ABSTRACT
The nosocomial pathogen Clostridioides difficile is a burden to the healthcare system. Gut microbiome disruption, most commonly by broad-spectrum antibiotic treatment, is well established to generate a state that is susceptible to CDI. A variety of metabolites produced by the host and/or gut microbiota have been shown to interact with C. difficile. Certain bile acids promote/inhibit germination while other cholesterol-derived compounds and amino acids used in the Stickland metabolic pathway affect growth and CDI colonization. Short chain fatty acids maintain intestinal barrier integrity and a myriad of other metabolic compounds are used as nutritional sources or used by C. difficile to inhibit or outcompete other bacteria in the gut. As the move toward non-antibiotic CDI treatment takes place, a deeper understanding of interactions between C. difficile and the host’s gut microbiome and metabolites becomes more relevant.
KEYWORDS: Clostridioides difficile, gut metabolites, microbiome, fecal microbial therapy, bile acids, Stickland, short-chain fatty acids
Introduction
Clostridioides difficile is a Gram-positive, pathogenic, spore forming, anaerobic bacterium that is considered the main cause of antibiotic-associated diarrhea, pseudomembranous colitis and toxic megacolon.1 According to the Centers for Disease Control and Prevention (CDC), C. difficile is a major nosocomial pathogen with more than 220,000 infections, 13,000 deaths, and nearly $5 billion in annual treatment associated costs that are predicted to increase in the future.2,3 This, combined with its inherent natural antibiotic resistance, led to the CDC classifying C. difficile as an ‘urgent threat’ to the United States healthcare system. The greatest risk factor for C. difficile infection (CDI) is prior treatment with broad-spectrum antibiotics. Antibiotics render the host susceptible to CDI by changing the ecology of the microbiota, which is known to provide ‘colonization resistance’ against invading pathogens, C. difficile-included.4,5 Interestingly, infants and newborns can be C. difficile carriers6 and C. difficile can be found in approximately 53% of healthy adults.7 Thus, C. difficile could be considered a part of the microbiome in humans, but this is mostly defined by age.8
Endospores are metabolically dormant forms of spore-forming bacteria produced in response to stress (e.g., nutrient limitation).9–11 For C. difficile, the spore form is essential for host-to-host transmission due to the strictly anaerobic nature of the vegetative form.12–14 Thus, in susceptible hosts, C. difficile spores must germinate into the active, vegetative form in order to multiply and cause disease. Germination by C. difficile spores is triggered upon recognition of certain bile acids and amino acids by germinant receptors.11,15,16
Dormant C. difficile spores are considered infectious agents, but the growing C. difficile vegetative cells secrete the TcdA and TcdB toxins that are responsible for the primary symptoms of disease (e.g., diarrhea and colitis);14,17–19 while some C. difficile strains secrete a third toxin, a binary toxin.20 The most common treatments for CDI are broad-spectrum antibiotics like vancomycin, although fidaxomicin (a narrower-spectrum antibiotic)is now recommended for initial and recurrent CDI . Unfortunately, the higher cost of fidaxomicin is limiting its use, and the continued alteration to the colonic microbiota by these antibiotics, lead to patients experiencing recurring disease due to the presence of C. difficile spores within the gastrointestinal tract or in the surrounding environment.21,22
Recently, fecal microbial transplantation (FMT) has emerged as an effective treatment against C. difficile, especially for patients with recurrent CDI. FMT is hypothesized to drive protection against CDI through the restoration of gut microbes, gut associated metabolites [e.g., secondary bile acids and short chain fatty acids (SCFA)]23 and/or microbial bile acid biotransformation . Nevertheless, the use of FMT is not without risk for the potential introduction of harmful microbes to the recipient.24–26 Thus, effective and safe treatments for CDI that result in the restoration of the protective metabolic effects provided by the microbiome still need to be developed. In this review, we summarize studies focusing on metabolites produced by the host or gut microbiota and how these molecules influence C. difficile pathogenesis.
Bile acid metabolites and their role in C. difficile pathogenesis
Bile acids are cholesterol-based molecules synthesized by hepatocytes in the liver that play key roles in regulating metabolic pathways and aiding in the absorption of fats and cholesterol during digestion.27,28 There are two pathways for bile acid synthesis: the classical pathway generates both cholic acid (CA) and chenodeoxycholic acid (CDCA) while the alternative bile acid synthesis pathway only forms CDCA.29 Before being secreted into the intestines, primary bile acids are conjugated with either taurine or glycine at C-24 to generate taurocholic acid (TA)/taurochenodeoxycholic acid (TCDCA) and glycocholic (GCA) acid/glycochenodeoxycholic acid (GCDCA) (Figure 1).30,31 Reabsorption of bile acids takes place throughout the intestines and the reabsorbed bile acids are recycled back to the liver for additional rounds of digestion.29,31 During this enterohepatic recirculation pathway, bile acid synthesis is regulated through the Farsenoid X receptor (FXR) transcriptional activator32 . Bile acid binding principally through CDCA to FXR generates a conformational change in FXR that leads to the synthesis of FGF19 in illeal enterocytes32,33 . FGF19 is secreted by the ileal enterocytes and binds to the FGFR4 receptor on the cell surface of hepatocytes. Binding of FGF19 to the receptor leads to a negative feedback regulation on bile acid synthesis.32,33 In an FMT model for recurrent CDI patients, levels of FGF19 were significantly increased post-FMT, thus suggesting upregulation of the FXR-FGF pathway after FMT that may aid in C. difficile clearance.34 Furthermore administration of obeticholic acid, an FXR agonist, to HFD CDI-infected mice resulted in decreases in disease severity, and the use of ursodeoxycholate (UCA) in a CDI mouse model showed increased levels of FXR as well as TGR5 that modulates the immune response against C. difficile.35,36 The mentioned studies on FXR suggest that FXR modulation may be important for CDI treatment but further research is necessary to move past the correlation observed in FXR that shows protection against CDI.
Figure 1.
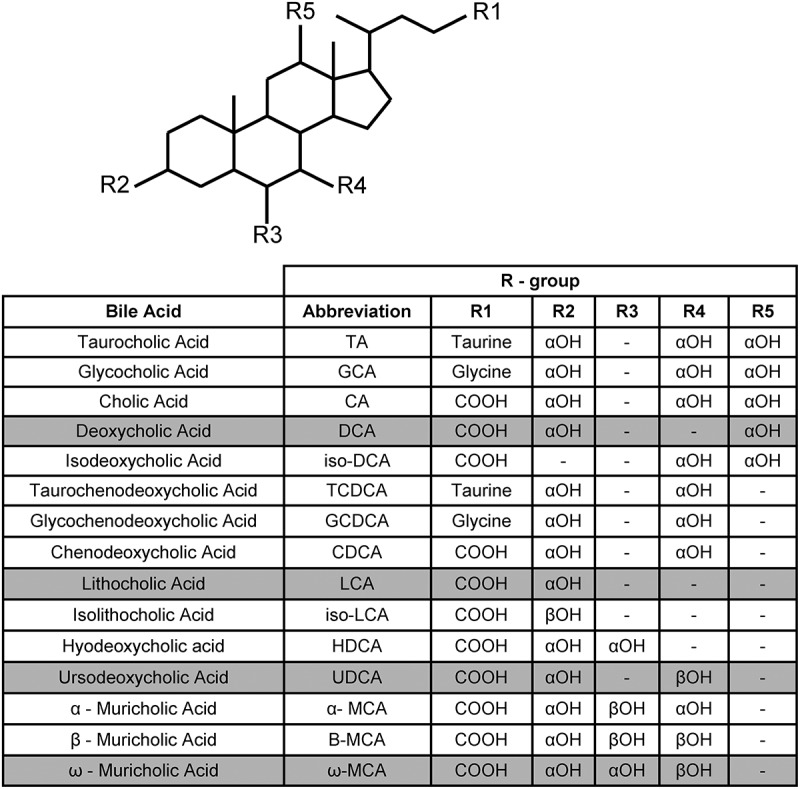
Structures of the bile acids discussed in this review.
Shaded rows indicate secondary bile acids. Unshaded rows indicate conjugated and deconjugated primary bile acids.
A small percentahe of bile acids escape enterohepatic recirculation, and can be deconjugated by the bile salt hydrolases that are expressed in many gut microbes. Subsequently, deconjugated bile acids can be 7α-dehydroxylated by a small subset of gut microbial species to generate secondary bile acids (deoxycholic acid (DCA), from CA, and lithocholic acid (LCA) from CDCA) (Figure 1).
