Abstract
Shortly after the onset of the COVID-19 pandemic, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has acquired numerous variations in its intracellular proteins to adapt quickly, become more infectious, and ultimately develop drug resistance by mutating certain hotspot residues. To keep the emerging variants at bay, including Omicron and subvariants, FDA has approved the antiviral nirmatrelvir for mild-to-moderate and high-risk COVID-19 cases. Like other viruses, SARS-CoV-2 could acquire mutations in its main protease (Mpro) to adapt and develop resistance against nirmatrelvir. Employing a unique high-throughput protein design technique, the hotspot residues, and signatures of adaptation of Mpro having the highest probability of mutating and rendering nirmatrelvir ineffective were identified. Our results show that ∼40% of the designed mutations in Mpro already exist in the globally circulating SARS-CoV-2 lineages and several predicted mutations. Moreover, several high-frequency, designed mutations were found to be in corroboration with the experimentally reported nirmatrelvir-resistant mutants and are naturally occurring. Our work on the targeted design of the nirmatrelvir-binding site offers a comprehensive picture of potential hotspot sites and resistance mutations in Mpro and is thus crucial in comprehending viral adaptation, robust antiviral design, and surveillance of evolving Mpro variations.
Keywords: Adaptability, Resistance mutations, Main protease, Nirmatrelvir, Protein design, SARS-CoV-2, Signatures of adaptation
1. Introduction
RNA virus evolution depends on regular recombination events, which are frequently accompanied by escape mutations and result in enhanced transmissibility and stronger adherence to host cell-surface receptors [1]. Moreover, certain mutations confer resistance to the available drugs and antibodies [2], which facilitate the evasion of the virus by the immune system, thereby developing uncertainty about the efficacy of the therapeutics [3]. Along with viral recombination, constant replication of the virus in immuno-compromised people or inter-species spill-over & spill-back events may also enable the accumulation of mutations to develop viral fitness and immune system evasion [4]. Thus, investigating the virus evolution is crucial to help comprehend the viral evolutionary mechanisms which control its pathogenicity and transmission.
Despite the development and widespread administration of several potent vaccines, the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) continues to infect people with Coronavirus disease 19 (COVID-19) and claim lives, making it possibly the most lethal virus of this century. Pfizer's Paxlovid (a combination of nirmatrelvir [PF-07321332] tablets and ritonavir tablets) has been approved as the first oral antiviral for the treatment of COVID-19 by the US Food and Drug Administration (FDA). In December 2021, Paxlovid was granted emergency use authorization (EUA) for adults and pediatric patients. Nirmatrelvir is emerging as a useful drug for treating mild-to-moderate and high-risk COVID-19 cases [5]. It is a potent inhibitor of the SARS-CoV-2 main protease (Mpro) with an inhibition constant (K i) of ∼1 nM [6], an EC50 value of ∼16 nM [7], and an IC50 value ranging between 22 and 225 nM [8]. Using a reversible covalent mechanism, nirmatrelvir, via its cyano group, reacts with the catalytic Cys145 residue of the Mpro. SARS-CoV-2 is continually evolving and generating new sequence variants globally. While the emergence of new variants is inevitable, predicting their evolution, spread, and impact through intense research may help improve countermeasures [9,10]. However, as current reports suggest the emergence of resistance mutations in SARS-CoV-2 against nirmatrelvir [[11], [12], [13], [14], [15]], we envisioned carrying out a cohesive and comprehensive study to identify all the potential resistance mutations and hotspots in the Mpro of SARS-CoV-2. In the current study, we used a unique high-throughput ligand-based interface protein design protocol to identify potential mutational hotspots in the nirmatrelvir-binding site in the SARS-CoV-2 Mpro. Notably, we correlate our findings with the globally circulating viral genomes and show that our design methodology can correctly predict the hotspot residues and adaptable mutations in the Mpro.
