Abstract
A chromosomal insertion of transposon Tn917 partially restores the expression of protease and alpha-toxin activities to PM466, a genetically defined agr-null derivative of the wild-type Staphylococcus aureus strain RN6390. In co-transduction experiments, transposon-encoded erythromycin resistance and a protease- and alpha-toxin-positive phenotype are transferred at high frequency from mutant strains to agr-null strains of S. aureus. Southern analysis of chromosomal DNA and sequence analysis of DNA flanking the Tn917 insertion site in mutant strains revealed that the transposon interrupted a 498-bp open reading frame (ORF). Similarity searches using a conceptual translation of the ORF identified a region of homology to the known staphylococcal global regulators AgrA and SarA. To verify that the mutant allele conferred the observed phenotype, a wild-type allele of the mutant gene was introduced into the genome of a mutant strain by homologous recombination. The resulting isolates had a restored agr-null phenotype. Virulence factor gene expression in mutant, restored mutant, and wild-type strains was quantified by measuring alpha-toxin activity in culture supernatant fluids and by Northern analysis of the alpha-toxin transcript. We named this ORF rot (for repressor of toxins) (GenBank accession no. AF189239) because of the activity associated with rot::Tn917 mutant strains.
In Staphylococcus aureus, the expression of many virulence factors is coordinately regulated. The genetics of this regulation is largely understood in terms of the function of agr and sar (30). Together, the components of these loci form part of a complex pathway that leads to decreased transcription of select cell surface virulence factor genes and increased transcription of regulated extracellular toxins and enzymes (see Fig. 1.) (6, 16, 17, 21, 31). Translational control of alpha-toxin by the agr-sar system has also been demonstrated (28, 32). In agr or sar mutant strains, regulated cell surface proteins (e.g., coagulase, fibronectin binding protein, and protein A) are produced throughout the exponential and postexponential phases of growth. This is in contrast to wild-type strains that only produce these proteins during exponential growth. Furthermore, in agr and sar mutant strains, many extracellular toxins and enzymes that are normally present in postexponential phase cultures (e.g., alpha-toxin, metalloprotease, and serine protease) are reduced to as low as 5% of their normal levels (17, 32).
FIG. 1.
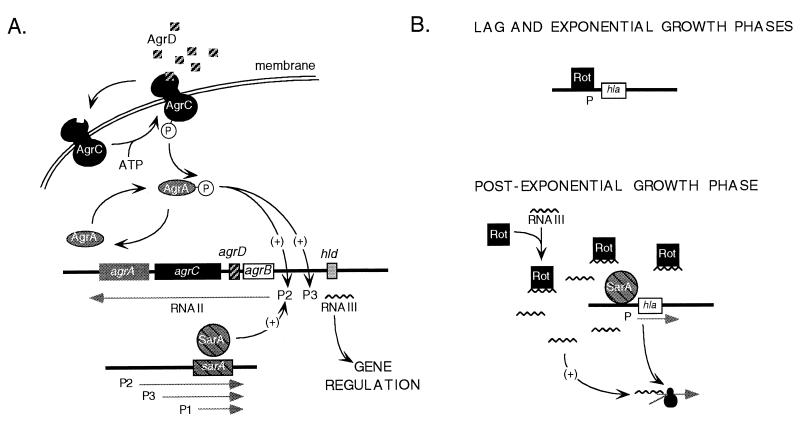
(A) Model of the function of the agr-sar system; (B) hypothetical model of the function of the rot gene product. Details can be found in the text. Chromosomal DNA is depicted as a thick black line. Promoters (P) are numbered. Genes (boxes) and their translation product are identically shaded. Phosphorylated proteins are associated with a circled letter P. The straight arrows and squiggly lines represent mRNA. Translation of hla mRNA is illustrated by the addition of a ribosome (black circles) to the message. The curved arrows show the relationships among the components of the system. Positive and negative effects are marked with (+) and (−), respectively.
The agr (accessory gene regulator) locus encodes a self-inducing, pheromone-sensing, signal transduction circuit (Fig. 1A). One of two divergent agr messages is transcribed from a promoter designated P2 (33). This message, RNAII, encodes four proteins, AgrA, AgrB, AgrC, and AgrD. Two of these agr-encoded proteins share sequence homology with elements of other bacterial two-component signal transduction systems: AgrC and AgrA function as sensor and regulator proteins, respectively. The activating signal of the agr system is a pheromone encoded within the prepeptide protein AgrD. AgrB is believed to be the enzyme responsible for the maturation and/or secretion of the 8-amino-acid peptide pheromone (18, 19).
The agr system up-regulates its function when AgrC binds the AgrD-derived signal (30). Like other bacterial sensor proteins, the binding of the signal results in the autophosphorylation of AgrC and, presumably, a concomitant activating conformational change (25). The phosphate group on AgrC is then thought to be transferred to the regulator protein AgrA, resulting in the activation of AgrA. Unlike other bacterial signal transduction systems in which the activated regulator protein directly initiates the transcription of target promoters, AgrA functions with the translation product of an unlinked locus named sar (staphylococcal accessory protein regulator) (7). The sar product, SarA, binds a region of DNA between the two agr promoters, and in conjunction with activated AgrA it up-regulates transcription of the agr messages (6, 16, 29). The result of the increased transcription is an amplification of the circuit encoded by RNAII and high-level production of a 510-ribonucleotide message known as RNAIII (17, 29, 32).
