at 250 nM protein (lanes 2 to 6). Positions of transcripts resulting from initiation of transcription from PRM and PR are indicated. (A) The 434 repressor was incubated with DNA template at 23°C for 10 min, followed by addition of RNA polymerase. The reaction mixture was transferred to 37°C for 10 min before the transcription reaction was initiated by the addition of nucleotides and heparin. (B) RNA polymerase was incubated with DNA at 37°C for 10 min before addition of the 434 repressor. After an additional 10-min incubation at 37°C, the transcription reaction was initiated by the addition of nucleotides and heparin. (C and D) Shown are the open complexes formed at PRM in the absence of the repressor (lanes 1) and the presence of increasing concentrations of the 434 repressor as detected by KMnO4 footprinting. Repressor concentrations were increased in 2.5-fold steps starting at 250 nM protein. Footprinting conditions are given in Materials and Methods. Positions of PR and PRM open complex are indicated. (C) Incubation conditions were as described for panel A. (D) Incubation conditions were as described for panel B. RNAP, RNA polymerase.
Having established the existence of promoter interference between PRM and PR, we wished to establish the precise mechanism of interference. One possible mechanism is that, similar to the related λ phage, PR interference with PRM function may occur by inhibiting the rate of transition of the RNA polymerase-PRM closed complex to an open complex (10, 26). If this hypothesis is correct, RNA polymerase should simultaneously form open complexes on both PR and PRM promoters. An alternative mechanism is that an open complex at PR may simply prevent the formation of any RNA polymerase-PRM complexes.
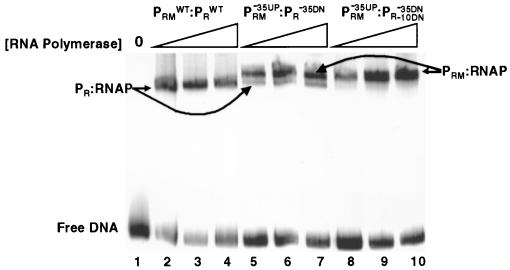
As the first step toward answering this question, we used gel mobility shift assays to determine how many open complexes can be formed on a single DNA template containing both 434 PR and PRM. We examined the ability of RNA polymerase to form open complexes on DNA templates bearing various arrays of wild-type and mutant PR and PRM promoters. Similar to Fig. 4, only one shifted band is observed when RNA polymerase is added to a template bearing both wild-type PR and PRM promoters (Fig. 6, lanes 2 to 4). This band is not observed with templates that bear a mutation that inhibits PR open complex formation (data not shown; also Fig. 6, lanes 8 to 10). This finding establishes that the band seen in Fig. 6, lanes 2 to 4, represents a heparin-resistant RNA polymerase-PR promoter complex.
FIG. 6.
Mutually exclusive binding of RNA polymerase to PR and PRM. Increasing concentrations of RNA polymerase were incubated with a DNA fragment containing wild-type PR and PRM (lanes 2 to 4), a fragment bearing a single mutation in the −35 region of PR and PRM that simultaneously decreases the strength of PR and increases the strength of PRM (lanes 5 to 7) (28), or a template bearing both the −35 mutation and a mutation in the −10 region of PR (lanes 8 to 10; see Fig. 1 for sequences). The concentration of RNA polymerase was increased in threefold steps starting from 10 nM protein (lanes 2 and 8). RNA polymerase was added to the labeled DNA fragment incubated for 10 min at 37°C before addition of heparin and loading on the gel. RNAP, RNA polymerase; WT, wild type.
We showed previously that a mutation that changes the sequence of the −35 region of 434 PRM toward the consensus sequence (Fig. 1) increases initiation from PRM and decreases transcription initiation from PR (28). When RNA polymerase is incubated with this template, the results in Fig. 6, lanes 5 to 7, show that two shifted bands are observed. Since KMnO4 footprinting data indicate that open complexes are formed at PR and PRM (data not shown), we suggest that each band represents an open complex formed at PRM or PR, respectively, on separate DNA molecules.