2. Methodology
The crystal structure of SARS-CoV-2 Mpro in complex with nirmatrelvir (PDB ID: 7TLL) was used to obtain the Mpro's interacting residues with nirmatrelvir [6] and then prepared using Molecular Operating Environment (MOE), v2022.03. Subsequently, a high-throughput protein design involving the resistance scan methodology of MOE was employed to identify single-point mutations of the nirmatrelvir-binding site in Mpro that exhibit the highest potential for adaptability and drug resistance [16,17]. To sample the mutations that are not lethal to the virus and more likely to evolve naturally, the nirmatrelvir-binding sites were mutated to single nucleotide polymorphisms (SNPs) of the native sequence only. From the nirmatrelvir-bound Mpro complex, 25 drug-binding residues of Mpro were designed, except the catalytic dyad (Cys145 and His41), because of their role in substrate cleave and processing [18,19]. For exhaustive sampling and designed site flexibility, the ensemble protein design protocol with the rotamer explorer option was utilized, followed by the root mean square deviation (RMSD) threshold of 0.5 Å. Other important parameters, including the “conformation limit”, “fix residues farther than,” and “energy window,” were set to 25 K, 4.5 Å, and 10 kcal/mol, respectively. A total of 210 designed mutations were generated, and the relative binding affinity of the mutation to the native Mpro (represented by dAffinity, which is the Boltzmann average of the relative affinities of the ensemble) was computed for the designed Mpro-nirmatrelvir complexes. The Arpeggio webserver was used to analyze the intermolecular interactions between nirmatrelvir and Mpro in the low-affinity and high-affinity designs of the Mpro-nirmatrelvir complex [20]. Finally, to validate the protein design protocol and correlate the designed Mpro mutations with available experimental data, the designed Mpro residues that bind to nirmatrelvir were compared with globally circulating SARS-CoV-2 sequences deposited in the Global Initiative on Sharing Avian Influenza Database (GISAID) enabled CoV-GLUE-Viz database, after which their frequencies of occurrence were obtained to demonstrate design accuracy [21,22].
3. Results
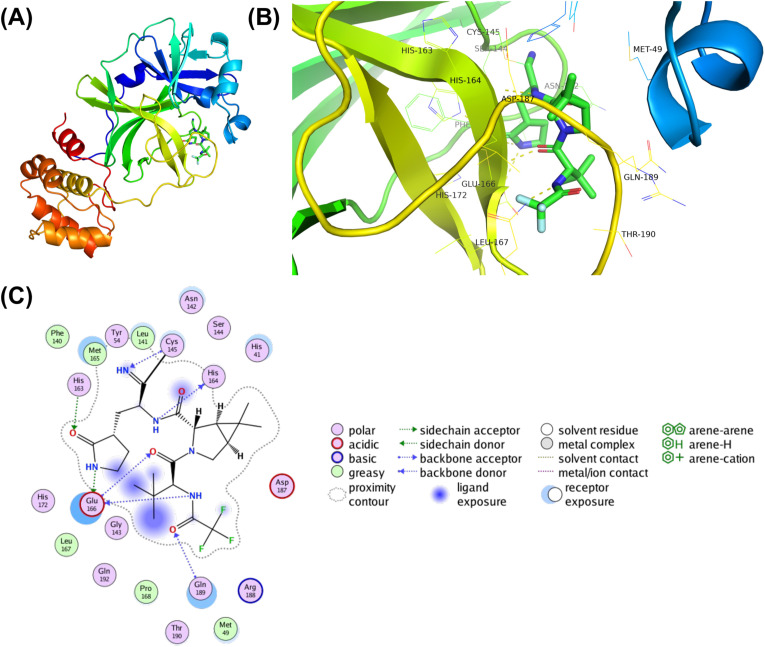
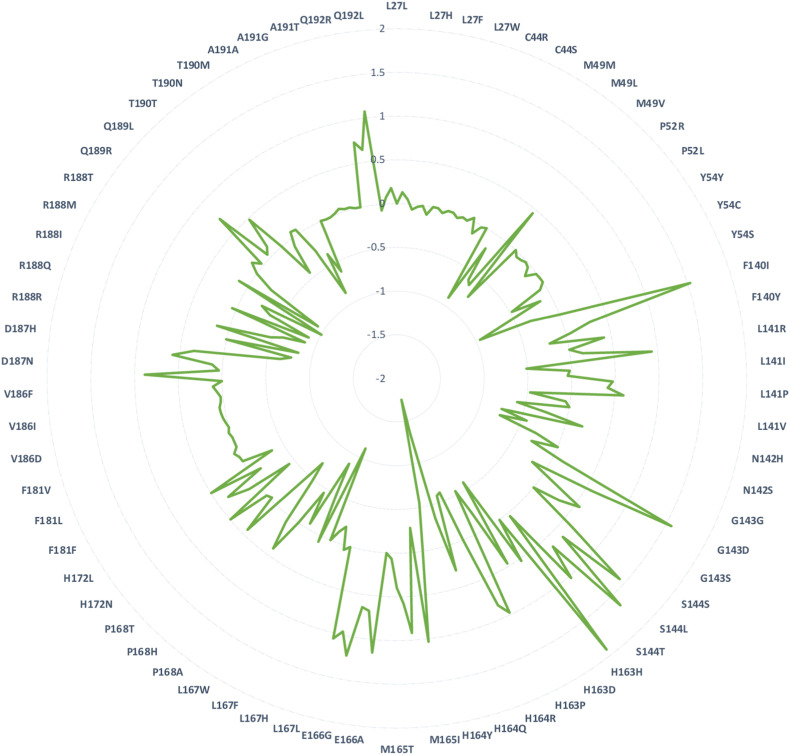
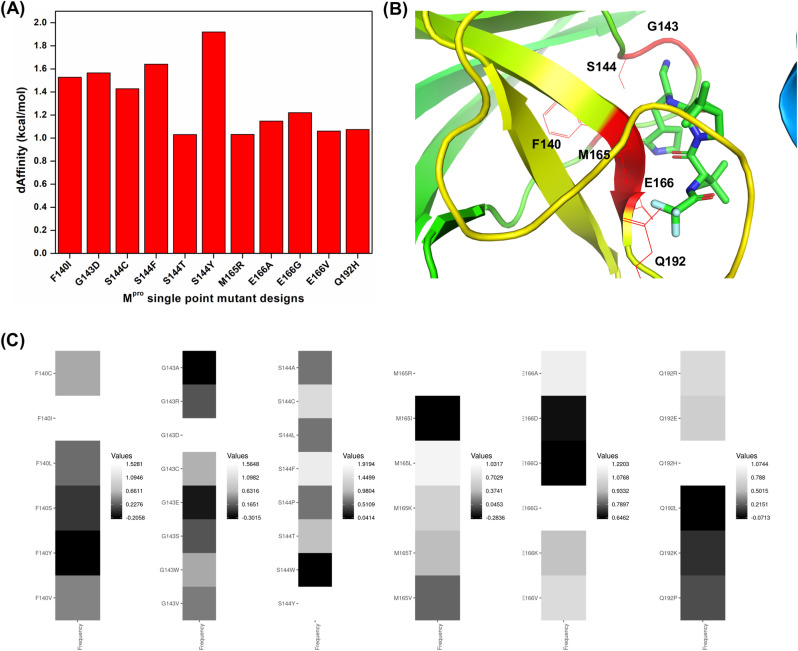