The current understanding is that RNAIII is involved in both repressing the transcription of cell surface protein genes and activating the transcription of extracellular protein genes (32, 33). In the case of alpha-toxin, a direct interaction between RNAIII and the alpha-toxin message is required for full translation (28, 29). When synthesized from a heterologous promoter in an agr-null mutant, RNAIII returns a wild-type pattern of virulence factor messages and translation products (28, 39; our unpublished data). Mutational analysis of RNAIII has shown that delta-toxin does not play a role in the regulation of virulence factor gene expression, suggesting that the message is the effector molecule of the agr-sar system (19, 32, 33).
An additional layer of complexity in this model is added by the observation that SarA is transcribed on three different overlapping messages known from largest to smallest as A, C, and B (3). These messages are initiated from distinct upstream promoters named P1, P3, and P2, respectively. Each message ends at a common terminator downstream of the SarA open reading frame. The P1 and P2 promoters are dependent on ςSA, the primary sigma factor in S. aureus, while the P3 promoter is dependent on ςSB, a multiple-stress-responsive sigma factor (23, 39). These data can be linked to the observation that virulence factor production is regulated by environmental conditions. For example, SarA message and levels of alpha-toxin are both elevated when the bacteria are exposed to oxidative stress (6). Additionally, SarA, its messages, or unidentified translation products encoded by the messages may play a direct role in virulence factor regulation. This hypothesis is based on the observation that decreases in transcription of the gene encoding cell protein A requires a different trans-encoded sar message in sar as compared to agr mutants of S. aureus (8).
Despite the progress made in understanding the agr-sar system, the best available evidence suggests that additional regulatory factors are required for virulence factor production (2, 11, 12, 39). One example of this difference is seen with alpha-toxin. This hemolysin is concomitantly transcribed, translated, and secreted 2 h after the appearance of RNAIII; however, RNAIII remains elevated while alpha-toxin production falls within 1 h of reaching peak production (39). In addition, unidentified regulatory molecules have also been invoked to explain the decrease in alpha-toxin message seen when S. aureus is treated with protein synthesis inhibitors (2). In the present study, we used transposon Tn917 mutagenesis to identify a gene that encodes a previously undescribed regulator of alpha-toxin.
MATERIALS AND METHODS
Bacterial strains, phage, plasmids, media, growth conditions, and virulence factor assays.
Bacteria, bacteriophage, and plasmids used in this study are described in Table 1. S. aureus was cultivated in tryptic soy broth (TSB) (Difco Laboratories, Detroit, Mich.) and incubated at 37°C with rotary agitation at 200 rpm or grown on tryptic soy agar plates (TSA). Escherichia coli was grown at 37°C in Luria-Bertani broth with agitation or on Luria-Bertani agar. Antibiotic-resistant staphylococci were selected and maintained in tetracycline (10 μg ml−1) or erythromycin or chloramphenicol (5 μg ml−1). Resistant E. coli were grown in media augmented with 100 μg of ampicillin ml−1. The method used for quantitative measurement of alpha-toxin has been previously described (15, 27). Assays for coagulase and protease have been described by Hart et al. (15).
TABLE 1.
Summary of bacterial strains, bacteriophage, and plasmids used in this study
| Strain, phage, or plasmid | Genotype, phenotype, description | Reference or source |
|---|---|---|
| E. coli DH5α | F− φ80lacZΔM15 Δ(lacZYA-argF)U196 endA1 recA1 hsdR17(rK− mK+) deoR thi-1 supE44 λ− gyrA96 relA1 | BRL |
| S. aureus | ||
| 8325-4 | Wild-type strain 8325 UV cured of phages φ11, φ12, and φ13 | National Culture Type Collection |
| PM466 | agr-null mutant of RN6390 | This study |
| PM614 | PM466 chr::Tn917::rot φ11 transductant | This study |
| PM615 | PM466 chr::Tn917::rot φ11 transductant | This study |
| PM616 | PM466 chr::Tn917::rot φ11 transductant | This study |
| PM702 | RN6390 chr::Tn917::rot φ11 transductant | This study |
| PM720 | PM614 with rot restored by allelic exchange | This study |
| RN4220 | 8325-4, nitrosoguanidine-induced restriction mutant used as primary recipient for plasmids propagated in E. coli | |
| RN6390 | 8325-4 | 32 |
| RN6911 | 8325-4 Δagr | 33 |
| Phage φ11 | S. aureus generalized transducing phage | |
| Plasmids | ||
| pBC SK | Cloning vector | Stratagene |
| pBluescript SK(+) | Cloning vector | |
| pCRII (+) | t-tail cloning vector | Invitrogen |
| pJM33 | pRN6650 with a 3.3-kb Agr-encoding ClaI-HpaI deletion | This study |
| pJM36 | pBluescript SK(+) with a 1.2-kb Tn551 transposase-encoding HindIII fragment | 27 |
| pJM37 | pBluescript KS with a 1.2-kb HindIII insert that encodes the transposase of Tn917 | This study |
| pJM42 | pT7::rnaiii; RNA III gene from RN6390 amplified using primers 5′-CACAGAGATGTGATGGAAAATAG and 5′-CATGACTAAACATAGATTTATGAG | This study |
| pJM48 | pSPT181(ts)::agr-null | This study |
| pJM202 | pSPT181(ts)::rot | This study |
| pJM531 | pCR-Script::hla; alpha-toxin gene from RN6390 amplified using primers 5′-GGAAGCTTAAACATCATTTCTGAAGTTATCGGC and 5′-GGGACTAGTGAAGGATGATGAAAATGAAAACACG | This study |
| pRN6650 | pUC18::agr; contains a 6.1-kb MboI Agr-encoding fragment | 35 |
| pSPT181(ts) | Temperature-sensitive S. aureus-E. coli shuttle vector | 17 |
| pT7Blue | t-tail vector | Novagen |
| pTV1 | E. coli-S. aureus shuttle vector with Tn917 | 41 |
DNA isolation.