Cholic acid-derived bile acids promote C. difficile germination, whereas bile acids of the CDCA family act as inhibitors of C. difficile germination.16,37,38 In addition to acting as activators/inhibitors of C. difficile spore germination, bile acids affect C. difficile vegetative growth. The host-derived, primary, bile acids TA and CDCA have different effects on C. difficile growth. Vegetative cell growth seems to be unaffected by TA, however C. difficile growth is strongly inhibited by CDCA.16 Interestingly, DCA, a product of the 7α-dehydroxylation of CA by the colonic microbiota, is also toxic to C. difficile growth in-vitro, suggesting that the 7α-dehydroxylation of CA by the colonic microbiota may prevent C. difficile from colonizing healthy hosts.16 When the minimal inhibitory concentrations (MIC) of various bile acids were determined, the MIC of CA was 10 mM for 2 different C. difficile strains, a concentration that is not found in the GI tract, thus not physiologically relevant.39 However, increasing the hydrophobicity of the base CA structure (which has three hydroxyl groups, Figure 1), by removing either the 12α hydroxyl (generating CDCA) or the 7α hydroxyl (generating DCA) resulted in the MIC decreasing from 10-fold to 1 mM.39 Moreover, the 7β-epimer of CDCA, UCA) (Figure 1), inhibits growth of C. difficile cells,40 although over a 24-h time period, C. difficile grew in the presence of 2 mM UCA.41 Because CDCA completely inhibited the growth of C. difficile vegetative cells over a 24-h period and UCA did not, this could imply that stereochemistry of the bile acid structure plays a role in toxicity.39,40 Importantly, though these two studies were done using different C. difficile isolates, which may have different MIC values for bile acids as observed for other C. difficile strains.39
Mice are one of the most common animal models used in CDI studies and their murine bile acid composition and their effects on CDI are well understood.39,42 In rodents, the presence of alternative hydroxylating enzymes yields other primary bile acids (α/β/ω-muricholic acids) (Figure 1).43 Muricholic acids have a MIC similar to that of CDCA and DCA.39 Similar to CA, muricholic acids are also trihydroxyl bile acids but the hydroxyls are in the 3, 6, and 7 positions instead of the 3, 7, and 12 positions that are found on CA-derived bile acids (Figure 1).39 A study found that in addition to the bile acids described above, isodeoxycholic acid (a 7α, 12α-hydroxy bile acid), lithocholic acid (a 3α-hydroxy bile acid), isolithocholic acid (a 3β-hydroxy bile acid), and hyodeoxycholic acid also inhibit the growth of C. difficile vegetative cells.44 Furthermore, isoallolithocholic acid (isoalloLCA), the isomer of isoLCA had an MIC90 of 2 µM in C. difficile CD630 (a laboratory strain) and in the highly toxigenic C. difficile VPI10463 strain.45
Due to the inherent toxicity of secondary bile acids for C. difficile growth, it was hypothesized that their production would toxify an environment and limit C. difficile growth.16,46 To test this hypothesis, Theriot and Young5 showed evidence that the presence of secondary bile acids is responsible for protection against CDI. Using a multi-omics approach, the authors identified C. difficile resistant and susceptible states in an antibiotic-treated murine model. A susceptible state correlated with high levels of conjugated primary bile acids (e.g., TA) whereas a resistance state correlated with higher levels of secondary bile acids (e.g., DCA). Later, Buffie et al.4 also provided compelling data in support of this hypothesis. Using mouse models and human subjects, the authors found that high levels of DCA or the presence of the genetic operon that is known to generate secondary bile acids (bai), strongly correlated with a protective environment. Additionally, mice treated with different antibiotics resulted in variable metabolic profiles and certain secondary bile acids (i.e., omega-muricholic acid, LCA and, to a lesser extent, hyodeoxycholic acid, and UCA) that could inhibit TA- and DCA-mediated colony formation by C. difficile spores.16,38,39,47
These landmark studies demonstrated that increased secondary bile acids and the genes encoding proteins responsible for their production strongly correlate with disease-resistant states and increased primary bile acids correlate with disease-susceptible states. A more recent article45 analyzing metabolites in centennials collected stool from centennials(~107 year olds), elderly adults (85–89 years old), and young adults (21–55 years old) and bile acids were measured. The authors observed higher levels of isoLCA, 3-oxoLCA, alloLCA, 3-oxoalloLCA, and isoalloLCA in the centennial samples, when compared to the other study population. Additionally, the authors characterized the biosynthetic pathways to generate these CDCA derivatives and suggested that further elucidation of microbial biotransformations may expand our understanding of intestinal homeostasis and, as a result, possible protection effects of additional bile acids against CDI45
Interestingly, a recent study using a cholate-deficient mouse strain showed that the cholate-derived secondary bile acid (DCA) is dispensable for protection against CDI, and that lack of the bile acid transcriptional activator FXR, does not show differences in CDI mice. Also, when germ free mice were monoassociated with bacteria known to generate secondary bile acids, no secondary bile acids were present in fecal samples, but the mice were still protected against CDI.48 This emerging evidence would suggest that although bile acids are important for C. difficile colonization to be established in the gut, these metabolites may not play such an important role in generating a protective environment against C. difficile. In support of this, a 2018 study49 showed that the levels of the conjugated primary bile acids TA and TCDCA decreased 2 days post-FMT. In contrast, there was an increase of the secondary bile acids DCA, LCA, and UDCA.49 Interestingly, the authors note that although levels of secondary bile acids increased, the presence of secondary bile acids was not indicative of FMT success or failure, thus suggesting that although bile acids may play a role in protection, there are other mechanisms driving CDI resistance.49,50
Regardless, C. difficile encounters bile acid during colonization and its growth in the gut may impact bile acid synthesis in the liver. Taking advantage of MALDI imaging mass spectrometry, Wexler et al.51 found the levels of primary bile acids in the mouse gut increased significantly as early as one-day post-CDI. Additionally, the authors found that the introduction of cholestyramine, a bile-acid sequestering drug, lead to delayed C. difficile colonization.51 Their results suggest that primary bile acids are required to efficiently establish C. difficile in a host.
Regarding secondary bile acids, in addition to its toxic effect against C. difficile vegetative cells, the secondary bile salt DCA induces C. difficile biofilm formation at low concentrations (240 µM) in a non-biofilm-producing C. difficile CD630∆erm strain.52 Thus, it is possible that a decrease in bile acid levels during late infection stages may generate favorable conditions for biofilm formation. Indeed, the authors hypothesized that the low concentrations of the bile salt encountered during the natural restoration of microbiome may allow the cells to transition toward biofilm formation. This would protect C. difficile vegetative cells against concentrations of antibacterial compounds that may normally inhibit growth (i.e., antibiotics or other bile acids).52
In addition to promoting germination and inducing biofilm formation, bile acids can alter C. difficile physiology in other ways. Low amounts of LCA (0.08 mM) resulted in more elongated vegetative cells and absence of flagella. Fewer flagella were also observed when vegetative cells were incubated in 0.8 mM DCA and 0.3 mM CDCA. The authors hypothesized that lack of flagella may allow for better adherence to intestinal lining during infection. Additionally, bile acids resulted in up-regulation of the chaperon proteins DnaK, DnaJ, GrpE, GroL, and GroS, but the mechanism of up-regulation for chaperones is still unknown.53
Finally, bile acids inhibit TcdB toxin activity by binding to the toxin. Both conjugated and deconjugated secondary bile acids (e.g., DCA, LCA, GDCA, TDCA, GLCA, and TLCA) have greater potency in inhibiting toxin activity than their primary bile acid counterparts, but toxin binding by bile acids is reversible. The reversible binding and inhibition of bile acids to C. difficile toxins may thus suggest modulation mechanisms taking place in the gut depending on bile acid composition in the host.54
The role of bile acids in relation to C. difficile physiology and pathogenesis has been studied and characterized, extensively, since the discovery of their function as germinants for C. difficile spores.16,46 Because of the many functions bile acids play in the host, it is not surprising that the interaction with C. difficile is complex and multifaceted, with both positive and negative interactions observed – depending on the conformation of, the abundance of, and the location of specific bile acids as well as the C. difficile life cycle stage in the host. More work should be done that move the field from data that correlate what bile acids are present in the host to potential causative interactions between bile acids and the host/C. difficile.
Stickland metabolites, amino acids, and the Wood Ljundahl pathway in C. difficile pathogenesis
Stickland metabolism was first identified in 1934 as the predominant pathway for Clostridium sporogenes energy production.55 Stickland metabolism couples pairs of amino acids that may act as electron donors or acceptors.56,57 The donor amino acid is oxidatively deaminated or decarboxylated to produce NADH, whereas the electron accepting amino acid is reduced to regenerate NAD+ (Figure 2(a)). Although a myriad of amino acids can be used in the oxidative branch, the reductive branch is fueled only by proline or glycine. In the reductive branch, proline is consumed by proline reductase (PrdB, PrdA), a selenium-containing enzyme (selenoenzyme) that generates 5-aminovalerate and NAD+, and glycine is consumed by glycine reductase (GrdA, GrdB), selenoproteins that generate acetate and NAD+ (Figure 2(b,c)).58,59
Figure 2.
