Abstract
The purple photosynthetic bacterium Rhodobacter sphaeroides has within its genome a cluster of photosynthesis-related genes approximately 41 kb in length. In an attempt to identify genes involved in the terminal esterification stage of bacteriochlorophyll biosynthesis, a previously uncharacterized 5-kb region of this cluster was sequenced. Four open reading frames (ORFs) were identified, and each was analyzed by transposon mutagenesis. The product of one of these ORFs, bchG, shows close homologies with (bacterio)chlorophyll synthetases, and mutants in this gene were found to accumulate bacteriopheophorbide, the metal-free derivative of the bacteriochlorophyll precursor bacteriochlorophyllide, suggesting that bchG is responsible for the esterification of bacteriochlorophyllide with an alcohol moiety. This assignment of function to bchG was verified by the performance of assays demonstrating the ability of BchG protein, heterologously synthesized in Escherichia coli, to esterify bacteriochlorophyllide with geranylgeranyl pyrophosphate in vitro, thereby generating bacteriochlorophyll. This step is pivotal to the assembly of a functional photosystem in R. sphaeroides, a model organism for the study of structure-function relationships in photosynthesis. A second gene, orf177, is a member of a large family of isopentenyl diphosphate isomerases, while sequence homologies suggest that a third gene, orf427, may encode an assembly factor for photosynthetic complexes. The function of the remaining ORF, bchP, is the subject of a separate paper (H. Addlesee and C. N. Hunter, J. Bacteriol. 181:7248–7255, 1999). An operonal arrangement of the genes is proposed.
A rigorous program of sequencing combined with mutational and biochemical analysis has revealed within the genome of the purple bacterium Rhodobacter sphaeroides the presence of a 41-kb gene cluster holding virtually all of the loci directly responsible for its photosynthetic ability (Fig. 1) (11, 12, 27), including those encoding enzymes of the bacteriochlorophyll (Bchl) biosynthesis pathway. The Bchla molecules of R. sphaeroides are esterified with phytol, a C20 isoprenoid alcohol, which constitutes 30% of the total molecular weight and also greatly influences the molecules' properties by investing them with much of their hydrophobicity. The structural data for bacterial reaction center and light-harvesting complexes (13, 15, 26) enable the conformations adopted by many of the phytol chains to be examined in atomic detail. In light-harvesting complex 2 (LH2), it appears that the close intertwining of the phytol tails of the B800 and B850 Bchls, along with the carotenoid chains, imparts a significant amount of stability to the complex. Indeed, it appears that the addition of the alcohol moiety, the final act of Bchl biosynthesis in this organism, is a crucial determinant of the assembly of the entire photosynthetic apparatus (references 9 and 31 and this study). The tails also play a major role in controlling the orientation of the transition dipoles of the tetrapyrrole rings, a function that is crucial for fast energy transfer. Despite the importance of this esterification, the genetics involved have previously been little studied. Coomber et al. (11) generated a nonphotosynthetic transposon mutant accumulating a pigment identified as bacteriochlorophyllide (Bchlide); this phenotype was attributed to insertion into a bchG locus, but no further analysis of the relevant region of the photosynthesis gene cluster was undertaken.
FIG. 1.
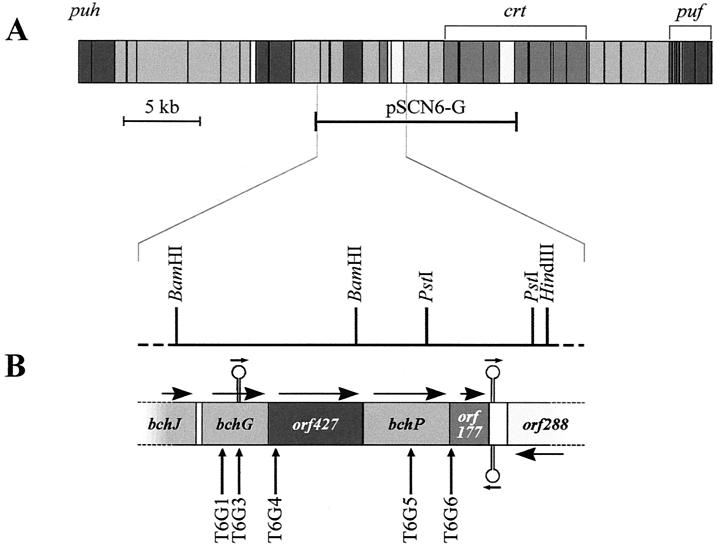
Physical map of the R. sphaeroides photosynthesis gene cluster, with expanded physical and restriction maps of the newly sequenced region containing the bchG locus. Structural genes, and those with putative regulatory or assembly functions, are shown in the darkest grey; the next darkest represents carotenoid and other isoprenoid biosynthesis genes; Bchl biosynthesis genes are paler again, while very pale grey indicates genes of no known function. For further information on the photosynthesis gene cluster of R. sphaeroides, see reference 27. The horizontal bar immediately below the full map (A) represents the insert of pSCN6-G, cloned in the pSUP202 vector. Horizontal arrows on the expanded physical map (B) indicate the direction of transcription of the labeled genes, while vertical arrows below this map mark the positions of Tn5 insertions within these genes; stem-loops indicate potential transcription terminators.