Chromosomal DNA was isolated from S. aureus using the method of Dyer and Iandolo (13). Staphylococcal plasmid DNA was purified using a Qiagen (Chatsworth, Calif.) Plasmid Mini Kit. The plasmid isolation procedure was modified by incubating the cell suspension in P1 buffer containing 100 μg of recombinant lysostaphin ml−1 (AMBI UK, Trowbridge, United Kingdom) for 30 min at 37°C. The procedure was further modified by removal of the precipitate formed after the addition of neutralization buffer by centrifugation for 30 min. Routine procedures were used to isolate DNA from E. coli (1).
Recombinant techniques.
Plasmids were constructed and amplified in E. coli strain DH5α using standard recombinant DNA techniques (1). Restriction endonucleases, DNA modification enzymes, and polymerases were obtained from Promega (Madison, Wis.) and used as recommended by the manufacturer.
For staphylococcal transductions, bacteriophage φ11 lysates were obtained from infected strains grown in overlaid soft agar (TSB, 0.5 mM CaCl2, 0.5% agar) and sterilized by passage through 0.2-μm-pore-size filters, and titers were determined on S. aureus strain RN6390. Transductions consisted of 5 × 1011 CFU ml−1 of exponentially grown bacteria in TSB containing 0.5 mM CaCl2 and 5 × 1010 PFU ml−1 of bacteriophage in a total of 0.6 ml. After 5 min at room temperature, 1.5 ml of TSB containing 0.5 mM CaCl2 was added, and the tubes were incubated for 20 min at 37°C. Following the addition of 1 ml of 0.2 mM sodium citrate, the cells were harvested by centrifugation at 4 × 103 × g for 20 min, resuspended in 1 ml of 0.2 mM sodium citrate, and plated on TSA supplemented with 2 mM sodium citrate and the appropriate antibiotic. Transductional frequencies, when reported, were based on scoring of at least 65 colonies.
Transformations of S. aureus were conducted using the electrotransformation procedure of Kraemer and Iandolo (22). All plasmid DNA initially isolated from E. coli was introduced into S. aureus RN4220 prior to introduction to other strains of S. aureus. Allelic exchange in S. aureus utilized pSPT181(ts)-based plasmids with the conditions for plasmid integration and cointegrate resolution that have been described in detail by Janzon and Arvidson (17).
Construction of S. aureus strain PM466.
To create PM466, the agr locus was deleted from strain RN6390 by allelic exchange using plasmid pJM48. Plasmid pJM48 was constructed in multiple steps. Initially, a 3.3-kb ClaI-HpaI Agr-encoding fragment was removed from plasmid pRN6650, creating pJM33. The 2.8-kb EcoRI-HindIII fragment from pJM33 that contains agr-flanking DNA was then cloned into similar sites in pBC SK (Stratagene, La Jolla, Calif.). This fragment was removed from pBC SK by digestion with PstI and transferred into similar sites in the temperature-sensitive shuttle vector pSPT181(ts), creating pJM48.
Transposon Tn917 mutagenesis and phenotypic screens.
Strain PM466 was subjected to mutagenesis with transposon Tn917 carried on plasmid pTV1 (41). To overcome the low transformation efficiency of S. aureus, a colony of PM466 harboring pTV1 was grown at 32°C, the permissive temperature, on TSA containing chloramphenicol, to create a pool of bacteria with pTV1. Mutant bacteria were selected at 42°C and screened for protease activity on nutrient agar (38) with 5% skim milk (Difco Laboratories) and hemolytic activity on blood agar base (Difco Laboratories) supplemented with 5% rabbit blood.
Southern and Northern hybridization.
Digested staphylococcal chromosomal DNA was subjected to electrophoresis through 0.7% agarose gels, transferred to nylon membranes (MagnaGraph; Micron Separators Inc., Westborough, Mass.), and probed using the Genius system (Boehringer Mannheim, Indianapolis, Ind.) as instructed by the manufacturer. Hybridizations used a randomly primed digoxigenin-labeled 6.1-kb BamHI fragment from pRN6650 that contains agr plus flanking DNA or a 1.2-kb HindIII probe from pIM36 that encodes the transposase of transposon Tn551, standard buffer plus 50% formamide for prehybridization and hybridization, and stringent washes performed at 68°C (27). Detection used the chemiluminescent substrate disodium 3-(4-methoxyspiro{1,2-dioxetane-3,2′-(5′-chloro)tricyclo[3.3.2.237]decan}-4-yl) phenyl phosphate (CSPD) (Boehringer Mannheim).
Total cellular RNA was isolated from 10-h cultures of S. aureus by the method of Hart et al. (15) and purified using RNAeasy (Qiagen). Electrophoresis of RNA was conducted in 1% LE agarose (FMC Bioproducts, Rockland, Maine) glyoxal gels. The RNA was transferred to a nylon membrane (MagnaGraph) and probed with a ClaI-XbaI fragment from pIM42 that encodes part of RNAIII or a SpeI-HindIII fragment from plasmid pJM531 that encodes alpha-toxin. The probes were digoxigenin labeled and hybridized using high-concentration sodium dodecyl sulfate buffer at 50°C. Stringent washes were performed at 65°C, and detection was carried out with CSPD. Levels of message were compared using Multi-Analyst Version 1.02 software (Bio-Rad Laboratories, Hercules, Calif.).
Inverse PCR and nucleotide sequencing.