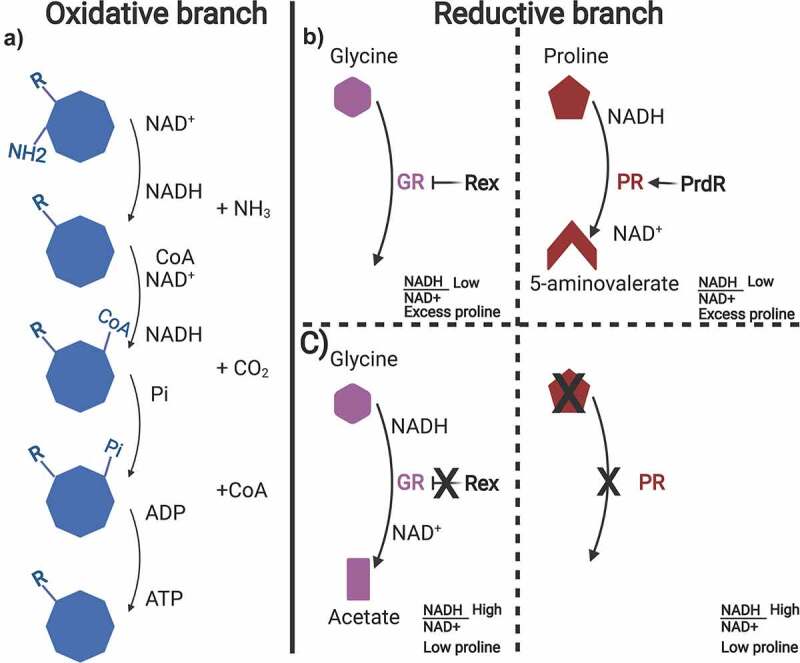
Oxidative and reductive branch of Stickland metabolism in C. difficile.
Graphical representation of the oxidative and reductive Stickland pathways. A) Oxidation of a myriad of amino acids takes place in the oxidative branch of Stickland metabolism resulting in 2 NADH molecules and 1 ATP molecule produced. B) Proline and glycine are used to regenerate NAD+ in the reductive branch. When excess proline is available in the surrounding environment and the NADH/NAD+ ratio in the cell is low, the PrdR activator promotes proline reductase (PR) expression to generate 5-aminovalerate and NAD+. Concurrently, Rex inhibits glycine reductase (GR). C) When proline levels are low and the NADH/NAD+ ratio is high, Rex is unable to inhibit glycine reductase allowing acetate formation. Proline reductase activity is not present. Created with BioRender.com.
Although Stickland metabolism is well-characterized in Clostridium sticklandii,60 this alternative metabolic pathway in other Clostridial species has just recently been elucidated. Bouillaut et al.59 analyzed the C. difficile glycine and proline reduction operons and their activity in the reductive pathway of Stickland metabolism. A mutation in the prdB subunit of the proline reductase enzyme resulted in a decrease in growth in rich media, but a mutation in a grdA mutant did not affect growth in rich medium.59 Additionally, the authors characterize the sigma-54 dependent activator, PrdR that acts as a mediator for PrdB-dependent activation and proline-dependent toxin repression.59 In addition, given the role the Stickland reductive branch has in regenerating NAD+ for the cell, the redox dependent transcriptional repressor, Rex, has a role in proline-dependent regulation and is controlled by PrdR in C. difficile.61 Using DNA binding assays and qRT-PCR, the authors found that proline seems to be the preferred amino acid for regeneration of NAD+. In presence of excess proline, PrdR stimulates proline reductase expression and simultaneously Rex represses the glycine reductase gene grdE. NADH is then oxidized and, as a result, the ratio of NADH/NAD+ is low. Alternatively, when levels of proline are low, the NADH/NAD+ ratio increases and the high levels of NADH prevents Rex from repressing glycine reductase expression (Figure 2(b,c)).61
Selenium is an essential component of the proline and glycine reductases. Selenium is incorporated into these proteins as selenocysteine. Selenocysteine is generated through a synthesis pathway where the product of the selD gene reacts inorganic phosphate with hydrogen selenide to generate selenophosphate.58,62,63 A selD mutant is unable to incorporate selenium into proteins and this results in a complete loss of the reductive branch of Stickland metabolism. The loss of selenophosphate generation results in a defect in the ability of C. difficile spores to outgrow following germination in peptide-rich media.63,64 Also, the absence of selenophosphate altered C. difficile physiology so that other NAD+ regeneration pathways were expressed.64
Several studies have correlated the depletion or increased levels of amino acids important for Stickland metabolism using both in vitro and in vivo approaches.48,65–68 In an in vitro, multi-omics approach, the metabolome of multiple C. difficile growth stages found that Stickland metabolites were dramatically depleted upon entry into stationary phase69 . Furthermore, in stool samples derived from hospitalized patients, amino acids used for Stickland metabolism were depleted in CDI patients, suggesting that these amino acids were consumed by C. difficile vegetative cells. In this study, branched chain amino acids, such as leucine, were hypothesized to be the preferred amino acid used by C. difficile; other amino acids, such as proline, tyrosine, and phenylalanine are possible sources of energy for both C. difficile and other gut microbes.67
Proline availability is important for C. difficile colonization in mice. A mutation in proline reductase (prdB) resulted in decreased colonization in a humanized microbiome mouse model.65 Moreover, C. difficile can take advantage of inflammation-induced collagen degradation. During this toxin-dependent process, the host cells respond to this inflammation by producing matrix metalloproteases. This results in the degradation of collagen – a protein rich in proline.70
Building upon this, in mice monoassociated with C. scindens, C. hiranonis, or C. leptum, proline was greatly depleted and mice were protected against CDI.48 This work is also supported by in vitro conditions where C. difficile and C. hiranonis compete for nutrients.71 Interestingly, Battagliogli et al.65 observed high levels of the amino acids glycine, proline, threonine, and alanine in a dysbiotic colon, whereas Aguirre et. al.48 observed depletion of proline and glycine in their monoassociated mice (amino acids essential for the reductive pathway of Stickland metabolism).48,65 Nevertheless, direct comparison should not be made since one study used a humanized microbiome mouse model and the other used germ-free mice that were monoassociated with individual bacteria.
Expanding on the hypothesis that depletion of amino acids is important for Stickland metabolism, and may drive protection against CDI, Girinathan et al.66 found that the gut commensal bacterium P. bifermentans protects against CDI, at least in part, through depleting nutrient sources used by C. difficile. Using P. bifermentans monoassociated mice, as well as carbon-source enrichment analysis of the gut-metabolomic environment, the authors found alterations in the substrates/products of Stickland metabolism (such as proline, glycine, threonine and 4-hydroxyproline) and production of 5-aminovalerate.66
Although in vivo FMT studies have reported levels of amino acids in FMT-recipients, such as increasing levels of valine, isoleucine, and leucine (amino acids able to be used in the oxidative branch), the presence of Stickland products in these recipients have not been reported [besides the SCFA (acetate, propionate)].72–74 In a chemostat model, levels of 5-aminovalerate decreased post-FMT and isobutyrate levels remain constant post-FMT. A plausible explanation for the observed 5-aminovalerate levels could be the lack of Stickland fermenters included in the stable communities introduced into the chemostat, such as P. bifermentans.75
Expressed at lower levels when other forms of energy production are present, the Wood-Ljungdahl Pathway (WLP) may need examining during CDI. Through the WLP, two molecules of CO2 are reduced to acetate.76 The WLP can be coupled with butyrate production to allow for increased efficiency of ATP formation by decreasing nutritional requirements. Thus, the WLP may prove useful when glucose or amino acid levels are low, and C. difficile may need to adapt to low nutrient conditions.77 Moreover, in a mouse model of CDI, the WLP increased expression twenty-four hours post-infection.78 This suggests more attention should be given to this pathway in animal models, or in patient cohort samples.
Stickland metabolism by C. difficile during host colonization is well-established.48,65,68 Nevertheless, the availability of the substrates needed for the oxidative and reductive branches of Stickland metabolism, and required compounds for WLP, during CDI likely shapes the ability of the bacterium to cause disease. Similarly, the presence of Stickland-using microbial competitors in the host microbiome also shapes how C. difficile colonizes a host.48,66,79 Therefore, a better understanding of specific conditions in which C. difficile takes advantage of alternative metabolic pathways is necessary for the study of novel therapeutics that may modulate these pathways.
Non bile acid metabolites that correlate or show protection against C. difficile pathogenesis
The gut microbiome is complex, with interactions occurring between resident and invading bacteria that result in the generation of secondary metabolites. One such metabolite is coprostanol. Coprostanol is generated through the reduction of the double bond between C5 and C6 of cholesterol.80 In the metabolome of healthy controls vs. CDI patients, 63 bacterial OTUs were identified that positively correlated with the presence of coprostanol, which in turn negatively correlated with CDI patients.81 The majority of phylotypes that correlated with coprostanol presence were members of the Lachnospiraceae and Ruminococcaceae families. Coprostanol may enhance resistance to CDI by decreasing the availability of cholesterol which could reduce the abundance of metabolites (i.e., primary bile acids) that are necessary for germination by C. difficile spores (Figure 3(a,b).81
Figure 3.
