Abstract
Intercellular transfer of plasmid DNA during bacterial conjugation initiates and terminates at a specific origin of transfer, oriT. We have investigated the oriT structure of conjugative plasmid R64 with regard to the initiation and termination of DNA transfer. Using recombinant plasmids containing two tandemly repeated R64 oriT sequences with or without mutations, the subregions required for initiation and termination were determined by examining conjugation-mediated deletion between the repeated oriTs. The oriT subregion required for initiation was found to be identical to the 44-bp oriT core sequence consisting of two units, the conserved nick region sequence and the 17-bp repeat A sequence, that are recognized by R64 relaxosome proteins NikB and NikA, respectively. In contrast, the nick region sequence and two sets of inverted repeat sequences within the 92-bp minimal oriT sequence were required for efficient termination. Mutant repeat A sequences lacking NikA-binding ability were found to be sufficient for termination, suggesting that the inverted repeat structures are involved in the termination process. A duplication of the DNA segment between the repeated oriTs was also found after mobilization of the plasmid carrying initiation-deficient but termination-proficient oriT and initiation-proficient but termination-deficient oriT, suggesting that the 3′ terminus of the transferred strand is elongated by rolling-circle-DNA synthesis.
Intercellular transfer of plasmid DNA during bacterial conjugation is accomplished by the function of transfer genes borne on each conjugative plasmid (for reviews, see references 4 and 15). All conjugative and mobilizable plasmids, such as R64, F, RP4, and R1162, contain oriT sites as cis elements which function as the origin of transfer of plasmid DNA. At the initiation stage of DNA transfer, a site- and strand-specific nick is introduced into the oriT site with a covalent attachment of the cognate relaxase protein to the 5′ terminus of the nicked strand. The nicked strand is transferred from donor to recipient cells with the 5′ terminus leading through a putative channel. In the donor cells, replacement strand DNA synthesis reconstitutes the double-stranded plasmid DNA. After one round of DNA transfer, the relaxase-attached 5′ terminus of the transferred-strand DNA is religated to its 3′ terminus to reconstitute the circular structure, and complementary-strand synthesis establishes double-stranded plasmid DNA in the recipient cells. Among these steps, the mechanisms by which initiation and termination of conjugative DNA transfer occur are important issues which remain to be elucidated.
Initiation and termination of conjugative DNA transfer at the oriT site were extensively studied using a small mobilizable plasmid, R1162 (RSF1010). R1162 oriT consists of a specific nick site and a 10-bp inverted repeat with one mismatch, which is situated 8 bp from the nick site (3, 27). Three R1162 proteins, MobA, MobB, and MobC, form a protein-DNA complex called the relaxosome at oriT (27). From the mobilization experiments with recombinant plasmids containing two tandemly repeated oriT sequences with or without mutations, the 10-bp inverted repeat structure as well as the sequence around the nick site was shown to be required for termination, while the nick site-distal arm of the inverted repeat was not required for initiation (1). From the analyses of mutations introduced into the 10-bp repeat, it has been postulated that during termination of DNA transfer, formation of a hairpin loop structure by the inverted repeat is required for the resealing of the transferred DNA by the R1162 MobA relaxase (32).
The initiation and termination of DNA transfer at oriT of the F plasmid, the fertility factor of Escherichia coli, were also studied. The F oriT sequence is located within an approximately 250-bp segment at one end of the transfer region (5, 10). Within the oriT sequence, the binding sites for F-encoded TraY and TraM and for integration host factor (IHF) are situated near the nick site that is recognized by F TraI protein. F TraI has both oriT-specific nicking activity and DNA helicase activity (17, 24). Although the 250-bp F oriT sequence is required for efficient conjugation, an approximately 100-bp sequence that contains the nick region sequence and IHF- and TraY-binding sites is required for efficient nicking in vivo by the TraI nickase activity (10). From the mobilization experiments with recombinant plasmids containing two tandemly repeated oriT sequences of various lengths, the 100-bp oriT sequence required for the efficient nicking was found to be essential for the initiation of DNA transfer (10). However, only a 36-bp F oriT sequence containing the nick site but missing all of the IHF-, TraY-, and TraM-binding sites was sufficient for termination (10).