Inverse PCRs contained chromosomal DNA from strains PM614, PM615, or PM616 digested with either EcoRI or PstI and self-ligated at a concentration of 5 ng of DNA μl−1. The Tn917-specific outward facing primers were 5′-GAGCATATCCACTTTTCTTGGAG-3′ and 5′-CACAATAGAGAGATGTCACGTC-3′ (GenBank accession no. M11180). DNA was amplified by the method of Coen (10). The nucleotide sequence for rot was obtained using an Applied Biosystems 373A or 377 DNA Sequencer with dye terminator cycle sequencing chemistry (Perkin-Elmer, Foster City, Calif.) on Qiagen purified DNA. Template DNA consisted of a pool of three independently amplified PCR products. Sequencing primers were designed to extend the newly acquired sequence. Additional S. aureus sequence data were obtained from The Institute for Genomic Research (website at http://www.tigr.org). Data were analyzed using the Wisconsin Genetics Computer Group sequence analysis software package Version 9.1.
Construction of S. aureus strain PM720.
PM720 was created by allelic exchange using S. aureus PM614 and plasmid pJM202. Plasmid pJM202 is plasmid pSPT181(ts) with a 1.3-kb PCR fragment generated from the wild-type S. aureus strain RN6390 using primers that correspond to sequence upstream and downstream of rot, 5′-CAAAGCCTGACACGACAATCC-3′ and 5′-CTGAAAGATGAGACAGTAGATG-3′, respectively. To construct pJM202, the rot-containing PCR fragment was cloned into plasmid pCRII (+) (Invitrogen, Carlsbad, Calif.) and verified by restriction endonuclease and sequence analysis. The rot fragment in pCRII was removed by digestion with EcoRI and moved into a similar site within the multiple-cloning site of pSPT181(ts).
RESULTS
Construction of S. aureus strain PM466.
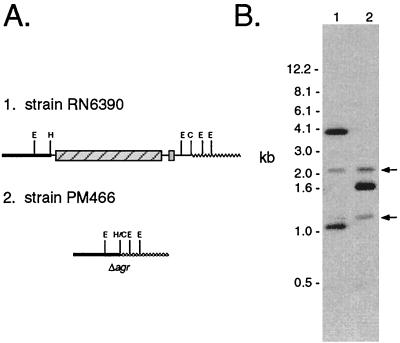
Strain PM466 is a new agr-null derivative of S. aureus RN6390 created by allelic exchange using plasmid pJM48. The deletion in PM466 encompasses the entire agr P2 operon and the first 379-bp of the P3 transcript. The expected chromosomal deletion was confirmed in strain PM466 by Southern analysis (Fig. 2). Measurements of virulence factor activity demonstrated that post-exponential-phase culture supernatant fluids from PM466 had less than 5% of the protease and alpha-toxin activities associated with RN6390. Coagulase activity was approximately 10-fold higher in PM466 than in the wild-type control. RNAIII in PM466 could not be detected by Northern analysis (data not shown).
FIG. 2.
(A) Restriction map of the wild-type allele of agr in RN6390 (top) and the agr-null allele (Δagr) in PM466 (bottom). The agr P2 operon and P3 transcript are represented by the striped and shaded boxes, respectively. Chromosomal DNA on the 3′ end of the P2 operon and 3′ end of the P3 transcript are represented by black and gray lines, respectively. Abbreviations: C, ClaI; E, EcoRI; H, HincII; H/C, ligation of a HincII site with a T4 polymerase blunt-ended ClaI site. (B) Chromosomal DNA from RN6390 (lane 1) and PM466 (lane 2) digested with EcoRI and analyzed by Southern hybridization using agr and flanking single-stranded DNA as the probe. DNA hybridizing to agr-flanking regions is marked with arrows.
Transposon Tn917 mutagenesis and transductional analysis.
Strain PM466 was subjected to mutagenesis with transposon Tn917. Approximately 2 × 104 bacteria with chromosomal insertions of the transposon were screened for proteolytic activity on skim milk agar, both with and without erythromycin. Eleven protease-positive strains were isolated. To rule out mutations in genes that only activate protease expression, the erythromycin-resistant, protease-positive strains were screened for hemolytic activity on rabbit blood agar plates. Nine of the original eleven isolates had alpha-toxin activity. The loss of plasmid pTV1 from these nine strains was confirmed by testing for vector-encoded antibiotic resistance on TSA supplemented with the MIC of chloramphenicol. The lack of growth of the nine strains in this medium suggested that the erythromycin resistance was mediated by a chromosomal insertion of the transposon.
To confirm the linkage between the transposon and the genetic lesion causing the restored phenotype, DNA surrounding the transposon from the presumptive mutant strains was back-transferred into the agr-null strains PM466 and RN6911 by transduction using bacteriophage φ11 in independent experiments. In independent experiments, the protease- and alpha-toxin-positive phenotype was shown to cotransfer with transposon-encoded erythromycin resistance in four of the nine isolates. No differences in phenotype were observed between mutations in the two agr-null genetic-backgrounds. Transduction of the erythromycin resistance marker into PM466 resulted in the isolation of strains PM614, PM615, and PM616. In these experiments, more than 98% of the transductants had a protease- and alpha-toxin-positive phenotype. In the remaining strains, genetic linkage could not be verified.
Southern analysis of chromosomal DNA from PM614, PM615, and PM616 using a Tn917-derived probe suggested that a single gene conferred the restored extracellular protein phenotype. Single digests of the chromosomal DNA using four different restriction endonucleases that do not cut within Tn917 resulted in an identical pattern of hybridizing DNA fragments (data not shown). These data suggest that the chromosomal insertion of the transposon in the three strains occurred within the same gene.