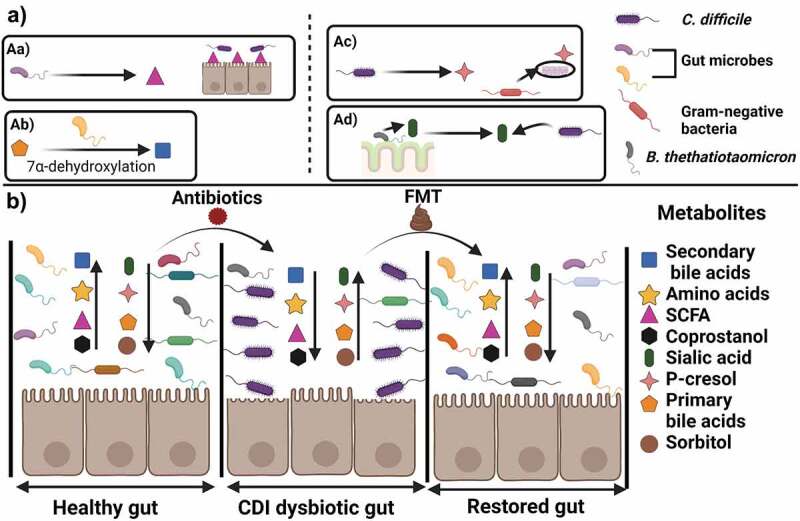
Gut metabolites effect in C. difficile growth and colonization.
A) Examples of mechanisms of inhibition or protection against CDI. Aa) Gut microbes produce SCFA that aid in intestinal barrier function. Ab) Bacteria that perform 7α-dehydroxylation generate secondary bile acids that correlate with CDI resistant states. On the other hand Ac) generation of p-cresol by C. difficile may affect barrier function in Gram-negative bacteria, Ad) release of sialic acid from the colon mucus layer is used as a nutrient source by C. difficile. B) In an undisrupted gut environment amino acid, SCFA and secondary bile acid levels are high, creating a resistant state against C. difficile through immune defenses and nutrient limitation. High levels of the cholesterol derivative, coprostanol, are observed indicative of limited availability of primary bile acids. Introduction of antibiotics compromises the gut environment, and C. difficile toxins disrupt the intestinal barrier. C. difficile also promotes increasing levels of p-cresol, conjugated primary bile acids (e.g. TA) and sorbitol as well as use sialic acid as a nutrient source. FMT treatment allows restauration of gut metabolites and restore a resistant state against CDI. Created with BioRender.com.
Three microbial-derived SCFAs (propionate, acetate, and butyrate) have long been associated with CDI resistance.82–85 In general, SCFAs are believed to play important roles in maintaining gut homeostasis.86 Several studies have shown that SCFAs are depleted during CDI5,81,86 Recently, butyrate, and acetate were found to protect against CDI by aiding in maintaining intestinal barrier integrity.87,88 Butyrate plays a role in maintaining intestinal barrier integrity through limiting the permeability of intestinal epithelial cells to C. difficile toxins by stabilizing the hypoxia-inducible factor 1 alpha (HIF-1α).87 Acetate acts during the early stages of C. difficile infection by activating the free fatty acid receptor 2 (FFAR2) signaling pathway by augmentation of IL-1B production from neutrophils and IL-1 R through signaling of ILC3. These two types of immune cells then induce production of IL-22 that is implicated in antimicrobial and repair mechanisms in intestinal epithelial cells.88
The mechanism by which propionate confers protection against CDI has yet to be elucidated. However, taking from data published on butyrate and acetate, a hypothesis could be that propionate acts upon specific host immune factors (Figure 3a). Interestingly, Gregory et al.89 suggested that increasing levels of SFCAs as the gut microbiome recovers from disruption, may be used by C. difficile vegetative cells as a triggering signal to upregulate toxin secretion and promote the inflammation that would allow the bacterium to maintain colonization in the gut.89
A recent study that used1H-NMR on stool samples derived from recurring CDI patients that received FMT, through capsule or colonoscopy, found increased levels of acetate, butyrate, and propionate in recipients 12-weeks post-FMT.85 Interestingly, valerate, another SCFA was depleted in a chemostat C. difficile infection model.75 Similar to the other SCFA, valerate levels were restored upon FMT. The study also found that valerate inhibited in vitro C. difficile vegetative growth in a dose-dependent manner, and that introducing valerate orally in the form of 15 mM glycerol trivalerate into a C. difficile mouse model decreased total viable counts.75,85 It is important to note, though, that the glycerol trivalerate given to the mice was above physiologically relevant concentration since the authors found that chemostat FMT cultures only reached 4 mM valerate.
In addition to SCFAs, the microbiome produces antimicrobial compounds. Two Clostridial species, C. scindens and C. sordellii secrete tryptophan-derived antibiotic compounds (i.e., 1-acetyl-β-carboline and turbomycin A) that inhibit C. difficile growth.90 Interestingly, the anti-C. difficile effect of the antibiotics was enhanced by DCA and LCA. These findings suggest that a combination of secondary bile acids as well as antibiotics is at play during prevention of disease.90 However, a recent study with mice that were monoassociated with C. scindens did not detect 1-acetyl-β-carboline in their metabolomics analysis,48 suggesting that the activity of 1-acetyl-β-carboline may require specific conditions to be produced.
Dynamic interactions between gut microbes are constantly taking place and involve the production of and the sensing of secondary metabolites.91 Although the mechanisms by which some of these metabolites protect against CDI are still unclear,81,87,88,90 their identification provides important data that can be directly tested as potential treatment options for CDI. Moreover, a microbiome-centric approach for CDI treatment will most likely find other metabolites that show protection against CDI and could be explored for therapeutic approaches.
Metabolites and their beneficial roles for C. difficile infection
In recent years, the research focus on the modulation of the microbiome by C. difficile and how the bacterium survives in the host has increased.92–96 C. difficile can produce products that promote inflammation and that have antibacterial effects. C. difficile generates p-cresol, a phenolic compound that affects the integrity of surface barriers in bacterial cells with a higher effect observed in Gram-negative bacteria.93 The hpdBCA operon is responsible for the fermentation of tyrosine to p-hydroxyphenylacetate to then generate p-cresol using the 4-hydroxyphenylacetate dehydrogenase enzyme. In a mouse model of recurrent CDI, mice infected with a hpdC mutant strain, had increased microbial diversity compared to mice infected with the wildtype strain, as well as lower C. difficile viable counts. Additionally, when exogenous p-cresol was introduced to healthy human fecal slurries, the number of viable total anaerobes increased, thus suggesting that generation of p-cresol by C. difficile may give the bacterium a competitive advantage over other gut microbial species (Figure 3(a,c)).93
Similar to the production of p-cresol, production of sorbitol by C. difficile may enhance colonization. A C. difficile strain that is unable to produce sorbitol is outcompeted 10-fold by its wildtype counterpart in a mouse model.94 Upon further investigation, the authors found that the inflammation induced by the C. difficile toxins results in an upregulation of aldose reductase which generates host-derived sorbitol. These results show how C. difficile is able to use a diet and host-derived nutrient to expand in the perturbed microbiome environment (Figure 3(b)).94
C. difficile also can use metabolites produced by the host and other gut microbes during colonization.92,95,96 The gut symbiont Bacteroides thetatiotaomicron encodes a sialidase enzyme that cleaves and releases sialic acid from mucosal glycoconjugates. C. difficile encodes a sialic acid catabolic operon. Using a transcriptional analysis of germ free and B. thetatiotaomicron monoassociated mice, the data showed that the ability of B. thetatiotaomicron to release sialic acid resulted in increased expression levels of the C. difficile sialic acid operon.95 Additionally, a spike in free sialic acid 1 day after antibiotic treatment of mice was observed but these levels reduced 3 days after antibiotic treatment.95 Moreover, mice monoassociated with a B. thetatiotaomicron sialidase mutant strain and then infected with C. difficile had lower C. difficile CFU counts (Figure 3(a-d)).95 These results suggest an important role of sialic acid during in vivo infection.