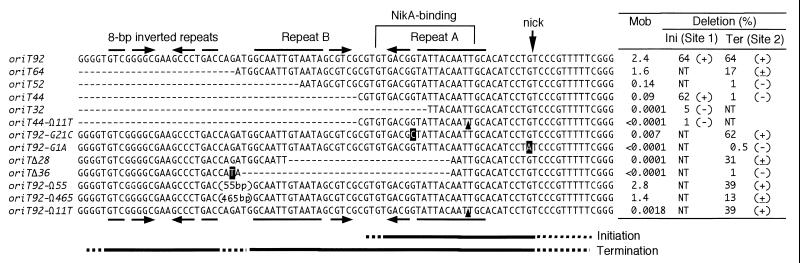
The IncI1 plasmid R64 carries a 54-kbp transfer region containing genes required for several steps of the conjugation process, such as the formation of two kinds of conjugative pili and processing of DNA during DNA transfer (12–14, 31). Located at one end of the R64 transfer region is an oriT operon consisting of an oriT site and two genes, nikA and nikB, encoding an oriT-binding protein and a putative relaxase, respectively (9). Deletion experiments have identified a 92-bp minimal oriT sequence (oriT92 [see Fig. 2]) which consists of a specific nick site, repeat A, repeat B, and 8-bp inverted repeat sequences and displays full oriT activity (8). The repeat A and repeat B sequences form a 17-bp inverted repeat with a 1-bp mismatch. The ATCCTG sequence from the 3′ end of the nick site is precisely conserved among the oriT sequences of various conjugative and mobilizable plasmids, such as RP4 (RK2), R751, and pTF-FC2, and the T-DNA border sequences of Ti and Ri plasmids (19, 29, 30). The specific relaxases, R64 NikB, RP4 and R751 TraI, and Ti VirD2, share three conserved amino acid sequence motifs that recognize the conserved nick region sequence (23). RP4 TraI and Ti VirD2 were actually shown to cleave and religate single-stranded oligonucleotides containing their respective nick region sequences (21–23), suggesting that the analogous R64 protein, NikB, also shares these activities.
FIG. 2.
Effects of various deletion, insertion, and substitution mutations on mobilization and on initiation or termination of DNA transfer. DNA segments deleted from the original sequence are indicated by dashes. Substitution mutations are indicated by white letters. One-base insertions of T in oriT44-Ω11T and oriT92-Ω11T are indicated by arrowheads. Insertions in oriT92-Ω55 and oriT92-Ω465 are indicated by “(55bp)” and “(465bp),” respectively. Mobilization frequencies (Mob) of plasmids carrying a single copy of the mutant oriT sequences are expressed as the ratio of transfer frequencies of the oriT plasmids to that of pKK607. Some data for mobilization frequencies were taken from reference 8. Initiation (Ini) or termination (Ter) activities of various oriT mutants are expressed as the ratio (percentage) of DNA of the plasmid that has been subjected to mobilization-mediated deletion to total mobilized plasmid DNA from the dual oriT plasmids carrying mutant oriT at site 1 (pKK542 to pKK544) or mutant oriT at site 2 (pKK545 to pKK554), respectively. The ratio was determined by measuring intensity of DNA bands in Fig. 3 using Southern hybridization with 32P-labeled pHSG398 DNA as a probe. NT, not tested. Inferred initiation or termination activities are indicated in parentheses (+, the efficiency is similar to that of oriT92; ±, weak activity; −, no activity). Solid lines at the bottom indicate the regions required for initiation and termination. Broken lines indicate the junctions bordering the essential sequences.
To analyze the relationship between the structure and function of the R64 oriT sequence, various deletion, insertion, and substitution mutants were constructed (see Fig. 2) (8). Removal of the 8-bp inverted repeats from the minimal oriT sequence resulted in a slight decrease in mobilization (oriT64 [see Fig. 2]). A 44-bp oriT core sequence containing the nick region and repeat A sequences (oriT44) exhibited a mobilization frequency 1/25 that of the minimal oriT (oriT92). The NikA protein was shown to specifically bind to the repeat A sequence but not to the repeat B sequence (see Fig. 2) (7). The NikA and NikB proteins form a relaxosome at the minimal or core oriT sequence. Upon sodium dodecyl sulfate or proteinase treatment of the relaxosome, a strand- and site-specific nick was introduced at the oriT nick site. The failure to form a functional relaxosome (oriT32, oriT92-G21C, and oriTΔ28) results in an incapability to mobilize. The NikA-binding sequence was required to be localized to a precise position relative to the nick site (oriT44-Ω11T).
To explain the differences in mobilization frequencies of the minimal oriT sequence and the oriT core sequence, we have previously predicted that the oriT core sequence is essential for the initiation of R64 DNA transfer and that the remaining sequence of the minimal oriT is involved in the termination (8). Here we present results which strongly support our previous prediction.