These findings do not definitively prove that each band represents an individual open complex at PRM and PR, which are formed on two different DNA molecules. It is formally possible that the higher-mobility band represents an RNA polymerase-DNA complex at a single promoter, while the complex having lower mobility represents an RNA polymerase complex at both PRM and PR on the same DNA fragment. Alternatively, it is possible that the two bands observed in Fig. 6, lanes 5 to 7, represent two forms of a complex formed at a single promoter. To begin to distinguish these possibilities, we examined the ability of RNA polymerase to form heparin-resistant complexes on a template bearing two mutations, the combined consequences of which increase the strength of PRM and decrease the strength of PR. The first mutation is located within the −35 region of PRM that simultaneously increases the match of its −35 region of 434 PRM with the consensus sequence but decreases the match of the −35 region of PR to the consensus sequence. The second mutation is in the −10 region of PR (see Fig. 1 for sequences). This change, combined with the −35 alteration, renders the PR promoter incapable of forming any complexes with RNA polymerase. KMnO4 probe experiments show that, on this template, RNA polymerase forms only open complexes at PRM (data not shown). The results in Fig. 6, lanes 8 to 10, show that only one shifted band is observed when RNA polymerase is incubated with this template. The mobility of this complex is identical to that of the lower-mobility species seen in Fig. 6, lanes 5 to 7. This finding indicates that the lower-mobility complex does not contain a DNA molecule bearing an RNA polymerase at both PRM and PR. Since only a single species is formed on this template, this finding also suggests that the two species that are observed in Fig. 6, lanes 5 to 7, do not represent two forms of the same RNA polymerase-promoter complex. Moreover, since only the lower-mobility complex forms on this template, and since this complex is observed only when RNA polymerase forms an open complex at PRM, we suggest that this complex is the RNA polymerase-PRM open complex. Hence, the lower- and higher-mobility complexes formed under the conditions of the experiment in Fig. 6, lanes 5 to 7, represent open complexes formed at PRM and PR, respectively, on separate DNA molecules. Most importantly, these observations indicate that formation of open complexes at PRM and PR promoters is mutually exclusive. Based on the analysis of the data in Fig. 6, if open complexes were allowed to simultaneously form on both PR and PRM promoters, a third, higher-molecular-weight species should be observed.
Role of PR sequence in inhibiting activation of PRM transcription.
The above results demonstrate that prior addition of RNA polymerase decreases the repressor's ability to stimulate open complex formation at PRM by prohibiting the access of RNA polymerase to the PRM promoter sequence. We wished to determine what kind of RNA polymerase-PR complex is capable of inhibiting repressor-mediated activation of PRM transcription. As a first step in this investigation, we examined whether the relative strength of the PR promoter plays a role in inhibiting the activation of PRM open complex formation by the 434 repressor. This experiment employs the −10 mutant PR promoter used in the experiments presented in Fig. 5 and 6 (see Fig. 1 for sequence). The −10 sequence change dramatically decreases the transcriptional activity of the PR promoter (Fig. 7A, lane 1). Similar to the results obtained using a template bearing the wild-type promoters (Fig. 2A), the 434 repressor is able to activate transcription from PRM when it is added to the template prior to RNA polymerase (Fig. 7A). However, in contrast to the results obtained with the wild-type promoters, the 434 repressor is also able to activate PRM transcription even if RNA polymerase is incubated with DNA template prior to adding the repressor (Fig. 7B; also compare these results to those shown in Fig. 2B). The failure of the mutant PR to inhibit initiation of transcription from PRM under these conditions may result from a decrease in the lifetime of the mutant PR-RNA polymerase open complex or an inability of the mutant promoter-RNA polymerase complex to interfere with repressor function (see Discussion). Nonetheless, these findings suggest that the RNA polymerase-mediated inhibition of PRM transcription requires a strong PR promoter.
KMnO4 footprinting experiments were performed to confirm that the loss of the inhibition, resulting from weakening PR by mutation, affects PRM open complex formation. The results in Fig. 7 show that, on a template bearing a mutant PR promoter, the maximal amount of PRM open complex is formed regardless of whether RNA polymerase is added after (Fig. 7C) or before (Fig. 7D) the 434 repressor. These results differ from those obtained on templates bearing the wild-type PR promoter (Fig. 2C and D) and confirm that decreasing the strength of PR relieves the RNA polymerase-mediated inhibition of the repressor-stimulated PRM open complex formation. Together with the results in Fig. 4 and 5, these results suggest that RNA polymerase-mediated inhibition of PRM transcription occurs at the level of RNA polymerase binding to PRM.