Another host-produced metabolite, heme, can be used by C. difficile.92 The heme-sensing membrane protein system, HsmRA, protects against redox damage generated from antibiotic treatment of CDI. C. difficile HsmA binds to heme that is released from the inflamed GI tract and shields the bacterium from redox-active molecules. Although the specific mechanism of protection by HsmA is still unknown, the authors hypothesized the use of HsmRA by other pathogens for protection against oxidative stress may be taking place.92
Finally, indole may play a role during C. difficile infections. Supernatant from C. difficile stationary phase cultures can induce the expression of tryptophanase (tnaA) in E. coli. The levels of indole increased in other indole-producing microbes in the gut (e.g., L. reuteri and E. faecalis) in co-culturing assays with C. difficile. Interestingly, the MIC (5 mM) of C. difficile strains were found to be higher than the MIC of multiple gastrointestinal bacteria tested, ranging from 2–4 mM. These results led the authors to the hypothesis that the ability of C. difficile to resist higher gut indole concentrations may provide the bacterium a competitive advantage.96 Importantly, these experiments were performed under in vitro conditions and future work with animal models is needed to test the proposed hypothesis.
Successful C. difficile colonization, occurs when the host gut environment is disrupted. As a result, the bacterium must adapt and take advantage of resources available to maintain colonization and exclude the reestablishment of competing microbes.97,98 The ability of the C. difficile bacterium to establish a niche in the gut environment by use of metabolites from intestinal epithelial cells, as well as secreted compounds to outcompete other gut microbes, suggests C. difficile is a bacterial generalist.68 Although the factors that correlate with an environment that excludes C. difficile are well documented, the factors that C. difficile uses to exclude the microbiome from reestablishing itself are comparatively less understood. By inhibiting these mechanism, the normally-protective microbiome may gain a foothold and return the GI to a colonization resistant environment.
Conclusions and future directions
This review highlights specific metabolic compounds that have been regarded as important during the life cycle and pathogenesis pathway of CDI. As the research field, moves toward a microbiome-centric study of gut diseases, molecules used as nutritional sources for both pathogen and resident gut bacteria are being identified. In Figure 3, we show graphical representations of microbiome stages that may allow or prevent the gut environment to succumb to a C. difficile disease stage, as well as promote recovery.
C. difficile pathogenesis is complex. Although much work has focused on the effect that bile acids and antimicrobials have on the bacterium, considering the physiology of C. difficile for developing new treatments is emerging in the field. Potential treatment options against CDI have slowly progressed toward approaches that focus on restoring the gut microbiome ecosystem as a whole and not what affects C. difficile. Although some progress has been made, there are still many aspects to uncover regarding gut microbiota niche growth and competition in the GI tract. The move from broad-spectrum antibiotic treatment to more specific antibiotics, like ridinilazole that showed fewer CDI recurrences in a phase 2 trial, is happening at present.99 Targeted molecules such as nanobodies or DARPins have also emerged and,100 will probably become the norm for treatment against CDI in the future. Alternatively, specific strategies to reintroduce defined microbial communities to combat CDI, in the form of targeted microbiome therapies (e.g., SER-109, composed of purified Firmicutes bacterial spores),101 might also work as an effective treatment strategy. Unraveling of additional bile salt biotransformations that can be taken advantage of to potentially modulate bile acid composition as well as bile acid analogs that do not undergo enterohepatic recirculation are yet other potential CDI treatments.45,102 Finally, introduction of probiotics specifically targeting CDI or synbiotics (probiotics combined with prebiotics) that may modify the gut microbiome by increasing engraftment through introduction of specific nutrient sources appear to have potential for treatment in the future.103,104 It is important to mention though, that as understanding of the complex CDI pathogenesis and life cycle expands, the realization that individual treatments options may not be the way forward and comprehensive treatment plans may provide the best results, especially in regards to relapsing C. difficile episodes.
Funding Statement
This project was supported by awards R01AI116895 and R21AI144454 from the National Institute of Allergy and Infectious Diseases. Andrea Martinez Aguirre is a recipient of a CONACYT-COECYT fellowship 2017–2022 scholar/scholarship 625561/472087. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIAID. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Disclosure statement
The authors report there are no competing interests to declare.
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
References
- 1.Finn E, Andersson FL, Madin-Warburton M.. Burden of Clostridioides difficile infection (CDI) - a systematic review of the epidemiology of primary and recurrent CDI. BMC Infect Dis. 2021;21:456. doi: 10.1186/s12879-021-06147-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clancy CJ, Buehrle D, Vu M, Wagener MM, Nguyen MH. Impact of revised infectious diseases Society of America and Society for Healthcare Epidemiology of America clinical practice guidelines on the treatment of Clostridium difficile Infections in the United States. Clin Infect Dis. 2021;72:1944–15. doi: 10.1093/cid/ciaa484. [DOI] [PubMed] [Google Scholar]
- 3.CDC . 2019. Antibiotic resistance threats in the United States, 2019. U.S. Department of Health and Human Services, CDC. [Google Scholar]
- 4.Buffie CG, Bucci V, Stein RR, McKenney PT, Ling L, Gobourne A, No D, Liu H, Kinnebrew M, Viale A, et al. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature. 2015;517:205–208. doi: 10.1038/nature13828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Theriot CM, Koenigsknecht MJ, Carlson PE Jr., Hatton GE, Nelson AM, Li B, Huffnagle GB, Li J, Young VB. Antibiotic-induced shifts in the mouse gut microbiome and metabolome increase susceptibility to Clostridium difficile infection. Nat Commun. 2014;5:3114. doi: 10.1038/ncomms4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shim JO. Clostridium difficile in children: to treat or not to treat? Pediatr Gastroenterol Hepatol Nutr. 2014;17:80–84. doi: 10.5223/pghn.2014.17.2.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bien J, Palagani V, Bozko P. The intestinal microbiota dysbiosis and Clostridium difficile infection: is there a relationship with inflammatory bowel disease? Therap Adv Gastroenterol. 2013;6:53–68. doi: 10.1177/1756283X12454590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Furuya-Kanamori L, Marquess J, Yakob L, Riley TV, Paterson DL, Foster NF, Huber CA, Clements ACA. Asymptomatic Clostridium difficile colonization: epidemiology and clinical implications. BMC Infect Dis. 2015;15:516. doi: 10.1186/s12879-015-1258-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Swick MC, Koehler TM, Driks A. Surviving between hosts: sporulation and transmission. Microbiol Spectr. 2016;4. doi: 10.1128/microbiolspec.VMBF-0029-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tocheva EI, Ortega DR, Jensen GJ. Sporulation, bacterial cell envelopes and the origin of life. Nat Rev Microbiol. 2016;14:535–542. doi: 10.1038/nrmicro.2016.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baloh M, Sorg JA. Clostridioides difficile spore germination: initiation to DPA release. Curr Opin Microbiol. 2022;65:101–107. doi: 10.1016/j.mib.2021.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deakin LJ, Clare S, Fagan RP, Dawson LF, Pickard DJ, West MR, Wren BW, Fairweather NF, Dougan G, Lawley TD, et al. The Clostridium difficile spo0A gene is a persistence and transmission factor. Infect Immun. 2012;80:2704–2711. doi: 10.1128/IAI.00147-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jump RL, Pultz MJ, Donskey CJ. Vegetative Clostridium difficile survives in room air on moist surfaces and in gastric contents with reduced acidity: a potential mechanism to explain the association between proton pump inhibitors and C. difficile-associated diarrhea? Antimicrob Agents Chemother. 2007;51:2883–2887. doi: 10.1128/AAC.01443-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barra-Carrasco J, Hernandez-Rocha C, Ibanez P, Guzman-Duran AM, Alvarez-Lobos M, Paredes-Sabja D. Clostridium difficile spores and its relevance in the persistence and transmission of the infection. Rev Chilena Infectol. 2014;31:694–703. doi: 10.4067/S0716-10182014000600010. [DOI] [PubMed] [Google Scholar]