MATERIALS AND METHODS
Bacterial strains and plasmids.
E. coli K-12 strains NF83 and NF84 are streptomycin-resistant (Smr) and nalidixic acid-resistant (Nalr) derivatives of E. coli CL83 recA (16), respectively. E. coli K-12 strain TN102 is a nalidixic acid-resistant derivative of the wild-type strain W3110 (13). E. coli K-12 strain JC7623 recB21 recC22 sbcB15 thr thi leu his pro arg rpsL (18) was used for the construction of mini-R64 plasmid pKK610a. Vector plasmid pHSG398 (28) was used for the construction of dual oriT plasmids.
Medium.
Luria-Bertani broth was prepared as previously described (26). The solid medium contained 1.5% agar. Antibiotics were added to liquid or solid medium at the following concentrations: chloramphenicol, 25 μg/ml; kanamycin, 50 μg/ml; nalidixic acid, 25 μg/ml; and streptomycin, 200 μg/ml.
Recombinant DNA techniques.
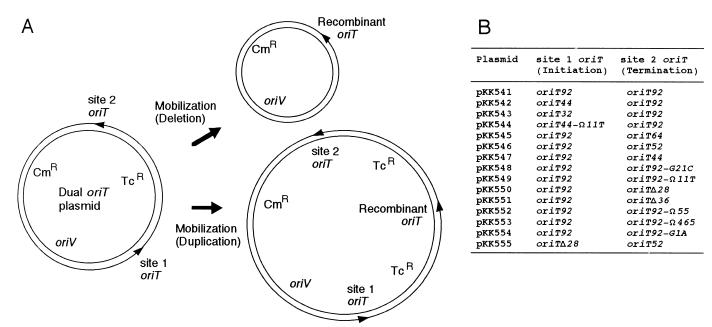
Recombinant DNA techniques and Southern blot hybridization were performed as previously described (26). Mini-R64 plasmid pKK610a, which contains the replication region and transfer region without the oriT core sequence of plasmid R64drd-11, as well as the kanamycin resistance (Kmr) gene, was constructed from pKK610 by the in vivo recombination method (13). Most of the mutant oriT sequences have been described previously (8). The oriT92-Ω55 and oriT92-Ω465 mutations were constructed by inserting 55- and 465-bp NdeI fragments, respectively, into an NdeI site generated between the 8- and 17-bp inverted repeat sequences by the PCR-mediated site-directed mutagenesis method (11). The inserted DNA fragments were obtained from the R64 traABCD region (nucleotide numbers 239 to 293 and 1358 to 1822 of the sequence under GenBank accession number AB027308 for the 55- and 465-bp fragments, respectively) (12). Dual oriT plasmids, pKK541 through pKK555 (see Fig. 1B), were constructed by inserting various mutant oriT sequences (see Fig. 2) and the tetracycline resistance (Tcr) gene cassette within the multicloning sites of pHSG398 as indicated in Fig. 1A.
FIG. 1.
(A) Schematic representation of a plasmid for the investigation of initiation and termination functions of the oriT sequence. The deletion of the Tcr segment occurs when the transfer is initiated and terminated at site 1 and site 2 oriTs, respectively, during mobilization. The duplication of the Tcr segment occurs when the transfer is initiated at site 2 oriT and terminated at site 1 oriT after passing through an entire round of transfer. The arrowheads indicate the 5′ ends of the nicked strand at oriT. Cmr, chloramphenicol resistance; oriV, origin of replication. (B) Structure of dual oriT plasmids. Mutant oriT sequences inserted into site 1 and site 2 of the dual oriT plasmids are described in Fig. 2.
Conjugal transfer.
To determine the mobilization frequency of plasmids with mutant oriTs, liquid mating was performed as described previously (12). Donor E. coli NF83 cells carrying both mini-R64 plasmid pKK607 and one of the mutant oriT plasmids were mated with recipient E. coli TN102 cells for 90 min at 37°C. The mobilization frequency was expressed as the ratio of the transfer frequency of the oriT plasmid to that of helper plasmid pKK607. For each mutant oriT plasmid, mobilization frequencies were determined from at least five independent experiments and their mean value was calculated.