Structural inspection of the nirmatrelvir-Mpro crystal structure revealed 25 interacting residues and were subjected to design (Fig. 1A & B). Employing the resistance scan module of MOE, the 25 nirmatrelvir-interacting Mpro residues were mutated to naturally evolving SNPs (Table 1 ). This led to 210 single-point mutants, of which the relative binding affinities of the mutations to the native Mpro (dAffinity) were computed, where a higher positive dAffinity denoted lower affinity with nirmatrelvir, thereby implying easy tolerance and resistance of the mutant to nirmatrelvir. The computed dAffinity of 210 single mutants ranged from 1.91 to −1.75 kcal/mol, of which ninety-four Mpro-designed mutants demonstrated positive dAffinity values. These results implied tolerated and resistant mutants (Fig. 2 ). Moreover, to shortlist the most potential resistant mutants, when a strict cut-off of dAffinity >1.0 kcal/mol was used, it resulted in the identification of eleven Mpro mutants. The hotspots were mostly from 140, 143, 144, 165, 166, and 192 positions of the Mpro. Fig. 3 shows the hotspot residues and potential resistant mutants against nirmatrelvir. Interestingly, this analysis suggested that positions 144 and 166 of Mpro are crucial and of high susceptibility for tolerance, adaptability, and conferring resistance during evolution.
Fig. 1.
Structure and interactions of the SARS-CoV-2 Mpro-nirmatrelvir bound complex. (A) The crystal structure of full-length SARS-CoV-2 Mpro is shown as a cartoon, and bound nirmatrelvir is shown as a stick. (B) The nirmatrelvir-interacting residues of Mpro are shown as lines along with hydrogen bond interactions as yellow dashed lines. (C) A 2D representation showing various types of interactions between Mpro and nirmatrelvir, where various types of intermolecular interactions are labeled in the legend. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Table 1.
Mpro-nirmatrelvir interacting residues that were designed with corresponding SNPs of the native sequence to emulate the mutations that are more likely to happen naturally over the evolution of Mpro.
| S. No | Mpro-nirmatrelvir interacting and designed native residuesa | Sampled SNPs in designs |
|---|---|---|
| 1 | Leu27 | RQHIMFPSWV |
| 2 | Cys44 | RGFSWY |
| 3 | Met49 | RILKTV |
| 4 | Pro52 | ARQHLST |
| 5 | Tyr54 | NDCHFS |
| 6 | Phe140 | CILSYV |
| 7 | Leu141 | RQHIMFPSWV |
| 8 | Asn142 | DHIKSTY |
| 9 | Gly143 | ARDCESWV |
| 10 | Ser144 | ACLFPTWY |
| 11 | His163 | RNDQLPY |
| 12 | His164 | RNDQLPY |
| 13 | Met165 | RILKTV |
| 14 | Glu166 | ADQGKV |
| 15 | Leu167 | RQHIMFPSWV |
| 16 | Pro168 | ARQHLST |
| 17 | His172 | RNDQLPY |
| 18 | Phe181 | CILSYV |
| 19 | Val186 | ADEGILMF |
| 20 | Asp187 | ANEGHYV |
| 21 | Arg188 | NCQGHILKMPSTW |
| 22 | Gln189 | REHLKP |
| 23 | Thr190 | ARNIKMPS |
| 24 | Ala191 | DEGPSTV |
| 25 | Gln192 | REHLKP |
The catalytic dyad residues His41 and Cys145 of Mpro were not designed, as they carry out the acylation-deacylation reaction and cleavage of the substrates.