DNA surrounding the insertion site of the transposon from strains PM614, PM615, and PM616 was amplified by inverse PCR, and the nucleotide sequence of approximately 2 kb of DNA flanking the transposon insertion site was determined. The size of the inverse PCR products was consistent with values predicted from Southern analysis of the protease- and alpha-toxin-positive transductants (data not shown). With each of these strains, the probe-hybridizing EcoRI fragment was 9 kb and the inverse PCR product, minus Tn917 DNA, was approximately 4 kb. Furthermore, in each of these strains, we found the nucleotide duplication that occurs upon the transposition of Tn917.
Nucleotide sequence analysis of the inverse PCR products indicated that the transposon insertion site in PM614 and PM616 was identical. In strain PM615, the transposon had inserted into a different site within the same gene. The open reading frame for the interrupted gene is 498 bp in length (GenBank accession no. AF189239). The predicted protein begins at an ATG translational start and terminates after 161 amino acid residues at a TAA stop. Alternatively, protein initiation from a downstream ATG start would result in a 141-amino-acid residue protein.
A BLASTP search using a conceptional translation of the predicted 161-amino-acid protein identified hypothetical proteins (GenBank accession no. U89914 and Swiss-protein accession no. P54182) and a region of homology to S. aureus AgrA and SarA as well as Staphylococcus epidermidis AgrA (Fig. 3). The transposon-inactivated gene was named rot (repressor of toxins) because loss of a wild-type allele results in the restoration of protease and alpha-toxin activities to S. aureus PM466 and to reflect the fact that it has homology to known transcriptional regulators.
FIG. 3.
BLASTP alignment of the rot gene product (Rot) and the S. aureus regulatory proteins AgrA (AgrA e value = 1.9; SarA e value = 5.9) and S. epidermidis AgrA (e value = 0.49). Rot was used as the query sequence against the nonredundant database at the National Center for Biotechnology Information. Identities are shown, + denotes similarity, and numbers at right refer to amino acids in the respective proteins.
Verification and initial characterization of the rot mutation.
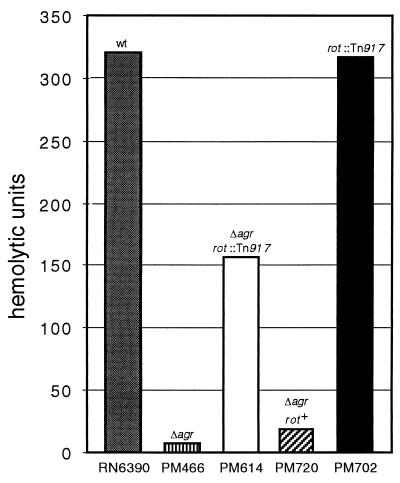
As viewed on indicator plates, inactivation of rot restores a post-exponential-phase protease- and alpha-toxin-positive phenotype to the agr-null strain of S. aureus PM466. To quantify the effect of the rot mutation on virulence factor production, we compared alpha-toxin activity in culture supernatant fluids from strains RN6390, PM466, PM614, and PM720 (Fig. 4). PM466, the agr-null strain, has approximately 4% of the activity associated with RN6390, its wild-type parental strain. Compared to the activity seen with PM466, the rot mutation in PM614 results in a 40-fold increase in alpha-toxin activity. This level is approximately half that associated with a wild-type strain. Similar results were seen with PM615 (data not shown).
FIG. 4.
Quantitative measurements of alpha-toxin activity in supernatant fluids from post-exponential-phase (10-h) cultures of S. aureus strains RN6390 (wild-type, gray bar), PM466 (vertically striped bar), PM614 (white bar), PM720 (diagonally striped bar), and PM702 (black bar). Relevant genotypes are shown (wt, wild-type; Δagr, agr-null allele, rot+, wild-type rot allele; rot::Tn917, insertionally inactivated rot).
Hemolytic activity in PM614 can be restored to PM466 levels by replacement of the chromosomal insertion of Tn917 with a wild-type copy of rot. PM614 was subjected to allelic exchange using plasmid pJM202. Unlike pSPT181(ts) alone, allele replacement using pJM202 resulted in the isolation of colonies that lacked protease and alpha-toxin on indicator plates. The genome of one resulting strain, PM720, was examined by Southern analysis. This strain both lacked DNA that hybridized with the transposase-encoding insert from plasmid pIM36 and had the expected 4-kb EcoRI rot-hybridizing fragment (data not shown). Measurement of hemolytic activity in culture supernatant fluids from PM720 revealed that the 1.3-kb rot-encoding fragment in pJM202 is sufficient to return PM466-like levels of activity to PM614 (Fig. 4).
Transposon-encoded erythromycin resistance was transduced from PM614 into the wild-type strain of S. aureus, RN6390. In the resulting transductants, no difference in protease activity could be visualized on indicator plates. One transductant, strain PM702 was shown by Southern analysis to have the expected rot mutation (data not shown). In this strain, alpha-toxin activity was similar to that seen in RN6390 (Fig. 4).
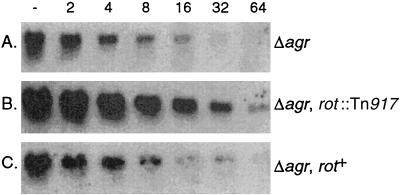
Post-exponential-phase alpha-toxin message from strains PM466, PM614, and PM720 was quantified by Northern analysis (Fig. 5). Consistent with our activity data, the alpha-toxin transcript in PM614 (rot agr double mutant) was elevated sixfold compared to the message found in the agr-null parental strain. Furthermore, in strain PM720 (PM614 with a wild-type copy of rot) the alpha-toxin message is returned to PM466 levels.
FIG. 5.
Northern analysis of the alpha-toxin message in RNA from post-exponential-phase cultures of S. aureus strains (A) PM466, (B) PM614, and (C) PM720. The leftmost lane contains 30 μg of total RNA serially diluted (1:2) in subsequent lanes. The transcript was identified by hybridization using digoxigenin-labeled probe specific for the gene encoding alpha-toxin with chemiluminescent chemistry.