To determine mobilization-mediated deletion and duplication, E. coli NF84 (Nalr) donor cells carrying both mini-R64 plasmid pKK610a and one of the dual oriT plasmids were mated with recipient E. coli NF83 (Smr) cells for 90 min at 37°C by the surface mating method (12). The mating mixture was directly inoculated into Luria-Bertani medium containing chloramphenicol and streptomycin for the selection of transconjugants and incubated overnight. DNAs were extracted from the transconjugants by the alkaline lysis method (26). After linearization at the unique EcoRI site, they were analyzed by 0.7% agarose gel electrophoresis. The DNA bands were visualized by staining with ethidium bromide. The relative amounts of DNA bands were measured by Southern blot analysis and probing with 32P-labeled pHSG398 DNA. The intensity of radiolabeled DNA bands was measured by the BAS-2000 bioimaging analyzer system (Fuji).
RESULTS
Experimental design.
To determine the oriT subregion(s) required for the initiation and termination of R64 DNA transfer, the dual oriT plasmid method originally developed by Bhattacharjee and collaborators was used (1). We have constructed plasmids carrying two oriT sequences in the same direction and a Tcr segment between the two oriTs (Fig. 1A). The dual oriT plasmids were mobilized from donor to recipient cells by mini-R64 plasmid pKK610a. Since pKK610a contains all the transfer genes except the oriT core sequence, it is able to mobilize the dual oriT plasmids but unable to transfer itself. If the initiation of DNA transfer occurs at site 1 oriT on the dual oriT plasmid and the termination occurs at site 2 oriT, the plasmids recovered from the transconjugants are predicted to lose the Tcr segment between the site 1 and site 2 oriTs (Fig. 1A). Such a deletion event is predicted to occur if site 1 and site 2 oriTs are initiation and termination proficient, respectively, whereas it may not occur if the site 2 oriT is termination deficient or the site 1 oriT is initiation deficient. Thus, it is possible to separately assess the initiation and termination abilities of mutant oriT sequences by examining the mobilization-mediated deletion of the dual oriT plasmids. Mutant oriT sequences used to construct dual oriT plasmids (Fig. 1B) are summarized in Fig. 2. Their mobilization frequencies when present once in each plasmid are also indicated in Fig. 2.
The R64 oriT segment required for the initiation of DNA transfer corresponds to the oriT core sequence.
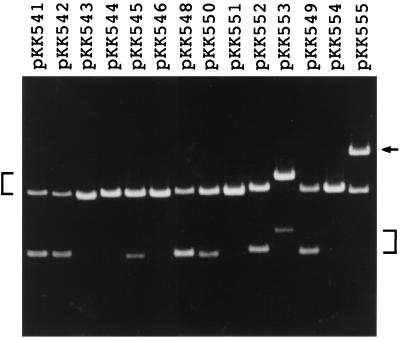
The minimal oriT sequence with full activity has been located within a 92-bp sequence of R64, as described in the introduction (oriT92 in Fig. 2). When pKK541 with dual oriT92 sequences was mobilized by pKK610a, transfer-mediated deletion was observed (Fig. 3). The fact that this deletion occurred precisely between the site 1 and site 2 oriTs was confirmed by restriction analysis and DNA sequencing. It was estimated that 64% of the mobilized plasmid carried the deletion (Fig. 2).
FIG. 3.
Mobilization-mediated recombination of dual oriT plasmids. Each plasmid was mobilized from donor cells by the mini-R64 plasmid pKK610a. Mobilized plasmids were recovered from the transconjugants, linearized at the unique EcoRI site, and subjected to agarose gel electrophoresis followed by staining with ethidium bromide. Plasmid bands of original size are indicated by a bracket on the left. Plasmid bands with the deletion or duplication of the Tcr segment are indicated by a bracket or an arrow, respectively, on the right.
The minimal R64 oriT sequence consists of four structural units: a nick region, repeat A, repeat B, and 8-bp inverted repeat sequences (8). To determine the oriT subregion(s) required for the initiation of DNA transfer, various lengths of oriT segments were introduced into site 1 with the oriT92 sequence retained at site 2, and then mobilization-mediated deletion was examined. When the oriT core sequence (oriT44) was used as a site 1 oriT, mobilization-mediated deletion was not affected (pKK542 in Fig. 3), indicating that the inverted repeat structures are not required for the initiation. However, when oriT32 and oriT44-Ω11T sequences without oriT activity were used as a site 1 oriT, mobilization-mediated deletion was not observed (pKK543 and pKK544 in Fig. 3). In pKK543 and pKK544, mobilization is thought to initiate and terminate at site 2 oriT. These results indicate that the R64 oriT segment required for the initiation of DNA transfer corresponds to the oriT core sequence.
The two inverted repeat structures within R64 oriT are required for efficient termination of DNA transfer.