- 15.Bhattacharjee D, McAllister KN, Sorg JA. Germinants and their receptors in Clostridia. J Bacteriol. 2016;198:2767–2775. doi: 10.1128/JB.00405-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sorg JA, Sonenshein AL. Bile salts and glycine as cogerminants for Clostridium difficile spores. J Bacteriol. 2008;190:2505–2512. doi: 10.1128/JB.01765-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Orrell KE, Zhang Z, Sugiman-Marangos SN, Melnyk RA. Clostridium difficile toxins A and B: receptors, pores, and translocation into cells. Crit Rev Biochem Mol Biol. 2017;52:461–473. doi: 10.1080/10409238.2017.1325831. [DOI] [PubMed] [Google Scholar]
- 18.Aktories K, Schwan C, Jank T. Clostridium difficile Toxin Biology. Annu Rev Microbiol. 2017;71:281–307. doi: 10.1146/annurev-micro-090816-093458. [DOI] [PubMed] [Google Scholar]
- 19.Di Bella S, Ascenzi P, Siarakas S, Petrosillo N, and Di Masi A. Clostridium difficile toxins A and B: insights into pathogenic properties and extraintestinal effects. Toxins (Basel). 2016;8(5);134. doi: 10.3390/toxins8050134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perelle S, Gibert M, Bourlioux P, Corthier G, Popoff MR. Production of a complete binary toxin (actin-specific ADP- ribosyltransferase) by Clostridium difficile CD196. Infect Immun. 1997;65:1402–1407. doi: 10.1128/iai.65.4.1402-1407.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eyre DW, Walker AS, Griffiths D, Wilcox MH, Wyllie DH, Dingle KE, Crook DW, Peto TE. Clostridium difficile mixed infection and reinfection. J Clin Microbiol. 2012;50:142–144. doi: 10.1128/JCM.05177-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shoff CJ, Spires SS, Yen C, Advani SD. Navigating the 2021 update to the IDSA/SHEA Clostridioides difficile guidelines: an ethical approach to equitable patient care. Antimicrob Stewardship Healthcare Epidemiol. 2022;2:e70. doi: 10.1017/ash.2022.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xiao Y, Angulo MT, Lao S, Weiss ST, Liu YY. An ecological framework to understand the efficacy of fecal microbiota transplantation. Nat Commun. 2020;11:3329. doi: 10.1038/s41467-020-17180-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.DeFilipp Z, Bloom PP, Torres Soto M, Mansour MK, Sater MRA, Huntley MH, Turbett S, Chung RT, Chen YB, Hohmann EL, et al. Drug-resistant E. coli bacteremia transmitted by fecal microbiota transplant. N Engl J Med. 2019;381:2043–2050. doi: 10.1056/NEJMoa1910437. [DOI] [PubMed] [Google Scholar]
- 25.Khoruts A. Is fecal microbiota transplantation a temporary patch for treatment of Clostridium difficile infection or a new frontier of therapeutics? Expert Rev Gastroenterol Hepatol. 2018;12:435–438. doi: 10.1080/17474124.2018.1465818. [DOI] [PubMed] [Google Scholar]
- 26.Jiang ZD, Ajami NJ, Petrosino JF, Jun G, Hanis CL, Shah M, Hochman L, Ankoma-Sey V, DuPont AW, Wong MC, et al. Randomised clinical trial: faecal microbiota transplantation for recurrent Clostridum difficile infection - fresh, or frozen, or lyophilised microbiota from a small pool of healthy donors delivered by colonoscopy. Aliment Pharmacol Ther. 2017;45:899–908. doi: 10.1111/apt.13969. [DOI] [PubMed] [Google Scholar]
- 27.Chiang JY. Regulation of bile acid synthesis: pathways, nuclear receptors, and mechanisms. J Hepatol. 2004;40:539–551. doi: 10.1016/j.jhep.2003.11.006. [DOI] [PubMed] [Google Scholar]
- 28.Molinaro A, Wahlstrom A, Marschall HU. Role of bile acids in metabolic control. Trends Endocrinol Metab. 2018;29:31–41. doi: 10.1016/j.tem.2017.11.002. [DOI] [PubMed] [Google Scholar]
- 29.John YL, and Chiang JMF. Bile acid biology, pathophysiology, and therapeutics. Clin Liver Dis. 2020;15(3):91–94. doi: 10.1002/cld.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ridlon JM, Kang DJ, Hylemon PB, Bajaj JS. Bile acids and the gut microbiome. Curr Opin Gastroenterol. 2014;30:332–338. doi: 10.1097/MOG.0000000000000057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ridlon JM, Harris SC, Bhowmik S, Kang DJ, Hylemon PB. Consequences of bile salt biotransformations by intestinal bacteria. Gut Microbes. 2016;7:22–39. doi: 10.1080/19490976.2015.1127483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kliewer SA, Mangelsdorf DJ. Bile acids as hormones: the FXR-FGF15/19 pathway. Dig Dis. 2015;33:327–331. doi: 10.1159/000371670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reue K, Lee JM, Vergnes L. Regulation of bile acid homeostasis by the intestinal Diet1-FGF15/19 axis. Curr Opin Lipidol. 2014;25:140–147. doi: 10.1097/MOL.0000000000000060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Monaghan T, Mullish BH, Patterson J, Wong GKS, Marchesi JR, Xu H, Jilani T, Kao D. Effective fecal microbiota transplantation for recurrent Clostridioides difficile infection in humans is associated with increased signalling in the bile acid-farnesoid X receptor-fibroblast growth factor pathway. Gut Microbes. 2019;10:142–148. doi: 10.1080/19490976.2018.1506667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jose S, Mukherjee A, Horrigan O, Setchell KDR, Zhang W, Moreno-Fernandez ME, Andersen H, Sharma D, Haslam DB, Divanovic S, et al. Obeticholic acid ameliorates severity of Clostridioides difficile infection in high fat diet-induced obese mice. Mucosal Immunol. 2021;14:500–510. doi: 10.1038/s41385-020-00338-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Winston JA, Rivera AJ, Cai J, Thanissery R, Montgomery SA, Patterson AD, Theriot, CW. Ursodeoxycholic acid (UDCA) mitigates the host inflammatory response during Clostridioides difficile infection by altering gut bile acids. Infect Immun. 2020;88:e00045–20. doi: 10.1128/IAI.00045-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wilson KH, Kennedy MJ, Fekety FR. Use of sodium taurocholate to enhance spore recovery on a medium selective for Clostridium difficile. J Clin Microbiol. 1982;15:443–446. doi: 10.1128/jcm.15.3.443-446.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sorg JA, Sonenshein AL. Chenodeoxycholate is an inhibitor of Clostridium difficile spore germination. J Bacteriol. 2009;191:1115–1117. doi: 10.1128/JB.01260-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Francis MB, Allen CA, Sorg JA. Muricholic acids inhibit Clostridium difficile spore germination and growth. PLoS One. 2013;8:e73653. doi: 10.1371/journal.pone.0073653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weingarden AR, Chen C, Zhang N, Graiziger CT, Dosa PI, Steer CJ, Shaughnessy MK, Johnson JR, Sadowsky MJ, Khoruts A, et al. Ursodeoxycholic acid inhibits Clostridium difficile spore germination and vegetative growth, and prevents the recurrence of ileal pouchitis associated with the infection. J Clin Gastroenterol. 2016;50:624–630. doi: 10.1097/MCG.0000000000000427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weingarden AR, Chen C, Bobr A, Yao D, Lu Y, Nelson VM, Sadowsky MJ, Khoruts A. Microbiota transplantation restores normal fecal bile acid composition in recurrent Clostridium difficile infection. Am J Physiol Gastrointest Liver Physiol. 2014;306:G310–9. doi: 10.1152/ajpgi.00282.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen X, Katchar K, Goldsmith JD, Nanthakumar N, Cheknis A, Gerding DN, Kelly CP. A mouse model of Clostridium difficile–associated disease. Gastroenterology. 2008;135:1984–1992. doi: 10.1053/j.gastro.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 43.Hofmann AF. The continuing importance of bile acids in liver and intestinal disease. Arch Intern Med. 1999;159:2647–2658. doi: 10.1001/archinte.159.22.2647. [DOI] [PubMed] [Google Scholar]
- 44.Thanissery R, Winston JA, Theriot CM. Inhibition of spore germination, growth, and toxin activity of clinically relevant C. difficile strains by gut microbiota derived secondary bile acids. Anaerobe. 2017;45:86–100. doi: 10.1016/j.anaerobe.2017.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sato Y, Atarashi K, Plichta DR, Arai Y, Sasajima S, Kearney SM, Suda W, Takeshita K, Sasaki T, Okamoto S, et al. Novel bile acid biosynthetic pathways are enriched in the microbiome of centenarians. Nature. 2021;599:458–464. doi: 10.1038/s41586-021-03832-5. [DOI] [PubMed] [Google Scholar]