Fig. 2.
Relative binding affinities (dAffinities) of all the designed mutations in the nirmatrelvir-binding site of Mpro. Radar plot showing the computed dAffinities for the most plausible resistant and adaptable single-point mutant designs of Mpro bound to nirmatrelvir. The mutated designs are labeled in the radar plot, and the corresponding dAffinity values are shown.
Fig. 3.
Relative binding affinities (dAffinities) and conservation of the most plausible resistance-eliciting designs from the Mpro-nirmatrelvir complex. (A) Bar graph highlighting the dAffinities for the most plausible resistance-eliciting mutants of Mpro against nirmatrelvir, where designs exhibiting dAffinity > 1.0 kcal/mol suggest a decrease in affinity towards nirmatrelvir and hence their ability to become resistant easily. (B) hotspot residues having the greatest tendency to undergo resistance are shown within the protein. (C) heatmaps showing the dAffinity values of every designed mutation in each hotspot, thus highlighting their conservation within the protein. Here, the frequencies of the designed mutations in the hotspot residues are shown in black to white colors as per their dAffinity values, respectively. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
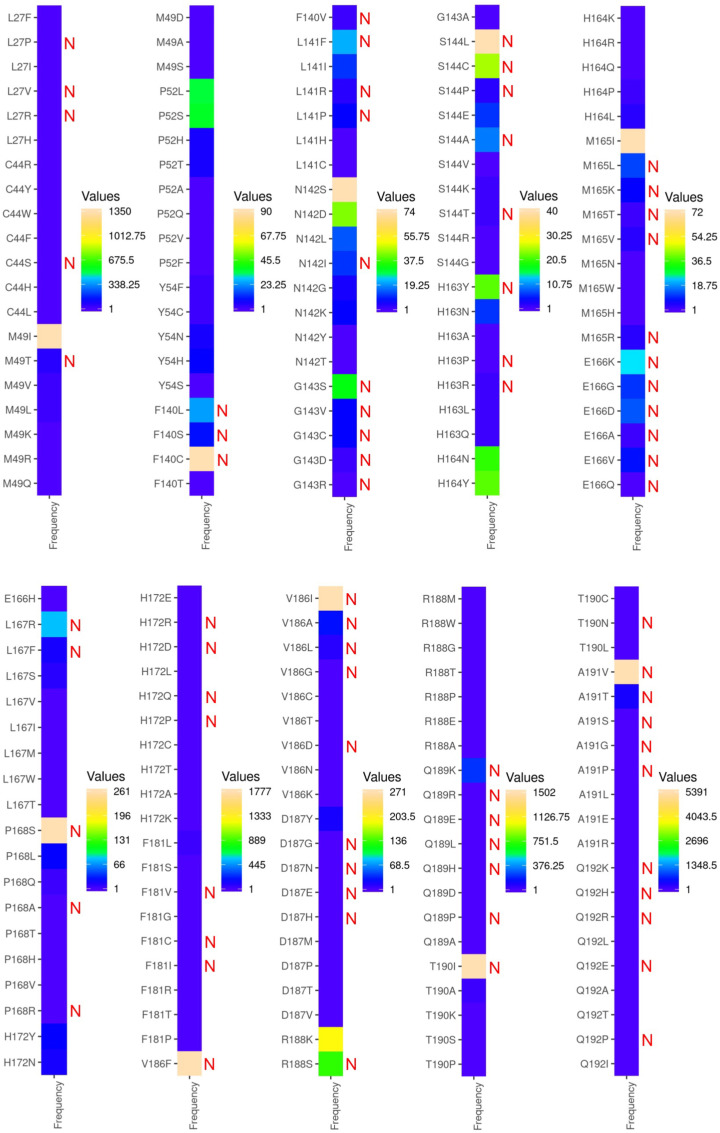
Next, a critical analysis was performed focusing on the accuracy of our ligand-based design protocol [17] and assessing how closely our designed single-point mutants of Mpro-nirmatrelvir corroborate with the experimentally determined SARS-CoV-2 sequences from the COVID-19 pandemic. As SARS-CoV-2 is actively mutating due to poor immunity, drug, and vaccine pressures, it resulted in the accumulation of >1000 unique mutations at the Mpro, thereby continuously being added into the GISAID and released through CoV-GLUE-Viz. We obtained all the deposited mutations and their frequencies of the nirmatrelvir-bound Mpro site and compared them with our designed mutants. The comparison showed that seventy-eight mutations (out of 199) were predicted as resistant-causing and hence crucial in conferring adaptability as obtained from the design calculations, thereby achieving ∼40% corroboration with the clinical-sequencing data (Fig. 4 ). Several of the globally circulating, frequently found mutants such as A191V, T190I, V186I, V186F, P168S, S144L, and F140C already have high dAffinity values as obtained from our computational design. This implies that these mutants are adaptive and resistant to nirmatrelvir, even without significant drug or selection pressure (Fig. 3, Fig. 4). However, it is important to comment that the dAffinities are not directly comparable with frequencies obtained from the databases and designed mutants (dAffinities >1.0 kcal/mol), for instance, S144C, which are being circulated as high-frequency mutants (Fig. 3, Fig. 4).