An open complex formed at PR is required for the inhibition of PRM activity.
The foregoing experiments demonstrate that changing the strength of PR can modulate the efficiency of repressor-mediated PRM activation. However, these experiments do not provide information regarding the molecular basis for this modulation. We took advantage of the fact that open complex formation is temperature dependent to examine whether an open complex engaged at the PR promoter is required to prevent repressor-mediated activation of PRM transcription. We have established that only closed, not open, complex formation can proceed at low (0 to 5°C) temperatures (data not shown) and that efficient open complex formation at PR requires temperatures in excess of 12°C. Thus, if formation of an open complex at PR is required for the inhibition of PRM activation, incubation of RNA polymerase at 0°C before adding the repressor should not prevent activation of PRM by the repressor. The results in Fig. 8 show that, in contrast to the results obtained at higher temperatures, incubating RNA polymerase and the DNA fragment at 0°C prior to adding the repressor does not inhibit transcription activation of PRM by the 434 repressor. This finding indicates that formation of open complex at the PR promoter is required for the inhibition of 434 repressor-mediated activation of PRM.
FIG. 8.
An open complex at PR is required for the inhibition of repressor-mediated activation of PRM. A DNA fragment containing wild-type PR and the PRM promoter was transcribed in vitro in the presence of the repressor. Transcription conditions are given in Materials and Methods. Repressor concentrations were increased in 2.5-fold steps starting at 250 nM protein. Positions of transcripts resulting from initiation of transcription from PRM and PR are indicated. RNAP, RNA polymerase.
DISCUSSION
Our data clearly demonstrate that open complex formation at the PR promoter prevents open complex formation on PRM. This finding indicates that, in 434 OR, 434 repressor-mediated activation of PRM transcription occurs through two mechanisms. First, the 434 repressor stimulates open complex formation at PRM by directly contacting RNA polymerase (2, 3, 28). Second, in agreement with the suggestions of others (1), the results shown in this paper indicate that the 434 repressor also activates PRM transcription by releasing an “inhibitory” effect of RNA polymerase bound at the strong promoter PR. The homologous λ repressor employs identical strategies in stimulating transcription from λ PRM (6, 10, 11, 12, 14, 16, 18). The congruence of mechanisms used in stimulating PRM in these two phages supports the assertion that these strategies may be used generally in all homologous phages (10).
Although the activities of the PRM promoters in bacteriophages λ and 434 are regulated by interference between PR and PRM, the specific mechanisms by which RNA polymerase at PR inhibits PRM transcription initiation appear to differ between the two phages. In the case of the λ repressor, an open complex at PR does not appear to affect binding of RNA polymerase to PRM (10). Instead, the RNA polymerase-PR complex inhibits the rate of isomerization of a closed complex at PRM to an open complex (6, 11, 26). Hence in λ OR, open complex formation at PR and that at PRM are not mutually exclusive. We are unable to detect a ternary complex in which both 434 PR and PRM are occupied (Fig. 6), indicating that in 434 OR open complex formation at PR prevents formation of a similar complex at PRM.
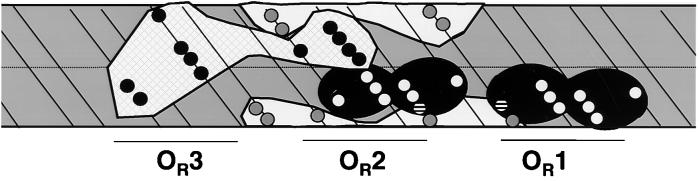
A comparison of sequences of the OR regions of the two phages suggests a reason for the difference in their promoter exclusion mechanisms. In λ OR, the −35 regions of PR and PRM are located to the right and left of OR2, respectively, and are separated from each other by 12 bp. In 434 OR, the −35 regions of PR and PRM virtually overlap on the left side of OR2. Hence, simultaneous occupancy of each promoter by RNA polymerase is forbidden by steric occlusion. To better understand the spatial relationships among the proteins that control the activities of PR and PRM, an unwrapped cylindrical projection of the OR region that identifies the positions of the phosphates actually or presumed to be contacted by the repressor and RNA polymerase is presented in Fig. 9 (2, 22). This DNA projection indicates that most of the phosphates contacted by the repressor bound at OR1 and OR2 are not contacted by RNA polymerase bound at the PR promoter. In addition, this model also indicates that RNA polymerase bound at the PR promoter and the repressor bound at OR1 and OR2 are essentially located on different faces of the DNA. These inferences are consistent with the results showing that the repressor at these sites and RNA polymerase at PR can coexist simultaneously on DNA. Examination of Fig. 9 also shows that RNA polymerase binding blocks the access of the repressor to phosphate at 3′ to the base at position 10 of OR2. This finding suggests that prior binding of RNA polymerase at PR may alter the structure of the repressor-OR2 complex. This putative structural alteration could also contribute to RNA polymerase-mediated inhibition of repressor-stimulated PRM transcription. Similarly, this model predicts that RNA polymerase may be able to form a nontranscriptionally active complex with PR in the presence of the repressor bound at OR2. Data obtained by our laboratory support both of these ideas (J. Xu and G. B. Koudelka, unpublished data).