DISCUSSION
We have identified a locus in S. aureus that encodes a regulator of virulence factors. This locus was named rot because our data suggests that the predicted gene product acts as a repressor of toxins. In an agr-null background, a mutation in rot increases the expression of protease and alpha-toxin. In addition, we showed increased transcription of the gene encoding alpha-toxin in an agr rot double mutant strain.
Based upon the fact that protein synthesis inhibitors can down-regulate transcription of virulence factor genes, mimicking RNAIII mutations, Balaban and Novick postulated that intermediary factors are required for transcription of virulence factor genes (2). Negative regulators of virulence factor genes constitute one possible class of these molecules. To identify loci that encode repressors, we subjected strain PM466 to mutagenesis with transposon Tn917 and screened for the restoration of two extracellular virulence factor activities.
S. aureus PM466, a genetically defined agr-null strain, was constructed for this study. PM466 is a derivative of RN6390, the wild-type strain used to define the molecular genetics of agr and sar (6, 34). In contrast to S. aureus RN6911, the published RN6390-derived agr-null mutant strain, PM466 has a specific deletion rather than an antibiotic marker and an accompanying deletion of unknown extent (33). Despite this genetic difference, quantitative measurements of cell surface and extracellular proteins suggest that the two agr-null strains have a common phenotype (data not shown). This observation supports previous findings in which the changes seen in RN6911 were interpreted as being solely due to the inactivation of agr.
Initially, we screened transposon Tn917 mutants for restored protease activity. Although the protein or proteins responsible for the zone of proteolysis surrounding single colonies of bacteria on skim milk agar have not been definitively identified, this activity has been shown to be RNAIII dependent (9). Wild-type strains produce clear zones of proteolysis on indicator plates, while the agr-null strains RN6911 and PM466 lack this activity (our unpublished data). To rule out mutations in genes that only up-regulate protease expression in the agr genetic background, the protease-positive strains were screened for hemolytic activity. S. aureus produces four different hemolysins (alpha-, beta-, delta- and gamma-toxins); however, rabbit erythrocytes suspended in agar are only susceptible to the action of alpha- and delta-toxins (14). Since the agr deletion in PM466 encompasses the gene encoding delta-toxin, the hemolytic activity associated with mutants created in the PM466 background is due to alpha-toxin (34). Despite the fact that RNAIII has been reported to be required for alpha-toxin translation, several of the proteolytic mutants displayed a hemolytic phenotype (28). Cotransductional analysis of the proteolytic- and alpha-toxin-positive mutants was used to verify genetic linkage between the extracellular protein phenotype and the erythromycin resistance encoded by the transposon. Finally, the phenotype associated with the rot allele in the agr-null strains was confirmed by demonstrating that wild-type rot is sufficient to restore an Agr− phenotype to PM614.
The rot locus has limited amino acid sequence homology with the known staphylococcal transcriptional regulators AgrA and SarA. The lack of structure and function data for AgrA and SarA precludes assigning a specific biological significance to the homologous region. Genetic experiments have suggested that AgrA is an activator of agr; however, AgrA has not been shown to bind DNA within the agr promoter region (29). Recent evidence suggests that SarA is a DNA-binding protein that activates both agr and individual virulence factor genes (4, 6). While it is not possible to exclude the hypothesis that the rot gene product is a component of a protein complex or the activator of a repressor, the limited homology among the regulatory genes and the phenotype conferred by the rot::Tn917 mutation suggests that the rot-encoded protein is a transcriptional repressor. In addition to those encoded by sar and agr, two uncharacterized proteins were identified by searches that used the predicted rot translation product. We have cloned the genes for these proteins and have produced knockout mutants in S. aureus.
Quantitative measurements of alpha-toxin activity and Northern analysis of the corresponding message were used to verify rot and, in part, to define its activity. Measurements of alpha-toxin activity indicated that restoration of rot in PM614 completely represses alpha-toxin to agr levels. Moreover, rot mutations were found to only partially restore alpha-toxin activity to agr-null strains. This observation may be explained by the translational effect of RNAIII on the alpha-toxin message, although the mirroring of alpha-toxin activity and message in PM614 suggests that regulation occurs at the level of transcription. Therefore, it is possible that the rot-encoded protein may up-regulate an activator that is necessary for full alpha-toxin expression.
Collectively, our data suggest that rot encodes a repressor of extracellular virulence factor transcription. We are testing a direct model (Fig. 1B) that predicts that the rot gene product (Rot) binds within the promoter region of regulated genes during the lag and exponential phase of bacterial growth, blocking their transcription. Transcription of Rot-regulated promoters occurs when levels of the bound repressor are decreased, thus exposing the promoter and allowing the binding of transcriptional activators and RNA polymerase. This model is analogous to the H-NS–DsrA-RNA pathway of E. coli (37). In the E. coli system, DrsA-RNA is part of a complex that binds the histone-like protein (H-NS), thus relieving DNA secondary structure that inhibits the transcription of regulated genes (24). A competing hypothesis is that rot and agr encode components of independent, yet partially redundant, pathways. Under this scenario, the rot translation product may act as either a repressor or an activator of factors necessary for virulence factor synthesis. In either case, the rot-associated activity appears to be altered by an agr product or factors that are regulated by agr, because the rot mutation does not alter alpha-toxin expression found in culture supernatant fluids from stationary cultures of wild-type strains.