To determine the oriT subregion(s) required for the termination of DNA transfer, various mutant oriT sequences were introduced into site 2 with the minimal oriT92 sequence retained at site 1, and then mobilization-mediated deletion was examined (Fig. 3). Deletion of the 8-bp inverted repeats (oriT64 in Fig. 2) resulted in a decrease in mobilization-mediated deletion (pKK545 in Fig. 3), indicating an involvement of the 8-bp inverted repeats in termination. A further decrease in mobilization-mediated deletion was observed for the oriT52 and oriT44 mutants, in which a portion and all of the repeat B sequence were removed, respectively (pKK546 in Fig. 3).
On the other hand, several oriT mutants without oriT activity displayed termination activity. The oriT92-G21C mutant without oriT activity was found to display normal termination activity (pKK548 in Fig. 3), suggesting that NikA binding to the repeat A sequence is not essential for termination. The oriT92-Ω11T mutation, which severely affects oriT activity but not NikA binding (8), also yielded efficient termination activity (pKK549 in Fig. 3), indicating that this mutation diminishes initiation activity but does not affect termination activity. Furthermore, the oriTΔ28 mutant, in which internal halves of the repeat A and B sequences were removed from the oriT92 sequence, was found to have less efficient but still significant termination activity (pKK550 in Fig. 3). In contrast, termination activity was severely reduced for oriTΔ36, in which the repeat B sequence was completely removed from oriTΔ28 (pKK551 in Fig. 3). These results indicate that both the 8- and 17-bp inverted repeat sequences are required for efficient termination of oriT-mediated DNA transfer, whereas NikA binding to repeat A is not essential for termination.
The 8-bp inverted repeats still function in termination when positioned at a more distant location.
The above-described results indicate that both the 8- and 17-bp inverted repeat structures are required for efficient termination. We next examined whether the distance between the two inverted repeats affects termination activity. The oriT92-Ω55 and oriT92-Ω465 mutants, containing 55- and 465-bp insertions between the two inverted repeats, respectively, were constructed (Fig. 2), and their termination activities were measured. pKK552, carrying oriT92-Ω55 at site 2, exhibited half the level of termination activity of the wild type (Fig. 3), indicating that the 8-bp inverted repeat is able to function at a distant location. On the other hand, pKK553, carrying oriT92-Ω465, exhibited the same level of termination activity as oriT64 lacking the 8-bp inverted repeat, indicating that the 8-bp repeat does not function beyond a certain distance. These results suggest that the 8-bp inverted repeat is functional in the termination of DNA transfer at a location separate from that of the 17-bp inverted repeat, but the effect gradually decreases with the length of distance between them.
The nick region sequence is required for the termination of DNA transfer.
Mutations introduced into the conserved nick region sequence resulted in severely decreased oriT activity (8). We examined whether this type of mutation affects the termination efficiency at site 2 oriT. The termination efficiency of pKK554 (with the oriT92-G1A mutation at site 2) was very low (Fig. 3), indicating the importance of the conserved nick region sequence in termination, as was expected from the established importance of the nick region sequence for recognition by relaxases (23).
Mobilization-mediated duplication of the Tcr segment.
It was found that mutant oriT sequences, such as oriT92Δ28 and oriT92-G21C, retain significant termination activities, although they do not display oriT activities due to the lack of a functional repeat A sequence. Such mutations are likely to be initiation deficient but termination proficient. When the initiation-deficient but termination-proficient oriT92Δ28 sequence and the initiation-proficient but termination-deficient oriT52 sequence are located at site 1 and site 2, respectively, transfer of the resultant plasmid, pKK555, is predicted to occur at oriT52 at site 2, proceed through the entire plasmid, pass site 2 oriT, and finally terminate at oriTΔ28 at site 1, resulting in the duplication of the Tcr segment (Fig. 1A, duplication). pKK555 was found to actually generate 62% of a plasmid larger than the unit length through mobilization (Fig. 3). The Tcr-duplicated structure of the large plasmid formed from pKK555 was confirmed by restriction analyses (data not shown). In addition, a low level (approximately 2%) of Tcr-duplicated plasmid was produced after mobilization of dual oriT plasmids carrying initiation-proficient oriT at site 2 and termination-proficient oriT at site 1, including pKK541, -545, -546, -547, -552, and -553 (data not shown). These results confirm the discrete functions of the R64 oriT sequence for the initiation and termination of DNA transfer.
DISCUSSION
In this study, we have analyzed the subregions of the R64 oriT sequence required for the initiation and termination of DNA transfer during conjugation. The initiation and termination activities of each mutant oriT sequence inferred from the present work are summarized in Fig. 2. From these results, we conclude that different portions within the R64 oriT sequence are required for the initiation and termination of DNA transfer as illustrated at the bottom of Fig. 2.