Fig. 4.
Heat maps of the nirmatrelvir-bound Mpro mutations with their frequencies of occurrence retrieved from GISAID-enabled CoV-GLUE-Viz databases. Mutations at the nirmatrelvir-binding site of Mpro obtained from the GISAID-enabled CoV-GLUE-Viz databases are shown. Frequencies of the mutations in SARS-CoV-2 sequences from the COVID-19 pandemic ranged from lower to higher numbers and from blue to orange colors, respectively. The designed mutants that potentially developed resistance and adaptation towards nirmatrelvir are denoted as 'N' in red. It was found that out of 199 mutations, 78 mutations were predicted as positively selected and resistant in the design computations, therefore attaining ∼40% correlation, already exist in the SARS-CoV-2 sequences and are currently circulating. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Notably, while this manuscript was being prepared, a preprint article experimentally reported 11 mutants at the drug-binding site of Mpro that are resistant to nirmatrelvir (K i > 10-fold increase) [13]. This offered us an opportunity to critically assess the designed Mpro mutants with that of the experimentally reported resistant mutants. Upon examination, it was found that several of the designed mutants, such as S144A, S144F, S144Y, M165T, E166Q, and H172Q, were already found to be resistant and naturally occurring. These mutants are reported to be nirmatrelvir resistant, with K i values increasing between 19.2 and 38.0-fold, therefore corroborating our findings and reflecting the accuracy of our design methodology [13]. Interestingly, the E166V mutant, experimentally shown to confer the strongest resistance (∼100-fold) [23], was found to be one of the most plausible resistance-causing mutants in our design experiments (Fig. 3). Moreover, our targeted design of the nirmatrelvir-binding site in Mpro provided a comprehensive picture of other potential hotspot sites and resistance mutations that may emerge if the drug pressure keeps mounting and the pandemic endures its devastating course.
Since the spike protein of SARS-CoV-2 has acquired some permanent mutations, such as the D614G, which continues to be a part of the new variants, a similar quest was conducted for potential clues in Mpro. It was found that there were a few mutations in the Mpro that were highly prevalent in different lineages (Table 2 ). Interestingly, these mutations were not in the nirmatrelvir-binding site of the SARS-CoV-2 Mpro and hence were not designed (Table 1). Nonetheless, it is possible that there could be a presence of (i) higher-order mutants (such as double, triple, quadruple mutants, and so on) which may harbor some of the mutations from the nirmatrelvir-binding site and/or (ii) compensatory mutations that are part of the nirmatrelvir-binding site and/or (iii) with mutations of other proteins, such as the spike protein. Although this information is not yet available to the best of our knowledge, resistance to nirmatrelvir may escalate by the simultaneous acquisition of two or more higher-order mutational combination events. A study is underway in this direction.
Table 2.
List of prevalent Mpro mutations and their corresponding SARS-CoV-2 lineages.
| S. No | Prevalent Mpro mutationsa | Lineage |
|---|---|---|
| 1 | G15S | C.37 (Lambda) |
| 2 | T21I | B.1.1.318 |
| 3 | L89F | B.1.2 |
| 4 | K90R | B.1.351 (Beta) |
| 5 | P132H | B.1.1.529 (Omicron) |
| 6 | L205V | P.2 (Zeta) |
The most prevalent Mpro mutations are not part of the nirmatrelvir-binding site.