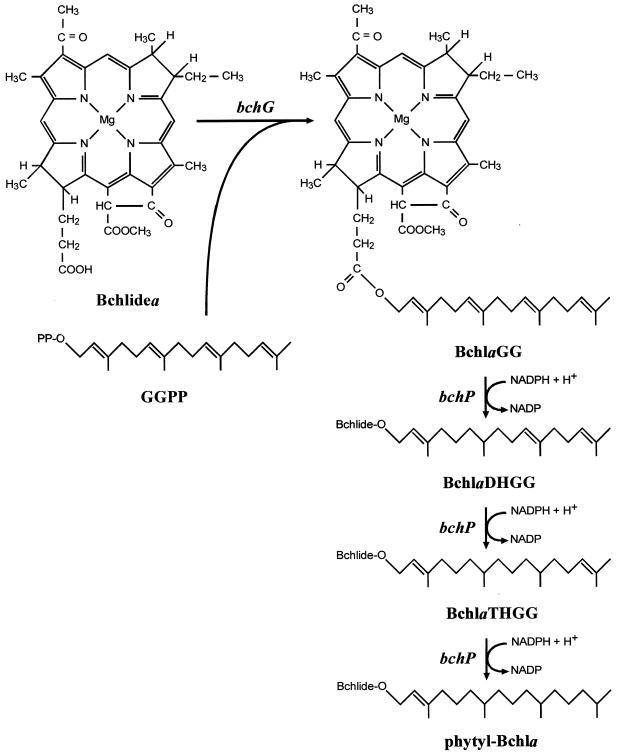
In Rhodobacter capsulatus, a closely related bacterium, sequencing of the entire photosynthesis gene cluster (3), followed by directed mutational analysis of selected loci (8), has identified two genes, bchG and bchP, as responsible for the phytylation of Bchlide. It was proposed that the product of the bchG gene performs an initial esterification of Bchlide with geranylgeranyl pyrophosphate (GGPP), an activated form of the C20 alcohol, geranylgeraniol (GG), and that this alcohol moiety is then reduced sequentially to phytol by the product of bchP (Fig. 2). In vitro assays using BchG protein of R. capsulatus, heterologously synthesized in Escherichia coli, have since confirmed its biosynthetic capability (28). Two open reading frames (ORFs) situated immediately downstream of bchG and bchP, respectively, orf428 and orf176, could be assigned no function by mutational analysis (7), but Hahn et al. (18) have since demonstrated orf176 to be an idi gene encoding isopentenyl diphosphate/pyrophosphate (IPP) isomerase. This enzyme catalyzes the conversion of IPP to its highly electrophilic isomer, dimethylallyl pyrophosphate, a crucial step in the isoprenoid pathway which generates, among other products, the GGPP required for Bchla and carotenoid biosynthesis.
FIG. 2.
Proposed pathway for the phytylation of Bchlidea in R. sphaeroides and R. capsulatus. The sequence of the hydrogenation reactions is based on that which occurs during chlorophyll biosynthesis in greening plants (32). The genes involved are in italics. DHGG, dihydro-GG; THGG, tetrahydro-GG.
We have now identified and localized bchG, bchP, and homologues of R. capsulatus orf428 and orf176 in the photosynthesis gene cluster of R. sphaeroides and subjected each to mutational analysis. The bchG gene has also been heterologously expressed in E. coli, and the resulting recombinant protein has been demonstrated to catalyze the in vitro esterification of Bchlidea with GGPP, thereby confirming the product of this gene as geranylgeranyl-Bchl synthetase. orf427 may encode an assembly factor for the photosynthetic complexes, while orf177 is a member of the idi gene family. Transposon insertion into this gene gave a mutant with significantly depleted levels of the photosynthetic complexes, particularly light-harvesting complex 1 (LH1), presumably as a result of a reduction in the supply of Bchl and carotenoids. Analysis of bchP, demonstrating its product to be geranylgeranyl-Bchl reductase, has been presented previously (2).
MATERIALS AND METHODS
Details of methods and equipment used are provided in reference 2. Unless otherwise stated, procedures employed were identical.
DNA sequencing and transposon Tn5 mutagenesis.
For sequencing purposes, a 5-kb region containing bchG was isolated in the form of its constituent 2.5-kb BamHI and 2.5-kb BamHI-HindIII fragments from R. sphaeroides genomic clones pSCN6-1 (19) and pSCN6-20 (12), respectively. Insertions of Tn5 were generated initially in plasmid pSCN6-G (2), prior to transfer into the genome of wild-type R. sphaeroides NCIB 8253.
Complementation of R. sphaeroides mutants.
The oligonucleotides 5′-CATCTAGAGGAGACGACCATATGAGTGTCAATCTATCCTTACAC-3′ and 5′-GGCTAAGCTTGGATCCTCACGGCAGCACCTCCAGCC-3′ were used to amplify the R. sphaeroides bchG gene by PCR and to introduce XbaI and HindIII sites, which enabled cloning into pRKEB (1), a vector which provides gene expression under the transcriptional control of the R. sphaeroides puf operon promoter, yielding plasmid pEBbchG. Plasmid pSK1bchP similarly provided expression of R. sphaeroides bchP via the R. sphaeroides puc operon promoter (2).
A 1,426-bp PstI fragment of the R. sphaeroides photosynthesis gene cluster, containing only orf177 as a full-length gene (Fig. 1B), was cloned into pBluescript II SK+ (Stratagene), from which it was subsequently excised using the XbaI and HindIII sites of the vector. The XbaI end of this fragment lay upstream of orf177, enabling it to be recloned into pRKEB, creating expression plasmid pEB177L.
Plasmids pEBbchG, pSK1bchP, and pEB177L were transferred by conjugation into different R. sphaeroides Tn5 mutants. In each case, the relevant parental plasmid was also independently introduced as a negative control. For pSK1bchP, this control plasmid was pRKSK1 (20).
HPLC of metal-free porphyrins.
Metal-free porphyrins were extracted into acetone-methanol (7:2 [vol/vol]) as for Bchl analysis but were found to give greater reproducibility of retention times when high-performance liquid chromatography (HPLC) analysis was performed using an analysis gradient designed for separation of carotenoids (2), rather than that used for the identification of Bchls.
Heterologous expression of bchG in E. coli.
In addition to XbaI and HindIII sites, the pair of primers used to amplify bchG for homologous expression in R. sphaeroides also contained restriction sites for NdeI and BamHI. These permitted cloning of the PCR product into vector pET9a (Novagen) for heterologous expression in E. coli. The resulting plasmid, pET9a-bchG, as well as the parental plasmid, pET9a, was transformed into E. coli BL21(DE3)pLysS (Novagen).
Bchl synthetase assays.
The assay procedure and buffer composition used were almost identical to those used by Addlesee and Hunter (2) for geranylgeranyl-Bchl reductase assays. Harvested E. coli cell pellets were resuspended to an optical density at 600 nm (OD600) of 50 in assay buffer consisting of 50 mM HEPES (pH 8.0), 120 mM potassium acetate, 10 mM magnesium acetate, 0.3 M glycerol, and 10 mM dithiothreitol; the cells were ruptured by sonication, and the soluble fraction was isolated by centrifugation. To 20 μl of E. coli soluble extract (representing the equivalent of 1 OD600 unit of sonicated cells) in an Eppendorf tube on ice were then added phenylmethanesulfonyl fluoride to a final concentration of 0.5 mM, GGPP to 20 μM, and Bchlidea (in acetone), prepared by the method of Fiedor et al. (16), to approximately 9 μM. The volume was made up to 300 μl with assay buffer. Incubations were carried out in the dark for 90 min at 30°C. The pigments were then extracted with acetone-methanol (7:2 [vol/vol]), and 50 μl was used for HPLC analysis.