- 46.Wilson KH. Efficiency of various bile salt preparations for stimulation of Clostridium difficile spore germination. J Clin Microbiol. 1983;18:1017–1019. doi: 10.1128/jcm.18.4.1017-1019.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Winston JA, Theriot CM. Impact of microbial derived secondary bile acids on colonization resistance against Clostridium difficile in the gastrointestinal tract. Anaerobe. 2016;41:44–50. doi: 10.1016/j.anaerobe.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aguirre AM, Yalcinkaya N, Wu Q, Swennes A, Tessier ME, Roberts P, Miyajima F, Savidge T, and Sorg JA. Bile acid-independent protection against Clostridioides difficile infection. PLoS Pathog. 2021;17:e1010015. doi: 10.1371/journal.ppat.1010015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Seekatz AM, Theriot CM, Rao K, Chang YM, Freeman AE, Kao JY, Young VB. Restoration of short chain fatty acid and bile acid metabolism following fecal microbiota transplantation in patients with recurrent Clostridium difficile infection. Anaerobe. 2018;53:64–73. doi: 10.1016/j.anaerobe.2018.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Staley C, Vaughn BP, Graiziger CT, Singroy S, Hamilton MJ, Yao D, Chen C, Khoruts A, Sadowsky MJ. Community dynamics drive punctuated engraftment of the fecal microbiome following transplantation using freeze-dried, encapsulated fecal microbiota. Gut Microbes. 2017;8:276–288. doi: 10.1080/19490976.2017.1299310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wexler AG, Guiberson ER, Beavers WN, Shupe JA, Washington MK, Lacy DB, Caprioli RM, Spraggins JM, Skaar EP. Clostridioides difficile infection induces a rapid influx of bile acids into the gut during colonization of the host. Cell Rep. 2021;36:109683. doi: 10.1016/j.celrep.2021.109683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dubois T, Tremblay YDN, Hamiot A, Martin-Verstraete I, Deschamps J, Monot M, Briandet R, Dupuy B. A microbiota-generated bile salt induces biofilm formation in Clostridium difficile. NPJ Biofilms Microbiomes. 2019;5:14. doi: 10.1038/s41522-019-0087-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sievers S, Metzendorf NG, Dittmann S, Troitzsch D, Gast V, Troger SM, Wolff C, Zuhlke D, Hirschfeld C, Schulter R, Riedel K, et al. Differential view on the bile acid stress response of Clostridioides difficile. Front Microbiol. 2019;10:258. doi: 10.3389/fmicb.2019.00258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tam J, Icho S, Utama E, Orrell KE, Gómez-Biagi RF, Theriot CM, Kroh HK, Rutherford SA, Lacy DB, Melnyk RA, et al. Intestinal bile acids directly modulate the structure and function of C. difficile TcdB toxin. Proc Natl Acad Sci. 2020;117:6792–6800. doi: 10.1073/pnas.1916965117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stickland LH. Studies in the metabolism of the strict anaerobes (genus Clostridium). Biochem J. 1934;28:1746–1759. doi: 10.1042/bj0281746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nisman BRM, Cohen GN, COHEN GN. Extension of the Stickland reaction to several bacterial species. Arch Biochem. 1948;16:473. [PubMed] [Google Scholar]
- 57.LH S. Studies in the metabolism of the strict anaerobes (genus Clostridium)- The oxidation of alanine by Cl. sporogenes. IV. The reduction of glycine by Cl. sporogenes. Biochem J. 1935;29:889–898. doi: 10.1042/bj0290889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jackson S, Calos M, Myers A, Self WT. Analysis of proline reduction in the nosocomial pathogen Clostridium difficile. J Bacteriol. 2006;188(24):8487–8495. doi: 10.1128/JB.01370-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bouillaut L, Self WT, Sonenshein AL. Proline-dependent regulation of Clostridium difficile Stickland metabolism. J Bacteriol. 2013;195:844–854. doi: 10.1128/JB.01492-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fonknechten Nuria CS, Sabine T, Aurélie L, Andreesen Jan R, Nadia P, Eric P, Michel G, Valérie B, Marcel S, Le Paslier D, et al. Clostridium sticklandii, a specialist in amino acid degradation:revisiting its metabolism through its genome sequence. BMC Genomics. 2010;11. doi: 10.1186/1471-2164-11-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bouillaut L, Dubois T, Francis MB, Daou N, Monot M, Sorg JA, Sonenshein AL, Dupuy B. Role of the global regulator Rex in control of NAD + -regeneration in Clostridioides (Clostridium) difficile. Mol Microbiol. 2019;111:1671–1688. doi: 10.1111/mmi.14245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Self WT. Specific and nonspecific incorporation of selenium into macromolecules. Comprehensive Natural Products Ii: Chemistry and Biology, Vol 5: Amino Acids, Peptides and Proteins. 2010;121–148. [Google Scholar]
- 63.McAllister KN, Bouillaut L, Kahn JN, Self WT, Sorg JA. Using CRISPR-Cas9-mediated genome editing to generate C. difficile mutants defective in selenoproteins synthesis. Sci Rep. 2017;7:14672. doi: 10.1038/s41598-017-15236-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.McAllister KN, Martinez Aguirre A, Sorg JA. The Selenophosphate Synthetase Gene, selD, Is Important for Clostridioides difficile physiology. J Bacteriol. 2021;203:e0000821. doi: 10.1128/JB.00008-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Battaglioli EJ, Hale VL, Chen J, Jeraldo P, Ruiz-Mojica C, Schmidt BA, Rekdal VM, Till LM, Huq L, Smits SA, Moor WJ, Hall YJ, Smyrk T, Khanna S, Pardi DS, Grover M, Patel R, Chia N, Nelson H, Sonnenburg JL, and Farrugia G, et al. Clostridioides difficile uses amino acids associated with gut microbial dysbiosis in a subset of patients with diarrhea. Sci Transl Med. 2018;10. doi: 10.1126/scitranslmed.aam7019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Girinathan BP, DiBenedetto N, Worley JN, Peltier J, Arrieta-Ortiz ML, Immanuel SRC, Lavin R, Delaney ML, Cummins CK, Hoffman M, et al. In vivo commensal control of Clostridioides difficile virulence. Cell Host Microbe. 2021;29:1693–708 e7. doi: 10.1016/j.chom.2021.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Robinson JI, Weir WH, Crowley JR, Hink T, Reske KA, Kwon JH, Burnham CAD, Dubberke ER, Mucha PJ, Henderson JP, et al. Metabolomic networks connect host-microbiome processes to human Clostridioides difficile infections. J Clin Invest. 2019;129:3792–3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jenior ML, JL L, Young BV, Schloss PD. Clostridium difficile colonizes alternative nutrient niches during infection across distinct murine gut microbiomes. mSystems. 2017;2. doi: 10.1128/mSystems.00063-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hofmann JD, Biedendieck R, Michel AM, Schomburg D, Jahn D, Neumann-Schaal M. Influence of L-lactate and low glucose concentrations on the metabolism and the toxin formation of Clostridioides difficile. PLoS One. 2021;16:e0244988. doi: 10.1371/journal.pone.0244988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fletcher JR, Pike CM, Parsons RJ, Rivera AJ, Foley MH, McLaren MR, Montgomery SA, Theriot CM, et al. Clostridioides difficile exploits toxin-mediated inflammation to alter the host nutritional landscape and exclude competitors from the gut microbiota. Nat Commun. 2021;12:462. doi: 10.1038/s41467-020-20746-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hromada S, Qian Y, Jacobson TB, Clark RL, Watson L, Safdar N, Amador-Noguez D, Venturelli OS, et al. Negative interactions determine Clostridioides difficile growth in synthetic human gut communities. Mol Syst Biol. 2021;17:e10355. doi: 10.15252/msb.202110355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bajaj JS, Kakiyama G, Savidge T, Takei H, Kassam ZA, Fagan A, Gavis EA, Pandak WM, Nittono H, Hylemon PB, Boonma P, Haag A, Heuman DM, Fuchs M, John B, Sikaroodi M, Gillevet PM, et al. Antibiotic-associated disruption of microbiota composition and function in cirrhosis is restored by fecal transplant. Hepatology. 2018;68:1549–1558. doi: 10.1002/hep.30037. [DOI] [PubMed] [Google Scholar]