A further inspection of the high-frequency mutants A191V, T190I, V186F, P168S, S144L, and F140C with respect to the lineages revealed that these mutants pertain to some of the globally distributed lineages that are found in nearly all the COVID-19 prevalent countries. More specifically, the F140C mutant is detected in six, S144L in 23, P168S in 69, V186F in 102, V186I in 41, T190I in 126, and A191V in 285 different lineages (a total of 652 lineages), out of which 421 lineages (64.5%) were found to be common across all the seven high-frequency mutants (Supplementary Table 1). This suggests that the SARS-CoV-2 strains with certain residue-specific mutations of Mpro are already circulating in humans, even without significant nirmatrelvir pressure. Remarkably, this result validated our ligand-based design protocol and signifies that several of our Mpro-designed mutants (having high dAffinity values) may materialize to be highly valuable in drug resistance and surveillance of newly evolving mutants and viral strains as SARS-CoV-2 encounters more immunogenicity, drug, and vaccine pressure, and as the high-throughput sequencing information keeps deposited.
Finally, the intermolecular interactions between nirmatrelvir and Mpro in the low-affinity and high-affinity designs were visualized and obtained. While the design with high affinity had 431 interactions, the design with lower affinity had only 407 interactions. The proximal and van der Waals interactions were decisive in influencing the affinity between nirmatrelvir and Mpro. The interaction diagrams between the high-affinity and low-affinity design demonstrated that losing a critical hydrogen bond between nirmatrelvir and Cys145 reduces the affinity and develops resistance and adaptability (Supplementary Figure 1).
4. Discussion
A recent phase 2–3 double-blind, randomized, controlled trial showed that the administration of Paxlovid to symptomatic COVID-19 patients resulted in an 89% lower risk of disease progression to severe conditions and quickly reduced viral load without evident safety issues [24,25]. However, the rapid and regular emergence of new SARS-CoV-2 variants highlights the fatal nature of the virus to mutate and illustrates that the COVID-19 pandemic will stay in the foreseeable future. Continuous surveillance and prediction of residues prone to mutations that can render therapies ineffective can emerge as a crucial repository for managing the spread of emerging mutants and controlling the pandemic. Currently, > 3 million SARS-CoV-2 genome sequences have been submitted to the GISAID website. The use of high-throughput technologies can considerably extend the insights gathered during drug development, improve drug efficacy and safety, predict the challenging multiple variants, and strengthen the countermeasures. Therefore, one may either wait for the new resistant and adaptable variants to develop and then battle it or anticipate it coming and expand the treatment arsenals in advance. With the currently available data and the emergence of new SARS-CoV-2 variants, the scientific community should be prepared to tackle the potential nirmatrelvir-resistant variants and intensify the development of additional interventions.
Nonetheless, we recognize certain limitations of the study, firstly, the requirement of the experimental and functional validation of the nirmatrelvir-resistant designs. Secondly, it is also reported that mutations distal from the catalytic or active site (e.g., HIV-1 protease) can also cause drug resistance, which is not addressed in this work. In conclusion, despite our recent results identified critical residues in the RNA-dependent RNA polymerase (RdRp) that could render SARS-CoV-2 resistant to remdesivir, molnupiravir, and favipiravir [26,27], a more systematic study to develop a unifying methodology is in progress to address some of these challenges. Nonetheless, our present study is significant with information on the potential structural and residue-specific sites in the Mpro that are susceptible to mutation under drug pressure and lead to viral adaptation with fitness advantages.
Author contributions
AKP carried out all the design experiments, data generation, and analysis. AKP and TT conceived the study, participated in its design and coordination, and drafted the manuscript. Both authors read and approved the final manuscript.
Data availability statement
Data will be made available upon reasonable request to the corresponding authors.
Declaration of competing interest
The authors declare no competing interests.