RESULTS
Nucleotide and deduced amino acid sequences of the bchG region of the R. sphaeroides photosynthesis gene cluster. (i) Translational organization.
A 5-kb region near the center of the R. sphaeroides photosynthesis gene cluster was sequenced (G. W. Naylor, EMBL accession no. AJ010302), and four complete ORFs conforming to a compiled codon usage table for R. sphaeroides photosynthesis genes were identified (Fig. 1B). The initial gene assignment of these ORFs is based on alignment with the R. capsulatus photosynthesis gene cluster (D. H. Burke, M. Alberti, G. A. Armstrong, and J. E. Hearst, GenBank accession no. Z11165). Putative translation starts are based on the codon usage and on the presence and position of a Shine-Dalgarno ribosome-binding site with homology to the 3′ end of the R. sphaeroides 16S rRNA (14). Analysis of one of these ORFs, bchP, is presented elsewhere (2).
(ii) Putative promoter sequences and transcription termination signals.
A putative promoter sequence was identified within the sequenced region, approximately 200 bp upstream of orf177. This promoter has four mismatches from the hexamers of the E. coli ς70 promoter consensus (25). Other possible promoter sequences also precede bchG and bchP, but each has five mismatches from the consensus hexamers, as do many other similar sequences within the 5 kb. A search for factor-independent transcription terminators (reviewed in reference 30) revealed one with a stable stem-loop (predicted ΔG [25°C] of −107.1 kJ [38]) 21 bp downstream of orf177. A second potential terminator incorporating a significantly weaker stem-loop (ΔG [25°C] of −86.2 kJ) was identified, within and running in the same direction as bchG, starting at bp 490 of the gene; the closest preceding stop codon is that of bchJ, 563 bp upstream. No terminators were found immediately following any of bchG, orf427, or bchP.
(iii) Properties of the putative gene products.
The putative gene products were analyzed principally using the Kyte-Doolittle hydropathy scale (23) and the Klein algorithm (22). These revealed both BchG and Orf427 to be highly hydrophobic, integral membrane proteins, with as many as 8 and 12 membrane spans, respectively. Orf177 is largely hydrophilic and is predicted to have no regions buried in the membrane bilayer.
(iv) Evolutionary conservation and structural similarities of deduced peptide sequences.
The predicted product of R. sphaeroides bchG has close homology to other Bchl synthetases from R. capsulatus and the green photosynthetic bacterium Chloroflexus aurantiacus (24). Lopez et al. (24) reported homologies with further Bchl and chlorophyll (Chl) synthetase enzymes, including a Synechocystis gene product, the activity of which has since been proven in vitro (28). All gave essentially identical hydropathy plots. Homologies were also found with three polyprenyltransferases, similarly involved in the attachment of an aliphatic alcohol pyrophosphate to an aromatic substrate, suggesting a broader relationship for these enzymes. Four domains appear to be particularly well conserved in the Bchl and Chl synthetases and are likely to protrude outside the membrane. These domains are shared to some extent by the polyprenyltransferases (Fig. 3). The second, present between amino acids 60 and 91 in R. sphaeroides BchG, is also found in many proteins that catalyze condensations involving a polyprenyl group.
FIG. 3.
Partial alignment of the R. sphaeroides bchG deduced amino acid sequence (Rsp_BchG) with sequences of other known and putative Bchl and Chl synthetases and of three polyprenyltransferases showing the four domains highly conserved between these sequences. Sequences (and, in parentheses, accession numbers): Rca_BchG, R. capsulatus BchG (Swissprot P26170); Cau_BchG, C. aurantiacus BchG (Swissprot P33326); Syn_ChlG, Synechocystis sp. strain PCC 6803 slr0056 gene product (DDBJ D64001); Ath_CHLG, Arabidopsis thaliana G4 (GenBank U19382); Cau_Orf2, C. aurantiacus orf2 gene product (EMBL Z34000). Polyprenyltransferase sequences and accession numbers: Sce_Coq2, Saccharomyces cerevisiae Coq2 (Swissprot P32378); Eco_UbiA, E. coli UbiA (Swissprot P32166); Eco_MenA, E. coli MenA (Swissprot P26601). Residues fully conserved within the synthetase enzymes are in bold type. A putative cation-binding DRXXD motif is marked by asterisks above the alignment (24). The figures above the alignment are amino acid positions within R. sphaeroides BchG.
R. sphaeroides orf427 shares close identity at the predicted polypeptide level, not only with R. sphaeroides orf428 but also with two other genes found in each of the Rhodobacter species, pucC and orf479/477, the latter of which is also present within the photosynthesis gene cluster (3, 27). Genes showing homology to orf427 were also identified in the cyanobacterium Synechocystis sp. strain PCC 6803 (sll1906; DDBJ accession no. D90910) and in E. coli (mhpT; EMBL accession no. AE000142). All of the gene products are extremely hydrophobic.
R. sphaeroides orf177 shares a high level of homology with known or putative idi genes from a wide range of organisms.
Production of a sequence-specific transposon Tn5 insertion map.
Random insertion of Tn5 was used to generate a set of mutants of wild-type R. sphaeroides in bchG, orf427, and orf177. The points of Tn5 insertion were precisely determined by sequencing from the left inverted repeat of Tn5 in the selected pSCN6-G derivatives, enabling an accurate Tn5 insertion map to be produced (Fig. 1B). Southern blot analysis verified that the locations of the Tn5 insertions in the mutant strains were the same as those in pSCN6-G.
Analysis of Tn5 mutants. (i) Photosynthetic growth analysis.
The two bchG mutants (T6G1 and T6G3) were nonphotosynthetic. The two remaining mutants, with insertions in orf427 and orf177, were capable of photosynthetic growth, but at significantly reduced rates (Table 1).
TABLE 1.
Maximal photosynthetic growth rates of R. sphaeroides strains under low light intensity (3 W/m2) at room temperature
| R. sphaeroides strain | Doubling time (h) |
|---|---|
| Wild-type | 15.9 |
| bchG insertion mutant T6G1 | No growth |
| bchG insertion mutant T6G3 | No growth |
| T6G1[pRKEB] | No growth |
| T6G1[pEBbchG] | 38.5 |
| orf427 insertion mutant T6G4 | 20 |
| T6G4[pRKSK1] | 27.8 |
| T6G4[pSK1bchP] | 23.8 |
| orf177 insertion mutant T6G6 | 31.3 |
| T6G6[pRKEB] | 58.8 |
| T6G6[pEB177L] | 23.3 |
(ii) Analysis of mutants by whole-cell absorbance spectroscopy.