ACKNOWLEDGMENTS
We thank Rae Ellen Syverson and Donna Bates for their helpful discussions and critical reading of this manuscript. This work was supported by the American Heart Association Beginning Grant-in-Aid (9960328Z) to P.J.M. and U.S. Public Health Service grant R01 AI42072-02 from the National Institute of Allergy and Infectious Diseases to R.A.P.
REFERENCES
- 1.Ausubel F A, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular Biology. New York, N.Y: Greene Publishing and Wiley-Interscience; 1996. [Google Scholar]
- 2.Balaban N, Novick R P. Translation of RNAIII, the Staphylococcus aureus agr regulatory RNA molecule, can be activated by a 3′-end deletion. FEMS Microbiol Lett. 1995;133:155–161. doi: 10.1111/j.1574-6968.1995.tb07877.x. [DOI] [PubMed] [Google Scholar]
- 3.Bayer M G, Heinrichs J H, Cheung A L. The molecular architecture of the sar locus in Staphylococcus aureus. J Bacteriol. 1996;178:4563–4570. doi: 10.1128/jb.178.15.4563-4570.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chan P F, Foster S J. The Staphylococcus aureus alternative sigma factor ςB controls the environmental stress response but not starvation survival or pathogenicity in a mouse abscess model. J Bacteriol. 1998;180:6232–6241. doi: 10.1128/jb.180.23.6082-6089.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheung A L, Koomey J M, Butler C A, Projan S J, Fischetti V A. Regulation of exoprotein expression in Staphylococcus aureus by a locus (sar) distinct from agr. Proc Natl Acad Sci USA. 1992;89:6462–6466. doi: 10.1073/pnas.89.14.6462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheung A L, Projan S J. Cloning and sequencing of sarA of Staphylococcus aureus, a gene required for the expression of agr. J Bacteriol. 1994;176:4168–4172. doi: 10.1128/jb.176.13.4168-4172.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheung A L, Woltz C, Yeaman M R, Bayer A S. Insertional inactivation of a chromosomal locus that modulates expression of potential virulence determinants in Staphylococcus aureus. J Bacteriol. 1995;177:3220–3226. doi: 10.1128/jb.177.11.3220-3226.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheung A L, Eberhardt K, Heinrichs J H. Regulation of protein A synthesis by the sar and agr loci of Staphylococcus aureus. Infect Immun. 1997;65:2243–2249. doi: 10.1128/iai.65.6.2243-2249.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chien Y, Manna A C, Projan S J, Cheung A L. SarA, a global regulator of virulence determinants in Staphylococcus aureus, binds to a conserved motif essential for sar-dependent gene regulation. J Biol Chem. 1999;24:37169–37176. doi: 10.1074/jbc.274.52.37169. [DOI] [PubMed] [Google Scholar]
- 10.Coen D. Enzymatic amplification of DNA by PCR: standard procedures and optimization. In: Ausubel F A, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Greene Publishing and Wiley-Interscience; 1992. pp. 15.1.1–15.1.7. [Google Scholar]
- 11.Compagnone-Post P, Malyankar U, Khan S A. Role of host factors in the regulation of the enterotoxin B gene. J Bacteriol. 1991;173:1827–1830. doi: 10.1128/jb.173.5.1827-1830.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dassy B, Hogan T, Foster T J, Fournier M. Involvement of the accessory gene regulator (agr) in expression of type 5 capsular polysaccharide by Staphylococcus aureus. J Gen Microbiol. 1993;139:1301–1306. doi: 10.1099/00221287-139-6-1301. [DOI] [PubMed] [Google Scholar]
- 13.Dyer D W, Iandolo J J. Rapid isolation of DNA from Staphylococcus aureus. Appl Environ Microbiol. 1983;46:283–285. doi: 10.1128/aem.46.1.283-285.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elek S, Levy E. Distribution of haemolysin in pathogenic and non-pathogenic staphylococci. J Pathol Bacteriol. 1950;62:541–554. doi: 10.1002/path.1700620405. [DOI] [PubMed] [Google Scholar]
- 15.Hart M E, Smeltzer M S, Iandolo J J. Phenotypic characterization of xpr, a global regulator of extracellular virulence factors in Staphylococcus aureus. Infect Immun. 1993;61:919–925. doi: 10.1128/iai.61.3.919-925.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heinrichs J H, Bayer M G, Cheung A L. Characterization of the sar locus and its interaction with agr in Staphylococcus aureus. J Bacteriol. 1996;178:418–423. doi: 10.1128/jb.178.2.418-423.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Janzon L, Arvidson S. The role of the delta-lysin gene (hld) in the regulation of virulence genes by the accessory gene regulator (agr) in Staphylococcus aureus. EMBO J. 1990;9:1391–1399. doi: 10.1002/j.1460-2075.1990.tb08254.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ji G, Beavis R C, Novick R P. Cell density control of staphylococcal virulence mediated by an octapeptide pheromone. Proc Natl Acad Sci USA. 1995;92:12055–12059. doi: 10.1073/pnas.92.26.12055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ji G, Beavis R C, Novick R P. Bacterial interference caused by autoinducing peptide variants. Science. 1997;276:2027–2030. doi: 10.1126/science.276.5321.2027. [DOI] [PubMed] [Google Scholar]
- 20.Jones R N. Impact of changing pathogens and antimicrobial susceptibility patterns in the treatment of serious infections in hospitalized patients. Am J Med. 1996;24:3S–12S. doi: 10.1016/s0002-9343(96)00102-7. [DOI] [PubMed] [Google Scholar]