The R64 oriT core sequence (8) turned out to be identical to the region required for the initiation of DNA transfer. However, the remaining region appears not to be required for the initiation process. It is likely that the decreased oriT activity of the R64 oriT core sequence compared to that of the minimum oriT sequence is due to deficiency of the termination activity.
The R64 oriT region required for efficient termination of DNA transfer was found to consist of the following three sequences: (i) the nick region sequence, (ii) the 17-bp inverted repeat sequences, and (iii) the 8-bp GC-rich inverted repeat sequences. The nick region sequence is essential for the termination process of DNA transfer, since the oriT92-G1A mutation caused severe defects in termination. In RP4, the TraI relaxase was found to cleave and religate single-stranded DNA containing the nick region sequence (20). Therefore, the R64 nick region sequence may be required for rejoining the 5′ terminus of the transferred strand by R64 NikB relaxase to the 3′ terminus at the termination stage of R64 DNA transfer.
Although the importance of the repeat A sequence for NikA binding and subsequent oriT activity has been established (8), the inverted repeat structure itself within the 17-bp inverted repeat sequences is sufficient for the termination of DNA transfer, since the mutations preventing NikA binding to the repeat A sequence (e.g., oriT92-G21C) did not abolish termination activity. Significant termination activities were detected when a portion of the 17-bp inverted repeat structure was removed (oriT92Δ28) or when a one-base insertion was introduced between the nick region sequence and the 17-bp inverted repeat sequences (oriT92-Ω11T). Thus, NikA binding to a precise location of oriT is not necessary for termination, although it is essential for R64 relaxosome formation at the initiation stage (8). The role of the 17-bp inverted repeat sequences in the termination of R64 DNA transfer is similar to that of the 10-bp inverted repeat sequences within R1162 oriT. In the case of R1162 oriT, it is postulated that, at the termination step of R1162 DNA transfer, a hairpin loop structure formed by the 10-bp inverted repeats located 8 bp upstream from the nick site is directly recognized by the R1162 relaxase protein MobA (2, 32).
In addition to the 17-bp inverted repeat sequences, the 8-bp inverted repeat sequences were also required for efficient termination. They might help the termination ability of 17-bp repeat sequences in an enhancer-like function. The 8-bp inverted repeats were still functional when moved to an upstream position within a certain distance (oriT92-Ω55). However, movement to a further location (oriT92-Ω465) diminished the stimulation effect.
It is noteworthy that the R64 core oriT sequence carries residual termination activity although the minimal oriT sequence is required for efficient termination, since most of pKK535 carrying a single copy of the R64 core oriT sequence was monomeric after mobilization (data not shown). On the other hand, the minimal oriT sequence did not exhibit 100% termination activity, since 36% of pKK541 was still in the original form after mobilization. The reasons for these phenomena are unknown.
There is a global similarity in the oriT structure between R64 and IncP plasmids RP4 and R751 (6). Such a similarity also exists within the oriT sequences of IncI2 plasmid R721 (data not shown) and the mobilizable plasmid pTF-FC2 (25). In these plasmids, there are long (17- to 19-bp) inverted repeats 8 bp apart from the nick site. The GC-rich short (6- to 8-bp) inverted repeats are situated 6 to 54 bp upstream of the long ones. Although the sequences of the inverted repeats themselves are not similar, the nick region sequence and most of the relaxosome proteins are conserved in these plasmids (R64 NikA and NikB, RP4 TraJ and TraI, and pTF-FC2 MobB and MobA). The RP4 TraJ protein was shown to specifically bind to the nick site-proximal arm of the 19-bp inverted repeat sequence (33). These observations suggest that similar termination mechanisms of DNA transfer exist among IncI, IncP, and pTF-FC2 plasmids. In particular, the termination proficiency of the R64 oriT92-Ω55 mutant suggests that short GC-rich inverted repeats function in efficient termination of IncP and pTF-FC2 plasmids even at a separate location.
It is an interesting question whether or not the 3′ terminus of the transferred strand is elongated by DNA synthesis in the donor cells to produce a transfer intermediate with a length greater than the unit length (15). Our present results suggest that this may be the case during R64 DNA transfer, in which the 3′ terminus of the transferred strand is elongated by rolling-circle DNA synthesis, since a plasmid greater than unit length was produced after mobilization of pKK555. If the 3′ terminus of the nicked strand remained free until completion of one round of DNA transfer, continuation of DNA transfer beyond the initiation site could not occur. It is thus obvious that the 3′ terminus of the nicked strand can be elongated before one round of DNA transfer is completed. It is possible that the 3′ terminus could be used directly as a primer for conjugative DNA synthesis.