Acknowledgments
The authors sincerely acknowledge the infrastructure facilities of IIT (BHU) Varanasi and DST funded I-DAPT Hub Foundation, IIT (BHU) [DST/NMICPS/TIH11/IIT(BHU)2020/02]. Further, the support and the computing resources provided by PARAM Shivay Facility under the National Supercomputing Mission, Government of India at the IIT (BHU), Varanasi, are gratefully acknowledged.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.bbrc.2022.09.010.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.Harvey W.T., Carabelli A.M., Jackson B., Gupta R.K., Thomson E.C., Harrison E.M., Ludden C., Reeve R., Rambaut A., Peacock S.J., Robertson D.L. SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021;19:409–424. doi: 10.1038/s41579-021-00573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Garcia-Beltran W.F., Lam E.C., St Denis K., Nitido A.D., Garcia Z.H., Hauser B.M., Feldman J., Pavlovic M.N., Gregory D.J., Poznansky M.C., Sigal A., Schmidt A.G., Iafrate A.J., Naranbhai V., Balazs A.B. Multiple SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity. Cell. 2021;184:2372–2383. doi: 10.1016/j.cell.2021.03.013. e2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen J., Gao K., Wang R., Wei G.W. Prediction and mitigation of mutation threats to COVID-19 vaccines and antibody therapies. Chem. Sci. 2021;12:6929–6948. doi: 10.1039/d1sc01203g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Solomon M., Liang C. Human coronaviruses: the emergence of SARS-CoV-2 and management of COVID-19. Virus Res. 2022;319 doi: 10.1016/j.virusres.2022.198882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Owen D.R., Allerton C.M.N., Anderson A.S., Aschenbrenner L., Avery M., Berritt S., Boras B., Cardin R.D., Carlo A., Coffman K.J., Dantonio A., Di L., Eng H., Ferre R., Gajiwala K.S., Gibson S.A., Greasley S.E., Hurst B.L., Kadar E.P., Kalgutkar A.S., Lee J.C., Lee J., Liu W., Mason S.W., Noell S., Novak J.J., Obach R.S., Ogilvie K., Patel N.C., Pettersson M., Rai D.K., Reese M.R., Sammons M.F., Sathish J.G., Singh R.S.P., Steppan C.M., Stewart A.E., Tuttle J.B., Updyke L., Verhoest P.R., Wei L., Yang Q., Zhu Y. An oral SARS-CoV-2 M(pro) inhibitor clinical candidate for the treatment of COVID-19. Science. 2021;374:1586–1593. doi: 10.1126/science.abl4784. [DOI] [PubMed] [Google Scholar]
- 6.Greasley S.E., Noell S., Plotnikova O., Ferre R., Liu W., Bolanos B., Fennell K., Nicki J., Craig T., Zhu Y., Stewart A.E., Steppan C.M. Structural basis for Nirmatrelvir in vitro efficacy against the Omicron variant of SARS-CoV-2. bioRxiv. 2022 doi: 10.1101/2022.01.17.476556. 2022.2001.2017.476556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rai D.K., Yurgelonis I., McMonagle P., Rothan H.A., Hao L., Gribenko A., Titova E., Kreiswirth B., White K.M., Zhu Y., Anderson A.S., Cardin R.D. Nirmatrelvir, an orally active Mpro inhibitor, is a potent inhibitor of SARS-CoV-2 Variants of Concern. bioRxiv. 2022 doi: 10.1101/2022.01.17.476644. 2022.2001.2017.476644. [DOI] [Google Scholar]
- 8.Rosales R., McGovern B.L., Rodriguez M.L., Rai D.K., Cardin R.D., Anderson A.S., group P.S.P.s, Sordillo E.M., van Bakel H., Simon V., García-Sastre A., White K.M. Nirmatrelvir, Molnupiravir, and Remdesivir maintain potent <em>in vitro</em> activity against the SARS-CoV-2 Omicron variant. bioRxiv. 2022 doi: 10.1101/2022.01.17.476685. 2022.2001.2017.476685. [DOI] [Google Scholar]
- 9.Padhi A.K., Tripathi T. Can SARS-CoV-2 accumulate mutations in the S-protein to increase pathogenicity? ACS Pharmacol Transl Sci. 2020;3:1023–1026. doi: 10.1021/acsptsci.0c00113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Padhi A.K., Tripathi T. High-throughput design of symmetrical dimeric SARS-CoV-2 main protease: structural and physical insights into hotspots for adaptation and therapeutics. Phys. Chem. Chem. Phys. 2022;24:9141–9145. doi: 10.1039/d2cp00171c. [DOI] [PubMed] [Google Scholar]