- 21.Kornblum J, Kreiswirth N, Projan S J, Ross H, Novick R P. agr: a polycistronic locus regulating exoprotein synthesis in Staphylococcus aureus. In: Novick R P, editor. Molecular biology of the staphylococci. New York, N.Y: VCH; 1990. pp. 373–402. [Google Scholar]
- 22.Kraemer G R, Iandolo J J. High-frequency transformation of Staphylococcus aureus by electroporation. Curr Microbiol. 1990;21:373–376. [Google Scholar]
- 23.Kullik I, Giachino P, Fuchs T. Deletion of the alternative sigma factor ςB in Staphylococcus aureus reveals its function as a global regulator of virulence genes. J Bacteriol. 1998;180:4814–4820. doi: 10.1128/jb.180.18.4814-4820.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lease R A, Cusick M E, Belfort M. Riboregulation in Escherichia coli: DsrA RNA acts by RNA:RNA interactions at multiple loci. Proc Natl Acad Sci USA. 1998;95:12456–12461. doi: 10.1073/pnas.95.21.12456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lina G, Jarraurd S, Ji G, Greenland T, Pedraza A, Etienne J, Novick R P, Vandenesch F. Transmembrane topology and histidine kinase activity of AgrC, the agr signal receptor in Staphylococcus aureus. Mol Microbiol. 1998;28:655–662. doi: 10.1046/j.1365-2958.1998.00830.x. [DOI] [PubMed] [Google Scholar]
- 26.Lyon B R, Skurray R. Antimicrobial resistance of Staphylococcus aureus: genetic basis. Microbiol Rev. 1987;51:88–134. doi: 10.1128/mr.51.1.88-134.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McNamara P J, Iandolo J J. Genetic instability of the global regulator agr explains the phenotype of the xpr mutation in Staphylococcus aureus KSI9051. J Bacteriol. 1998;180:2609–2615. doi: 10.1128/jb.180.10.2609-2615.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morfeldt E, Taylor D, von Gabain A, Arvidson S. Activation of alpha-toxin translation in Staphylococcus aureus by the trans-encoded antisense RNA, RNAIII. EMBO J. 1995;14:4569–4577. doi: 10.1002/j.1460-2075.1995.tb00136.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morfeldt E, Tegmark K, Arvidson S. Transcriptional control of the agr-dependent virulence gene regulator, RNAIII, in Staphylococcus aureus. Mol Microbiol. 1996;21:1227–1237. doi: 10.1046/j.1365-2958.1996.751447.x. [DOI] [PubMed] [Google Scholar]
- 30.Novick R. Signal transduction in staphylococci and other gram-positive cocci. In: Rappuoli R, Scarlato V, Aricò B, editors. Signal transduction and bacterial virulence. R. G. Austin, Tex: Landes Company; 1995. pp. 39–53. [Google Scholar]
- 31.Novick R, Kornblum J, Kreiswirth B, Projan S, Ross H. Regulation of postexponential phase exoprotein synthesis in Staphylococcus aureus. In: Ayout E M, Cassell G H, Branche W C, Henery T J, editors. Microbial determinants of virulence and host responses. Washington, D.C.: American Society for Microbiology; 1990. pp. 9–18. [Google Scholar]
- 32.Novick R P, Ross H F, Projan S J, Kornblum J, Kreiswirth B, Moghazeh S. Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J. 1993;12:3967–3975. doi: 10.1002/j.1460-2075.1993.tb06074.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Novick R P, Projan S J, Kornblum J, Ross H F, Ji G, Kreiswirth B, Vandenesch F, Moghazeh S. The agr P2 operon: an autocatalytic sensory transduction system in Staphylococcus aureus. Mol Gen Genet. 1995;248:446–458. doi: 10.1007/BF02191645. [DOI] [PubMed] [Google Scholar]
- 34.Peng H-I, Novick R P, Kreiswirth B, Kornblum J, Schlievert P. Cloning, characterization, and sequence of an accessory gene regulator (agr) in Staphylococcus aureus. J Bacteriol. 1988;170:4365–4372. doi: 10.1128/jb.170.9.4365-4372.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Regassa L B, Novick R P, Betley M J. Glucose and nonmaintained pH decrease expression of the accessory gene regulator (agr) in Staphylococcus aureus. Infect Immun. 1992;60:3381–3388. doi: 10.1128/iai.60.8.3381-3388.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Skurray R A, Firth N. Molecular evolution of multiply-antibiotic-resistant staphylococci. Ciba Found Symp. 1997;207:167–183. doi: 10.1002/9780470515358.ch11. [DOI] [PubMed] [Google Scholar]
- 37.Sledjeski D D, Gupta A, Gottesman S. The small RNA, dsrA, is essential for the low temperature expression of rpoS during exponential growth in Escherichia coli. EMBO J. 1996;15:3993–4000. [PMC free article] [PubMed] [Google Scholar]
- 38.Smibert R M, Krieg N R. General characterization. In: Krieg N, editor. Manual of methods for general bacteriology. Washington, D.C.: American Society for Microbiology; 1981. p. 435. [Google Scholar]
- 39.Vandenesch F, Kornblum J, Novick R P. A temporal signal, independent of agr, is required for hla but not spa transcription in Staphylococcus aureus. J Bacteriol. 1991;173:6313–6320. doi: 10.1128/jb.173.20.6313-6320.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu S, de Lancastre H, Tomaz A. Sigma-B, a putative operon encoding alternate sigma factor of Staphylococcus aureus RNA polymerase: molecular cloning and DNA sequencing. J Bacteriol. 1996;178:6036–6942. doi: 10.1128/jb.178.20.6036-6042.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Youngman P. Plasmid vectors for recovering and exploiting Tn917 transpositions in Bacillus and other gram-positive bacteria. In: Hardy K G, editor. Plasmids: a practical approach. Oxford, United Kingdom: IRL Press Limited; 1987. pp. 79–103. [Google Scholar]