FIG. 9.
Disposition of RNA polymerase and the 434 repressor bound at the 434 OR region. Ethylation interference data are mapped onto an unwrapped cylindrical projection of the surface of a DNA double helix, assuming 10.5 bp per turn. Ethylation interference patterns for RNA polymerase bound at the PR promoter are deduced from data obtained for the T7A3 and PlacUV5 promoters (22). Ethylation interference patterns for RNA polymerase at the 434 PRM promoter and the 434 repressor at OR1 and OR2 are based on the data in reference 2. The black area represents the 434 repressor. The white area represents RNA polymerase at PR, and the hatched area denotes the position of RNA polymerase at PRM. Phosphate contacts for each protein are denoted by white circles (repressor at OR1 and OR2), black circles (RNA polymerase at PRM), and gray circles (RNA polymerase at PR). The hatched circles denote phosphates that could be contacted by RNA polymerase and/or the 434 repressor.
Since inactivating PR allows RNA polymerase to initiate transcription at the weak PRM promoter (Fig. 5), we are interested in assessing the relative contribution of the indirect effect of relieving promoter competition to the overall efficiency of the 434 repressor activation of PRM transcription. The work of Gussin and coworkers (26) with λ phage suggests that interference between λ PRM and λ PR does not limit the rate of open complex formation at λ PRM in the cell. Apparently, rapid transcription initiation clears both the λ PR and λ PRM promoters rapidly enough that neither is occupied for a significant fraction of the time, thereby minimizing the effects of promoter interference in vivo. Although we do not have direct evidence, correlation between the in vivo and in vitro studies of 434 promoter utilization and the data presented in this paper suggest that promoter interference may have a role in vivo in the 434 bacteriophage. Overall, adding the repressor increases the amount of open complexes and transcripts from PRM by 10-fold (2, 28). The effect of mutating PR increases the amount of runoff transcripts by about threefold and the amount of open complexes by a similar amount (Fig. 5). This analysis indicates that repressor-mediated relief of promoter competition contributes nearly as much to the 434 repressor's activation of PRM transcription as does direct stimulation of RNA polymerase. Consistent with the relative importance of the indirect stimulation mechanism, positive control mutants that are presumably defective in directly contacting RNA polymerase stimulate PRM transcription at least half as well as does the wild-type repressor in vivo (2). This finding also indicates that relief of promoter interference may have a significant role in regulating PRM transcription in the bacteriophage. Further support for this view comes from the finding that eliminating RNA polymerase binding at PR by 434 Cro binding to OR1 and/or OR2 also stimulates transcription from PRM (1). Similarly, deletion of PR increased transcription from PRM threefold in vitro (2).
Kinetic assays indicate that the direct stimulation of λ PRM by the λ repressor occurs by increasing the rate of isomerization of RNA polymerase from a closed to an open complex at λ PRM (7, 8). Relief of promoter interference by the λ repressor also leads to an increase in the rate of isomerization (6, 11, 26). Although the precise kinetic mechanism by which the 434 repressor stimulates transcription from 434 PRM has not yet been determined, several lines of evidence indicate that the 434 repressor enhances the formation of closed complexes by recruiting RNA polymerase to the PRM promoter (28). Similarly, the 434 repressor-mediated relief of promoter competition would also be expected to result in an increase in the number of closed complexes at PRM. The variance in overall mechanism of activation in these two phages is likely related to promoter sequence-dependent differences in the identity of the rate-limiting steps between the two PRM promoters and not to a difference in the stimulatory properties of the two repressors (21). This idea is supported by the observation that the λ repressor can stimulate closed complex formation by a mutant RNA polymerase (15). Moreover, the λ repressor is also able to activate transcription simply by providing an arbitrary protein-protein contact with RNA polymerase (5).