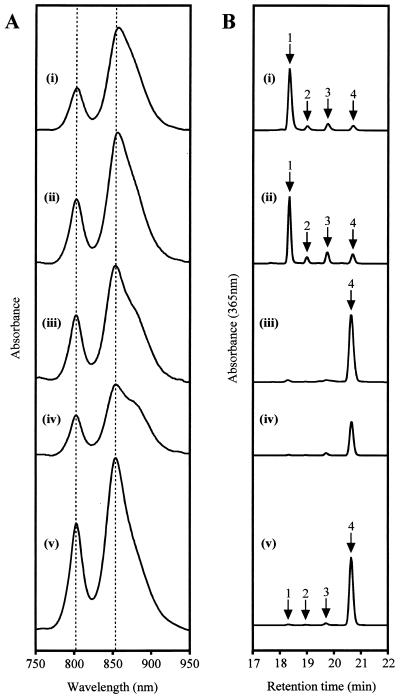
Insertion into orf427 (T6G4) gave a mutant with reductions in the height of its B800 and B850 peaks of approximately 40 and 24%, respectively [Fig. 4A, trace (ii)]. The B850 peak was also red shifted by approximately 3 nm relative to the wild type.
FIG. 4.
Spectral analyses of R. sphaeroides strains: (A) Whole-cell absorbance spectra of nonphotosynthetic cultures, balanced according to cell density; (B) HPLC traces of acetone-methanol extracts. Strains: (i), complemented bchG mutant T6G1[pEBbchG]; (ii), orf427 mutant T6G4; (iii), T6G4[pSK1bchP]; (iv), orf177 mutant T6G6; (v), wild-type R. sphaeroides. Labeled peaks on HPLC traces: 1, BchlaGG; 2, BchlaDHGG; 3, BchlaTHGG; 4, BchlaP.
Insertion T6G6 into orf177 led to a substantial reduction, of over 50%, in the overall level of light-harvesting complexes, particularly LH2 [Fig. 4A, trace (iv)]. This was reduced by approximately two-thirds relative to the wild type, while LH1 appeared to be reduced by less than one-third. No shifts in peak positions were apparent.
No photosynthetic pigment-protein complexes were detectable in cells of either of the bchG mutants. All peaks present in absorbance spectra of cultures were found to result from excretion of pigments into the growth medium; none of these peaks was at a wavelength above 750 nm (data not shown).
(iii) Porphyrin analysis.
Analysis by HPLC showed that the principal Bchl pigment of T6G6 was unchanged from that of wild-type R. sphaeroides, with a retention time of approximately 20.7 min under the conditions used [Fig. 4B, trace (iv)]. This pigment is BchlaP. It was noted, however, that it was present in a significantly reduced amount on a cell-for-cell basis compared to the wild type.
The Bchl content of orf427 insertion mutant T6G4 was very different from that of wild-type R. sphaeroides, with four different Bchla species present in significant quantities, even in stationary-phase cells [Fig. 4B, trace (ii)], where only approximately 11.5% of the Bchl present was BchlaP. The majority (approximately 68%) was BchlaGG, which occurs as a minor constituent in wild-type R. sphaeroides cells, particularly those undergoing rapid growth and pigment biosynthesis [Fig. 4B, trace (v)] (33), and is also the principal Bchl pigment of Rhodospirillum rubrum (21). The remainder consisted of BchlaDHGG (7.3%) and BchlaTHGG (13.2%), which are also present as minor constituents in wild-type R. sphaeroides [Fig. 4B, trace (v)] (33).
Insertion mutants T6G1 and T6G3 in bchG were found to be completely lacking in Bchl. Instead, they appeared to be accumulating bacteriopheophorbide a (Bpheida) or bacteriopheophytin a (Bphea)-type pigments, giving two distinct, but very closely associated, early-eluting HPLC peaks. The first eluted pigment was presumed to have been the 132-hydroxy form of the second, as Chl and associated pigments are particularly labile at this position. These pigments were also excreted into the growth medium, particularly when this was supplemented with Tween 80 (0.2% [vol/vol]). Their presence in the medium demonstrated that they were water soluble, a feature of pheophorbides but not of the more hydrophobic pheophytins. The identity of the putative Bpheid peaks was confirmed by the preparation of authentic samples of Bphea and Bpheida from BchlaP by the method of Perkins and Roberts (29) and the determination of their HPLC retention times. The main elution peaks of the Bpheid sample came after 3.9 and 4.1 min, exactly as for a cell extract of mutant T6G1, whereas the Bphe eluted only after 14.8 min. In the presence of Tween, Bchl- or Bchlide-type pigments remained undetectable.
(iv) Carotenoid analysis.
The carotenoid pigments of the transposon insertion mutants were also analyzed by HPLC to determine whether these too had been affected by any of the insertions. Analyses were performed on nonphotosynthetically grown cells; if unaltered in carotenoid biosynthesis or accumulation, these cells were expected to contain principally spheroidenone. This pigment, which had a major absorbance peak at 485.5 nm in the solvents used, eluted after approximately 13.8 min. It was found to be by far the most abundant carotenoid in all of the mutants, including bchG insertion mutants T6G1 and T6G3, despite their apparent lack of photosynthetic complexes.
Complementation analysis of transposon mutants. (i) Porphyrin analysis.
Upon introduction of plasmid pEBbchG into bchG insertion mutant T6G1, Bchla became detectable by HPLC [Fig. 4B, trace (i)]. However, very little of the pigment was found to be BchlaP (less than 10%), the vast majority (approximately 77%) being BchlaGG, with much smaller proportions of BchlaDHGG and BchlaTHGG present. No Bchl was detectable in the presence of control vector pRKEB alone (data not shown).
It was noted that the phenotype of orf427 insertion mutant T6G4 was similar to that of bchP insertion mutant T6G5 of R. sphaeroides (2), in that both accumulated excess nonphytylated Bchla; T6G5 accumulates only BchlaGG. Furthermore, the bchP locus has been identified as the geranylgeranyl-Bchl reductase of this organism (2). Therefore, it was determined that a plasmid-borne copy of bchP should be introduced into T6G4. Plasmid pSK1bchP had previously given complete complementation of the T6G5 lesion (2); upon introduction of this plasmid into T6G4, the Bchl content was returned to normal [Fig. 4B, trace (iii)]. The presence alone of the vector control pRKSK1 had no detectable effect on the pigment composition.