ACKNOWLEDGMENTS
We are grateful to K. Takayama for critical reading of the manuscript.
This work was supported in part by a grant from the Ministry of Education, Science, Sports and Culture of Japan.
REFERENCES
- 1.Bhattacharjee M, Rao X-M, Meyer R J. Role of the origin of transfer in termination of strand transfer during bacterial conjugation. J Bacteriol. 1992;174:6659–6665. doi: 10.1128/jb.174.20.6659-6665.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bhattacharjee M K, Meyer R J. Specific binding of MobA, a plasmid-encoded protein involved in the initiation and termination of conjugal DNA transfer, to single-stranded oriT DNA. Nucleic Acids Res. 1993;21:4563–4568. doi: 10.1093/nar/21.19.4563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brasch M A, Meyer R J. A 38 base-pair segment of DNA is required in cis for conjugative mobilization of broad host-range plasmid R1162. J Mol Biol. 1987;198:361–369. doi: 10.1016/0022-2836(87)90286-5. [DOI] [PubMed] [Google Scholar]
- 4.Clewell D B, editor. Bacterial conjugation. New York, N.Y: Plenum Press; 1993. [Google Scholar]
- 5.Fu Y-H F, Tsai M-M, Luo Y N, Deonier R C. Deletion analysis of the F plasmid oriT locus. J Bacteriol. 1991;173:1012–1020. doi: 10.1128/jb.173.3.1012-1020.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Furuya N, Komano T. Determination of the nick site at oriT of IncI1 plasmid R64: global similarity of the oriT structure of IncI1 and IncP plasmids. J Bacteriol. 1991;173:6612–6617. doi: 10.1128/jb.173.20.6612-6617.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Furuya N, Komano T. Specific binding of the NikA protein to one arm of 17-base-pair inverted repeat sequences within the oriT region of plasmid R64. J Bacteriol. 1995;177:46–51. doi: 10.1128/jb.177.1.46-51.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Furuya N, Komano T. Mutational analysis of the R64 oriT region: requirement for precise location of the NikA-binding sequence. J Bacteriol. 1997;179:7291–7297. doi: 10.1128/jb.179.23.7291-7297.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Furuya N, Nisioka T, Komano T. Nucleotide sequence and functions of the oriT operon in IncI1 plasmid R64. J Bacteriol. 1991;173:2231–2237. doi: 10.1128/jb.173.7.2231-2237.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gao Q, Luo Y, Deonier R C. Initiation and termination of DNA transfer at F plasmid oriT. Mol Microbiol. 1994;11:449–458. doi: 10.1111/j.1365-2958.1994.tb00326.x. [DOI] [PubMed] [Google Scholar]
- 11.Ito W, Ishiguro H, Kurosawa Y. A general method for introducing a series of mutations into cloned DNA using the polymerase chain reaction. Gene. 1991;102:67–70. doi: 10.1016/0378-1119(91)90539-n. [DOI] [PubMed] [Google Scholar]
- 12.Kim S-R, Funayama N, Komano T. Nucleotide sequence and characterization of the traABCD region of IncI1 plasmid R64. J Bacteriol. 1993;175:5035–5042. doi: 10.1128/jb.175.16.5035-5042.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Komano T, Funayama N, Kim S-R, Nisioka T. Transfer region of IncI1 plasmid R64 and role of shufflon in R64 transfer. J Bacteriol. 1990;172:2230–2235. doi: 10.1128/jb.172.5.2230-2235.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Komano T, Yoshida T, Narahara K, Furuya N. The transfer region of IncI1 plasmid R64: similarities between R64 tra and Legionella icm/dot genes. Mol Microbiol. 2000;35:1348–1359. doi: 10.1046/j.1365-2958.2000.01769.x. [DOI] [PubMed] [Google Scholar]
- 15.Lanka E, Wilkins B M. DNA processing reactions in bacterial conjugation. Annu Rev Biochem. 1995;64:141–169. doi: 10.1146/annurev.bi.64.070195.001041. [DOI] [PubMed] [Google Scholar]
- 16.Lerner C G, Inouye M. Low copy number plasmids for regulated low-level expression of cloned genes in Escherichia coli with blue/white insert screening capability. Nucleic Acids Res. 1990;18:4631. doi: 10.1093/nar/18.15.4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matson S W, Morton B S. Escherichia coli DNA helicase I catalyzes a site- and strand-specific nicking reaction at the F plasmid oriT. J Biol Chem. 1991;266:16232–16237. [PubMed] [Google Scholar]