We have shown that open complex formation at the PRM promoter is inhibited by open complex formation on PR. With this observation in mind, one question still remains. Given that an open complex on PR appears to be required to inhibit transcription from PRM, how can 15 to 20% of DNA that forms an open RNA polymerase-PR complex cause the 80% decrease in PRM transcription (Fig. 5)? One possible answer is that these complexes do not represent all of the heparin-resistant DNA-RNA polymerase complexes. It is possible that these intermediate complexes account for a reasonably large portion of the population and enforce an inhibitory effect on PRM transcription.
ACKNOWLEDGMENT
This work was supported by PHS grant GM42138 from the National Institutes of Health, National Institute of General Medical Sciences.
REFERENCES
- 1.Bushman F D. The bacteriophage 434 right operator. Roles of OR1, OR2 and OR3. J Mol Biol. 1993;230:28–40. doi: 10.1006/jmbi.1993.1123. [DOI] [PubMed] [Google Scholar]
- 2.Bushman F D, Ptashne M. Activation of transcription by the bacteriophage 434 repressor. Proc Natl Acad Sci USA. 1986;83:9353–9357. doi: 10.1073/pnas.83.24.9353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bushman F D, Ptashne M. Turning lambda Cro into a transcriptional activator. Cell. 1988;54:191–197. doi: 10.1016/0092-8674(88)90551-x. [DOI] [PubMed] [Google Scholar]
- 4.Choy H E, Adhya S. RNA polymerase idling and clearance in gal promoters: use of supercoiled minicircle DNA template made in vivo. Proc Natl Acad Sci USA. 1993;90:472–476. doi: 10.1073/pnas.90.2.472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dove S L, Joung J K, Hochschild A. Activation of prokaryotic transcription through arbitrary protein-protein contacts. Nature. 1997;386:627–630. doi: 10.1038/386627a0. [DOI] [PubMed] [Google Scholar]
- 6.Fong R S C, Woody S, Gussin G N. Modulation of PRM activity by the lambda PR promoter in both the presence and absence of repressor. J Mol Biol. 1993;232:792–804. doi: 10.1006/jmbi.1993.1432. [DOI] [PubMed] [Google Scholar]
- 7.Hawley D K, McClure W R. Mechanism of activation of transcription initiation from the lambda PRM promoter. J Mol Biol. 1982;157:493–525. doi: 10.1016/0022-2836(82)90473-9. [DOI] [PubMed] [Google Scholar]
- 8.Hawley D K, McClure W R. The effect of a lambda repressor mutation on the activation of transcription initiation from the lambda PRM promoter. Cell. 1983;32:327–333. doi: 10.1016/0092-8674(83)90452-x. [DOI] [PubMed] [Google Scholar]
- 9.Heltzel A, Lee I W, Totis P A, Summers A O. Activator-dependent preinduction binding of sigma-70 RNA polymerase at the metal-regulated mer promoter. Biochemistry. 1990;29:9572–9584. doi: 10.1021/bi00493a011. [DOI] [PubMed] [Google Scholar]
- 10.Hershberger P A, DeHaseth P L. RNA polymerase bound to the PR promoter of bacteriophage lambda inhibits open complex formation at the divergently transcribed PRM promoter. Implications for an indirect mechanism of transcriptional activation by lambda repressor. J Mol Biol. 1991;222:479–494. doi: 10.1016/0022-2836(91)90491-n. [DOI] [PubMed] [Google Scholar]
- 11.Hershberger P A, DeHaseth P L. Interference by PR-bound RNA polymerase with PRM function in vitro. Modulation by the bacteriophage λ cI protein. J Biol Chem. 1993;268:8943–8948. [PubMed] [Google Scholar]
- 12.Hochschild A, Irwin N, Ptashne M. Repressor structure and the mechanism of positive control. Cell. 1983;32:319–325. doi: 10.1016/0092-8674(83)90451-8. [DOI] [PubMed] [Google Scholar]