(ii) Photosynthetic growth analysis.
Maximal growth rates of complemented transposon mutants, recorded under low light intensity, are presented in Table 1. The maximal growth rate of orf177 insertion mutant T6G6 containing pEB177L was approximately 2.5 times greater than that of the mutant containing the insert-free plasmid pRKEB. Mutant T6G1 with an insertion in bchG remained nonphotosynthetic in the presence of plasmid pRKEB. However, with pEBbchG present, this mutant regained the ability to grow photosynthetically, though the maximal growth rate was still rather slow.
(iii) Whole-cell absorbance spectroscopy.
The absorbance spectrum of T6G6[pEB177L] showed that it contained approximately wild-type levels of the photosynthetic pigment-protein complexes (data not shown). In contrast, the control plasmid pRKEB barely affected the spectrum of the orf177 mutant.
The introduction of plasmid pSK1bchP into orf427 insertion mutant T6G4 produced an absorbance spectrum very similar to that of wild-type R. sphaeroides. The red shift of T6G4 was fully reversed, and the B800/B850 ratio returned closer to the wild-type level [Fig. 4A, trace (iii)]. In the presence of pRKSK1, or a derivative of this vector, the overall levels of complexes appeared to be lowered proportionately. The reason for this is unknown. The shape of the T6G4 spectrum was not, however, altered significantly by the presence of pRKSK1 alone (data not shown).
The absorbance spectrum of T6G1[pEBbchG] [Fig. 4A, trace (i)], unlike that of the mutant containing the parental vector, displayed peaks indicating the presence of photosynthetic pigment-protein complexes. The spectrum of T6G1[pEBbchG] was noted to be somewhat similar to that of orf427 insertion mutant T6G4, although the total amount of complexes was lower, and proportionately even less B800 was present. Additionally, the B850 peak of LH2 was red shifted by about 4 nm relative to the wild type, rather than by 3 nm as in T6G4.
Heterologous expression of bchG in E. coli.
The R. sphaeroides bchG gene was cloned into vector pET9a for overexpression in E. coli. Protein production from the construct was induced in E. coli BL21(DE3)pLysS by the addition of isopropyl-β-d-thiogalactopyranoside; cultures of cells transformed with the parental vector pET9a were similarly treated for use as a negative control. The cells were subsequently harvested and fractionated by sonication and centrifugation, and the fractions were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Additional protein bands were not detected.
Activity of BchG in vitro.
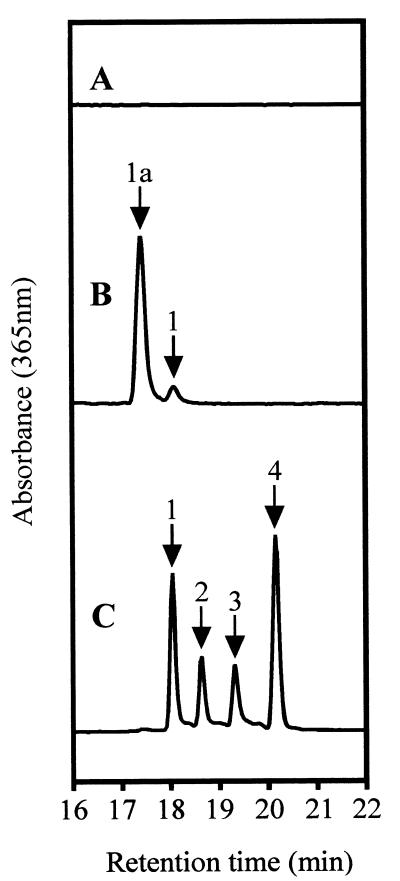
HPLC analysis of in vitro assays utilizing induced cells of BL21(DE3)pLysS carrying expression plasmid pET9a-bchG as the source of enzyme activity demonstrated that BchG was capable of catalyzing the esterification of Bchlidea with GGPP. When approximately 2.7 nmol of Bchlidea was incubated with 6 nmol of GGPP in the presence of the E. coli cell extract, at least 30% esterification of Bchlidea was achieved (Fig. 5B). It should be noted that only a relatively small proportion of the resulting Bchl actually had the expected retention time for BchlaGG and that the majority had a slightly reduced retention time. However, this was, as for the Bpheid pigments of the bchG mutants, almost certainly a result of hydroxylation at the 132 position, either before or after esterification. No esterified pigments were detectable when pET9a-containing cells were used as the source of enzyme activity (Fig. 5A).
FIG. 5.
HPLC analysis of Bchl synthetase assays. Chromatograms are of acetone-methanol extracts of incubations containing porphyrin substrates and E. coli extracts as follows: Bchlidea plus pET9a control (trace A) and Bchlidea plus pET9a-bchG (trace B). Trace C represents a mixture of Bchlidea esters. Labeled peaks: 1, BchlaGG; 2, BchlaDHGG; 3, BchlaTHGG; 4, BchlaP. Peak 1a is likely to be 132-hydroxy-BchlaGG.
DISCUSSION
The aim of this work was to identify and analyze the ORFs present in a previously uncharacterized 5-kb region near the center of the R. sphaeroides photosynthesis gene cluster, in particular that responsible for the esterification of Bchlide in the final stage of Bchl biosynthesis. Mutational analysis of the cluster by Coomber et al. (11) had suggested that this region contained a bchG locus involved in the reaction. Here we describe the results of sequencing and further transposon Tn5 mutagenesis of this region and the establishment of an in vitro assay demonstrating the activity of the product of the bchG gene.
Analysis and overexpression of bchG.
Sequence analysis identified four complete ORFs. The first of these, bchG, encodes an integral membrane protein possessing strong homology with Bchl and Chl synthetases, enzymes catalyzing esterification of Bchlide and chlorophyllide, respectively. R. sphaeroides BchG shares with all of these polypeptides, and with many other enzymes that catalyze condensations involving polyprenyl groups, a domain hypothesized to be the binding site of a polyprenyl pyrophosphate group (24). This domain may also contain a motif responsible for the binding of a divalent cation (either Mg2+ or Mn2+), required for the catalytic activity of at least some polyprenyltransferases (4, 10). This motif was originally proposed to be DDXXD, but Lopez et al. (24) suggested that it was actually DRXXD; R. sphaeroides BchG contains the DRXXD motif (Fig. 3). The requirement of Bchl and Chl synthetases for divalent cations has not yet been investigated.