- 18.Oishi M, Cosloy S D. The genetic and biochemical basis of the transformability of Escherichia coli K12. Biochem Biophys Res Commun. 1972;49:1568–1572. doi: 10.1016/0006-291x(72)90520-7. [DOI] [PubMed] [Google Scholar]
- 19.Pansegrau W, Lanka E. Common sequence motifs in DNA relaxases and nick regions from a variety of DNA transfer systems. Nucleic Acids Res. 1991;19:3455. doi: 10.1093/nar/19.12.3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pansegrau W, Lanka E. Mechanisms of initiation and termination reactions in conjugative DNA processing. Independence of tight substrate binding and catalytic activity of relaxase (TraI) of IncPα plasmid RP4. J Biol Chem. 1996;271:13068–13076. doi: 10.1074/jbc.271.22.13068. [DOI] [PubMed] [Google Scholar]
- 21.Pansegrau W, Schoumacher F, Hohn B, Lanka E. Site-specific cleavage and joining of single-stranded DNA by VirD2 protein of Agrobacterium tumefaciens Ti plasmids: analogy to bacterial conjugation. Proc Natl Acad Sci USA. 1993;90:11538–11542. doi: 10.1073/pnas.90.24.11538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pansegrau W, Schröder W, Lanka E. Relaxase (TraI) of IncPα plasmid RP4 catalyzes a site-specific cleaving-joining reaction of single-stranded DNA. Proc Natl Acad Sci USA. 1993;90:2925–2929. doi: 10.1073/pnas.90.7.2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pansegrau W, Schröder W, Lanka E. Concerted action of three distinct domains in the DNA cleaving-joining reaction catalyzed by relaxase (TraI) of conjugative plasmid RP4. J Biol Chem. 1994;269:2782–2789. [PubMed] [Google Scholar]
- 24.Reygers U, Wessel R, Müller H, Hoffmann-Berling H. Endonuclease activity of Escherichia coli DNA helicase I directed against the transfer origin of the F factor. EMBO J. 1991;10:2689–2694. doi: 10.1002/j.1460-2075.1991.tb07812.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rohrer J, Rawlings D E. Sequence analysis and characterization of the mobilization region of a broad-host-range plasmid, pTF-FC2, isolated from Thiobacillus ferrooxidans. J Bacteriol. 1992;174:6230–6237. doi: 10.1128/jb.174.19.6230-6237.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 27.Scherzinger E, Lurz R, Otto S, Dobrinski B. In vitro cleavage of double- and single-stranded DNA by plasmid RSF1010-encoded mobilization proteins. Nucleic Acids Res. 1992;20:41–48. doi: 10.1093/nar/20.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takeshita S, Sato M, Toba M, Masahashi W, Hashimoto-Gotoh T. High-copy-number and low-copy-number plasmid vectors for lacZα-complementation and chloramphenicol- or kanamycin-resistance selection. Gene. 1987;62:63–74. doi: 10.1016/0378-1119(87)90365-9. [DOI] [PubMed] [Google Scholar]
- 29.Waters V L, Guiney D G. Processes at the nick region link conjugation, T-DNA transfer and rolling circle replication. Mol Microbiol. 1993;9:1123–1130. doi: 10.1111/j.1365-2958.1993.tb01242.x. [DOI] [PubMed] [Google Scholar]
- 30.Waters V L, Hirata K H, Pansegrau W, Lanka E, Guiney D G. Sequence identity in the nick regions of IncP plasmid transfer origins and T-DNA borders of Agrobacterium Ti plasmids. Proc Natl Acad Sci USA. 1991;88:1456–1460. doi: 10.1073/pnas.88.4.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yoshida T, Kim S-R, Komano T. Twelve pil genes are required for biogenesis of the R64 thin pilus. J Bacteriol. 1999;181:2038–2043. doi: 10.1128/jb.181.7.2038-2043.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang S, Meyer R J. Localized denaturation of oriT DNA within relaxosomes of the broad-host-range plasmid R1162. Mol Microbiol. 1995;17:727–735. doi: 10.1111/j.1365-2958.1995.mmi_17040727.x. [DOI] [PubMed] [Google Scholar]
- 33.Ziegelin G, Fürste J P, Lanka E. TraJ protein of plasmid RP4 binds to a 19-base pair invert sequence repetition within the transfer origin. J Biol Chem. 1989;264:11989–11994. [PubMed] [Google Scholar]