- 13.Jacob F, Monod J. Genetic regulatory mechanisms in the synthesis of proteins. J Mol Biol. 1961;3:318–356. doi: 10.1016/s0022-2836(61)80072-7. [DOI] [PubMed] [Google Scholar]
- 14.Joung J K, Le L U, Hochschild A. Synergistic activation of transcription by Escherichia coli cAMP receptor protein. Proc Natl Acad Sci USA. 1993;90:3083–3087. doi: 10.1073/pnas.90.7.3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim Y I, Hu J C. Oriented DNA binding by one-armed lambda repressor heterodimers and contacts between repressor and RNA polymerase at P(RM) Mol Microbiol. 1997;25:311–318. doi: 10.1046/j.1365-2958.1997.4651831.x. [DOI] [PubMed] [Google Scholar]
- 16.Kuldell N, Hochschild A. Amino acid substitutions in the −35 recognition motif of ς70 that result in defects in phage λ repressor-stimulated transcription. J Bacteriol. 1994;176:2991–2998. doi: 10.1128/jb.176.10.2991-2998.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee J, Goldfarb A. lac repressor acts by modifying the initial transcribing complex so that it cannot leave the promoter. Cell. 1991;66:793–798. doi: 10.1016/0092-8674(91)90122-f. [DOI] [PubMed] [Google Scholar]
- 18.Li M, Moyle H, Susskind M M. Target of the transcriptional activation function of phage lambda cI protein. Science. 1994;263:75–77. doi: 10.1126/science.8272867. [DOI] [PubMed] [Google Scholar]
- 19.Mikaelian I, Sergeant A. A general and fast method to generate multiple site directed mutations. Nucleic Acids Res. 1992;20:376. doi: 10.1093/nar/20.2.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rojo F, Mencia M, Monsalve M, Salas M. Transcription activation and repression by interaction of a regulator with the alpha subunit of RNA polymerase: the model of phage phi 29 protein p4. Prog Nucleic Acid Res Mol Biol. 1998;60:29–46. doi: 10.1016/s0079-6603(08)60888-0. [DOI] [PubMed] [Google Scholar]
- 21.Roy S, Garges S, Adhya S. Activation and repression of transcription by differential contact: two sides of a coin. J Biol Chem. 1998;273:14059–14062. doi: 10.1074/jbc.273.23.14059. [DOI] [PubMed] [Google Scholar]
- 22.Siebenlist U, Simpson R B, Gilbert W. E. coli RNA polymerase interacts homologously with two different promoters. Cell. 1980;20:269–281. doi: 10.1016/0092-8674(80)90613-3. [DOI] [PubMed] [Google Scholar]
- 23.Wharton R P, Ptashne M. An α-helix determines the DNA specificity of a repressor. Trends Biochem Sci. 1986;11:71–73. [Google Scholar]
- 24.Wharton R P. Determinants of 434 repressor binding specificity. Ph.D. thesis. Cambridge, Mass: Harvard University; 1986. [Google Scholar]
- 25.Wharton R P, Brown E L, Ptashne M. Substituting an α-helix switches the sequence specific DNA interactions of a repressor. Cell. 1985;38:361–369. doi: 10.1016/0092-8674(84)90491-4. [DOI] [PubMed] [Google Scholar]
- 26.Woody S T, Fong R S C, Gussin G N. Effects of a single base-pair deletion in the bacteriophage lambda PRM promoter. Repression of PRM by repressor bound at OR2 and by RNA polymerase bound at PR. J Mol Biol. 1993;229:37–51. doi: 10.1006/jmbi.1993.1006. [DOI] [PubMed] [Google Scholar]
- 27.Wu L, Koudelka G B. Sequence-dependent differences in DNA structure influence the affinity of P22 operator for P22 repressor. J Biol Chem. 1993;268:18975–18981. [PubMed] [Google Scholar]
- 28.Xu J, Koudelka G B. DNA-based positive control mutants in the binding site sequence of 434 repressor. J Biol Chem. 1998;273:24165–24172. doi: 10.1074/jbc.273.37.24165. [DOI] [PubMed] [Google Scholar]