Two independent transposon Tn5 insertions into bchG (T6G1 and T6G3) generated mutants which were devoid of photosynthetic pigment-protein complexes. Spectral analysis of the pigments present in acetone-methanol cell extracts suggested that the mutants lacked Bchla, and this was confirmed by HPLC analysis. Instead they were found to accumulate principally Bpheida, the metal-free derivative of Bchlidea. The absence of esterified porphyrins suggested that the lesion in these mutants was at this stage of Bchl biosynthesis. Complementation of T6G1 with bchG in vector pRKEB restored both the ability to synthesize Bchl and photosynthetic growth.
Although both sequence homologies and Tn5 mutagenesis suggested that the product of R. sphaeroides bchG was a Bchla synthetase, confirmation required the performance of an in vitro assay utilizing the gene product. The bchG gene was successfully overexpressed in E. coli using the pET expression system, and although the BchG protein was not detectable in SDS-PAGE analysis of cell extracts, its enzymatic activity was detectable, and these extracts were capable of catalyzing the attachment of GG to Bchlidea, producing BchlaGG.
orf427.
R. sphaeroides orf427 is situated immediately downstream of bchG and encodes an extremely hydrophobic polypeptide, which shows significant homology with the products of two other R. sphaeroides genes, orf479 and pucC. orf479 is also sited within the photosynthesis gene cluster (27), while pucC is a member of the puc operon which lies just 18 kb from the cluster (35). The similarity between the products of the R. capsulatus equivalents of these three genes, orf428, orf477, and pucC, has been noted previously (7). No biochemical functions have yet been assigned to Orf479/477 or PucC, but mutational analyses have indicated that they may be involved in the assembly or stability of LH1 and LH2, respectively (5, 17, 36, 37); mutations resulted in total absence or greatly reduced levels of complexes. One of the factors supporting the interpretation that PucC may have a role in photosynthetic membrane assembly was its extreme hydrophobicity (37), a feature shared by both Orf479 and Orf427. Considering the close proximity of orf427 to the genes encoding the terminal enzymes of Bchla biosynthesis, and its probable coexpression with them, it seems likely that its gene product may also be involved in the assembly of a functional photosystem, possibly at the stage of Bchl insertion. Homologues of these Rhodobacter genes have been identified in Synechocystis sp. strain PCC 6803 and E. coli. The presence of such homologues in these distantly related organisms suggests that these genes must play an important role and are unlikely to be purely regulatory. The E. coli mhpT gene product belongs to the sugar transporter family and is hypothesized to be a transporter for 3-hydroxyphenylpropionic acid: the bearing of this on orf427 is as yet unclear.
Transposon mutagenesis of orf427 failed to provide any further insight into the function of this gene. The accumulation of significant quantities of nonphytylated Bchla suggested a disruption of the downstream bchP gene, encoding geranylgeranyl-Bchl reductase (2). This was tested by trans complementation of the mutant, T6G4, by plasmid-borne expression of bchP, the effects of which strongly suggested that the observed mutant phenotype resulted solely from disruption of the downstream bchP gene. Similarly, trans complementation of an R. capsulatus interposon mutant in orf428 has previously been achieved with an expression plasmid bearing the downstream bchP and orf176 loci (7).
The assignment of the observed effects of insertion T6G4 to bchP leaves orf427 apparently having no observable phenotype of its own. One possible explanation for this is that mutations within the gene may be compensated for by the products of the homologous orf479 and pucC genes. An equivalent scenario has previously been proposed for R. capsulatus orf428 (7).
orf177 encodes a putative IPP isomerase.
The fourth gene, lying immediately downstream of bchP, orf177, is a member of a family of idi genes which have been sequenced from a wide range of organisms, from eubacteria to humans. The strongest homology between the IPP isomerases encoded by these genes is in a catalytic core of between 152 (both R. sphaeroides and R. capsulatus) and 181 (Saccharomyces cerevisiae) amino acids. This core constitutes almost the entire enzyme in bacteria but is flanked by amino- and carboxy-terminal extensions in eukaryotes. The R. sphaeroides enzyme shares with all of the known and putative idi gene products, a cysteine residue and a glutamate residue: C63 and E110, respectively, in R. sphaeroides. In S. cerevisiae, affinity labeling and site-directed mutagenesis have shown these conserved residues to be essential for catalysis (34), and they are presumably of equal importance in the enzymes of other organisms. It is thought that they are located in the active site of the enzyme, on opposite faces of the allyl moiety of IPP, and assist with the addition and elimination of protons.
Transposon insertion T6G6 in orf177 generated a mutant strain with a substantial reduction in its population of photosynthetic complexes and a corresponding reduction in its photosynthetic growth rate. Both the Bchl and carotenoid pigments were unchanged from those of wild-type R. sphaeroides, although it was noted that the quantity of Bchl, at least, was considerably reduced. These features are consistent with the identification of orf177 as an idi gene, since mutation within such a gene would be expected to reduce or halt the supply of GGPP for both Bchl and carotenoid biosynthesis, leading to a reduction in the availability of these pigments for photosynthetic complex assembly. LH2 was reduced approximately twofold relative to LH1, suggesting that, if scarce, available pigments may be channeled to the more central LH1 complex and away from the antenna LH2 complex. The fact that neither Bchl nor carotenoid biosynthesis was totally halted indicates that, as was suggested by Southern blot analysis in R. capsulatus (18), there may well be more than one idi gene in R. sphaeroides. In addition to photosynthetic pigment biosynthesis, bacteria also require isoprenoids for respiration (ubiquinones) and cell wall biosynthesis (dolichols). It may be that different IPP isomerase enzymes generally function in different biosynthetic pathways in wild-type R. sphaeroides cells. Expression of orf177 in T6G6 restored both photosynthetic growth and photosynthetic complex levels to at least those of wild-type R. sphaeroides.
The likely role of orf177 in R. sphaeroides means that, as in R. capsulatus, candidates for genes encoding all of the enzymes required for the incorporation of IPP into the final Bchl and carotenoid molecules of the organism have now been found within its photosynthesis gene cluster.
Transcriptional coupling in this region of the photosynthesis gene cluster.