- 11.Jochmans D., Liu C., Donckers K., Stoycheva A., Boland S., Stevens S.K., De Vita C., Vanmechelen B., Maes P., Trüeb B., Ebert N., Thiel V., De Jonghe S., Vangeel L., Bardiot D., Jekle A., Blatt L.M., Beigelman L., Symons J.A., Raboisson P., Chaltin P., Marchand A., Neyts J., Deval J., Vandyck K. The substitutions L50F, E166A and L167F in SARS-CoV-2 3CLpro are selected by a protease inhibitor <em>in vitro</em> and confer resistance to nirmatrelvir. bioRxiv. 2022 doi: 10.1101/2022.06.07.495116. 2022.2006.2007.495116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou Y., Gammeltoft K.A., Ryberg L.A., Pham L.V., Fahnøe U., Binderup A., Hernandez C.R.D., Offersgaard A., Fernandez-Antunez C., Peters G.H.J., Ramirez S., Bukh J., Gottwein J.M. Nirmatrelvir resistant SARS-CoV-2 variants with high fitness in vitro. bioRxiv. 2022 doi: 10.1101/2022.06.06.494921. 2022.2006.2006.494921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu Y., Lewandowski E.M., Tan H., Morgan R.T., Zhang X., Jacobs L.M.C., Butler S.G., Mongora M.V., Choy J., Chen Y., Wang J. Naturally occurring mutations of SARS-CoV-2 main protease confer drug resistance to nirmatrelvir. bioRxiv. 2022:2022. doi: 10.1101/2022.06.28.497978. 2006.2028.497978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang L., Volkow N.D., Davis P.B., Berger N.A., Kaelber D.C., Xu R. COVID-19 rebound after Paxlovid treatment during Omicron BA.5 vs BA.2.12.1 subvariant predominance period. medRxiv. 2022 doi: 10.1101/2022.08.04.22278450. 2022.2008.2004.22278450. [DOI] [Google Scholar]
- 15.Ou J., Lewandowski E.M., Hu Y., Lipinski A.A., Morgan R.T., Jacobs L.M.C., Zhang X., Bikowitz M.J., Langlais P., Tan H., Wang J., Chen Y., Choy J.S. A yeast-based system to study SARS-CoV-2 Mpro structure and to identify nirmatrelvir resistant mutations. bioRxiv. 2022:2022. doi: 10.1101/2022.08.06.503039. 2008.2006.503039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vilar S., Cozza G., Moro S. Medicinal chemistry and the molecular operating environment (MOE): application of QSAR and molecular docking to drug discovery. Curr. Top. Med. Chem. 2008;8:1555–1572. doi: 10.2174/156802608786786624. [DOI] [PubMed] [Google Scholar]
- 17.Padhi A.K., Tripathi T. A comprehensive protein design protocol to identify resistance mutations and signatures of adaptation in pathogens. Brief Funct Genomics. 2022 doi: 10.1093/bfgp/elac020. [DOI] [PubMed] [Google Scholar]
- 18.Hilgenfeld R. From SARS to MERS: crystallographic studies on coronaviral proteases enable antiviral drug design. FEBS J. 2014;281:4085–4096. doi: 10.1111/febs.12936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ramos-Guzmán C.A., Ruiz-Pernía J.J., Tuñón I. Unraveling the SARS-CoV-2 main protease mechanism using multiscale methods. ACS Catal. 2020;10:12544–12554. doi: 10.1021/acscatal.0c03420. [DOI] [PubMed] [Google Scholar]
- 20.Jubb H.C., Higueruelo A.P., Ochoa-Montaño B., Pitt W.R., Ascher D.B., Blundell T.L. Arpeggio: a web server for calculating and visualising interatomic interactions in protein structures. J. Mol. Biol. 2017;429:365–371. doi: 10.1016/j.jmb.2016.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Elbe S., Buckland-Merrett G. Data, disease and diplomacy: GISAID's innovative contribution to global health. Glob Chall. 2017;1:33–46. doi: 10.1002/gch2.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Singer J., Gifford R., Cotten M., Robertson D. CoV-GLUE: a web application for tracking sars-CoV-2 genomic variation. Preprints. 2020 doi: 10.20944/preprints202006.0225.v1. [DOI] [Google Scholar]
- 23.Iketani S., Mohri H., Culbertson B., Hong S.J., Duan Y., Luck M.I., Annavajhala M.K., Guo Y., Sheng Z., Uhlemann A.-C., Goff S.P., Sabo Y., Yang H., Chavez A., Ho D.D. Multiple pathways for SARS-CoV-2 resistance to nirmatrelvir. bioRxiv. 2022 doi: 10.1101/2022.08.07.499047. 2022.2008.2007.499047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hammond J., Leister-Tebbe H., Gardner A., Abreu P., Bao W., Wisemandle W., Baniecki M., Hendrick V.M., Damle B., Simón-Campos A., Pypstra R., Rusnak J.M. Oral nirmatrelvir for high-risk, nonhospitalized adults with covid-19. N. Engl. J. Med. 2022 doi: 10.1056/NEJMoa2118542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ullrich S., Ekanayake K.B., Otting G., Nitsche C. Main protease mutants of SARS-CoV-2 variants remain susceptible to nirmatrelvir. Bioorg. Med. Chem. Lett. 2022;62 doi: 10.1016/j.bmcl.2022.128629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Padhi A.K., Dandapat J., Saudagar P., Uversky V.N., Tripathi T. Interface-based design of the favipiravir-binding site in SARS-CoV-2 RNA-dependent RNA polymerase reveals mutations conferring resistance to chain termination. FEBS Lett. 2021;595:2366–2382. doi: 10.1002/1873-3468.14182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Padhi A.K., Shukla R., Saudagar P., Tripathi T. High-throughput rational design of the remdesivir binding site in the RdRp of SARS-CoV-2: implications for potential resistance. iScience. 2021;24 doi: 10.1016/j.isci.2020.101992. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available upon reasonable request to the corresponding authors.