- 73.Cheng S, M X, Geng S, Jiang X, Yuan L, Luansha H, Jianrong L, Wang Y, and Han X. Fecal microbiota transplantation beneficially regulates intestinal mucosal autophagy and alleviates gut barrier injury. mSystems. 2018;3(5):e00137–18. doi: 10.1128/mSystems.00137-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Littmann ER, Lee JJ, Denny JE, Alam Z, Maslanka JR, Zarin I, Matsuda R, Carter RA, Susac B, Saffern MS, et al. Host immunity modulates the efficacy of microbiota transplantation for treatment of Clostridioides difficile infection. Nat Commun. 2021;12:755. doi: 10.1038/s41467-020-20793-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.McDonald JAK, Mullish BH, Pechlivanis A, Liu Z, Brignardello J, Kao D, Holmes E, Li JV, Clarke TB, Thursz MR, et al. Inhibiting growth of Clostridioides difficile by restoring valerate, produced by the intestinal microbiota. Gastroenterology. 2018;155:1495–507 e15. doi: 10.1053/j.gastro.2018.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Neumann-Schaal M, Jahn D, Schmidt-Hohagen K. Metabolism the difficile way: the key to the success of the pathogen Clostridioides difficile. Front Microbiol. 2019;10:219. doi: 10.3389/fmicb.2019.00219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gencic S, and Grahame DA. Diverse energy-conserving pathways in Clostridium difficile: growth in the absence of amino acid Stickland acceptors and the role of the wood-Ljungdahl pathway. J Bacteriol. 2020:202(20):e00233–20. doi: 10.1128/JB.00233-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Koenigsknecht MJ, Theriot CM, Bergin IL, Schumacher CA, Schloss PD, Young VB. Dynamics and establishment of Clostridium difficile infection in the murine gastrointestinal tract. Infect Immun. 2015;83:934–941. doi: 10.1128/IAI.02768-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lopez CA, McNeely TP, Nurmakova K, Beavers WN, Skaar EP. Clostridioides difficile proline fermentation in response to commensal clostridia. Anaerobe. 2020;63:102210. doi: 10.1016/j.anaerobe.2020.102210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kriaa A, Bourgin M, Mkaouar H, Jablaoui A, Akermi N, Soussou S, Maguin E, Rhimi M. Microbial reduction of cholesterol to Coprostanol: an old concept and new insights. Catalysts. 2019;9. doi: 10.3390/catal9020167. [DOI] [Google Scholar]
- 81.Antharam VC, McEwen DC, Garrett TJ, Dossey AT, Li EC, Kozlov AN, Mesbah Z, Wang GP. An integrated metabolomic and microbiome analysis identified specific gut microbiota associated with fecal cholesterol and coprostanol in Clostridium difficile infection. PLoS One. 2016;11:e0148824. doi: 10.1371/journal.pone.0148824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jump RL, Polinkovsky A, Hurless K, Sitzlar B, Eckart K, Tomas M, Deshpande A, Nerandzic MM, Donskey CJ. Metabolomics analysis identifies intestinal microbiota-derived biomarkers of colonization resistance in clindamycin-treated mice. PLoS One. 2014;9:e101267. doi: 10.1371/journal.pone.0101267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kellingray L, Gall GL, Defernez M, Beales ILP, Franslem-Elumogo N, Narbad A. Microbial taxonomic and metabolic alterations during faecal microbiota transplantation to treat Clostridium difficile infection. J Infect. 2018;77:107–118. doi: 10.1016/j.jinf.2018.04.012. [DOI] [PubMed] [Google Scholar]
- 84.Lavelle A, Sokol H. Gut microbiota-derived metabolites as key actors in inflammatory bowel disease. Nat Rev Gastroenterol Hepatol. 2020;17:223–237. doi: 10.1038/s41575-019-0258-z. [DOI] [PubMed] [Google Scholar]
- 85.Martinez-Gili L, McDonald JAK, Liu Z, Kao D, Allegretti JR, Monaghan TM, Barker GF, Blanco JM, Williams HRT, Holmes E, Thursz MR, Marchesi JR, Mullish BH, et al. Understanding the mechanisms of efficacy of fecal microbiota transplant in treating recurrent Clostridioides difficile infection and beyond: the contribution of gut microbial-derived metabolites. Gut Microbes. 2020;12:1810531. doi: 10.1080/19490976.2020.1810531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Garland M, Hryckowian AJ, Tholen M, Bender KO, Van Treuren WW, Loscher S, Sonnenburg JL, Bogyo M. The clinical drug ebselen attenuates inflammation and promotes microbiome recovery in mice after antibiotic treatment for CDI. Cell Rep Med. 2020;1(1):100005. doi: 10.1016/j.xcrm.2020.100005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Fachi JL, Felipe JS, Pral LP, da Silva BK, Correa RO, de Andrade MCP, da Fonseca DM, Basso PJ, Câmara NOS, de Sales E Souza ÉL, et al. Butyrate protects mice from Clostridium difficile-induced colitis through an HIF-1-dependent mechanism. Cell Rep. 2019;27:750–61 e7. doi: 10.1016/j.celrep.2019.03.054. [DOI] [PubMed] [Google Scholar]
- 88.Fachi JL, Secca C, Rodrigues PB, Mato FCP, Di Luccia B, Felipe JS, Pral LP, Rungue M, Rocha VDM, Sato FT, et al. Acetate coordinates neutrophil and ILC3 responses against C. difficile through FFAR2. J Exp Med. 2020;217. doi: 10.1084/jem.20190489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gregory AL, Pensinger DA, Hryckowian AJ. A short chain fatty acid-centric view of Clostridioides difficile pathogenesis. PLoS Pathog. 2021;17:e1009959. doi: 10.1371/journal.ppat.1009959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kang JD, Myers CJ, Harris SC, Kakiyama G, Lee IK, Yun BS, Matsuzaki K, Furukawa M, Min HK, Bajaj JS, Zhou H, and Hylemon PB, et al. Bile acid 7alpha-dehydroxylating gut bacteria secrete antibiotics that inhibit Clostridium difficile: role of secondary bile acids. Cell Chem Biol. 2019;26:27–34 e4. doi: 10.1016/j.chembiol.2018.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Saa P, Urrutia A, Silva-Andrade C, Martín AJ, Garrido D. Modeling approaches for probing cross-feeding interactions in the human gut microbiome. Comput Struct Biotechnol J. 2022;20:79–89. doi: 10.1016/j.csbj.2021.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Knippel RJ, Wexler AG, Miller JM, Beavers WN, Weiss A, de Crecy-Lagard V, Edmonds KA, Giedroc DP, Skaar EP. Clostridioides difficile senses and hijacks host heme for incorporation into an oxidative stress defense system. Cell Host Microbe. 2020;28:411–21 e6. doi: 10.1016/j.chom.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Passmore IJ, Letertre MPM, Preston MD, Bianconi I, Harrison MA, Nasher F, Kaur H, Hong HA, Baines SD, Cutting SM, Swann JR, Wren BW, Dawson LF. Para-cresol production by Clostridium difficile affects microbial diversity and membrane integrity of gram-negative bacteria. PLoS Pathog. 2018;14:e1007191. doi: 10.1371/journal.ppat.1007191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Pruss KM, Sonnenburg JL. C. difficile exploits a host metabolite produced during toxin-mediated disease. Nature. 2021;593:261–265. doi: 10.1038/s41586-021-03502-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ng KM, Ferreyra JA, Higginbottom SK, Lynch JB, Kashyap PC, Gopinath S, Naidu N, Choudhury B, Weimer BC, Monack DM, Sonnenburg JL. Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature. 2013;502:96–99. doi: 10.1038/nature12503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Darkoh C, Plants-Paris K, Bishoff D, and DuPont HL. Clostridium difficile modulates the gut microbiota by inducing the production of indole, an interkingdom signaling and antimicrobial molecule. mSystems. 2019;4(2):e00346–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Donskey CJ, Kundrapu S, Deshpande A. Colonization versus carriage of Clostridium difficile. Infect Dis Clin North Am. 2015;29:13–28. doi: 10.1016/j.idc.2014.11.001. [DOI] [PubMed] [Google Scholar]
- 98.Janoir C, Deneve C, Bouttier S, Barbut F, Hoys S, Caleechum L, Chapeton-Montes D, Pereira FC, Henriques AO, Collignon A, Monot M, Dupuy B, et al. Adaptive strategies and pathogenesis of Clostridium difficile from in vivo transcriptomics. Infect Immun. 2013;81:3757–3769. doi: 10.1128/IAI.00515-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Qian X, Yanagi K, Kane AV, Alden N, Lei M, Snydman DR, Vickers RJ, Lee K, Thorpe CM. Ridinilazole, a narrow spectrum antibiotic for treatment of Clostridioides difficile infection, enhances preservation of microbiota-dependent bile acids. Am J Physiol Gastrointest Liver Physiol. 2020;319:G227–G37. doi: 10.1152/ajpgi.00046.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Simeon RA, Zeng Y, Chonira V, Aguirre AM, Lasagna M, Baloh M, Sorg JA, Tommos C, Chen Z. Protease-stable DARPins as promising oral therapeutics. Protein Eng Des Sel. 2021;34. doi: 10.1093/protein/gzab028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Feuerstadt P, Louie TJ, Lashner B, Wang EEL, Diao L, Bryant JA, Sims M, Kraft CS, Cohen SH, Berenson CS, et al. SER-109, an oral microbiome therapy for recurrent Clostridioides difficile Infection. N Engl J Med. 2022;386:220–229. doi: 10.1056/NEJMoa2106516. [DOI] [PubMed] [Google Scholar]
- 102.Stoltz KL, Erickson R, Staley C, Weingarden AR, Romens E, Steer CJ, Khoruts A, Sadowsky MJ, Dosa PI. Synthesis and biological evaluation of bile acid analogues inhibitory to Clostridium difficile spore germination. J Med Chem. 2017;60:3451–3471. doi: 10.1021/acs.jmedchem.7b00295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hryckowian AJ, Van Treuren W, Smits SA, Davis NM, Gardner JO, Bouley DM, Sonnenburg JL. Microbiota-accessible carbohydrates suppress Clostridium difficile infection in a murine model. Nat Microbiol. 2018;3:662–669. doi: 10.1038/s41564-018-0150-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Rätsep M, Kõljalg S, Sepp E, Smidt I, Truusalu K, Songisepp E, Stsepetova J, Naaber P, Mikelsaar RH, Mikelsaar M, et al. A combination of the probiotic and prebiotic product can prevent the germination of Clostridium difficile spores and infection. Anaerobe. 2017;47:94–103. doi: 10.1016/j.anaerobe.2017.03.019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.