Both the sequence and complementation data presented in this paper support an operonal arrangement for bchG, orf427, bchP, and, in part, orf177. Complementation of bchG mutant T6G1 with bchG generated a strain (T6G1 [pEBbchG]) in which the Bchlide esterification reaction was restored but the hydrogenation of the resulting BchlaGG was mainly impeded. Insertion into orf427 (T6G4) also led to a disruption of the hydrogenation steps. In each case, disruption of the expression of the downstream ORF, bchP, was implicated as the cause of the partial loss of Bchl hydrogenation activity, and for T6G4, it was shown that this disruption could indeed be abolished by plasmid-borne expression of bchP. The only transcription terminator identified for the four sequenced genes is located just downstream of orf177; this may function as the terminator for the entire operon. Naylor et al. (27) suggested that the operon involving these four ORFs also included the upstream genes, bchE and bchJ, since the nearest potential promoter sequence lies upstream of bchE. We have identified a potential terminator within the coding region of bchG, which may function as a terminator for these upstream genes, but the relatively low stability of the stem-loop and the low number of uridines immediately following it suggest that transcription may frequently, if not always, continue beyond it; this must, in any case, occur for bchG to be expressed.
It actually appears that a second, shorter transcript probably also exists for orf177 and is responsible for the majority of the expression of this gene. A putative ς70-type promoter is situated approximately 200 bp upstream of orf177. Furthermore, neither insertion into orf427 (T6G4) nor into bchP (2) led to as substantial a reduction in the overall levels of the light-harvesting complexes as did insertion into orf177 (T6G6), despite the fact that Tn5 insertions generally have strongly polar effects on downstream genes in multigene operons (6). Low-level GG-Bchl reductase activity apparent in T6G4 may also be an indication that the bchP gene has a weak endogenous promoter, a putative candidate for which was located during sequence analysis.
ACKNOWLEDGMENT
This work was supported by a grant from the BBSRC (United Kingdom).
REFERENCES
- 1.Addlesee H A, Gibson L C D, Jensen P E, Hunter C N. Cloning, sequencing and functional assignment of the chlorophyll biosynthesis gene, chlP, of Synechocystis sp. PCC 6803. FEBS Lett. 1996;389:126–130. doi: 10.1016/0014-5793(96)00549-2. [DOI] [PubMed] [Google Scholar]
- 2.Addlesee H A, Hunter C N. Physical mapping and functional assignment of the geranylgeranyl-bacteriochlorophyll reductase gene, bchP, of Rhodobacter sphaeroides. J Bacteriol. 1999;181:7248–7255. doi: 10.1128/jb.181.23.7248-7255.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alberti M, Burke D H, Hearst J E. Structure and sequence of the photosynthesis gene cluster. In: Blankenship R E, Madigan M T, Bauer C E, editors. Anoxygenic photosynthetic bacteria. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1995. pp. 1083–1106. [Google Scholar]
- 4.Ashby M N, Edwards P A. Elucidation of the deficiency in two yeast coenzyme Q mutants: characterization of the structural gene encoding hexaprenyl pyrophosphate synthetase. J Biol Chem. 1990;265:13157–13164. [PubMed] [Google Scholar]
- 5.Bauer C E, Buggy J J, Yang Z, Marrs B L. The superoperonal organization of genes for pigment biosynthesis and reaction center proteins is a conserved feature in Rhodobacter capsulatus: analysis of overlapping bchB and puhA transcripts. Mol Gen Genet. 1991;228:433–444. doi: 10.1007/BF00260637. [DOI] [PubMed] [Google Scholar]
- 6.Berg D E, Weiss A, Crossland L. Polarity of Tn5 insertion mutations in Escherichia coli. J Bacteriol. 1980;142:439–446. doi: 10.1128/jb.142.2.439-446.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bollivar D W, Suzuki J Y, Beatty J T, Dobrowolski J M, Bauer C E. Directed mutational analysis of bacteriochlorophyll a biosynthesis in Rhodobacter capsulatus. J Mol Biol. 1994;237:622–640. doi: 10.1006/jmbi.1994.1260. [DOI] [PubMed] [Google Scholar]
- 8.Bollivar D W, Wang S, Allen J P, Bauer C E. Molecular genetic analysis of terminal steps in bacteriochlorophyll a biosynthesis: characterization of a Rhodobacter capsulatus strain that synthesizes geranylgeraniol-esterified bacteriochlorophyll a. Biochemistry. 1994;33:12763–12768. doi: 10.1021/bi00209a006. [DOI] [PubMed] [Google Scholar]
- 9.Brown A E, Eiserling F A, Lascelles J. Bacteriochlorophyll synthesis and the ultrastructure of wild type and mutant strains of Rhodopseudomonas sphaeroides. Plant Physiol. 1972;50:743–746. doi: 10.1104/pp.50.6.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carattoli A, Romano N, Ballario P, Morelli G, Macino G. The Neurospora crassa carotenoid biosynthetic gene (Albino 3) reveals highly conserved regions among prenyltransferases. J Biol Chem. 1991;266:5854–5859. [PubMed] [Google Scholar]
- 11.Coomber S A, Chaudri M, Connor A, Britton G, Hunter C N. Localized transposon Tn5 mutagenesis of the photosynthetic gene cluster of Rhodobacter sphaeroides. Mol Microbiol. 1990;4:977–989. doi: 10.1111/j.1365-2958.1990.tb00670.x. [DOI] [PubMed] [Google Scholar]
- 12.Coomber S A, Hunter C N. Construction of a physical map of the 45 kb photosynthetic gene cluster of Rhodobacter sphaeroides. Arch Microbiol. 1989;151:454–458. [Google Scholar]
- 13.Deisenhofer J, Epp O, Miki K, Huber R, Michel H. Structure of the protein subunits in the photosynthetic reaction centre of Rhodopseudomonas viridis at 3 Å resolution. Nature. 1985;318:618–624. doi: 10.1038/318618a0. [DOI] [PubMed] [Google Scholar]
- 14.Dryden S C, Kaplan S. Localization and structural analysis of the ribosomal RNA operons of Rhodobacter sphaeroides. Nucleic Acids Res. 1990;18:7267–7277. doi: 10.1093/nar/18.24.7267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ermler U, Fritzsch G, Buchanan S K, Michel H. Structure of the photosynthetic reaction centre from Rhodobacter sphaeroides at 2.65 Å resolution: cofactors and protein-cofactor interactions. Structure. 1994;2:925–936. doi: 10.1016/s0969-2126(94)00094-8. [DOI] [PubMed] [Google Scholar]
- 16.Fiedor L, Rosenbach-Belkin V, Scherz A. The stereospecific interaction between chlorophylls and chlorophyllase. J Biol Chem. 1992;267:22043–22047. [PubMed] [Google Scholar]
- 17.Gibson L C D, McGlynn P, Chaudri M, Hunter C N. A putative anaerobic coproporphyrinogen III oxidase in Rhodobacter sphaeroides. II. Analysis of a region of the genome encoding hemF and the puc operon. Mol Microbiol. 1992;6:3171–3186. doi: 10.1111/j.1365-2958.1992.tb01773.x. [DOI] [PubMed] [Google Scholar]
- 18.Hahn F M, Baker J A, Poulter C D. Open reading frame 176 in the photosynthesis gene cluster of Rhodobacter capsulatus encodes idi, a gene for isopentenyl diphosphate isomerase. J Bacteriol. 1996;178:619–624. doi: 10.1128/jb.178.3.619-624.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hunter C N, Coomber S A. Cloning and oxygen-regulated expression of the bacteriochlorophyll biosynthesis genes bch E, B, A and C of Rhodobacter sphaeroides. J Gen Microbiol. 1988;134:1491–1497. [Google Scholar]
- 20.Hunter C N, Hundle B S, Hearst J E, Lang H P, Gardiner A T, Takaichi S, Cogdell R J. Introduction of new carotenoids into the bacterial photosynthetic apparatus by combining the carotenoid biosynthetic pathways of Erwinia herbicola and Rhodobacter sphaeroides. J Bacteriol. 1994;176:3692–3697. doi: 10.1128/jb.176.12.3692-3697.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Katz J J, Strain H H, Harkness A L, Studier M H, Svec W A, Janson T R, Cope B T. Esterifying alcohols in the chlorophylls of purple photosynthetic bacteria. A new chlorophyll, bacteriochlorophyll(gg), all-trans-geranylgeranyl bacteriochlorophyllide a. J Am Chem Soc. 1972;94:7938–7939. doi: 10.1021/ja00777a054. [DOI] [PubMed] [Google Scholar]
- 22.Klein P, Kanehisa M, Delisi C. The detection and classification of membrane-spanning proteins. Biochim Biophys Acta. 1985;815:468–476. doi: 10.1016/0005-2736(85)90375-x. [DOI] [PubMed] [Google Scholar]
- 23.Kyte J, Doolittle R F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 24.Lopez J C, Ryan S, Blankenship R E. Sequence of the bchG gene from Chloroflexus aurantiacus: relationship between chlorophyll synthase and other polyprenyltransferases. J Bacteriol. 1996;178:3369–3373. doi: 10.1128/jb.178.11.3369-3373.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McClure W R. Mechanism and control of transcription initiation in prokaryotes. Annu Rev Biochem. 1985;54:171–204. doi: 10.1146/annurev.bi.54.070185.001131. [DOI] [PubMed] [Google Scholar]
- 26.McDermott G, Prince S M, Freer A A, Hawthornthwaite-Lawless A M, Papiz M Z, Cogdell R J, Isaacs N W. Crystal structure of an integral membrane light-harvesting complex from photosynthetic bacteria. Nature. 1995;374:517–521. [Google Scholar]
- 27.Naylor G W, Addlesee H A, Gibson L C D, Hunter C N. The photosynthesis gene cluster of Rhodobacter sphaeroides. Photosyn Res. 1999;62:121–139. [Google Scholar]
- 28.Oster U, Bauer C E, Rüdiger W. Characterization of chlorophyll a and bacteriochlorophyll a synthases by heterologous expression in Escherichia coli. J Bacteriol. 1997;272:9671–9676. doi: 10.1074/jbc.272.15.9671. [DOI] [PubMed] [Google Scholar]
- 29.Perkins H J, Roberts D W A. Purification of chlorophylls, pheophytins and pheophorbides for specific activity determinations. Biochim Biophys Acta. 1962;58:486–498. doi: 10.1016/0006-3002(62)90059-8. [DOI] [PubMed] [Google Scholar]
- 30.Platt T. Transcription termination and the regulation of gene expression. Annu Rev Biochem. 1986;55:339–372. doi: 10.1146/annurev.bi.55.070186.002011. [DOI] [PubMed] [Google Scholar]
- 31.Richards W R, Lascelles J. The biosynthesis of bacteriochlorophyll. The characterization of latter stage intermediates from mutants of Rhodopseudomonas spheroides. Biochemistry. 1969;8:3473–3482. doi: 10.1021/bi00836a051. [DOI] [PubMed] [Google Scholar]
- 32.Schoch S, Schäfer W. Tetrahydrogeranylgeraniol, a precursor of phytol in the biosynthesis of chlorophyll a: localization of the double bonds. Z Naturforsch. 1978;33c:408–412. [Google Scholar]
- 33.Shioi Y, Sasa T. Terminal steps of bacteriochlorophyll a phytol formation in purple photosynthetic bacteria. J Bacteriol. 1984;158:340–343. doi: 10.1128/jb.158.1.340-343.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Street I P, Coffman H R, Baker J A, Poulter C D. Identification of Cys139 and Glu207 as catalytically important groups in the active site of isopentenyl diphosphate: dimethylallyl diphosphate isomerase. Biochemistry. 1994;33:4212–4217. doi: 10.1021/bi00180a014. [DOI] [PubMed] [Google Scholar]
- 35.Suwanto A, Kaplan S. Physical and genetic mapping of the Rhodobacter sphaeroides 2.4.1 genome: genome size, fragment identification and gene localization. J Bacteriol. 1989;171:5840–5849. doi: 10.1128/jb.171.11.5840-5849.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tichy H V, Oberle B, Stiehle H, Schiltz E, Drews G. Genes downstream from pucB and pucA are essential for formation of the B800-850 complex of Rhodobacter capsulatus. J Bacteriol. 1989;171:4914–4922. doi: 10.1128/jb.171.9.4914-4922.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tichy H V, Albien K-U, Gad'on N, Drews G. Analysis of the Rhodobacter capsulatus puc operon: the pucC gene plays a central role in the regulation of LHII (B800-850 complex) expression. EMBO J. 1991;10:2949–2955. doi: 10.1002/j.1460-2075.1991.tb07845.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tinoco I, Borer P N, Dengler B, Levine M D, Uhlenbeck O C, Crothers D M, Gralla J. Improved estimation of secondary structure in ribonucleic acids. Nature. 1973;246:40–41. doi: 10.1038/newbio246040a0. [DOI] [PubMed] [Google Scholar]